ABSTRACT
The implementation of artemisinin (ART) combination therapies (ACTs) has greatly decreased deaths caused by Plasmodium falciparum malaria, but increasing ACT resistance in Southeast Asia and Africa could reverse this progress. Parasite population genetic studies have identified numerous genes, single-nucleotide polymorphisms (SNPs), and transcriptional signatures associated with altered artemisinin activity with SNPs in the Kelch13 (K13) gene being the most well-characterized artemisinin resistance marker. However, there is an increasing evidence that resistance to artemisinin in P. falciparum is not related only to K13 SNPs, prompting the need to characterize other novel genes that can alter ART responses in P. falciparum. In our previous analyses of P. falciparum piggyBac mutants, several genes of unknown function exhibited increased sensitivity to artemisinin that was similar to a mutant of K13. Further analysis of these genes and their gene co-expression networks indicated that the ART sensitivity cluster was functionally linked to DNA replication and repair, stress responses, and maintenance of homeostatic nuclear activity. In this study, we have characterized PF3D7_1136600, another member of the ART sensitivity cluster. Previously annotated as a conserved Plasmodium gene of unknown function, we now provide putative annotation of this gene as a Modulator of Ring Stage Translation (MRST). Our findings reveal that the mutagenesis of MRST affects gene expression of multiple translation-associated pathways during the early ring stage of asexual development via putative ribosome assembly and maturation activity, suggesting an essential role of MRST in protein biosynthesis and another novel mechanism of altering the parasite’s ART drug response.
IMPORTANCE
Plasmodium falciparum malaria killed more than 600,000 people in 2021, though ACTs have been critical in reducing malaria mortality as a first-line treatment for infection. However, ACT resistance in Southeast Asia and emerging resistance in Africa are detrimental to this progress. Mutations to Kelch13 (K13) have been identified to confer increased artemisinin tolerance in field isolates, however, genes other than K13 are implicated in altering how the parasite responds to artemisinin prompts additional analysis. Therefore, in this study we have characterized a P. falciparum mutant clone with altered sensitivity to artemisinin and identified a novel gene (PF3D7_1136600) that is associated with alterations to parasite translational metabolism during critical timepoints for artemisinin drug response. Many genes of the P. falciparum genome remain unannotated, posing a challenge for drug–gene characterizations in the parasite. Therefore, through this study, we have putatively annotated PF3D7_1136600 as a novel MRST gene and have identified a potential link between MRST and parasite stress response mechanisms.
KEYWORDS: malaria, antimalarial agents, drug resistance mechanisms, gene annotation
INTRODUCTION
Malaria afflicted about 247 million people and killed about 619,000 people in 2021, with increasing Plasmodium falciparum artemisinin resistance and artemisinin combination therapy (ACT) failure in Southeast (SE) Asia threatening to exacerbate its clinical burden (1). Alarmingly, studies have revealed the emergence of ACT resistance in Africa prompting the need to annotate novel genetic factors associated with altered artemisinin sensitivity to better track resistance in the field (2). Understanding ART resistance is complicated by the multi-pronged artemisinin mechanism of action, which includes damage to DNA, disruption of proteasome activity, accumulation of damaged/misfolded proteins leading to disruption of proteostasis, disruption to mitochondrial activity, free-radical-mediated oxidative stress, and disruption of hemoglobin digestion (3 - 9).
Mutations in the Kelch13 (K13) gene are the most well-characterized marker for artemisinin resistance, with the highest prevalence in SE Asia and recent emergence in Africa. Mutations to K13 have been linked to reduced binding to and polyubiquitination of phosphatidylinositol 3-kinase, increasing kinase levels and PI3P product, a key mediator of artemisinin resistance (6). Additionally, mutations to K13 are associated with disrupted nuclear metabolism, augmented antioxidant and damage-sensing properties of the mitochondria, and inactivation of K13 has led to cell cycle arrest at the ring stage promoting cell quiescence under ART exposure (3, 7, 8, 10 - 13). Recent studies have also identified links between K13 and altered endocytosis of host red blood cell cytosol and hemoglobin that can alter the rate of hemoglobin digestion and artemisinin activation, mediated by the K13 endocytosis complex (14). Several key mutations to the K13 propeller domain have been identified in the field to cause partial artemisinin resistance, including C580Y, Y493H, R539T, and I543T (6, 15).
While distinct single-nucleotide polymorphisms (SNPs) in K13 have previously been identified and studied, ongoing research posits a more interactive network between genetic makeup, transcriptional responses, and metabolic activities to play a combined role in the introduction, spread, and maintenance of artemisinin treatment failure in Asia and Africa (16 - 20). P. falciparum has demonstrated an ability to elongate its ring stage of development during the intraerythrocytic development cycle (IDC), thus leading to a longer period of no hemoglobin uptake and digestion, thereby decreasing artemisinin activation (7, 10, 11, 16). Recent studies from our laboratory and other research groups have also shown that P. falciparum appears to utilize essential and conserved pathways associated with its innate fever response mechanism to withstand artemisinin exposure, including heat shock, proteasome activity, protein folding, and apicoplast-derived isoprenoid biosynthesis (5, 9, 21 - 23). Proper balance of proteostasis under artemisinin-mediated oxidative stress is necessary to respond to a buildup of damaged, misfolded, and unfolded proteins, with artemisinin-tolerant parasites upregulating chaperones to stabilize proteins and upregulating the core proteasome and vesicular trafficking to degrade or eliminate non-reparable proteins (5, 9, 21 - 23).
Previous genome-wide association studies (GWAS) and transcriptome-wide association studies (TWAS) have shown that K13 mutation-mediated resistance is more likely to occur with upregulated unfolded protein response progression (to maintain a proper balance of proteostasis under artemisinin exposure) and delayed IDC progression (elongated ring stage) (18, 24). GWAS and TWAS studies have additionally noted high transcriptional diversity of P. falciparum isolates in areas with high artemisinin resistance and that parasite physiology, and thereby phenotype, is controlled by specific transcriptional profiles and genetic patterns, including genes regulating metabolic growth (glycolysis), response to proteotoxic stress, redox metabolism, vesicular trafficking, and host cytoplasm remodeling (16, 25). Lastly, GWAS/TWAS studies have identified novel genetic and transcriptional variations of genes not related to K13 that confer artemisinin tolerance in vivo and in vitro, including ubiquitin carboxyl-terminal hydrolase 1 (UBP-1), autophagy-related protein 18 (ATG18), and coronin (26 - 30). Though ongoing studies have hypothesized a link between the interactions of these genes with K13 to illicit a complex artemisinin-resistant response, the ability of P. falciparum to develop artemisinin tolerance independent of K13 mutations stresses additional transcriptomic studies.
With increasing resistance to artemisinin and the variety of mechanisms not linked to K13 that P. falciparum can undergo to become artemisinin tolerant, characterization of novel artemisinin-linked genes and the transcriptomic patterns associated with these genes are needed to address the need for novel antimalarials targeting key vulnerable parasite-specific processes. Chemogenomic profiling of single-insertion piggyBac transposon mutants has been integral in defining drug–gene interactions based on piggyBac mutant drug response profiles (31, 32). We previously validated the usefulness of this process in identifying novel artemisinin-linked genes through annotation of the artemisinin sensitivity cluster (ASC), a group of seven genes with associated piggyBac mutants exhibiting an increased sensitivity to artemisinin, including a piggyBac mutant of K13 (31). Transcriptomic analysis of this K13 piggyBac mutant showed significant dysregulation of K13 gene expression, correlating to dysregulation of DNA replication and repair processes, consistent with Kelch13’s role as a stress response regulator (12). In this study, we further define the transcriptome of an ASC piggyBac mutant of a Conserved Plasmodium Gene of Unknown Function, which we have now annotated as a Modulator of Ring Stage Translation (MRST, PF3D7_1136600). We show significant dysregulation of a variety of translation-associated pathways in the MRST mutant due to MRST downregulation, linking this gene to proper translational gene expression in P. falciparum.
RESULTS
Artemisinin-sensitive phenotype of the MRST piggyBac mutant
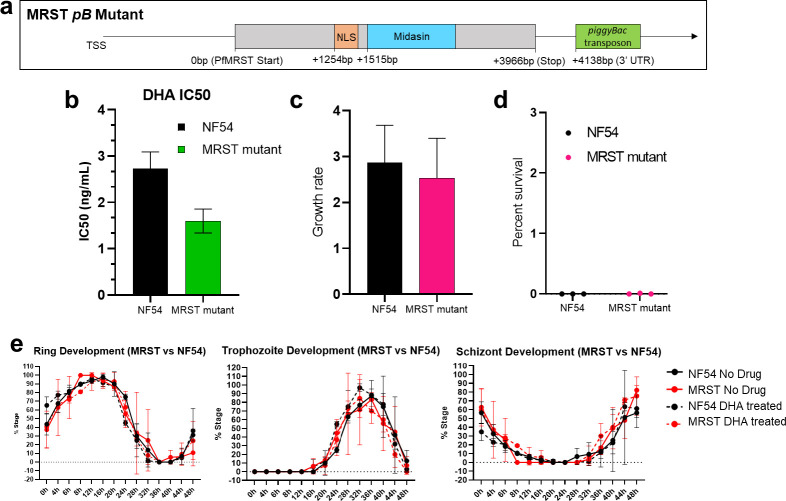
The mutant selected for this study contains a 3′ UTR piggyBac transposon insertion +4,138 bp downstream of the annotated start codon of a currently annotated conserved Plasmodium gene of unknown function, PF3D7_1136600 (Fig. 1a) (33, 34). WGS was performed using the Illumina NextSeq 550 system for the NF54 parent clone and the PF3D7_1136600 piggyBac mutant to both confirm the piggyBac insertion site location in the mutant and to ensure that additional mutations were not present in the NF54 and PF3D7_1136600 mutants used in this study (WGS data available via the NCBI Sequence Read Archive [SRA] with the BioProject ID PRJNA947639). We confirmed no additional mutations in our NF54 and mutant and identified the correct piggyBac transposon insertion in the annotated 3′ UTR of PF3D7_1136600 in the mutant, +4,138 bp downstream of the start codon (33, 34).
