Summary
The pancreatic islets are composed of discrete hormone producing cells that orchestrate systemic glucose homeostasis. Here we identify subsets of β cells using a single-cell transcriptomic approach. One subset of β cells marked by high CD63 expression is enriched for the expression of mitochondrial metabolism genes and exhibit higher mitochondrial respiration compared to CD63lo β cells. Human and murine pseudo-islets derived from CD63hi β cells demonstrate enhanced glucose-stimulated insulin secretion compared to pseudo-islets from CD63lo β cells. We show that CD63hi β cells are diminished in mouse models of and in humans with type 2 diabetes. Finally, transplantation of pseudo-islets generated from CD63hi but not CD63lo β cells into diabetic mice restores glucose homeostasis. These findings suggest that loss of a specific subset of β cells may lead to diabetes. Strategies to reconstitute or maintain CD63hi β cells may represent a potential anti-diabetic therapy.
Keywords: beta cell, type 2 diabetes, insulin, CD63, single cell RNA-Seq, beta cell subtype, beta cell heterogeneity, insulin secretion, CD81, islet transplantation
Introduction
Type 2 diabetes (T2D) is characterized by a significant decline in pancreatic β cell mass or function which results in hyperglycemia1. Whether and when certain individuals progress to T2D is multifactorial with genetic predispositions and acquired metabolic abnormalities playing a role2-5. In T2D, insulin resistance promotes metabolic stress leading to β cell failure through multiple cellular and molecular mechanisms such as mitochondrial dysfunction, endoplasmic reticulum (ER) stress, glucolipotoxicity, dedifferentiation, and death6-10. Recent technological advances in unbiased single cell analyses are reshaping our molecular understanding of the diversity of β cell outcomes in T2D.
Heterogeneity in β cell function and morphology11,12 have previously been appreciated13,14. Several studies reported differences in insulin secretion and biosynthesis and abnormal glucose metabolism15-18. It was noted that ~75% of β cells were glucose responsive and consistently secreted insulin in response to multiple glucose challenges19. NAD(P)H, a marker of metabolic activity discriminates between β cells with a lower glucose threshold and higher insulin secretion20, 21. Many of these “unresponsive” β cell populations had normal electrophysiologic responses and properties22. Interestingly, glucagon-like peptide 1 (GLP-1) pathway agonists were largely effective at rendering glucose competent previously glucose unresponsive β cells23.
Single cell RNA sequencing (scRNA-Seq) has been applied to pancreatic islet cells from human donors with or without T2D24-30 and mice24, 27, 31. Overall, these scRNA-Seq studies did not arrive at a consistent conclusion regarding the determination and extent of β cell heterogeneity32. This could be due to interindividual heterogeneity, relatively low number of β cells per donor, and/or low sequencing depth. However, a meta-analysis pooling data from multiple studies did indeed show two subtypes of β cells33. Camunas-Soler et al. coupled scRNA-Seq and patch-clamp electrophysiology analyses and found RBP4 on β cells to be inversely correlated with exocytosis and ETV1 elevated in donors with T2D34. While there has been much recent progress, our understanding of the cellular and molecular basis for functional β cell heterogeneity and how perturbations thereof contribute to normal physiology and T2D is still poorly defined.
We have assessed the single-cell transcriptomes of pancreatic islets in a mouse model of obesity and glucose intolerance to characterize the cellular and molecular perturbations induced by obesity that lead to T2D. Here we identify 4 subsets of β cells each with distinct molecular signatures. One β cell subset expresses high levels of mitochondrial and metabolic genes and is reduced in a mouse model of T2D. We reveal CD63 as a cell surface marker to enrich for this population of β cells and show that CD63hi β cells have augmented glucose metabolism and insulin secretion in humans and mice. Transplantation of pseudo-islets derived from CD63hi but not CD63lo β cells effectively corrects hyperglycemia in diabetic mice.
Results
Islet cell transcriptomes from lean and obese mice
To interrogate the transcriptional perturbations induced by mild diet-induced obesity (DIO) across specific endocrine pancreatic islet cell types and to determine the heterogeneity of the response within a cell type, we evaluated the transcriptomes of isolated pancreatic islet cells at single cell resolution. High fat diet (HFD) feeding for 6 weeks resulted in mild increases in body weight and fat mass and a decrease in lean mass (Extended Data Fig. 1a-c) in male mice. HFD-fed male mice also exhibited fasting hyperglycemia and hyperinsulinemia, glucose intolerance, and insulin resistance without significant changes in β cell mass compared to Regular diet (RD)-fed controls (Extended Data Fig. 1d-n). However, female mice did not show significant differences in those parameters after HFD feeding (Extended Data Fig. 1o-t). Thus, we performed scRNA-Seq on pancreatic islet cells from 10-week old male C57BL/6J mice fed RD or HFD (Fig. 1a). We sequenced the transcriptomes of 10,582 pancreatic islet cells (RD=6284, HFD=4298). Unsupervised graph-based clustering and marker gene expression identified the expected endocrine cell types, similarly represented in mice from both conditions, with β cells comprising more than 70% of the total (Fig. 1b and Extended Data Fig. 1u,v). The percentage of macrophages doubled from 1% in RD-fed mice to 2% with HFD, suggesting that DIO promotes a proinflammatory environment within the islets (Extended Data Fig. 1u).
Figure 1. scRNA-seq analysis reveals changes in β cell heterogeneity promoted by diet-induced obesity.
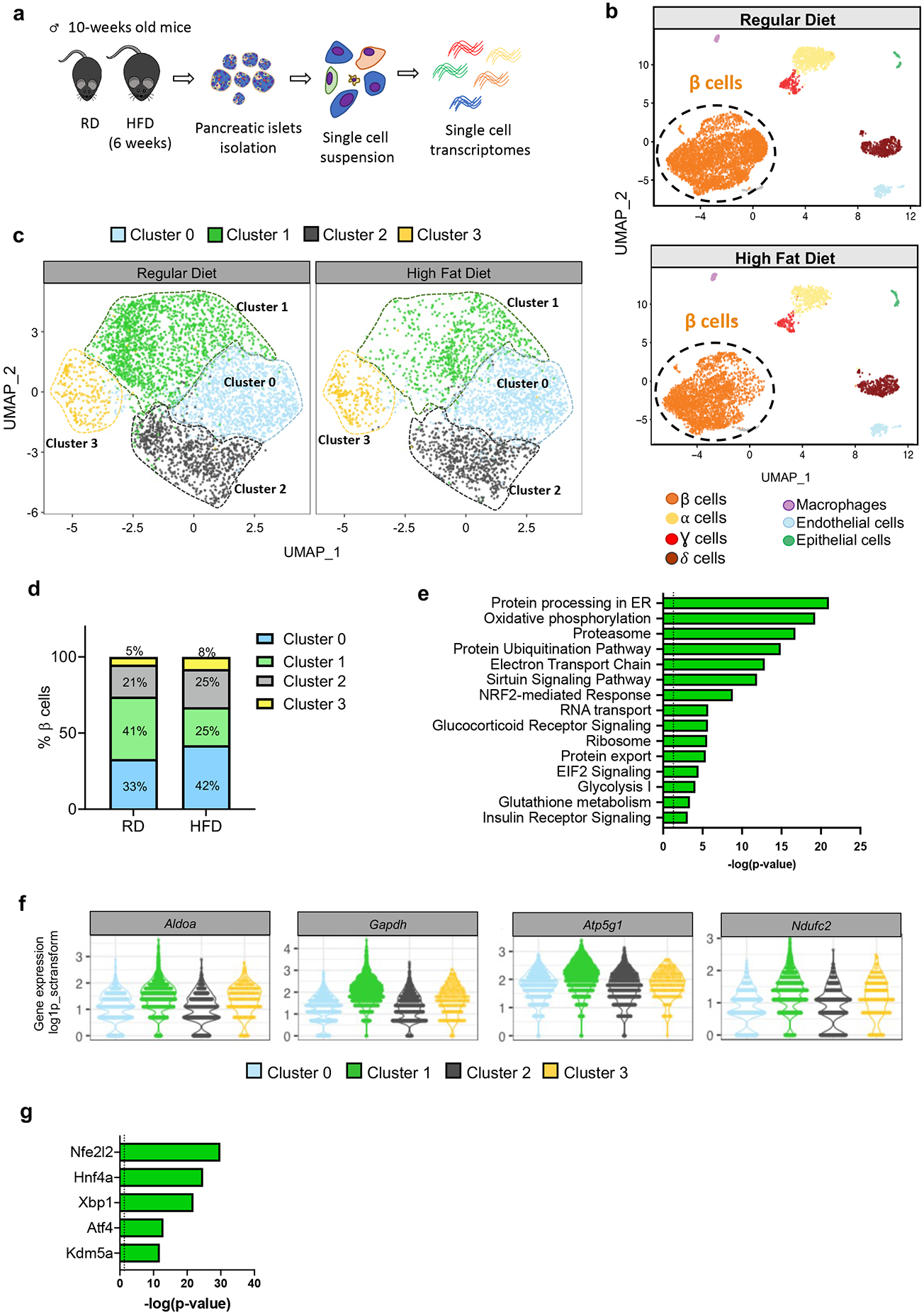
a, Schematic representation of the workflow used for single cell RNA-sequencing (scRNA-Seq) of pancreatic islet cells. b, Unsupervised clustering of single cell transcriptome visualized with uniform manifold approximation and projection (UMAP) analysis. Data represent pancreatic islet cells (N=10,582) from mice fed with Regular Diet (RD, N=6,284 cells pooled from 4 mice) or High Fat Diet (HFD, N=4,298 cells pooled from 5 mice) for 6 weeks after weaning. Annotated cell types are assigned based on known marker gene expression. c, Projection of β cells (N=7,564) from both RD and 6-weeks HFD-fed mice using UMAP analysis. β cell groups are defined according to kNN-based clusters. d, Distribution of β cell clusters in RD- and HFD-fed mice. e, Pathway enrichment analysis of differentially expressed genes in Cluster 1 β cells (p-value was calculated using two-tailed Benjamini-Hochberg). f, Violin plots representing the expression levels (log2(TPM+1)) of genes involved in glycolysis (Aldoa and Gapdh) and electron transport chain (Atp5g1 and Ndufc2) in each β cell cluster. g, Top 5 most statistically significant predicted transcription factors in Cluster 1 β cells (p-value was calculated using two-tailed Benjamini-Hochberg). Source numerical data are available in source data.