Fig 1.
Phenotype of the MRST mutant. (a) The MRST mutant contains a piggyBac insertion in the annotated 3′ UTR of PF3D7_1136600, which we have termed a Modulator of Ring Stage Translation (MRST) (33, 34). This insertion lies +4138 bp downstream of the start codon. Because MRST lacks functional annotation, bioinformatic analysis identified a putative nuclear localization signal +1,254 bp downstream of the start codon, along with a predicted midasin protein linker and disordered region homology, associated with ribosome nuclear export and maturation, +1,515 bp downstream of start codon. NLS and midasin domain were predicted in Prosite ExPasy and NCBI Blast, respectively. (b) SYBR Green I malaria drug sensitivity assay was performed to validate the artemisinin-sensitive phenotype of the mutant. IC50 values averaged across six bio-replicates of the NF54 parent and MRST mutant plotted via bar graph and analyzed via Mann–Whitney test (P-value: 0.002). Data were analyzed and plotted via GraphPad Prism. Ring stage survival assay was also performed to validate MRST mutant DHA sensitivity, demonstrating that NF54 and MRST mutant exhibited similar growth rates under normal conditions (c). Both the mutant and NF54 had decreased survival under a DHA drug pulse (0.004% survival for mutant and 0% survival for NF54) (d). Progression of ring, trophozoite, and schizont cell cycle progression was analyzed with and without a 6-hour DHA drug pulse (e). Line graphs show average percentages of each stage every 4 hours during the 48-hour life cycle in drug- and non-drug-treated NF54 and MRST mutant clones. We observed no significant changes in asexual cell cycle development between NF54 and mutant with and without DHA drug exposure, pointing to metabolic changes playing a role in sensitive phenotype (significant P-value < 0.05). Stage progression was analyzed via microscopy, with data analysis and statistics (Fisher’s exact test of cell counts) performed in GraphPad Prism and R, respectively. NLS, nuclear localization signal; IC50, half maximal inhibitory concentration; DHA, dihydroartemisinin.
PF3D7_1136600 remains a conserved Plasmodium gene with a lack of functional annotation, thus prompting our decision to perform a preliminary bioinformatic analysis of the PF3D7_1136600 protein sequence to determine putative conserved domains and protein characteristics. We first identified via NCBI Blast partial similarity to the linker and disordered region of the Saccharomyces cerevisiae midasin protein (MDN1p, YLR106p), a 4,910 amino acid-long AAA+ ATPase family nuclear chaperone protein required for the maturation and nuclear export of ribosome subunits (Fig. 1a) (35, 36). Similarity spanned the 505-951aa positions in PF3D7_1136600 and the 3874-4305aa position in the midasin gene (E-value: 3.97e-03). This ~440-residue similarity in the 4,910-residue midasin protein represents only a portion of the midasin linker domain (which provides a structural role in the midasin protein while also predicted to act as a hinge or protein-binding site) and disordered domain, thus partially linking PF3D7_1136600 to putative ribosome maturation processes and requiring further investigation (36). A nuclear localization signal was also bioinformatically predicted via ExPasy Prosite located 418aa–432aa downstream of the start codon, bioinformatically supporting a nuclear-associated function for PF3D7_1136600 (Fig. 1a) (37). We herein refer to PF3D7_1136600 as an MRST based on our preliminary bioinformatic partial characterization and our downstream analysis of the MRST mutant asexual blood stage transcriptome.
We next sought to further characterize the ART response phenotype of the MRST mutant through additional drug sensitivity assays. SYBR Green I malaria drug sensitivity assay was used to validate the dihydroartemisinin IC50 of the MRST mutant clone as previously published (mutant IC50 value: 1.6 ng/mL; NF54 IC50 value: 2.7 ng/mL, P-value: 0.002165, averaged across six bio-replicates) (Fig. 1b; Data Set S1 Tab 1) (31, 38, 39). Further, RSA identified an average percent survival rate of 0.004% for the MRST mutant under a DHA drug pulse and an average percent survival rate of 0% for the NF54 parent clone, indicating DHA drug sensitivity of the mutant and NF54 (Fig. 1c and d; Data Set S1 Tab 2) (40). Tolerance to artemisinin particularly occurs during the ring stage of the IDC, with artemisinin-resistant (ART-R) parasite clones being shown to elongate its ring stage progression (10, 11). As ring stage is the least metabolically active stage and thereby the least sensitive to DHA exposure compared to trophozoite, schizont, and merozoite stages, we sought to determine whether ring stage progression is altered in the artemisinin-sensitive (ART-S) MRST mutant by characterizing IDC development under a 6-hour DHA drug pulse, similar to the RSA (10, 11, 19). We observed no significant changes in MRST development of ring, trophozoite, and schizont stages with and without DHA drug pulse compared to the NF54 parent clone, indicating that the ART-S phenotype may not be directly attributed to the intraerythrocytic ring stage development (P-value > 0.05) (Fig. 1e; Fig. S1; Data Set S1 Tab 3).
Early ring stage gene expression is dysregulated due to the MRST gene disruption
To determine the molecular pathways associated with the ART-S phenotypic changes in the MRST mutant, we investigated the transcriptomic differences between the mutant and NF54 WT during the IDC. Mutant and NF54 parent clone samples were harvested at 6 hours post-merozoite invasion (hpi) (early ring), 12 hpi (ring), 24 hpi (early trophozoite), 36 hpi (late trophozoite), and 48 hpi (schizont) for subsequent RNA sequencing and analysis (Data Set S1 Tab 4 and 5). Normalization, calculation of fold change expression, and statistical assessment of differential gene expression in the mutant were performed via DEseq2 (version 1.34.0). Significant downregulation of the MRST gene in the mutant compared to the NF54 parent occurred at 6 hpi and 48 hpi (6 hpi FC value: −2.342, 6 hpi P-value: 0.00095; 48 hpi FC value: −2.939; 48 hpi P-value: 0.00014) (Fig. 2a). We observed high transcriptional concordance between the MRST mutant and NF54 at the same IDC stages (Spearman correlation: 6 hours: 0.88; 12 hours: 0.96; 24 hours: 0.94; 36 hours: 0.96; 48 hours: 0.95) (Fig. 2b; Fig. S2; Data Set S1 Tab 6). A relatively moderate correlation was shown at 6 hours (Spearman correlation: 6 hours: 0.88), corresponding to the highest number of DEGs in the mutant (6-hour DEGs: 493 upregulated, 608 downregulated), compared to the other IDC stages at 12 hours (95 total DEGs), 24 hours (223 total DEGs), 36 hours (111 total DEGs), and 48 hours (328 total DEGs) (Fig. 2c).
Fig 2.
MRST disruption alters the early ring stage gene expression. (a) MRST gene FPKM expression analysis in NF54 and the mutant shows significant downregulation of MRST at 6 hpi (P-value: 0.00095). (b) Analysis of gene expression correlation of each harvested timepoint (6 hpi, 12 hpi, 24 hpi, 36 hpi, 48 hpi) between NF54 and MRST mutant. The lowest correlation between matched timepoint is seen at 6 hpi, with a Spearman correlation value of 0.88. Spearman correlation values and heatmap generated in R. (c) The number of significantly differentially expressed genes (DEGs) per timepoint in the MRST mutant was analyzed and plotted via bar graph. The highest number of DEGs were observed at 6 hpi, corresponding to the timepoint of MRST gene dysregulation, along with the identical timepoint of the lowest correlation between NF54 and mutant.
We also counted the proportion of different IDC stages during parasite development via microscopy for our RNAseq samples, showing no significant difference in RNAseq samples stages and demonstrating the stage alignment between the MRST mutant and NF54 (Fisher’s exact test, P-value > 0.05) (Fig. S3; Data Set S1 Tab 7). Additionally, the transcriptional profiles in our study are highly consistent with the previously published NF54 and K13 piggyBac mutant transcriptome data (12). High correlation values were observed at identical timepoints between the NF54 parasites in the different studies (Spearman correlation: 6 hours: 0.87; 12 hours: 0.87; 24 hours: 0.85; 36 hours: 0.85; 48 hours: 0.87) (Fig. S4; Data Set S1 Tab 8) (12). Furthermore, the transcriptional correlation of housekeeping pathways (protein biosynthesis, DNA replication, transcriptional machinery, ribosome structure, fatty acid synthesis, redox metabolism, and genes coding for proteasome-related pathways) further demonstrates the life cycle consistency and supports metabolic similarity between NF54 and MRST mutant (Fig. S5; Data Set S1 Tab 9 and 10).