Diet-induced obesity and beta cell heterogeneity
Although there is evidence for β cell heterogeneity in both mouse and human, little is known about whether DIO affects β cell heterogeneity. To answer this question, we reperformed the clustering analysis, focusing only on β cells and defined 4 clusters, each present in RD- and HFD-fed mice (Fig. 1c,d).
To gain insight into the molecular differences that underly the four β cell clusters and infer function, we performed pathway analysis on the differentially expressed genes (DEGs) (FDR<0.001, fold change>1.5) for each cluster (Extended Data Fig. 2a-d). Cluster 0 cells are enriched for genes involved in proliferation and β cell development35-37 (Extended Data Fig. 2a-d). Four of the top five transcription factors (TFs) predicted by Upstream Analysis of Ingenuity Pathway Analysis Studio (IPA)38 are associated with β cell development39,40 (Extended Data Fig. 2e). β cells from Cluster 1 were enriched for genes that regulate glucose metabolism, mitochondrial metabolism, and protein synthesis/processing, which are all critical components for glucose-stimulated insulin secretion (GSIS) (Fig. 1e,f). Other notable pathways found in Cluster 1 β cells relate to insulin synthesis such as RNA transport, ribosomes, elongation of protein, eIF2 signaling, protein folding, protein processing in the ER, glutathione metabolism41, sirtuin signaling42, 43, and Nrf2 antioxidant pathways44 (Fig. 1e and Extended Data Fig. 2a). Analysis of the upstream TFs that might regulate Cluster 1 β cells revealed Nrf2 (Nfe2l2), Hnf4a, Xbp1, Atf4, and Kdm5a (Fig. 1g). Notably, Nrf2 regulates antioxidant defenses, mitochondrial activity, and β cell function, key processes observed in Cluster 1. Loss of function mutations in HNF4A result in impaired GSIS and cause maturity-onset diabetes of the young 1 (MODY1)45-47. Finally, XBP1 and ATF4 play critical roles in β cell function and survival by regulating the ER stress response48, 49. These data suggest that Cluster 1 β cells may represent a β cell subset with enhanced metabolic activity and play an important role in insulin secretion and handling metabolic stress.
In addition to Clusters 0 and 1 that represent ~75% of total β cells, β cells in clusters 2 and 3 comprised the remaining β cells and showed lower expression of β cell signature genes such as Pdx1, Nkx2-2, Nkx6-1, and Neurod1 compared to Clusters 0 and 1 (Extended Data Fig. 2b). Cluster 2 showed the highest expression of Ins1, Ins2, Pam and Iapp (Extended Data Fig. 2c). The top pathways in Cluster 2 β cells indicated the enrichment of many genes with important roles in the pathogenesis of diabetes (Extended Data Fig. 2d). Upstream pathway analysis identified TFs involved in neuroendocrine development (Extended Data Fig. 2e). Cluster 3 β cells expressed higher levels of Cd81, a marker of immature/stressed β cells50, and were enriched in pathways related to ribosomal activity and oxidative and ER-stress, while showing a dramatic reduction in the expression of β cell maturation genes, suggesting that Cluster 3 cells might represent dysfunctional or less mature β cells (Extended Data Fig. 2b-d).
Trajectory analyses by pseudo-time ordering suggested that clusters 2, 0, and 1 track along a linear molecular continuum whereas cluster 3 branched out on another path, indicating marked differences in their transcriptome content (Extended Data Fig. 2f). Taken together, our single cell analysis allowed us to describe four clusters of β cells with gene sets from each cluster suggesting distinct biological functions. Cluster 1 may represent β cells in a fate/state enriched for genes associated with increased metabolic and mitochondrial activity as well as protection against byproducts produced in the process of insulin synthesis and secretion. On the other hand, Cluster 0 exhibited higher expression of genes related to β cell proliferation and differentiation. Cluster 3 may represent immature/stressed β cells. Furthermore, our data suggested there may be a shift in the frequencies of these β cell subsets in T2D with a decline in the proportion of Cluster 1 cells and an increase in Clusters 0 and 3 with mild DIO.
Identification of a metabolically active β cell subset
Given the transcriptomic profile of Cluster 1 β cells with genes playing important roles in glucose metabolism and insulin secretion, we hypothesized that these β cells are robust insulin secretors. To molecularly characterize and functionally assess Cluster 1 β cells, we searched for cell surface markers that were enriched in this population and identified Cd63 as the most significant DEG (Fig. 2a and Extended Data Fig. 2c). Immunofluorescence staining for CD63 on pancreatic tissue sections demonstrated that CD63 protein is mainly expressed on β cells and that some β cells express higher levels than others (Fig. 2b and Extended Data Fig. 3a). Cd63 is also expressed on other pancreatic islet cells (Extended Data Fig. 3b). Next, we found that ~40% of β cells are CD63hi with no major differences in frequency across the head, body, and tail of the pancreas (Extended Data Fig. 3c). Flow cytometry of islet cells from MIP-GFP mice and revealed robust membrane expression of CD63 on β cells (Extended Data Fig. 3d). We next FAC-sorted β cells from WT mice according to high and low levels of CD63 expression (top and bottom ~1/3) and indeed detected higher Cd63 mRNA expression in CD63hi β cells compared to CD63lo β cells (Fig. 2c-e and Extended Data Fig. 3e)
Figure 2. Identification of a metabolically active β cell cluster.
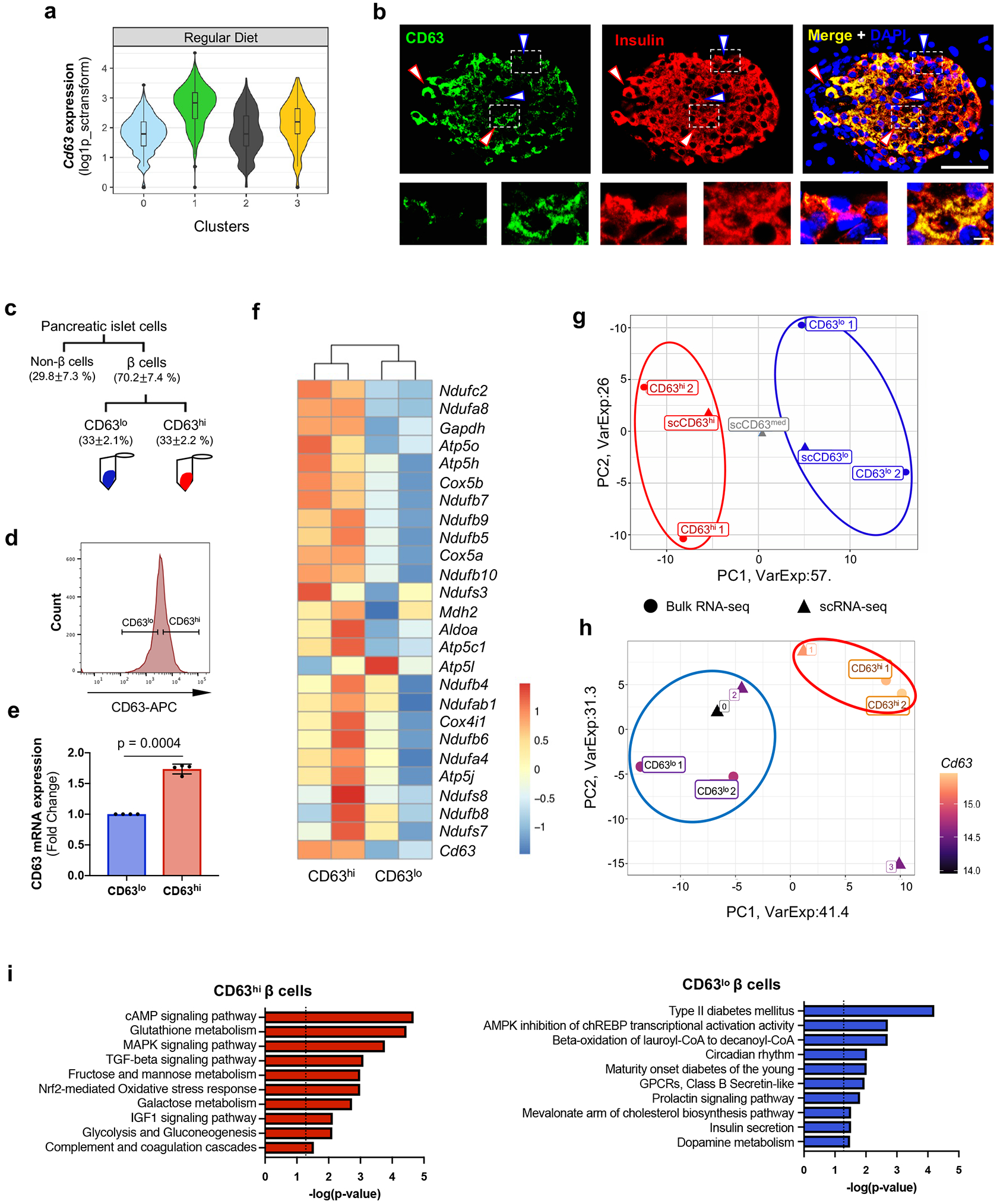
a, Violin plots showing Cd63 expression across β cell clusters in Regular Diet (RD) fed mice. Minimum to maximum values are shown by the whiskers, the bounds of boxes represent the first and third quartiles and the center line indicates the median. b, CD63 protein expression in β cells determined by immunofluorescence staining for CD63 (green), Insulin (red), and DAPI (blue) from RD-fed mice (10-weeks old) (Scale bar, 50 μm) (Magnified insets shown below. Scale bar, 5 μm). This is representative of more than 20 fields (N = 10 mice). Blue arrows mark CD63lo β cells and red arrows mark CD63hi β cells. c, Schematic for the FACS analysis to isolate CD63hi and CD63lo β cells from mouse pancreatic islets. d, Histogram of CD63 expression in β cells from mouse β cells. e, Cd63 mRNA expression in sorted CD63hi and CD63lo β cells determined by RT-qPCR (N=4 independent experiments per group). Data are presented as mean± S.E.M. Paired two-tailed t test is used for comparison. ***p < 0.001. f, Heatmap of glycolysis, TCA cycle, and mitochondrial genes from FAC-sorted CD63hi and CD63lo β cells obtained through bulk RNA-Seq (N=2 per group). The significance of DEGs between pairwise comparisons were identified by Wald test using DESeq2 v1.26.0. Benjamini–Hochberg corrected two-tiled p-values<0.05 were considered statistically significant g, PCA scatter plot comparing the transcriptomes of FAC-sorted CD63hi and CD63lo β cells obtained through bulk RNA-Seq to scRNA-Seq analyses of CD63hi, CD63med, and CD63lo β cells. (N=2, each replicate is pooled from 5 mice).. h, PCA scatter plot comparing the transcriptomes of FAC-sorted CD63hi and CD63lo β cells obtained through bulk RNA-Seq to β cell Clusters 0, 1, 2 and 3 obtained by scRNA-Seq. i, Pathway enrichment analysis of differentially expressed genes in CD63hi and CD63lo β cells ordered by statistical significance (p-value was calculated using two-tailed Benjamini-Hochberg). Source numerical data are available in source data.
Similar to Cluster 1 β cells, bulk RNA-Seq analyses of CD63hi β cells showed elevated expression of glycolysis, TCA cycle, and mitochondrial genes compared to CD63lo β cells (Fig. 2f). This raised the possibility that CD63hi β cells represent Cluster 1 β cells and are amenable to FACS-isolation using CD63 as a marker. We compared the transcriptomes of FAC-sorted CD63hi and CD63lo β cells obtained through bulk RNA-Seq to CD63hi, CD63med, and CD63lo β cells procured by scRNA-Seq. Principal component analysis (PCA) revealed that CD63hi β cells from scRNA-Seq and FAC-sorted experiments have similar gene expression patterns (Fig. 2g). Likewise, FAC-sorted CD63lo β cells and CD63lo β cells from scRNA-Seq clustered closely together with CD63med β cells falling between the CD63hi and CD63lo populations. Next, we compared the FAC-sorted CD63hi and CD63lo β cells (bulk RNA-Seq) to the four β cell clusters (scRNA-Seq). Unsupervised clustering revealed a similar gene expression pattern between FAC-sorted CD63hi β cells and Cluster 1 β cells whereas FAC-sorted CD63lo β cells exhibited a similar gene signature compared to β cells from Clusters 0 and 2 (Fig. 2h). Because Cd63 is also expressed on alpha, delta, and gamma cells (Extended Data Fig. 3b), we assessed the purity of FAC-sorted CD63hi and CD63lo β cells and found they contained about 97-98% β cells (Supplementary table 1). Furthermore, Arx and Hhex were detectable at very low levels and 20-50 fold lower than β cell genes that were not significantly different between CD63hi and CD63lo sorted β cells.
Having shown that FAC-sorted CD63hi β cells and Cluster 1 β cells are similar, we used the more sensitive bulk RNA-Seq results to gain further molecular insight into the transcriptomic profile. We found 667 DEGs between CD63hi and CD63lo β cells with an enrichment of immune-related genes. Since CD63 is highly expressed on macrophages (Extended Data Fig. 3b), we cannot exclude minor immune cell contamination in the CD63hi population. To assess the transcriptome of β cells in the CD63hi population, we excluded those genes that are highly expressed on macrophages 51 (Extended Data Fig. 4a,b). CD63hi β cells are enriched in genes related to cAMP-mediated signaling, glutathione metabolism, MAPK and TGF-β signaling pathways, Nrf2-mediated response, monosaccharide metabolism, glycolysis, and complement and coagulation cascades (Fig. 2i). CD63lo β cells were enriched in genes associated with T2D, AMPK inhibition of ChREBP transcriptional activity, beta-oxidation, circadian rhythm, and maturity onset diabetes of the young (MODY) (Fig. 2i). Next, we compared the pathways enriched in FAC-sorted β cells and in the different clusters. Both Cluster 1 and CD63hi β cells showed an enrichment for pathways related to glycolysis, TCA cycle and Nrf2-mediated response, while CD63lo β cells and β cells from Clusters 0 and 2 exhibited an enrichment in MODY signaling, hepatic fibrosis, and T2D and NF-κB signaling pathways (Extended Data Fig. 4c). Since the read depth in scRNA-Seq is relatively shallow, we used bulk RNA-Seq data to identify differentially expressed TFs (DET) in CD63hi β cells. We found 18 DETs upregulated and 9 DETs downregulated in CD63hi compared to CD63lo β cells (Extended Data Fig. 4d). 16 of the top 20 TF pathways identified in CD63hi β cells were also significantly enriched in Cluster 1 β cells (Extended Data Fig. 4e,f). Together, these results demonstrate a shared transcriptional signature between CD63hi and Cluster 1 β cells, highlighting mitochondrial activity and glucose metabolism pathways. Importantly, FACS isolation of CD63hi β cells enables purification of Cluster 1 β cells, targeting CD63 as a surface marker.
Next, we assessed CD63 expression across β cell populations characterized by differential maturation from published datasets52, 53 and found that mature β cells exhibited higher expression of Cd63 in comparison to immature cells (Extended Data Fig. 5a,b). Furthermore, we found that Cd63 expression on β cells increases postnatally (Extended Data Fig. 5c,d)53, 54. These data suggest that CD63 represents a marker for mature β cells.
CD63hi β cells exhibit enhanced insulin secretion
The gene expression suggested that Cluster 1 and CD63hi β cells may have enhanced metabolic and mitochondrial activity compared to other β cell populations. We evaluated mitochondrial mass and mitochondrial membrane potential in CD63hi and CD63lo β cell subsets and found that CD63hi β cells exhibited elevated mitochondrial mass as measured by MitoTracker Green and transmission electron microscopy (TEM) compared to CD63lo β cells (Fig. 3a-d). To evaluate mitochondrial activity, we stimulated β cells with a high concentration of glucose and assessed mitochondrial membrane potential using MitoTracker Red CMXRos. β cells with higher mitochondrial membrane potential displayed 5-fold higher CD63 expression compared to β cells with lower mitochondrial membrane potential (Extended Data Fig. 6a). After glucose stimulation, CD63hi β cells showed augmented NAD(P)H autofluorescence, as a real time measure of glucose metabolism, whereas CD63lo β cells remained at levels close to basal (Extended Data Fig. 6b). Insulin expression in β cells is known to be heterogenous with high and low insulin cell subsets14, 55. CD63hi β cells exhibited increased intracellular granularity compared to CD63lo β cells (Extended Data Fig. 6c). CD63lo β cells showed increased density of immature insulin granules in comparison with CD63hi β cells (Fig. 3e).
Figure 3. Functional characterization of a metabolically active β cell cluster.
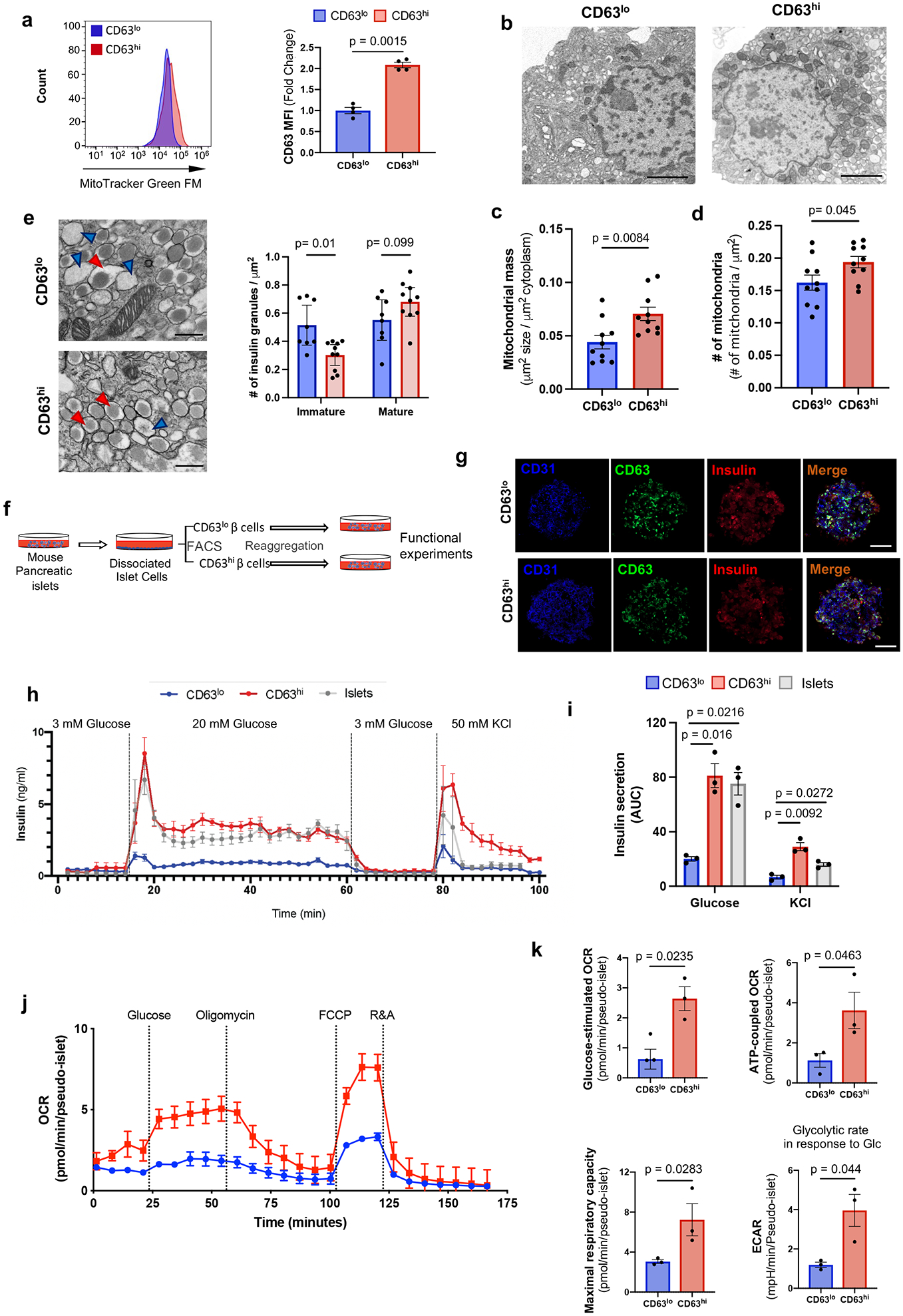
a, Mitochondrial mass in CD63hi and CD63lo β cells assessed by Mitotracker green (MTG). Results are representative of 4 independent experiments. Data are presented as mean±S.E.M. Paired two-tailed t test is used for comparison. b-d, Transmission electron microscopy (TEM) of FAC-sorted CD63hi and CD63lo β cells (Scale bars, 2 μm) (b). Mitochondrial mass (c) and numbers (d) were counted in a blinded manner from 10 fields per group with about 3 cells per field. Welch’s unpaired two-tailed t test is used for comparison. e, Representative TEM images of FAC-sorted CD63hi and CD63lo β cells from mice fed with RD. Blue arrows point to immature secretory granules. Red arrows indicate mature insulin secretory granules. Magnification 30,000x (Scale bars, 500 nm). Quantification of insulin granules normalized by area (right panel). Welch’s unpaired two-tailed t test is used for comparison. Results of granules from CD63lo β cells (8 fields) and CD63hi (10 fields) were pooled from 5mice. f, Schematic illustration of strategy to assemble pseudo-islets from FAC-sorted CD63hi and CD63lo β cells prior to functional assays. g, Representative immunofluorescence images of pseudo-islets showing Insulin (Red), CD63 (Green) and CD31 (Blue) (Scale bars, 50 μm). N=5 pseudo-islets per group. h, Dynamic glucose-stimulated insulin secretion assay of perifused CD63hi and CD63lo pseudo-islets, and whole islets (20 pseudo-islets or whole islets per replicate). Results are representative of 3 independent experiments with 3 replicates. i, Area under the curve (AUC) for insulin secretion in response to glucose and KCl from 3 different perifusion experiments. Data are presented as mean±S.E.M. Paired two-tailed t test is used for comparison. j, Oxygen consumption rates (OCR) of CD63hi and CD63lo pseudo-islets normalized to the number of pseudo-islets per well (20 pseudo-islets). Results are representative of 3 independent experiments. k, Quantification of glucose-stimulated OCR, ATP-coupled OCR, maximal respiratory capacity, and glycolytic rate in response to 20 mM glucose (at least 4 replicates in 3 independent experiments). Data are presented as mean±S.E.M. Unpaired two-tailed t test is used for comparison. Source numerical data are available in source data.