Gene ontology enrichment of MRST mutant early ring stage expression reveals dysregulation in RNA metabolic processes
We next interrogated the biological pathway alterations behind the high number of DEGs at 6 hpi between the MRST mutant and NF54. We performed GO enrichment analysis, via the PlasmoDB GO enrichment tool, for the dysregulated genes at 6 hpi (Fig. 3a), highlighting pathways such as RNA processing, RNA splicing, translational initiation, tRNA aminoacylation for protein translation, and ribonucleoprotein complex biogenesis dysregulation (P-value cutoff < 0.05) (Data Set S1 Tab 11). To further support these findings, additional GO enrichment analysis via the pfGO enrichment package in R was also performed on 6 hpi DEGs of the mutant (41, 42). Upregulated GO terms included terms associated with intracellular protein transport (intracellular protein transport P-value: 0.00054, vesicle-mediated transport P-value: 0.02788) and terms associated with the mitochondrial cellular compartment (P-value: 4.80E-07) (Data Set S1 Tab 12), while downregulated GO terms were significantly enriched in genes related to RNA binding (P-value: 6.20E-06), mRNA binding (P-value: 0.00018), ribonucleoprotein complex (P-value: 0.0151), and tRNA aminoacylation for protein translation (P-value: 0.002) (Fig. 3b; Data Set S1 Tab 12). Fold change expression values of genes associated with these top upregulated and downregulated GO terms are shown in Fig. 3c (Data Set S1 Tab 5 and 13).
Fig 3.
Gene ontology (GO) enrichment of 6 hpi expression in the MRST mutant. (a) GO enrichment of significantly dysregulated genes at 6 hpi in the mutant as shown in word cloud plot. Word color and size correspond to the P-value, and word cloud plot is generated via PlasmoDB GO enrichment tool. (b) In addition to PlasmoDB database GO enrichment, 6 hpi expression in the mutant was subjected to GO enrichment via the pfGO package in R. Downregulated GO terms were highly enriched in translational metabolism terms such as mRNA binding, ribonucleoprotein complex, and tRNA aminoacylation for protein translation. GO terms are plotted via bubble plot, with the x-axis corresponding to the fraction of annotated genes present in significant DEGs, and the y-axis corresponding to −log(P-value) of each term. Bubble size corresponds to the number of significant DEGs associated with the term in the mutant, and color corresponds to the GO category (green: molecular function, blue: biological process, red: cellular compartment). (c) Log(foldchange) plots of genes associated with select upregulated and downregulated GO terms at 6 hpi in the mutant. Plot is generated in GraphPad Prism.
Protein biosynthesis pathways are significantly dysregulated at the early ring stage in the MRST mutant
Using the Malaria Parasite Metabolic Pathways database (https://mpmp.huji.ac.il/maps/all), we chose to investigate the mutant gene expression of a select number of established housekeeping pathways in P. falciparum to identify the significance of dysregulated RNA processing and translation-related pathways at 6 hpi (21, 43 - 45). Gene sets were obtained corresponding to the following housekeeping pathways: protein biosynthesis, DNA replication, transcriptional machinery, ribosome structure, fatty acid synthesis, redox metabolism, and genes coding for the proteasome (Data Set S1 Tab 9). High correlations of these housekeeping pathways were seen between NF54 and the MRST mutant at identical timepoints (Fig. S5; Data Set S1 Tab 10). Additionally, we analyzed fold change expression values of genes corresponding to each pathway at 6 hours, with downregulation of protein biosynthesis–related genes seen compared to the other housekeeping pathways (Fig. 4a; Data Set S1 Tab 9). Analysis of fold change expression of the same protein biosynthesis–related genes in the mutant shows widespread dysregulation only at 6 hours, with no widespread change in gene expression at 12 hours, 24 hours, 36 hours, and 48 hours (Fig. 4b; Data Set S1 Tab 14).
Fig 4.
Protein biosynthesis–related pathways are significantly dysregulated at 6 hpi in the mutant. (a) Housekeeping pathway expression at 6 hpi was analyzed to determine the significance of translation-associated GO term dysregulation, with gene lists obtained from Malaria Parasite Metabolic Pathways database for protein biosynthesis, DNA replication, transcriptional machinery, ribosome structure, fatty acid synthesis, redox metabolism, and genes coding for the proteasome. Widespread downregulation was only observed for genes associated with protein biosynthesis. (b) Analysis of fold change expression of these same protein biosynthesis–related genes in the MRST mutant show downregulation at only 6 hpi, compared to the other timepoints tested. (c) Log2(foldchange) across all sampled timepoints was analyzed for gene sets corresponding to the various translation-associated GO terms as determined by pfGO analysis in R and displayed via heatmap. The majority of these translation-associated genes and pathways are solely downregulated at 6 hpi. Graph and statistics are generated in GraphPad Prism.
To further validate significantly enriched gene ontologies at 6 hours in the mutant, as identified by PlasmoDB GO Enrichment analysis (Fig. 3a; Data Set S1 Tab 11), we analyzed fold change expression of genes associated with select 6- hour GO terms. As shown in Fig. 4c, we highlight the following pathways that were dysregulated at 6 hours: RNA splicing (P-value: 2.83E-05), mRNA processing (P-value: 0.00124), regulation of translation (P-value: 0.00062), translation initiation (P-value: 0.00281), ribonucleoprotein complex biogenesis (P-value: 0.00304), transcription by RNA polymerase III (P-value: 0.00419), and tRNA aminoacylation for protein translation (P-value: 0.00437). Fold change expression of genes in these pathways show dysregulation at 6 hours with a lack of gene dysregulation at the other harvested timepoints (Data Set S1 Tab 15). Because we observed significant dysregulation of the MRST gene also at 6 hours (P-value: 0.00095) (Fig. 2a) and identified dysregulation of translation associated genes at only 6 hpi in the mutant (Fig. 4c), this points to a potential link between MRST and translation gene dysregulation.
MRST disruption leads to the dysregulation of ribosome biogenesis pathways causing downstream effects on translation
MRST contains partial predicted similarity to the linker and disordered domains of the midasin domain of the AAA+ ATPase family, involved in the maturation and nuclear export of ribosome subunits (Fig. 1a). To further investigate the link between this predicted domain and the MRST mutant transcriptome, we chose to analyze differential expression of gene lists obtained from the MPMP database for ribosomal subunit genes and for genes coding for components involved in ribosomal assembly at 6 hpi. The majority of ribosomal subunit genes (79 out of 97 genes) lacked significant dysregulation at 6 hpi (P-value > 0.05), with only 13 out of 97 genes (~13%) being significantly downregulated (Fig. 5a; Data Set S1 Tab 16). In comparison, we observed more significant downregulation (P-value cutoff < 0.05) of genes coding for components involved in ribosome assembly (51 out of 133 genes, ~38%), including ATP-dependent RNA helicases, eukaryotic translation initiation factors, ribosome biogenesis proteins, and small nucleolar RNA-associated proteins (Fig. 5a; Data Set S1 Tab 16). This points to the initial assembly of ribosomes, instead of the ribosomal subunits themselves, as the primary dysregulated process of ribosomal maturation due to the MRST disruption.
Fig 5.
MRST disruption leads to the dysregulation of ribosome biogenesis pathways, causing downstream effects on translation expression. (a) Due to the bioinformatically predicted homology of the MRST protein to the midasin protein linker domain and disordered regions, ribosome metabolism pathway expression was analyzed at 6 hpi. While only 13 out of 97 genes (~13%) related to ribosome structure were downregulated at 6 hpi, 51 out of 133 genes (~38%) of the ribosome assembly pathway were downregulated, primarily pointing to the dysregulation of ribosomal assembly/maturation, and not the ribosome subunits themselves, being affected by MRST disruption. (b) Several tRNA ligases were differentially expressed at 6 hpi, with the majority lacking significant dysregulation at the other harvested timepoints in the mutant (P-value > 0.05). (c) Seven genes associated with RNA polymerase III activity (PF3D7_0609900, PF3D7_0708100, PF3D7_1104700, PF3D7_1206600, PF3D7_1210400, PF3D7_1329000, PF3D7_1449300) were dysregulated at 6 hpi compared to the other timepoints. Graphs and statistics are generated in GraphPad Prism.
We have shown that several protein biosynthesis pathways other than ribosome biogenesis are downregulated at 6 hpi in the mutant, including translation initiation, mRNA processing, transcription by RNA polymerase III, and tRNA aminoacylation (Fig. 4c). We identified 16 tRNA ligases, the majority of which (12 out of 16) are considered essential for asexual blood stage development (Fig. S7A), that have significant downregulation solely at the early ring stages (P-value cutoff < 0.05) (Fig. 5b; Fig. S7b and c; Data Set S1 Tab 15) (46). Additionally, several genes associated with RNA polymerase III, which transcribes DNA known to synthesize tRNA, were significantly dysregulated at 6 hpi including DNA-directed RNA polymerases I, II, and III subunit RPABC5 (PF3D7_0708100), DNA-directed RNA polymerase III subunit RPC8 (PF3D7_1104700), DNA-directed RNA polymerase III subunit RPC2 (PF3D7_1206600), and DNA-directed RNA polymerase III subunit RPC1 (PF3D7_1329000) (6 hpi P-value cutoff < 0.05) (Fig. 5c; Data Set S1 Tab 15) (47). Additionally, MAF1 (PF3D7_0416500), a negative regulator of RNA polymerase III, is not dysregulated at any stage of development in the MRST mutant, pointing to downstream effects associated with MRST dysregulation on ribosome assembly and other regulatory pathways of translation (47).