Analysis of β cells from MIP-GFP mice revealed that β cells with higher insulin content (MIP-GFPmed/high) were predominantly CD63hi rather than CD63lo β cells (Extended Data Fig. 6d). In contrast, CD63lo β cells represented the majority of β cells with lower insulin content (MIP-GFPlo). To directly assess insulin secretion in CD63hi and CD63lo β cells, we FAC-sorted CD63hi and CD63lo β cells and reaggregated them with human umbilical vein endothelial cells (HUVEC) to obtain CD63hi and CD63lo pseudo-islets (Fig. 3f,g and Extended Data Fig. 6e). We performed GSIS assays and found that CD63hi pseudo-islets robustly secreted 2.5-fold more insulin and displayed a modestly higher but non-significant trend in insulin content compared to CD63lo pseudo-islets (Extended Data Fig. 6f,g). In a higher resolution and dynamic GSIS, both CD63hi pseudo-islets and whole islets demonstrated 3-4-fold higher insulin secretion in response to both glucose and KCl than CD63lo pseudo-islets (Fig. 3h,i). Additionally, both phase I and phase II of GSIS were augmented in CD63hi pseudo-islets. CD63hi pseudo-islets also exhibited increased oxygen consumption rates (OCR) (ATP-coupled respiration, maximal respiratory capacity, and glycolytic rates) and extracellular acidification rates (ECAR) compared to CD63lo pseudo-islets (Fig. 3j,k). CD63hi pseudo-islets expressed higher levels of GAPDH, a key glycolytic enzyme involved in GSIS compared to CD63lo pseudo-islets (Extended Data Fig. 6h). Together, these data demonstrate that CD63hi β cells exhibit elevated mitochondrial respiratory and glycolytic activity and GSIS compared to CD63lo β cells.
To test whether the insulin secretion defect in CD63lo β cells was due to diminished cell viability or alternatively differences in the insulin secretion machinery, we assessed cell death and found no differences between CD63hi and CD63lo β cells (Extended Data Fig. 7a). Additionally, we performed GSIS with the addition of the cAMP phosphodiesterase inhibitor 3-isobutyl-1-methylxanthine (IBMX). IBMX strongly potentiated insulin secretion in both CD63hi and CD63lo β cells resulting in similar insulin secretion (Extended Data Fig. 7b). These results demonstrate that CD63lo β cells are viable and functional β cells that can secrete insulin mediated by cAMP signaling. Next, we searched for genes associated with the regulation of insulin secretion in CD63lo and Cluster 0 β cells and found that Glp1r expression was higher in CD63lo compared to CD63hi β cells (Extended Data Fig. 7c,d). To evaluate whether GLP-1 receptor (GLP1R) may play a role in regulation of insulin secretion in CD63lo β cells, we performed GSIS assays with the GLP1R agonist Exendin-4. Akin to IBMX stimulation, Exendin-4 enhanced insulin secretion in both CD63hi and CD63lo β populations, indicating that reduced insulin secretion observed in CD63lo β cells can be modulated by activation of GLP1R (Extended Data Fig. 7e).
CD63 marks human β cells with augmented insulin secretion
To test whether CD63 also marks human β cells with increased glucose metabolism and GSIS, we assessed non-diabetic human β cells by immunofluorescence and found human CD63hi and CD63lo β cells (Fig. 4a, Extended Data Fig. 8a, Supplementary Table 2). Flow cytometry from islets demonstrated a variance in cell surface CD63 expression among human β cells (Fig. 4b). Then we FAC-sorted human β cells according to high and low CD63 expression (top and bottom ~1/3) and observed that CD63hi β cells displayed higher Cd63 mRNA expression compared to CD63lo β cells (Fig. 4c and Supplementary Tables 3-4). Furthermore, human CD63hi β cells exhibited higher NAD(P)H levels and intracellular granularity, suggesting increased glucose metabolism and insulin secretion compared to CD63lo β cells (Fig. 4d,e). GSIS assays demonstrated that human CD63hi pseudo-islets robustly secreted insulin in the presence of 20 mM glucose. In contrast, GSIS was markedly lower in CD63lo pseudo-islets (Fig. 4f). We found no significant difference in insulin content between human CD63hi and CD63lo pseudo-islets (Fig. 4g). Dynamic cell perifusion revealed that CD63hi pseudo-islets, similar to whole islets, robustly secreted 3-fold more insulin than CD63lo pseudo-islets in response to glucose and 2-fold more to KCl (Fig. 4h,i). In addition, we found that islet CD63 mRNA is positively correlated with insulin secretion in 14 non-diabetic donors (Fig. 4j and Supplementary Table 5).
Figure 4. CD63 marks human beta cells with enhanced insulin secretion.
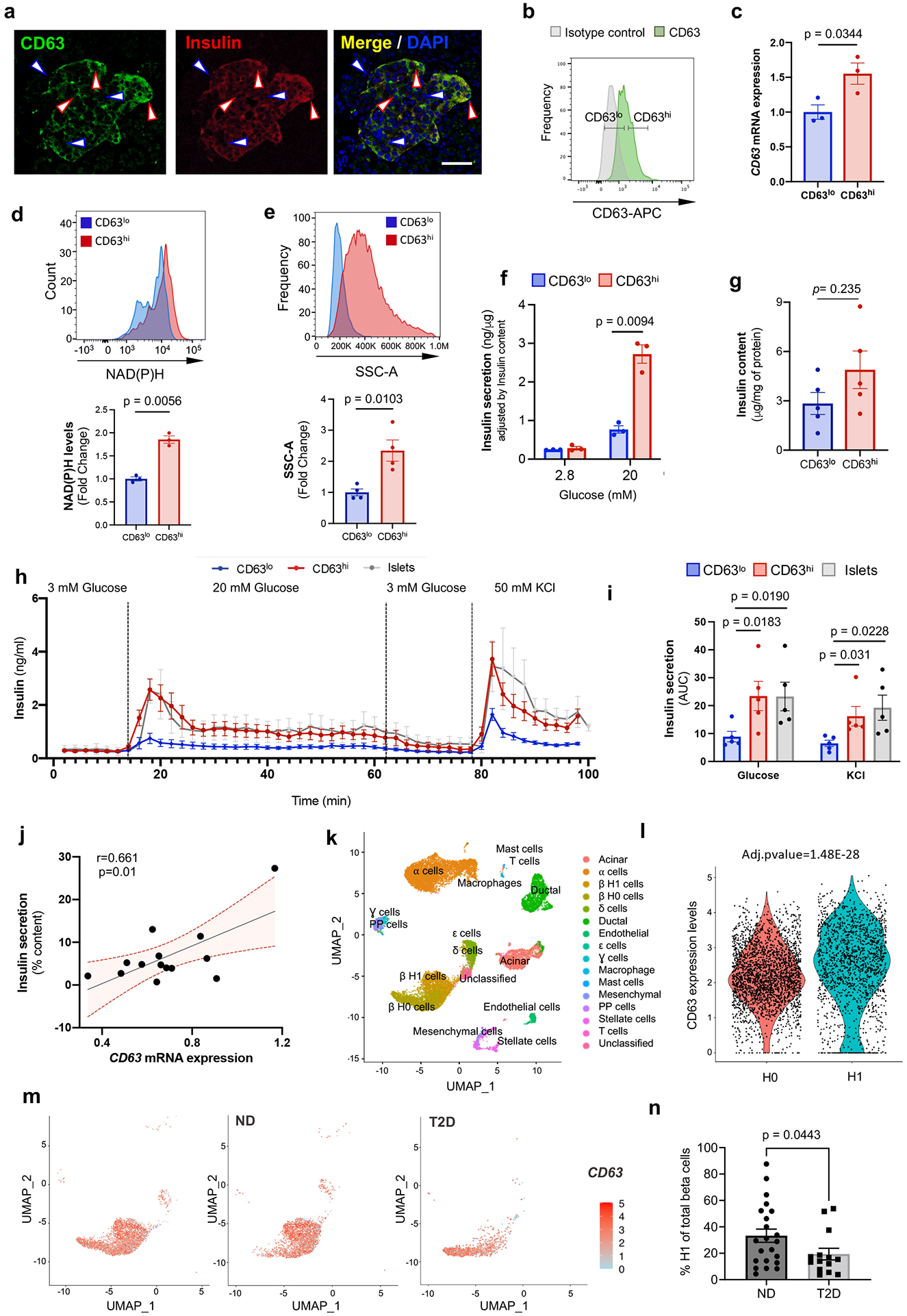
a, Immunofluorescence for CD63 (green), Insulin (red), and DAPI (blue) in human β cells (Scale bar, 50 μm). Blue arrows (CD63lo β cells). Red (CD63hi β cells). Representative of more than 10 fields. b, Flow cytometry analysis of CD63 in human β cells. Representative of 5 independent experiments. c, CD63 expression in sorted human CD63hi and CD63lo β cells determined by RT-qPCR (N=3 independent experiments) d, NAD(P)H levels in human CD63hi and CD63lo β cells. (N=3 independent experiments). e, Analysis of cellular granularity in human CD63hi and CD63lo β cells assessed by side scatter (SSC) (N=4 independent experiments). f, Static glucose-stimulated insulin secretion (GSIS) assay in CD63hi and CD63lo human pseudo-islets at 2.8 or 20 mM glucose (N=3 independent experiments). g, Insulin content of CD63hi and CD63lo human pseudo-islets normalized by intracellular protein (N=5 samples per group). h, Dynamic GSIS in CD63hi and CD63lo human pseudo-islets subjected to indicated concentrations of glucose and KCl (20 pseudo-islets or whole islets per replicate. i, Area under the curve (AUC) for insulin secretion in response to 20mM glucose and 50mM KCl shown in h (N = 5 independent experiments [5 donors]). j, Correlation between human islet CD63 expression and insulin secretion to 16.7mM glucose (N=14 donors). Linear regression with Pearson correlation coefficient analysis (bars indicate the 95% confidence intervals). k, UMAP projection of human pancreatic cells from integrated analysis of five human pancreatic islet scRNA-Seq studies across non-diabetic (ND, N=29) and type 2 diabetes (T2D, N=15) donors. l, Violin plots for CD63 in H0 and H1 β cell subclusters. m, CD63 expression in β cells in ND and T2D donors using UMAP analysis. All β cells on left panel. n, Frequency of Cluster H1 cells in ND (N=22) and T2D (N=14) donors with at least 13 β cells. Data are presented as mean±S.E.M and paired two-tailed t test is used for comparison in c, d, e, f, g, and i. Welch’s unpaired two-tailed t test is used for comparison in m. Source numerical data are available in source data.