DISCUSSION
With the urgent and potentially disastrous issue of artemisinin resistance and ACT failure in Asia and Africa, combined with the various molecular mechanisms of P. falciparum resistance, it is imperative to identify and characterize novel genes that can alter the artemisinin stress response. GWAS and TWAS have made substantial gains in detecting mutations associated with delayed parasite clearance in the field, along with characterizing transcriptional signatures linked to artemisinin resistance and response (15 - 18 - 25). However, while these studies focus on the large-scale analysis of field isolates and data sets to identify shared molecular mechanisms, they lack the comprehensive metabolic and transcriptomic characterization of unique individual genes in P. falciparum, many of which lack functional annotation, that can alter artemisinin response (17, 25, 46). Therefore, in this study we have transcriptionally characterized the piggyBac transposon mutant of the novel, unannotated gene PF3D7_1136600 (conserved Plasmodium gene, unknown function) that we have termed a modulator of ring stage translation (MRST) based on its bioinformatically predicted domains and associated transcriptional profile of the mutant.
Our study has several key findings and observations. First, we observed that piggyBac disruption in the 3′ untranslated region of the MRST gene led to the downregulation of expression at 6 hpi. Our data point to the significant dysregulation of MRST (P-value: 0.000953) as a factor in the differential gene expression seen at 6 hpi in the mutant. Genes related to mRNA binding, translation initiation, and ribosome assembly pathways were dysregulated at 6 hpi. Additionally, we identified the downregulation of seven genes associated with RNA polymerase III activity, along with the downregulation of 16 tRNA ligases. tRNA ligases are highly conserved housekeeping genes of the P. falciparum protein synthesis pathway, known for the charging of tRNAs for protein translation, and the majority are classified as essential for asexual stage development (46). The inhibition of tRNA ligase activity leads to an accumulation of uncharged tRNAs, associated with decreased protein synthesis and cell death, making tRNA ligases potent and widely studied antimicrobial drug targets (48 - 50). Additionally, dysregulation of RNA polymerase III proteins, also essential for asexual development, notably impacts tRNA ligase expression, ribosome biogenesis, RNA processing, and protein transport (47). Overall, we observe a multi-pronged dysregulation of protein biosynthesis regulatory pathways at the early ring stage that we suspect plays a role in the artemisinin-sensitive phenotype of the MRST mutant.
MRST is currently annotated as a conserved Plasmodium gene of unknown function (PF3D7_1136600); however, MRST was bioinformatically predicted to have putative partial similarity to the linker and disordered regions of the midasin protein, involved in the nuclear export and maturation of ribosomes (36). Supporting this is the identification of a predicted nuclear localization signal in MRST and the observed dysregulation in translational gene expression at 6 hpi, the same timepoint of MRST downregulation. While the majority of ribosomal subunit genes lacked significant dysregulation at 6 hpi, genes involved in the regulation of assembly of the ribosome are downregulated. Considering the annotated role of the midasin domain in ribosome nuclear export and maturation and the timepoints of MRST gene, translation pathways, and ribosome assembly dysregulation, our finding suggest that MRST may be mildly associated with ribosome maturation processes at the early ring stage. As our bioinformatic characterization of the MRST protein was preliminary to identify putative domains and characteristics to elucidate gene function, it will be important to perform additional analysis to determine how the moderate similarity with midasin is associated with MRST gene function.
Our predicted model is that MRST may be responsible for the early regulation of ribosome maturation processes, via moderate similarity to the midasin protein, at the early ring stage and, due to piggyBac transposon-mediated disruption, downregulation of MRST activity negatively impacts downstream effects on protein translation and the ability to respond to artemisinin-mediated stress at the ring stage. While we observed significant dysregulation of translational, tRNA ligase, and ribosomal genes, we did not observe significant impacts on cell cycle progression in the mutant compared to NF54 (Fig. 1e). Under normal growth, translation increases as the parasite matures, notably ramping up at the trophozoite/schizont border compared to the ring stage parasites (19, 51). Protein biosynthesis is already considerably low at the ring stage compared to other stages and, under normal conditions that the parasite samples in this study were subjected to, dysregulation may not affect parasite growth (19). However, as the ring stage is a critical timepoint for ART phenotypic response, dysregulation of translational activity may lead to decreased protein synthesis capacity under ART exposure, having lasting effects on later stages of parasite development.
MRST is considered essential for the asexual development and combined with the putative domains and transcriptome of the MRST mutant, supports MRST as an essential modulator for the ring stage translation. Due to the non-mutability of MRST during asexual stage, future studies will need to involve targeted mutagenesis of specific regions of the gene, such as a knockout of the predicted midasin homology regions or nuclear localization signal sites, or epitope tagging of MRST to confirm protein localization throughout the IDC with and without DHA stress. Lastly, it will be important to measure protein production throughout the life cycle, achievable through liquid chromatography with tandem mass spectrometry, to confirm MRST’s role in altering protein translation and activity. Combined, these future studies will give us more clarification on the function of MRST and the essential role this gene may play in ribosome metabolism and translation in P. falciparum.
In conclusion, we have performed an initial characterization of the novel PF3D7_1136600 gene, which we have termed MRST, via the analysis of the transcriptome of its piggyBac mutant, predicting that this gene plays a role in ribosome assembly and translational activity during the ring stage. With many genes of the P. falciparum genome lacking functional annotation, our putative annotation of the MRST gene contributes to the critical goal of characterizing novel drug–gene interactions. There is a continuing need to comprehensively characterize genetic factors that can alter artemisinin response in P. falciparum to understand mechanisms of resistance along with identifying novel drug targets. With this being the first characterization of how MRST can alter the transcriptome of P. falciparum, we hope our findings can provide an additional step into the characterization of this gene and other novel molecular interactions between P. falciparum and artemisinin.
MATERIALS AND METHODS
Parasite culturing methods
Standard in vitro P. falciparum culturing protocols were used to freeze, thaw, and maintain culture (52). Parasite cultures were maintained in human O+ red blood cells (RBCs) (obtained from commercial supplier Interstate Blood Bank, Inc.) at 4% hematocrit in culture media containing RPMI 1640 supplemented with HEPES, hypoxanthine, sodium bicarbonate, Albumax II, and gentamicin. A 37°C mixed gas incubator was used with O2, CO2, and N2. piggyBac mutants were created in the wild-type (WT) isogenic NF54 parent (including a WT K13), which is the parent line of the 3D7 clone, and validated via thermal asymmetric interlaced (TAIL)-PCR (32). Asexual phenotypic growth of mutant clones was first characterized (53). The NF54 line, containing no known K13 ART-R mutations and sensitive to antimalarials, was re-cloned and sequenced to serve as the reference genome for insertion mapping (46, 54). The MRST mutant was first described as piggyBac mutant 52 (PB52) in a previous study via TAIL-PCR (32). piggyBac mutants, including the MRST mutant and the previously characterized K13 piggyBac mutant PB58 (12), contain a NF54 genetic background except for a single piggyBac transposon insertion (an MRST 3′ untranslated region [UTR] insertion and K13 5′ UTR insertion for PB52 and PB58, respectively).
Bioinformatic characterization of MRST protein
The protein sequence of MRST was input into the National Center for Biotechnology Information (NCBI) Protein Blast database (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to determine conserved protein sequences and putative domains (accessed July 2022) using the MRST protein sequence available on the PlasmoDB database (35, 55). ExPasy Prosite Database was used (August 2022) to identify putative localization signals, amino acid-rich regions, and other bioinformatically determined protein characteristics (37).
SYBR Green I malaria drug sensitivity assay
The standardized SYBR Green I drug sensitivity assay was performed to determine altered sensitivity to ART drugs and other antimalarials (38, 39). Thawed parasite clones were allowed to reach ~2% parasitemia in culture before being synchronized with 10% sorbitol at the majority ring stage. Cultures were sorbitol synchronized at the next cycle of ring stage and allowed to recover for another 48-hour life cycle before starting the assay. A 1 mg/mL stock solution of DHA was diluted to the appropriate mother plate starting concentration, then diluted two-fold in 96-well Greiner black bottom well plates, leaving the last column for positive and negative drug controls. Ten microliters of mother plate drugs were added to the 96-well daughter plate containing 90 μL of parasite culture (2% hematocrit, 0.5% parasitemia) and incubated in a mixed gas incubator for 72 hours. Plates were then removed, frozen at −20°C for 24 hours, removed, and allowed to thaw to room temperature (RT), and 100 μL of 4× SYBR Green I dye solution (from an initial 10,000× SYBR Green stock) in 1:1 lysis buffer:water solution was added to each plate well. For the lysis buffer solution, dissolve 15.8 g Tris-HCl in 964-mL culture water (adjust pH to 7.5). Add 20 mL of 0.5M EDTA, 160 mg of saponin (0.016% wt/vol), and 16 mL of Triton X-100 (1.6% vol/vol) before mixing and sterilizing via vacuum filtration. After incubating plates at RT for 1 hour, SYBR Green fluorescence was read using the ClarioStar plate reader and analyzed via Excel and GraphPad Prism. IC50 values were determined by non-linear regression (sigmoidal dose–response/variable slope equation) of plotted normalized % inhibition values versus log(drug concentrations). Statistical analysis of IC50 values performed via Mann–Whitney test in GraphPad Prism. The assay was performed with six biological replicates.
Ring stage survival assay
Ring stage survival assay (RSA) was conducted for NF54 and MRST mutant to validate mutant artemisinin sensitivity (40). Parasite clones were thawed and maintained in culture until reaching 2%–5% majority ring stage for sorbitol synchronization. Sorbitol synchronization was again performed 30–48 hours later depending on parasite development and stages at the time of sorbitol treatment. After 30 hours of culture (majority late schizont stages), cultures were subjected to Percoll synchronization for late-stage schizont enrichment (70%:40% gradient). Thin smear slides were taken, and parasites returned to culture with the appropriate volume of complete media and uninfected RBC added. The culture was allowed to grow as normal in a mixed gas incubator for exactly 3 hours. Thin smear slides were then taken to evaluate the proportion of ring stages (>0.5%), cultures were incubated with sorbitol for 10 minutes, parasitemia was made to 0.5%–1% with a 2% hematocrit, and thin smear slides were taken again as initial slides “INI.”
Previously prepared DHA test solution (700 nM concentration) and dimethyl sulfoxide (DMSO) control solution were added to a 24-well culture plate under the following parameters: 100 μL of DMSO solution to the “non-exposed” wells and 100 μL of DHA solution to the “DHA-exposed” well. Infected RBC suspension of 900 μL was added to each well, with the 24-well plate being maintained in a mixed gas incubator for exactly 6 hours. Following the 6 hours, the contents of each well were washed with RPMI. The contents of each well were added to 15-mL centrifuge tubes, pelleted at 1,750 rpm for 3 minutes, the supernatant discarded, and 10 mL of RPMI added. Cultures were again pelleted, the supernatant discarded, and washed again with 10 mL of RPMI. Cultures were pelleted for a final time before the supernatant was discarded, 1 mL of complete culture media added, and returned to culture at 2% hematocrit in a new 24-well plate. Thin smear slides were taken for each well, and cultures were maintained in a mixed gas incubator for 66 hours. Following incubation, thin smear slides were taken for each well for microscopic analysis.
Slides were classified as “INI” smear to define the initial parasitemia, “NE” smear to define the non‐exposed parasitemia at 72 hours, and “DHA” smear to define the DHA‐exposed parasitemia at 72 hours. The number of infected RBCs containing viable parasites were counted (in a total of 10,000 RBCs estimated by the number of RBCs per field). The percent survival (% survival) values in NE and DHA slides are calculated using the following equation:
Percentage survival values are interpretable if the growth rate is ≥1.5 for the in vitro RSA.
Intraerythrocytic development cycle growth assay
The MRST mutant was analyzed for altered IDC progression under DHA exposure compared to NF54. Clones were thawed via standard protocols and synchronized at the same timepoint using 10% sorbitol for ring stage enrichment. After reaching ~2% majority ring, clones were sorbitol synchronized three times every 48 hours and allowed an additional 48 hours for recovery to obtain highly synchronized rings. When the NF54 parent clone reached 0 hpi (about half schizonts and half rings) (56), microscopy slides were taken for each culture every 4 hours for 48 consecutive hours, with culture media and fresh RBCs being replenished when needed. For the DHA drug pressure group, DHA at NF54 IC50 (concentration was added to each culture starting at 0 hpi, pulsed for 6 hours (with slides being taken at 0 hpi and 4 hpi) after which cultures were washed with RPMI twice, and returned to culture. Slides were taken post-washing of RBC cultures (~6 hpi) and continued to be taken every 4 hpi (8 hpi, 12 hpi, 16 hpi, etc.). Following 48 hours, all assay slides were counted to determine parasitemia and percentages of ring, trophozoite, and schizont per slide (dividing the total number of rings, trophozoite, or schizonts counted by the total number of parasites counted, multiplied by 100 to get a percentage of each stage). Data were analyzed using Excel spreadsheet, GraphPad Prism, and R. Percentage ring, trophozoite, or schizont were plotted against the timepoint post-merozoite invasion per clone. GraphPad Prism 9.3.1 was used to generate graphs, and R was used to perform Fisher’s exact test of cell counts per mutant and per timepoint for drug and no drug treatment.
Whole-genome sequencing and analysis
The MRST mutant and NF54 WT parent clone were subjected to whole-genome sequencing (WGS) to confirm a lack of additional mutations and to confirm the insertion site sequence in the mutant. Clones were thawed via standard thawing protocols and maintained in a 20-mL culture at 4% hematocrit in complete RPMI media. Cultures were allowed to reach ~5% parasitemia of the majority of schizonts before being subjected to lysis and DNA extraction. Briefly, RBCs and parasitized RBCs in culture were pelleted at 1,750 rpm for 3 minutes in a 50-mL centrifuge tube. The supernatant was removed, and cells were resuspended in 20-mL 1× PBS (phosphate-buffered saline). One hundred microliters of 10% saponin was added to the solution to lyse RBCs, and the tube was inverted three to four times to mix. Crude lysed cells were then pelleted, and the supernatant was removed. Lysed cells were washed three times with 10 mL of 1× PBS and resuspended in 2 mL of 1× PBS before continuing with DNA extraction via the Qiagen QIAamp DNA Blood Mini Kit, with DNA concentration determined via Qubit 4 Fluorometer. The NEBNext Ultra II DNA Library Preparation Kit was used to prepare samples for sequencing, and samples were sequenced using NextSeq 550 at 300 cycles. The sequencing reads’ quality was assessed using FastQC (v0.11.5) and were trimmed to remove adapter and low-quality bases using Trimgalore (v0.4.4). Raw reads were mapped to the P. falciparum 3D7 reference genome (PlasmoDB version 58) using mapped reads from Burrows-Wheeler aligner supermaximal exact matches (sequence aligner for reads > 70 bp) (BWA-MEM ) (v0.7.17). The sequence alignment data were then post-processed using SAMtools (v1.3.1). The Integrative Genomics Viewer was used to visualize reads mapped at the piggyBac insertion site. Bedtools (version 2.28.0) was used for comparative analysis to evaluate the presence of the mutations, such as SNPs and indels.
RNA extraction and sequencing
Five timepoints during the 48-hour IDC were harvested for RNAseq analysis in this study: 6 hours post-invasion (hpi), 12 hpi, 24 hpi, 36 hpi, and 48 hpi. Tightly synchronous isogenic WT NF54 and MRST mutant were obtained via three rounds of sorbitol synchronization for ring stage isolation and one round of Percoll synchronization for schizont stage isolation, maintained in a 200-mL culture at 4% hematocrit and ~5% parasitemia. 0 hpi of the culture was estimated via microscopy and samples were harvested at the correct timepoints, with morphological stages analyzed similar to growth screen analysis via Excel, GraphPad Prism, and Fisher’s exact test of microscopy cell counts in R. The standard TRIzol extraction method was used to extract RNA from crude saponin–lysed samples. Cultures were transferred from culture flasks to conical tubes, centrifuged at 1,750 rpm for 3 minutes, and supernatant was removed. RBC pellets were resuspended in cold 1× PBS, and RBCs were lysed via saponin, followed by washing three times afterward with cold PBS. The pellet was resuspended in 1-mL TRIzol for every 50 mL of culture and stored at –80°C for RNA extraction. Before extraction, samples were allowed to reach RT, and 2 µL of glycogen was added. The samples were incubated at 4°C overnight, removed, and allowed to reach RT. After adding chloroform, the samples were vortexed vigorously and incubated at RT for 5 minutes, followed by centrifugation at 12,000× g at 4°C for 10 minutes, supernatant discarded, and 1 mL of 75% ethanol was added. This was again followed by centrifugation at 10,000× g for 5 minutes, the supernatant was discarded, and the RNA pellet was allowed to dry. RNA pellets were then dissolved in 20–50 µL of diethyl pyrocarbonate (DEPC)-treated water while being incubated at 55°C for 10 minutes. RNA was quantified using a Qubit fluorometer RNA HS Assay kit, and quality was determined via RNA ScreenTape Analysis. Using the Illumina TruSeq Stranded mRNA Library Prep Kit, mRNA was sequenced using Illumina Nextseq Mid-output X5 kit at 300 cycles.
Gene expression determination
Gene expression data were obtained using previously described protocols (12). Reads from RNA sequencing (RNAseq) were aligned with the P. falciparum 3D7 reference clone via the HISAT2 program (Hierarchical indexing for spliced alignment of transcripts). FeatureCounts read summarization program was used to obtain raw counts. The expression of each gene was normalized and calculated using Cuffnorm as fragments per kilobase per million mapped reads (FPKM).