There may be differences in the frequencies of CD63hi and CD63lo β cells between human donors. We reperformed a meta-analysis of 5 scRNA-Seq studies on human pancreatic islets from 29 donors without diabetes and included 15 donors with T2D (Fig. 4k and Extended Data Fig. 8b-e)33. We identified two subsets of β cells, H0 and H1, that are both present in donors with or without T2D (Extended Data Fig. 8c). CD63 is expressed at significantly higher levels in H1 compared to H0 β cells (Fig. 4l and Extended Data Fig. 8c,d). Importantly, H1 β cells represent about 35% of all β cells in donors without diabetes compared to only 20% in donors with T2D, though the variation within each group was large (Fig. 4m,n and Extended Data Fig. 8d,e).
Next, we evaluated for the potential impact of biological sex and did not find differences in islet CD63 expression, frequency of H1 β cells, and BMI between male and female donors (Extended Data Fig. 8f-h).
H1 β cells were also enriched for genes related to ribosome, VEGF, oxidative phosphorylation, mRNA processing, and actin cytoskeleton (Extended Data Fig. 8i). Next, we compared the DEGs in human H1 β cells with our two main murine clusters (0 and 1). Remarkably, mouse cluster 1 and human H1 β cells shared 196 genes associated with ribosome, electron transport chain, oxidative phosphorylation, proteasome degradation, and glycolysis and gluconeogenesis (Extended Data Fig. 8j). Evaluation of DEGs in H1 cells between ND and T2D donors revealed upregulation of nuclear receptor and AGE/RAGE pathway and downregulation of ribosomal, VEGF, and electron transport chain pathways (Extended Data Fig. 8k-m). Collectively, these results suggest human β cells with higher CD63 expression represent a subset with enhanced metabolic activity and insulin secretion that is reduced in T2D.
CD63hi beta cells are diminished in mouse models of T2D
We hypothesized that CD63 expression on β cells might be useful to track this β cell subset with increased insulin secretion in vivo. To determine the effects of obesity on expression of CD63 in β cells, we evaluated our scRNA-Seq data and found that Cd63 is downregulated in β cells from HFD compared to RD-fed mice (Fig. 5a). Using MIP-GFP mice, we confirmed a reduction in CD63hi β cells and a concomitant rise in CD63lo β cells in male mice fed with HFD for 6 weeks compared to RD (Fig. 5b). In addition, we co-stained pancreatic sections with CD63 and insulin and found a 2-3 fold reduction in the percentage of CD63hi β cells in mice fed a HFD for 6 and 12 weeks compared to RD controls (Fig. 5c). A separate genetic model of obesity and T2D that does not depend on HFD feeding showed that CD63hi β cells are dramatically reduced to less than 5% of all β cells in male db/db mice (Fig. 5d). Furthermore, we stained β cells for CD63 and CD81 to distinguish CD63hi and CD81+ β cells, which could be used as markers for Cluster 1 and 3 β cells, respectively. We found that CD63hi β cells are decreased while CD81+ β cells are increased with HFD (Extended Data Fig. 9a,b) similar to our scRNA-Seq analyses (Fig. 1d). HFD did not increase body weight, blood glucose levels, nor induce changes in the CD63hi β cell population in female mice (Extended Data Fig. 1f-j and 9c-e).
Figure 5. CD63 expression on beta cells in mouse models of T2D.
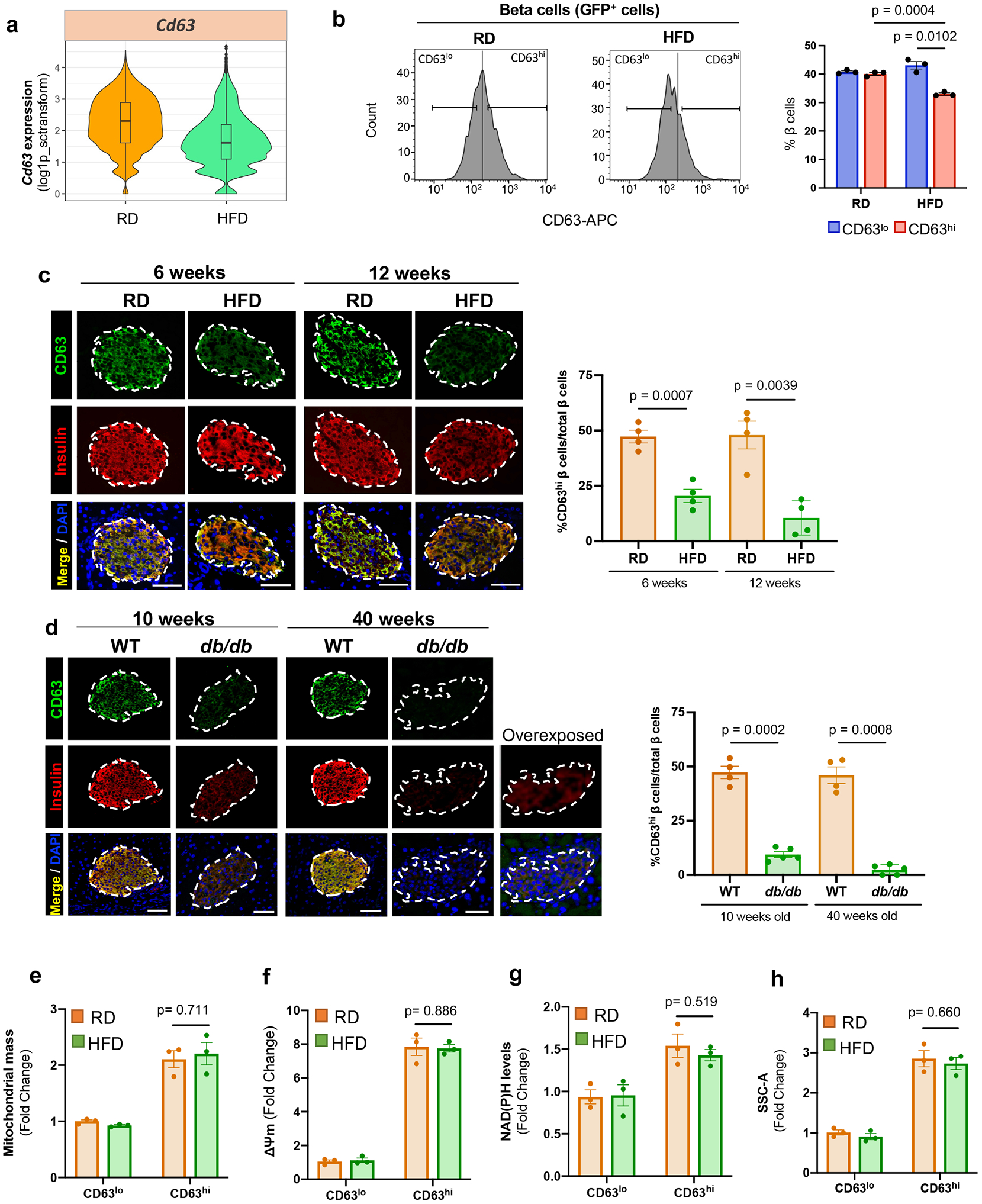
a, Violin plots showing Cd63 expression in β cells from regular diet (RD) and high fat diet (HFD) fed mice (scRNA-Seq data). Minimum to maximum values are shown by the whiskers, the bounds of boxes represent the first and third quartiles and the center line indicates the median. RD=4514 cells; HFD=3050 cells. b, Percentage of CD63hi and CD63lo β cells in MIP-GFP mice fed RD or HFD for 6 weeks assessed by flow cytometry (N=3 independent experiments). c, Representative immunofluorescence images showing CD63 (green) and insulin (red) in pancreas sections from WT mice fed with RD or HFD (6 and 12 weeks) (left panel) (Scale bars, 50 μm). Quantification of the percentage of β cells that are CD63hi (right panel) (N=4 mice per group) d, Representative immunofluorescence showing CD63 (green) and insulin (red) in pancreas sections from WT and db/db mice (10 and 40 weeks old) (left panel) (Scale bars, 50 μm). Quantification of the percentage of β cells that are CD63hi (right panel). WT (N=4 mice per group) and db/db (N=5 mice per group) mice (10 and 40 weeks old) . e, Mitochondrial mass in CD63hi and CD63lo β cells from RD and HFD-fed mice assessed by Mitotracker green (MTG). Results are representative of 3 independent experiments (5 mice per experiment). f, Mitochondrial membrane potential in CD63hi and CD63lo β cells from RD and HFD-fed mice previously incubated with 20 mM glucose. Results are representative of 3 independent experiments (5 mice per experiment). g, NAD(P)H fluorescence patterns of CD63hi and CD63lo β cells from RD and HFD-fed mice after incubation with 20 mM glucose. Results are representative of 3 independent experiments (5 mice per experiment). h, Analysis and quantification of cellular granularity in CD63hi and CD63lo β cells from RD and HFD-fed mice assessed by side scatter (SSC). Results are representative of 3 independent experiments (5 mice per experiment). Data are presented as mean ± S.E.M and Welch’s unpaired two-tailed t test is used for comparison unless otherwise stated. Source numerical data are available in source data.