DEseq2 analysis to determine gene dysregulation
Normalization, fold change (FC) calculation (MRST mutant/NF54), and statistical assessment of differential gene expression (DE) were performed via DEseq2 (version 1.34.0). A log2FC > 1.4 and false discovery rate (FDR)-corrected P-value < 0.05 classified genes are significantly upregulated in the mutant. log2FC < −1.4 and FDR-corrected P-value < 0.05 determined genes are significantly downregulated in the mutant. DEseq2 analysis performed in R.
Transcriptional alignment and correlation assessment
To determine the similarity in gene expression patterns between NF54 and the MRST mutant, FPKM gene expression values per timepoint were analyzed for expression correlation. Using R and cor.test function, FPKM values of genes for NF54 and mutant were correlated to determine Spearman’s rank correlation coefficient. To support that our samples are correctly transcriptionally aligned, NF54 and MRST mutant data sets from this study were compared to sequencing data from our previously reported K13 piggyBac mutant study from Gibbons et al. (12). Spearman’s rank correlation coefficient was determined between sample timepoints of both clones from both data sets as previously described in R. Genes were disregarded unless expression data were present in samples from both studies. Housekeeping pathways were then analyzed for correlated gene expression between NF54 and MRST clones in this study to determine transcriptional alignment at the metabolic pathway level. Gene lists corresponding to seven housekeeping pathways (protein biosynthesis, DNA replication, transcriptional machinery, ribosome structure, fatty acid synthesis, redox metabolism, and genes coding for proteasome) were obtained from the Malaria Parasite Metabolic Pathways database (https://mpmp.huji.ac.il/maps/all). FPKM values for each pathway for all timepoints sampled were analyzed for Spearman correlation between NF54 and MRST mutants in R. Correlation coefficients were plotted via heatmap.2 function in R in a heatmap.
Gene ontology enrichment analysis
To analyze DEGs, gene ontology (GO) enrichment was performed for 6 hpi DEGs in the mutant. Significantly upregulated and downregulated gene IDs (upregulated > 1.4 log2FC, downregulated < −1.4 log2FC, significant P-value < 0.05) were input into the PlasmoDB database GO enrichment tool and analyzed for biological process of GO terms (55). GO terms were plotted with word cloud plot in PlasmoDB, with a 0.005 P-value cutoff to highlight the top significantly determined GO terms in the figure (55).
Additional GO enrichment assessment of 6 hpi expression in the mutant was performed by analyzing GO terms mapped to genes in a category of interest compared to a background of GO terms mapped to all other genes in the analysis using the R package pfGO (v 1.1) (41, 42). A GO-term database was created from the latest curated P. falciparum ontology available from PlasmoDB (accessed June 2022). GO terms are enriched in dysregulated gene categories in the mutant versus NF54 by timepoint and ontology, with “Up” and “Down” representing upregulation in the mutant or downregulation in the mutant compared to NF54, respectively. Enrichment was assessed via weighted Fisher’s/elim-hybrid P-value ≤ 0.05. Upregulated and downregulated GO terms were plotted in bubble plot, and/or box and whisker graphs were made in GraphPad Prism.
Metabolic pathway gene expression analysis
Select housekeeping pathways were chosen for gene expression analysis and gene expression correlation in the mutant. Gene lists corresponding to housekeeping pathways were obtained from the Malaria Parasite Metabolic Pathways database (https://mpmp.huji.ac.il/maps/all) and downloaded (June 2022) into Excel. Gene lists corresponding to each pathway were used to obtain log2FC (mutant/WT) expression values (generated via DEseq) in the mutant across the sampled timepoints. Violin plots of log2FC gene expression were generated and statistically analyzed in GraphPad Prism. Heatmaps of various protein biosynthesis pathways (obtained from pfGO GO enrichment analysis [41, 42]) was generated with log2FC values in GraphPad Prism. Significantly differentially expressed tRNA ligase gene dot plots and line graphs, along with RNA polymerase III–related genes bar graphs, were also generated and analyzed in GraphPad Prism.
ACKNOWLEDGMENTS
We thank the USF Genomics Equipment Core and Omics Hub for RNAseq and analysis support. We also thank Matthew Mercurio for RNAseq library preparation assistance and Swamy Rakesh Adapa for WGS analysis assistance.
This work is supported by National Institute of Allergy and Infectious Diseases grants R01AI117017 and R01AI130171.
Experimental design: C.S., J.G., M.Z., A.S., and J.H.A.; methodology: C.S., J.G., M.Z., F. S., C.V.P., D.C., and J.H.A.; software: J.G., J.O., and C.W.; formal analysis: C.S., J.G., and C.W.; data curation: C.S., J.G., C.W., F. S., C.V.P., T.D.O., and J.H.A.; writing—original draft: C.S.; writing—review and editing: C.S., J.G., C.W., C.V.P., and J.H.A.; supervision: A.S., L.C., T.D.O., and J.H.A.; all authors have read and agreed to the published version of the manuscript.
The authors declare no competing interests.
Contributor Information
John H. Adams, Email: ja2@usf.edu.
Laura A. Kirkman, Weill Cornell Medicine, New York, New York, USA
DATA AVAILABILITY
RNAseq data generated for NF54 and MRST mutants in this study have been deposited to the NCBI Gene Expression Omnibus (GEO) database with the accession number GSE223497. Processed RNAseq data are provided in Data Set S1 Tab 4 and Data Set S1 Tab 5. The IDC reference data can be obtained from Bozdech Z, Llinás M, Pulliam BL, Wong ED, Zhu J, DeRisi JL. 2003. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol 1:e5.
https://doi.org/10.1371/journal.pbio.0000005
WGS data for NF54 and MRST mutants in this study are available via the NCBI SRA with the BioProject ID PRJNA947639. The Plasmodium falciparum NF54 reference genome can be obtained from ftp://ftp.sanger.ac.uk/pub/project/pathogens/Plasmodium/falciparum/NF54/Assembly/V1_morphed/
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/msphere.00152-23.
Growth screen.
RNAseq correlations.
RNAseq sample stage analysis.
Correlation of RNAseq samples.
Correlation to housekeeping pathways.
GO 6 hrs.
tRNA ligase expression.
Tables of quantitative data sets.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. World Health Organization . 2022. World malaria report 2022. World Health Organization; 293. [Google Scholar]
- 2. Balikagala B, Fukuda N, Ikeda M, Katuro OT, Tachibana S-I, Yamauchi M, Opio W, Emoto S, Anywar DA, Kimura E, Palacpac NMQ, Odongo-Aginya EI, Ogwang M, Horii T, Mita T. 2021. Evidence of artemisinin-resistant malaria in Africa. N Engl J Med 385:1163–1171. doi: 10.1056/NEJMoa2101746 [DOI] [PubMed] [Google Scholar]
- 3. Xiong A, Prakash P, Gao X, Chew M, Tay IJJ, Woodrow CJ, Engelward BP, Han J, Preiser PR. 2020. K13-mediated reduced susceptibility to artemisinin in Plasmodium falciparum is overlaid on a trait of enhanced DNA damage repair. Cell Rep 32:107996. doi: 10.1016/j.celrep.2020.107996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang J, Huang L, Li J, Fan Q, Long Y, Li Y, Zhou B, Bereswill S. 2010. Artemisinin directly targets malarial mitochondria through its specific mitochondrial activation. PLoS ONE 5:e9582. doi: 10.1371/journal.pone.0009582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rocamora F, Zhu L, Liong KY, Dondorp A, Miotto O, Mok S, Bozdech Z. 2018. Oxidative stress and protein damage responses mediate Artemisinin resistance in malaria parasites. PLoS Pathog 14:e1006930. doi: 10.1371/journal.ppat.1006930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mbengue A, Bhattacharjee S, Pandharkar T, Liu H, Estiu G, Stahelin RV, Rizk SS, Njimoh DL, Ryan Y, Chotivanich K, Nguon C, Ghorbal M, Lopez-Rubio J-J, Pfrender M, Emrich S, Mohandas N, Dondorp AM, Wiest O, Haldar K. 2015. A molecular mechanism of artemisinin resistance in Plasmodium falciparum malaria. Nature 520:683–687. doi: 10.1038/nature14412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Klonis N, Crespo-Ortiz MP, Bottova I, Abu-Bakar N, Kenny S, Rosenthal PJ, Tilley L. 2011. Artemisinin activity against Plasmodium falciparum requires hemoglobin uptake and digestion. Proc Natl Acad Sci U S A 108:11405–11410. doi: 10.1073/pnas.1104063108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gupta