Obesity diminishes Cd63 expression in the whole β cell population and specifically in the different β cell clusters (Extended Data Fig. 9f-k). We also compared our findings with previously published bulk RNA-Seq data from β cells obtained from mice fed with HFD56. Cd63 expression was severely reduced (log2FoldChange= −5.71e-01; p-value=7.21e-05) in β cells with 16 weeks HFD feeding. In addition, HFD induced a reduction in the expression of genes associated with pathways related to GSIS in the whole β cell population from our scRNA-Seq data and published bulk RNA-Seq data (Extended Data Fig. 9l). Next, we isolated CD63hi β cells from HFD-fed male mice to interrogate their functional capacity. CD63hi β cells from HFD-fed mice had similar mitochondrial mass, mitochondrial membrane potential, NAD(P)H autofluorescence, and granularity compared to those from RD-fed mice (Fig. 5e-h). In HFD-fed mice, there was a trend towards increased GSIS in CD63hi compared to CD63lo β cells (Extended Data Fig. 9m). However, CD63hi β cells from HFD-fed mice had diminished insulin secretion than those from RD-fed mice. Together, these results suggest that obesity has two effects on CD63hi β cells: 1) reduces the frequency of CD63hi β cells that are similar to Cluster 1 β cells, and 2) impairs insulin secretion but not all markers of β cell function.
We compared the transcriptomes of each β cell cluster between RD and HFD conditions and performed pathway enrichment analysis to determine how obesity alters gene expression within each cluster of β cells. All β cell clusters from HFD-fed mice showed decreased expression of genes in the oxidative phosphorylation pathway (Extended Data Fig. 9h-k), consistent with mitochondrial dysfunction in obesity. In cluster 1 β cells, HFD feeding led to a reduction in pathways involved in GSIS as well as an enrichment for genes involved in ferroptosis57 and FoxO signaling pathways58, 59 (Extended Data Fig. 9i). Collectively, these results suggest that DIO disrupts β cell homeostasis by both perturbing the ratio of β cell subsets and dysregulating genes critical for β cell function.
CD63hi beta cells restore euglycemia to diabetic mice
To evaluate the insulin secretory capacity of CD63hi β cells in vivo, we transplanted CD63hi and CD63lo pseudo-islets into NOD/SCID mice rendered diabetic by streptozotocin (STZ) (Fig. 6a). Diabetic mice transplanted with CD63hi pseudo-islets exhibited a marked reduction in blood glucose levels and reversal of weight loss compared to those receiving CD63lo pseudo-islets (Fig. 6b-d). Hyperglycemia reemerged after graft removal in CD63hi pseudo-islet transplanted mice, indicating that the engrafted CD63hi pseudo-islets were responsible for the improved blood glucose control. β cells from the explanted CD63hi pseudo-islets approximately two months later retained augmented CD63 expression compared to CD63lo pseudo-islets (Fig. 6e-g). Moreover, CD63hi pseudo-islets showed increased levels of Cd63 and genes involved in glycolysis and mitochondrial activity compared to CD63lo pseudo-islets (Fig. 6f). In addition, CD63hi pseudo-islets exhibited higher expression of core β cell genes such as Ins2, Pdx1, and Mafa in comparison to CD63lo pseudo-islets (Fig. 6h). Remarkably, CD63lo pseudo-islets displayed elevated levels of genes found in dedifferentiated60 and dysfunctional β cells3, 34 (Fig. 6i,j). These data indicate that transplantation of CD63hi but not CD63lo β cells can restore normoglycemia in mice with insulin-deficient diabetes. After 60 days, CD63hi β cells continue to exhibit a heightened metabolic gene expression program in stark contrast to CD63lo β cells, which show higher expression of dedifferentiation and stress genes.
Figure 6. CD63hi beta cells restores euglycemia to diabetic mice.
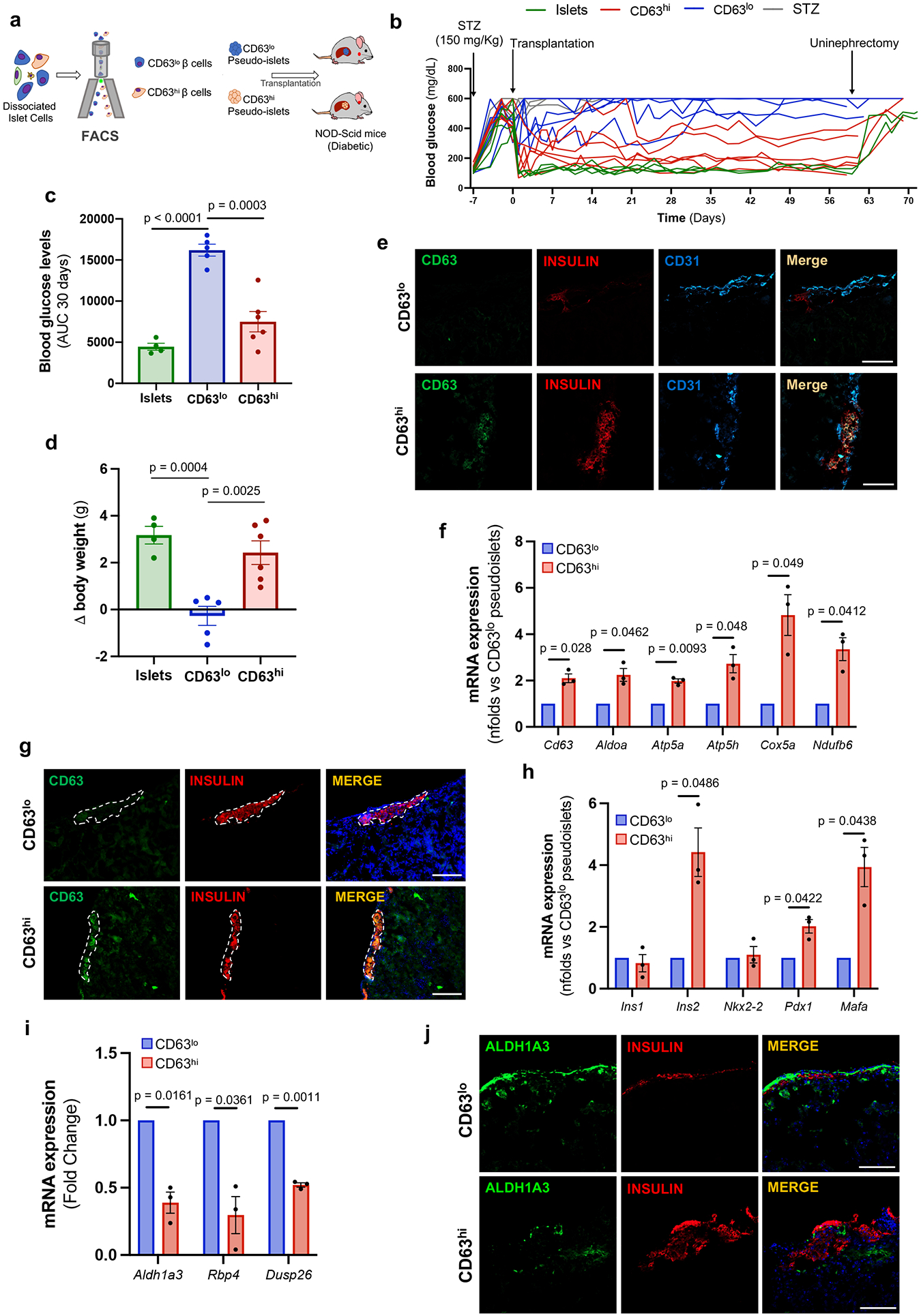
a, Schematic illustration of CD63hi and CD63lo pseudo-islet transplantation into diabetic NOD-SCID mice previously treated with streptozotocin (STZ). b, Random-fed blood glucose levels in mice transplanted with 150 whole islets (N=4; 2 were followed for 30 days), CD63lo pseudo-islets (N=5), or CD63hi pseudo-islets (N=6). Untransplanted STZ mice remained hyperglycemic (N=3). c, Area under the curve (AUC) of blood glucose levels for 30 days after transplantation until uninephrectomy. Whole islets (N=4), CD63lo pseudo-islets (N=5), or CD63hi pseudo-islets (N=6). Welch’s unpaired two-tailed t test is used for comparison. d, Change in body weight 30 days after transplantation in diabetic NOD-SCID transplanted with 150 whole islets (N=4), CD63lo pseudo-islets (N=5), or CD63hi pseudo-islets (N=6). Data are presented as mean±S.E.M. Welch’s unpaired two-tailed t test is used for comparison. e, Representative immunofluorescence images showing CD63 (green) and insulin (red) and CD31 (blue) in grafted kidney sections from NOD-SCID mice 60 days post-transplantation (N=3 per group) (Scale bars, 100 μm). f, mRNA expression of Cd63 and indicated metabolic genes in CD63hi and CD63lo pseudo-islets 60 days after transplantation determined by RT-qPCR. Paired two-tailed t test is used for comparison (N=3 per group). g, Representative immunofluorescence images of CD63 (green) and insulin (red) in grafted kidney sections 60 days post-transplantation (Scale bars, 100 μm) (N=3 independent experiments). h, mRNA expression ofβ cell signature genes in CD63hi and CD63lo pseudo-islets 60 days after transplantation determined by RT-qPCR. Paired two-tailed t test is used for comparison (N=3 per group). i, mRNA expression of indicated genes in grafted CD63hi and CD63lo pseudo-islets determined by RT-qPCR. Paired two-tailed t test is used for comparison (N=3 per group). j, Representative immunofluorescence images of Aldh1a3 (green) and insulin (red) in grafted kidney sections 60 days post-transplantation (Scale bars, 100 μm). Paired two-tailed t test is used for comparison (N=3 per group). Data are presented as mean±S.E.M. Source numerical data are available in source data.
Discussion
Our study identifies four subclusters of β cells each with their distinct molecular signature. We focus on one β cell subset marked by high CD63 expression, which we find to be reduced in mouse models of and in humans with T2D. Notably, this subset of β cells demonstrate enhanced GSIS and is enriched for genes in pathways that promote GSIS. In contrast, this subset of β cells shows lower transcript levels of genes associated with immature or dysfunctional β cells34, 50, 52. To ascertain the function of this β cell subset, we use CD63 as a molecular handle to functionally assess these β cells in T2D and further molecularly characterize them. It is unknown whether cell surface CD63 may be more than a molecular marker of β cells with enhanced glucose metabolism and insulin secretion. One report found that CD63 can target insulin granules to the lysosomes as part of in vitro stress-induced nascent granule degradation61. In contrast, our analyses focused on cell surface expression of CD63 and its mRNA levels in specific primary beta cell subclusters. It is possible that CD63 has multiple functions determined by its subcellular localization (late endosomes, lysosomes, secretory vesicles, and at the plasma membrane). Interestingly, other proteins from this family such as CD9 or CD81 have been used as markers to determine β cell populations50, 62, suggesting that tetraspanins may play a role in the detection of β cell subpopulations.
Functionally, CD63hi β cells have enhanced glucose metabolism and increased mitochondrial activity. Furthermore, CD63hi β cells show higher cellular granularity, lower numbers of immature insulin granules, and enhanced insulin secretory capacity. Through in silico approaches, we identify numerous TF pathways that are active in Cluster 1 and CD63hi β cells but have yet to determine which of these or other molecular pathways endow these β cells with their robust insulin secretory properties. It will be of considerable future interest to establish the essential molecular elements that ‘wire’ these Cluster 1 and CD63hi β cells as this knowledge could inform other ongoing approaches of cell-based therapies for T2D such as stem cell-derived islet/β cell transplantation.
In our study, male but not female B6 mice fed HFD for 6 weeks developed hyperglycemia. However, we did not observe increases in β cell mass, which has been variably reported with this duration of HFD feeding63, 64. This time period is also not sufficient to observe β cell dedifferentiation64. Additionally, engraftment of CD63hi but not CD63lo β cells into diabetic mice restores normoglycemia further demonstrating the critical role of CD63hi β cells. Of translational relevance to human physiology and disease, we confirm the presence of CD63hi β cells in human pancreatic islets and find they exhibit increased glycometabolic activity and insulin secretion. Finally, we demonstrate that humans with T2D have lower frequencies of CD63hi β cells compared to those without diabetes.
Previous studies have shown that β cells exhibit differential insulin secretory responses65. Subsequently different cell surface markers or fluorescent reporters have been used to describe β cell functional heterogeneity14, 53, 62, 66, 67. We find that Cluster 1 cells express high levels of Ucn3 and Fltp suggesting this subset of β cells may contain Ucn3+ and Fltp+ β cells53, 66. However, unlike the high insulin secretors found by Dorrell et al., cluster 1 β cells express high levels of both Cd9 and St8sia1. Several groups have performed scRNA-Seq in human pancreatic islets28, 30, 68, 69; however, only a few have described β cell heterogeneity and mostly in regards to select marker genes or UPR responses24-26. Pancreatic islets from human donors with T2D have only been assessed by scRNA-Seq in a few studies26, 28, 30 and no changes in β cell heterogeneity were found. Several reasons could account for these discrepancies such as shallow sequencing depth, inter-individual genetic and environmental differences, or differing stage of T2D30, 33, 70. A common caveat associated with scRNA-Seq analyses is that this technology does not provide a functional output. A Patch-Seq study by MacDonald and colleagues has integrated single cell functional secretion analysis followed by transcriptomic data acquisition and observed a population of dysfunctional β cells with high RBP4 and decreased exocytosis34. Here we performed our analysis in pancreatic islet cells from genetically identical mice and confirmed our findings in human samples. This was followed by combining our scRNA-Seq data with functional analyses, integrating β cell physiology and single cell transcriptomics.
In addition, we find three other clusters (0, 2 and 3) of β cells. Although β cells from Cluster 0 show high expression of β cell markers, these cells are enriched for genes involved in proliferation and development of β cells36, 71. Interestingly, both Cluster 0 and CD63lo β cells show higher expression of Glp1r. GLP-1 increases insulin secretion through a pleiotropic mechanism involving increased mitochondrial membrane potential and ATP production, improves access of insulin-secreting granules to the plasma membrane, and enhances β cell excitability72. Intriguingly, GLP-1 confers glucose sensitivity to glucose-resistant β cells, allowing membrane depolarization and subsequent insulin release23. Similarly, we find that Exendin-4 erases the difference in insulin secretion between CD63hi and CD63lo β cells , indicating a potential therapeutic approach for individuals with decreased frequency of CD63hi β cells. Farack et al. showed that a small fraction of “extreme” β cells with elevated Ins1, Ins2, and Iapp mRNA levels may contribute to insulin secretion in fasted states when blood glucose levels are normal55. We find that Cluster 2 β cells (21% of total β cells), highly express Ins1, Ins2, and Iapp in comparison with the rest of the clusters. Altogether, this data led us to postulate that this subset of β cells may partially contain the extreme β cell population. However, further studies are needed to test this hypothesis, since there is not currently a specific marker for extreme β cells.
Finally, we find that β cells from Cluster 3, increased in HFD-fed mice, are enriched for genes associated with immature and-stressed β cells such as Cd81 and show lower expression of β cell signature genes 34, 50, 52. Interestingly, Lickert and colleagues recently identified a population of immature, stressed, and dedifferentiated β cells with high levels of CD8150. Cluster 3 β cells and CD81+ β cells may have a high degree of overlap or represent the same population.
It is possible that there may be more than 4 subsets of β cells in mice that are either fewer in frequency or arise in higher numbers with age, advanced diabetes, or different strains of mice. Similarly, there may be more than two human β cell subsets that we show. Deeper sequencing and sequencing of more human β cells from donors of different ages, medical conditions, and ethnic backgrounds in the future may identify additional β cell subsets. In humans, this dataset cannot differentiate between the possibility that some individuals may naturally have lower numbers of H1 β cells at birth which predisposes them to T2D or that loss of H1 β cells with aging or metabolic stress triggers T2D. An alternative scenario is that the frequency of H1 β cells declines after T2D, marking the progression towards β cell failure.
Pseudo-time analysis suggests a trajectory of transcriptional phenotypes or states within the β cell clusters, progressing or maturing from clusters 2 to 0 to 1 whereas cluster 3 may represent a stressed state of β cells. However, whether the variations we observed in β cell clusters in T2D models are due to the presence of transient states or fixed subpopulations of β cells is currently unknown and represent a general challenge for the field. Trajectory analysis relies on the assumption that each subset is connected to one another. It is possible that this distinction between states or lineages may not be mutually exclusive. For example, one subset may be a distinct subtype/lineage with no interaction/crossover with others whereas other subsets may denote transient cellular states. A reduction in Cluster 1 β cells with DIO but without an appreciable change in β cell mass may signify plasticity. Our data show that CD63hi β cells remain CD63hi for two months after transplantation, suggesting a persistent identity or state. However, it is also possible that CD63hi β cells maintain their transcriptional profile because the recipient mice are no longer hyperglycemic. Lineage tracing analysis of each putative β cell cluster will ultimately be needed to test if those are transient states or fixed subpopulations.
After transplantation, CD63lo pseudo-islets displayed increased levels of genes associated with β cell dedifferentiation . Although the dedifferentiation of β cells has been observed in 10-week-old db/db mice, 6 weeks of HFD in C57BL/6 mice may not be sufficient to induce this process64. Future studies with longer duration of HFD or other mouse models of β cell failure integrating scRNA-Seq and lineage tracing will be a valuable approach to elucidate dedifferentiation. The cellular and molecular mechanisms leading to loss of Cluster 1 β cells in mice and H1 β cells in humans will be an important question to explore and has the potential to impact cell-based transplantation therapy in patients with T2D.
In summary, this study demonstrates that perturbations in β cell heterogeneity are associated with the development of T2D in mouse models and in humans. The identification of CD63 as a marker provides a valuable tool to target β cells with enhanced metabolic activity and insulin secretion. Thus, the number of CD63hi β cells may serve as a useful index to predict the risk of T2D or used to assess the success of therapeutic interventions. We do not yet know if T2D is correlated with the loss of human CD63hi β cells or that this β cell subset directly protects against T2D in people. Our data also suggest that treatment with GLP1R agonists may be a strategy to improve insulin secretion in CD63lo β cells. Lastly, it is possible that CD63hi and CD63lo β cells are present to coordinate optimal GSIS at low and high blood glucose concentrations, respectively.
Methods
All animal experiments performed in this study were approved by the Institutional Animal Care and Use Committee (IACUC) and Research Animal Resource Center at Weill Cornell Medical College. The experiments in this manuscript are in compliance with relevant guidelines and ethical regulations.
Animals.
C57BL/6J wild-type (WT) mice (Stock No. 000664), B6.BKS(D)-Leprdb/J (db/db) (Stock No. 000697), B6.Cg-Tg(Ins1-EGFP)1Hara/J (MIP-GFP) (Stock No. 006864), and NOD.Cg-PrkdcScid/J (NOD-Scid) (Stock No. 001303) were obtained from the Jackson Laboratory. Male and female mice were group-housed under a standard rodent chow at ambient temperature (22°C), 48% humidity in average and a 12 hour light-dark cycle in individually ventilated cages. Food and water were provided ad libitum. Body composition was analyzed using EchoMRI. Samples were obtained from female mice at 10 weeks of age and male mice at 10, 18 or 40 weeks of age. WT and MIP-GFP transgenic mice were fed a regular chow diet (RD; 5053, LabDiet) or a 60% high fat diet (HFD; D12492i, Research Diets) for 6 and 12 weeks after weaning. C57BL/6J wild-type fed with RD and HFD for 6 weeks were fasted 6 hours for glucose tolerance tests (GTT) and given an i.p. injection of D-Glucose (1g/kg). For insulin tolerance tests (ITT), mice were fasted for 4 hours and given an i.p. injection of insulin (0.75 units/kg body weight). Blood samples were taken at different times from the tail vein and measured with a glucometer.