DK, Patra AT, Zhu L, Gupta AP, Bozdech Z. 2016. DNA damage regulation and its role in drug-related phenotypes in the malaria parasites. Sci Rep 6:23603. doi: 10.1038/srep23603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bridgford JL, Xie SC, Cobbold SA, Pasaje CFA, Herrmann S, Yang T, Gillett DL, Dick LR, Ralph SA, Dogovski C, Spillman NJ, Tilley L. 2018. Artemisinin kills malaria parasites by damaging proteins and inhibiting the proteasome. Nat Commun 9:3801. doi: 10.1038/s41467-018-06221-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Klonis N, Xie SC, McCaw JM, Crespo-Ortiz MP, Zaloumis SG, Simpson JA, Tilley L. 2013. Altered temporal response of malaria parasites determines differential sensitivity to artemisinin. Proc Natl Acad Sci U S A 110:5157–5162. doi: 10.1073/pnas.1217452110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hott A, Casandra D, Sparks KN, Morton LC, Castanares G-G, Rutter A, Kyle DE. 2015. Artemisinin-Resistant Plasmodium falciparum parasites exhibit altered patterns of development in infected erythrocytes. Antimicrob Agents Chemother 59:3156–3167. doi: 10.1128/AAC.00197-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gibbons J, Button-Simons KA, Adapa SR, Li S, Pietsch M, Zhang M, Liao X, Adams JH, Ferdig MT, Jiang RHY. 2018. Altered expression of K13 disrupts DNA replication and repair in Plasmodium falciparum. BMC Genomics 19:849. doi: 10.1186/s12864-018-5207-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gnädig NF, Stokes BH, Edwards RL, Kalantarov GF, Heimsch KC, Kuderjavy M, Crane A, Lee MCS, Straimer J, Becker K, Trakht IN, Odom John AR, Mok S, Fidock DA. 2020. Insights into the intracellular localization, protein associations and artemisinin resistance properties of Plasmodium falciparum K13. PLoS Pathog 16:e1008482. doi: 10.1371/journal.ppat.1008482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Birnbaum J, Scharf S, Schmidt S, Jonscher E, Hoeijmakers WAM, Flemming S, Toenhake CG, Schmitt M, Sabitzki R, Bergmann B, Fröhlke U, Mesén-Ramírez P, Blancke Soares A, Herrmann H, Bártfai R, Spielmann T. 2020. A Kelch13-defined endocytosis pathway mediates artemisinin resistance in malaria parasites. Science 367:51–59. doi: 10.1126/science.aax4735 [DOI] [PubMed] [Google Scholar]
- 15. Rawat M, Kanyal A, Choubey D, Deshmukh B, Malhotra R, Mamatharani DV, Rao AG, Karmodiya K. 2022. Identification of co-existing mutations and gene expression trends associated with K13-mediated artemisinin resistance in Plasmodium falciparum. Front Genet 13:824483. doi: 10.3389/fgene.2022.824483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhu L, van der Pluijm RW, Kucharski M, Nayak S, Tripathi J, White NJ, Day NPJ, Faiz A, Phyo AP, Amaratunga C, Lek D, Ashley EA, Nosten F, Smithuis F, Ginsburg H, von Seidlein L, Lin K, Imwong M, Chotivanich K, Mayxay M, Dhorda M, Nguyen HC, Nguyen TNT, Miotto O, Newton PN, Jittamala P, Tripura R, Pukrittayakamee S, Peto TJ, Hien TT, Dondorp AM, Bozdech Z. 2022. Artemisinin resistance in the malaria parasite, Plasmodium falciparum, originates from its initial transcriptional response. Commun Biol 5:274. doi: 10.1038/s42003-022-03215-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Oberstaller J, Zoungrana L, Bannerman CD, Jahangiri S, Dwivedi A, Silva JC, Adams JH, Takala-Harrison S. 2021. Integration of population and functional genomics to understand mechanisms of artemisinin resistance in Plasmodium falciparum. Int J Parasitol Drugs Drug Resist 16:119–128. doi: 10.1016/j.ijpddr.2021.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mok S, Stokes BH, Gnädig NF, Ross LS, Yeo T, Amaratunga C, Allman E, Solyakov L, Bottrill AR, Tripathi J, Fairhurst RM, Llinás M, Bozdech Z, Tobin AB, Fidock DA. 2021. Artemisinin-Resistant K13 mutations rewire Plasmodium falciparum’s intra-erythrocytic metabolic program to enhance survival. Nat Commun 12:530. doi: 10.1038/s41467-020-20805-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mok S, Imwong M, Mackinnon MJ, Sim J, Ramadoss R, Yi P, Mayxay M, Chotivanich K, Liong K-Y, Russell B, Socheat D, Newton PN, Day NPJ, White NJ, Preiser PR, Nosten F, Dondorp AM, Bozdech Z. 2011. Artemisinin resistance in Plasmodium falciparum is associated with an altered temporal pattern of transcription. BMC Genomics 12:391. doi: 10.1186/1471-2164-12-391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mok S, Ashley EA, Ferreira PE, Zhu L, Lin Z, Yeo T, Chotivanich K, Imwong M, Pukrittayakamee S, Dhorda M, Nguon C, Lim P, Amaratunga C, Suon S, Hien TT, Htut Y, Faiz MA, Onyamboko MA, Mayxay M, Newton PN, Tripura R, Woodrow CJ, Miotto O, Kwiatkowski DP, Nosten F, Day NPJ, Preiser PR, White NJ, Dondorp AM, Fairhurst RM, Bozdech Z. 2015. Drug resistance. population transcriptomics of human malaria parasites reveals the mechanism of artemisinin resistance. Science 347:431–435. doi: 10.1126/science.1260403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang M, Wang C, Oberstaller J, Thomas P, Otto TD, Casandra D, Boyapalle S, Adapa SR, Xu S, Button-Simons K, Mayho M, Rayner JC, Ferdig MT, Jiang RHY, Adams JH. 2021. The apicoplast link to fever-survival and artemisinin-resistance in the malaria parasite. Nat Commun 12:4563. doi: 10.1038/s41467-021-24814-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tintó-Font E, Michel-Todó L, Russell TJ, Casas-Vila N, Conway DJ, Bozdech Z, Llinás M, Cortés A. 2021. A heat-shock response regulated by the PfAP2-Hs transcription factor protects human malaria parasites from febrile temperatures. Nat Microbiol 6:1163–1174. doi: 10.1038/s41564-021-00940-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lu K-Y, Pasaje CFA, Srivastava T, Loiselle DR, Niles JC, Derbyshire E. 2020. Phosphatidylinositol 3-phosphate and Hsp70 protect Plasmodium falciparum from heat-induced cell death. Elife 9:e56773. doi: 10.7554/eLife.56773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Daily JP, Scanfeld D, Pochet N, Le Roch K, Plouffe D, Kamal M, Sarr O, Mboup S, Ndir O, Wypij D, Levasseur K, Thomas E, Tamayo P, Dong C, Zhou Y, Lander ES, Ndiaye D, Wirth D, Winzeler EA, Mesirov JP, Regev A. 2007. Distinct physiological states of Plasmodium falciparum in malaria-infected patients. Nature 450:1091–1095. doi: 10.1038/nature06311 [DOI] [PubMed] [Google Scholar]
- 25. Zhu L, Tripathi J, Rocamora FM, Miotto O, van der Pluijm R, Voss TS, Mok S, Kwiatkowski DP, Nosten F, Day NPJ, White NJ, Dondorp AM, Bozdech Z, Tracking Resistance to Artemisinin Collaboration I . 2018. The origins of malaria artemisinin resistance defined by a genetic and transcriptomic background. Nat Commun 9:5158. doi: 10.1038/s41467-018-07588-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hunt P, Afonso A, Creasey A, Culleton R, Sidhu ABS, Logan J, Valderramos SG, McNae I, Cheesman S, do Rosario V, Carter R, Fidock DA, Cravo P. 2007. Gene encoding a deubiquitinating enzyme is mutated in artesunate- and chloroquine-resistant rodent malaria parasites. Mol Microbiol 65:27–40. doi: 10.1111/j.1365-2958.2007.05753.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Demas AR, Sharma AI, Wong W, Early AM, Redmond S, Bopp S, Neafsey DE, Volkman SK, Hartl DL, Wirth DF. 2018. Mutations in Plasmodium falciparum actin-binding protein coronin confer reduced artemisinin susceptibility. Proc Natl Acad Sci U S A 115:12799–12804. doi: 10.1073/pnas.1812317115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Henrici RC, Sutherland CJ. 2018. Alternative pathway to reduced artemisinin susceptibility in Plasmodium falciparum. Proc Natl Acad Sci U S A 115:12556–12558. doi: 10.1073/pnas.1818287115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mukherjee A, Bopp S, Magistrado P, Wong W, Daniels R, Demas A, Schaffner S, Amaratunga C, Lim P, Dhorda M, Miotto O, Woodrow C, Ashley EA, Dondorp AM, White NJ, Wirth D, Fairhurst R, Volkman SK. 2017. Artemisinin resistance without pfkelch13 mutations in Plasmodium falciparum isolates from Cambodia. Malar J 16:195. doi: 10.1186/s12936-017-1845-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Breglio KF, Amato R, Eastman R, Lim P, Sa JM, Guha R, Ganesan S, Dorward DW, Klumpp-Thomas C, McKnight C, Fairhurst RM, Roberts D, Thomas C, Simon AK. 2018. A single nucleotide polymorphism in the Plasmodium falciparum Atg18 gene associates with artemisinin resistance and confers enhanced parasite survival under nutrient deprivation. Malar J 17:391. doi: 10.1186/s12936-018-2532-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pradhan A, Siwo GH, Singh N, Martens B, Balu B, Button-Simons KA, Tan A, Zhang M, Udenze KO, Jiang RHY, Ferdig MT, Adams JH, Kyle DE. 2015. Chemogenomic profiling of Plasmodium falciparum as a tool to aid antimalarial drug discovery. Sci Rep 5:15930. doi: 10.1038/srep15930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Balu B, Chauhan C, Maher SP, Shoue DA, Kissinger JC, Fraser MJ, Adams JH. 2009. Piggybac is an effective tool for functional analysis of the Plasmodium falciparum genome. BMC Microbiol 9:83. doi: 10.1186/1471-2180-9-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Böhme U, Otto TD, Sanders M, Newbold CI, Berriman M. 2019. Progression of the canonical reference malaria parasite genome from 2002–2019. Wellcome Open Res 4:58. doi: 10.12688/wellcomeopenres.15194.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chappell L, Ross P, Orchard L, Russell TJ, Otto TD, Berriman M, Rayner JC, Llinás M. 2020. Refining the transcriptome of the human malaria parasite Plasmodium falciparum using amplification-free RNA-seq. BMC Genomics 21:395. doi: 10.1186/s12864-020-06787-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 36. Garbarino JE, Gibbons IR. 2002. Expression and genomic analysis of midasin, a novel and highly conserved AAA protein distantly related to dynein. BMC Genomics 3:18. doi: 10.1186/1471-2164-3-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. de Castro E, Sigrist CJA, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A, Hulo N. 2006. ScanProsite: detection of PROSITE signature matches and proRule-associated functional and structural residues in proteins. Nucleic Acids Res 34:W362–5. doi: 10.1093/nar/gkl124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Smilkstein M, Sriwilaijaroen N, Kelly JX, Wilairat P, Riscoe M. 2004. Simple and inexpensive fluorescence-based technique for high-throughput antimalarial drug screening. Antimicrob Agents Chemother 48:1803–1806. doi: 10.1128/AAC.48.5.1803-1806.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bennett TN, Paguio M, Gligorijevic B, Seudieu C, Kosar AD, Davidson E, Roepe PD. 2004. Novel, rapid, and inexpensive cell-based quantification of antimalarial drug efficacy. Antimicrob Agents Chemother 48:1807–1810. doi: 10.1128/AAC.48.5.1807-1810.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Witkowski B. n.d. Ring‐stage survival assays (RSA) to evaluate the in‐vitro and ex‐vivo susceptibility of Plasmodium falciparum to artemisinins, . In Institute Pasteur Du cambodge National Institutes of health procedure RSAv1 [Google Scholar]
- 41. Alexa A, Rahnenfuhrer J. 2016. TopGO: enrichment analysis for gene ontology
- 42. Oberstaller J. 2022. pfGO (version 1.1.0). doi: 10.5281/zenodo.5931520 [DOI]
- 43. Goodman CD, Su V, McFadden GI. 2007. The effects of anti-bacterials on the malaria parasite Plasmodium falciparum. Mol Biochem Parasitol 152:181–191. doi: 10.1016/j.molbiopara.2007.01.005 [DOI] [PubMed] [Google Scholar]
- 44. Goodman CD, McFadden GI. 2014. Ycf93 (orf105), a small apicoplast-encoded membrane protein in the relict plastid of the malaria parasite Plasmodium falciparum that is conserved in Apicomplexa. PLoS One 9:e91178. doi: 10.1371/journal.pone.0091178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bauer H, Fritz-Wolf K, Winzer A, Kühner S, Little S, Yardley V, Vezin H, Palfey B, Schirmer RH, Davioud-Charvet E. 2006. A fluoro analogue of the menadione derivative 6- [2'-(3'-methyl) -1',4'-naphthoquinolyl ] hexanoic acid is a suicide substrate of glutathione reductase. crystal structure of the alkylated human enzyme. J Am Chem Soc 128:10784–10794. doi: 10.1021/ja061155v [DOI] [PubMed] [Google Scholar]
- 46. Zhang M, Wang C, Otto TD, Oberstaller J, Liao X, Adapa SR, Udenze K, Bronner IF, Casandra D, Mayho M, Brown J, Li S, Swanson J, Rayner JC, Jiang RHY, Adams JH. 2018. Uncovering the essential genes of the human malaria parasite Plasmodium falciparum by saturation mutagenesis. Science 360:eaap7847. doi: 10.1126/science.aap7847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vannini A, Ringel R, Kusser AG, Berninghausen O, Kassavetis GA, Cramer P. 2010. Molecular basis of RNA polymerase III transcription repression by Maf1. Cell 143:59–70. doi: 10.1016/j.cell.2010.09.002 [DOI] [PubMed] [Google Scholar]
- 48. Xie SC, Metcalfe RD, Dunn E, Morton CJ, Huang S-C, Puhalovich T, Du Y, Wittlin S, Nie S, Luth MR, Ma L, Kim M-S, Pasaje CFA, Kumpornsin K, Giannangelo C, Houghton FJ, Churchyard A, Famodimu MT, Barry DC, Gillett DL, Dey S, Kosasih CC, Newman W, Niles JC, Lee MCS, Baum J, Ottilie S, Winzeler EA, Creek DJ, Williamson N, Parker MW, Brand S, Langston SP, Dick LR, Griffin MDW, Gould AE, Tilley L. 2022. Reaction hijacking of tyrosine tRNA synthetase as a new whole-of-life-cycle antimalarial strategy. Science 376:1074–1079. doi: 10.1126/science.abn0611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nyamai DW, Tastan Bishop Ö. 2019. Aminoacyl tRNA synthetases as malarial drug targets: a comparative bioinformatics study. Malar J 18:34. doi: 10.1186/s12936-019-2665-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chhibber-Goel J, Yogavel M, Sharma A. 2021. Structural analyses of the malaria parasite aminoacyl-tRNA synthetases provide new avenues for antimalarial drug discovery. Protein Sci 30:1793–1803. doi: 10.1002/pro.4148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ng CS, Sinha A, Aniweh Y, Nah Q, Babu IR, Gu C, Chionh YH, Dedon PC, Preiser PR. 2018. Trna epitranscriptomics and biased codon are linked to proteome expression in Plasmodium falciparum. Mol Syst Biol 14: e8009. doi: 10.15252/msb.20178009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kirsten Moll AK, Scherf A, Wahlgren M. 2013. EVIMalaR and MR4/ATCC, . In Methods in malaria research. 6th ed. [Google Scholar]
- 53. Balu B, Singh N, Maher SP, Adams JH. 2010. A genetic screen for attenuated growth identifies genes crucial for intraerythrocytic development of Plasmodium falciparum. PLoS One 5:e13282. doi: 10.1371/journal.pone.0013282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bronner IF, Otto TD, Zhang M, Udenze K, Wang C, Quail MA, Jiang RHY, Adams JH, Rayner JC. 2016. Quantitative insertion-site sequencing (QIseq) for high throughput phenotyping of transposon mutants. Genome Res 26:980–989. doi: 10.1101/gr.200279.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Amos B, Aurrecoechea C, Barba M, Barreto A, Basenko EY, Bażant W, Belnap R, Blevins AS, Böhme U, Brestelli J, Brunk BP, Caddick M, Callan D, Campbell L, Christensen MB, Christophides GK, Crouch K, Davis K, DeBarry J, Doherty R, Duan Y, Dunn M, Falke D, Fisher S, Flicek P, Fox B, Gajria B, Giraldo-Calderón GI, Harb OS, Harper E, Hertz-Fowler C, Hickman MJ, Howington C, Hu S, Humphrey J, Iodice J, Jones A, Judkins J, Kelly SA, Kissinger JC, Kwon DK, Lamoureux K, Lawson D, Li W, Lies K, Lodha D, Long J, MacCallum RM, Maslen G, McDowell MA, Nabrzyski J, Roos DS, Rund SSC, Schulman SW, Shanmugasundram A, Sitnik V, Spruill D, Starns D, Stoeckert CJ, Tomko SS, Wang H, Warrenfeltz S, Wieck R, Wilkinson PA, Xu L, Zheng J. 2022. VEuPathDB: the eukaryotic pathogen, vector and host bioinformatics resource center. Nucleic Acids Res 50:D898–D911. doi: 10.1093/nar/gkab929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lambros C, Vanderberg JP. 1979. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J Parasitol 65:418–420. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Growth screen.
RNAseq correlations.
RNAseq sample stage analysis.
Correlation of RNAseq samples.
Correlation to housekeeping pathways.
GO 6 hrs.
tRNA ligase expression.
Tables of quantitative data sets.
Data Availability Statement
RNAseq data generated for NF54 and MRST mutants in this study have been deposited to the NCBI Gene Expression Omnibus (GEO) database with the accession number GSE223497. Processed RNAseq data are provided in Data Set S1 Tab 4 and Data Set S1 Tab 5. The IDC reference data can be obtained from Bozdech Z, Llinás M, Pulliam BL, Wong ED, Zhu J, DeRisi JL. 2003. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol 1:e5.
https://doi.org/10.1371/journal.pbio.0000005
WGS data for NF54 and MRST mutants in this study are available via the NCBI SRA with the BioProject ID PRJNA947639. The Plasmodium falciparum NF54 reference genome can be obtained from ftp://ftp.sanger.ac.uk/pub/project/pathogens/Plasmodium/falciparum/NF54/Assembly/V1_morphed/