Human Samples.
Formalin-fixed paraffin-embedded pancreas sections from non-diabetic donors were obtained from the Human Pancreas Analysis Program (HPAP; https://hpap.pmacs.upenn.edu) (Supplementary Table 2). Fresh and frozen human pancreatic islets were obtained through the Integrated Islet Distribution Program (IIDP) at City of Hope (Duarte, CA) (https://iidp.coh.org) (Supplementary Tables 3, 5). The HPAP and IIDP approved the use of human islets and required informed consent for research purpose from the donor relatives prior to offering the de-identified islets to established laboratories engaged in islet research and registered with IIDP. De-identified specimens are not considered human subjects research (NHSR) by the NIH. For the human pancreatic tissue slides, the organs were processed in the Human Islet Core at University of Pennsylvania following the guidelines of Clinical Islet Transplantation consortium protocol. The tissue biopsies were harvested from the pancreatic body and fixed in 10% formalin solution. The paraffin tissue blocks were cut into 5-mm sections for the immunofluorescence studies. This study used data from the Organ Procurement and Transplantation Network (OPTN). The OPTN data system includes data on all donor, wait-listed candidates, and transplant recipients in the US, submitted by the members of the Organ Procurement and Transplantation Network (OPTN). The Health Resources and Services Administration (HRSA), U.S. Department of Health and Human Services provides oversight to the activities of the OPTN contractor. The data reported here have been supplied by UNOS as the contractor for the Organ Procurement and Transplantation Network (OPTN). The content, interpretation, and reporting of these data is solely the responsibility of the authors and does not necessarily represent the official views of the OPTN or the National Institutes of Health.
Mouse pancreatic islet isolation and culture.
Mouse pancreatic islets were isolated by perfusion of the pancreases with CIzyme (VitaCyte) through the common hepatic duct and digested at 37°C for 17 minutes. After gradient-based separation using Histopaque-1077 (Sigma), islets were then hand-picked to avoid exocrine contamination and processed for different applications. For flow cytometry and FAC-sorted islet cells, dissociation was performed with 0.05% trypsin.
Human islet culture.
Human islets were cultured in islet medium overnight after delivery. After 1 day of recovery, human islets were hand-picked and processed for different applications. For FAC-sorted islet cells, dissociation was performed with 0.05% trypsin.
Single cell RNA sequencing.
Pancreatic islets were isolated and cultured overnight in RPMI 1640 supplemented with 10% FBS and penicillin/streptomycin. Then, islets were dissociated with 0.05% Trypsin (Corning) for 10 minutes at 37°C. Dissociated cells were passed through a 35 μm nylon mesh and resuspended in 1x PBS + 0.05% BSA. Cellular viability was higher than 95% as determined by using Trypan Blue assay and doublets represented less than 5% of total cells. Single cell suspensions were sequenced by the Chromium Single Cell 3’ Reagent Kit v3 (10x Genomics) and 10X Genomics’ Chromium Controller at the Weill Cornell Medicine Genomics Core Facility.
scRNA-Seq analysis.
The raw reads were aligned and processed with the CellRanger pipeline (v. 2.1.0) using the mouse transcriptome and genome version mm10. Subsequent analyses were performed in R following the recommendations of Amezquita et al. (https://osca.bioconductor.org/) using numerous functions provided in the R packages scater and scran73, 74. Based on the calculation of outliers, we removed cells with fewer than 1,000. In addition, cells with more than 7.5% mitochondrial reads were removed as well as genes that were expressed in fewer than 5 cells of the same sample type. We then processed and integrated the different samples using Seurat version 3.1 following the recommendations from the Satija Lab’s vignette (https://satijalab.org/seurat/v3.1/integration.html). More specifically, read counts were first normalized using SCTransform for each sample individually75. The different samples were then integrated using the top 3,000 most informative genes76 before performing various dimensionality reduction steps including PCA and UMAP77. A shared nearest neighbor graph was constructed using Seurat’s FindNeighbors function with default settings (e.g. k=20) using the first 20 principal components. Subsequent clustering was performed with Seurat’s FindClusters function. In order to find major subclusters of β cells that are transcriptionally distinct and not rare (<1% of β cells), we set the resolution parameter set to 0.278. For visualizations and assessments of normalized expression values, the SCTransform-normalized (log-transformed) expression values were used unless noted otherwise. To remove putative doublets that may have resulted from the simultaneous capture of cells representing two different cell types within the same GEM, we additionally employed normalized expression thresholds for each of the marker genes and cells that expressed more than one marker gene above the given threshold were removed from the downstream analyses focusing on β cells. To determine genes that show significant expression differences that can be attributed to the condition (RD or HFD), we first extracted all the β cells from both conditions and re-calculated the most variable genes, PCA and UMAP coordinates as well as clusters (resolution parameter = 0.2). For each cluster, we then used the findMarkers function of the scran package73, comparing cells of the RD condition to those of the HFD condition and performing gene-wise one-tailed t-tests testing for up-regulation. Differentially expressed genes (DEG) were defined by an FDR threshold of 1%. Gene enrichment analyses to investigate canonical pathways, biological processes or transcription factors were performed using the Ingenuity Pathway Analysis tool (Qiagen).
RNA extraction and real-time qPCR analysis.
Total RNA from β cells were obtained using the RNAeasy Micro and Mini kits (Qiagen). cDNA was synthesized from the RNA by reverse transcription using the High Capacity cDNA Archive Kit (Invitrogen) and analyzed by QuantStudio 6 Flex Real-Time PCR Systems (Invitrogen) according to the DeltaDelta Ct method. Expression levels were normalized to Ribosomal Protein S18 (Rps18). Primer sequences are shown in Supplementary Table 6.
Bulk RNA-seq library generation.
After RNA isolation, total RNA integrity was analyzed using an Agilent 2100 Bioanalyzer and concentrations were measured using the NanoDrop Spectrophotometer (Thermo Fisher Scientific). Preparation of the RNA sample library were performed by the Genomics Core Laboratory at Weill Cornell Medicine. cDNA was generated using the SMARTer v4 Ultra Low Input RNA Kit (Clontech, # 63488) and Nextera XT kit (Nextera XT DNA Library Preparation Kit (Illumina, San Diego, CA) was used to make cDNA libraries suitable for Illumina sequencing. The normalized cDNA libraries were pooled and sequenced on an Illumina HiSeq4000 sequencer at 50 pair-end cycles. Transcriptome Data Analysis Sequencing reads were mapped with STAR v2.6.0c with default parameters to the mouse reference genome79 (GRCm38.p6). Fragments per gene were counted with featureCounts v1.6.2 with respect to Ensembl annotations 33137190 (Liao et al., 2014). Differentially expressed genes between pairwise comparisons were identified by Wald tests using DESeq2 v1.26.080 and only Benjamini–Hochberg corrected two-tailed p-values < 0.05 were considered statistically significant. Biological analyses, including canonical pathways, biological processes or transcription factors were performed using the Ingenuity Pathway Analysis tool (Qiagen). Base-2 log-transformed counts per million (CPM) values were used for heatmap plots of bulk RNA-seq data, which were centered and scaled by row. More code details can be found at https://github.com/abcwcm/RubioNavarro_Pancreas
β cell reaggregation into pseudo-islets.
Both mice and human FAC-sorted β cells were reaggregated along HUVECs cells to generate pseudo-islets. Briefly, 2 x 103 β cells and 2 x 103 HUVECs cells were cultured at a 1:1 ratio in culture media using 96-well plates with V-bottom conical wells (Thomas Scientific) pre-treated with Anti-Adherence Rinsing Solution (STEMCELL Technologies Inc.) at 37°C in a humidified 5% CO2 incubator. Pseudo-islets formed autonomously after approximately 24 hour of culture and were used for functional experiments or transplanted under the left kidney capsule of the mouse.
Diabetes Induction in Recipient Mice.
Acute diabetes induction in the mice was achieved via single intraperitoneal injection of streptozotocin (STZ; 150 mg/kg). Mice were considered diabetic when three consecutive readings of non-fasting blood glucose were over 300 mg/dL.
Islet Transplantation.
Islet transplantation into the kidney capsule of diabetic mice was performed on 10-11-weeks old diabetic male NOD.Cg-PrkdcScid/J recipient mice. For transplantation, we transferred 150 islets or pseudo-islets to sterile PBS and aspirated them into a sterile Polyethylene PE50 Tubing (BD INTRAMEDIC) connected to a 50 μl Hamilton syringe (Hamilton). Recipient mice were anesthetized using isoflurane (Abott), shaved, and scrubbed. A small incision was made on the left flank of the mouse to expose the kidney. Left kidney was kept moist with sterile PBS swab. Using a 27-gauge needle, we punctured the kidney capsule to carefully insert the beveled end of the PE50 tubing. Then, islets and pseudo-islets were slowly injected into the kidney capsule and the kidney was placed back into the cavity. Finally, the peritoneum and skin were sutured and stapled, and mice placed in a cage on a heating pad until recovery. To determine gene expression after 60 days of transplantation, islet grafts were dissected out of the kidneys for RNA isolation.
Unilateral nephrectomy.
To study the effect of graft removal, we performed uninephrectomy in transplanted mice. Mice were anesthetized using isoflurane (Abbott), shaved, and scrubbed. A midline abdominal incision was made on the left flank of the mouse. Then, we exposed the left ureter as well as then renal artery and vein by displacing the intestines towards the right side of the abdominal cavity and cover them with moistened drapes. We ligated the vessels and ureter with a single knot and cut to remove the kidney. Finally, we carefully replaced the kidney and the intestines into the peritoneal cavity and the peritoneum and skin were sutured and stapled.
Blood chemistry and serum insulin analysis.
For measuring blood glucose levels, mice were fasted for 6 hours and fasting blood samples were taken from the tail vein and assayed with a glucometer (OneTouch). For measuring mouse serum insulin levels, mice were fasted for 6 hours and blood was collected from the tail vein, the serum was separated by centrifugation and insulin levels were determined by ELISA with a standard curve (Mercodia).
Antibodies.
The following antibodies were used for immunohistochemistry and immunofluorescence: Guinea pig anti-insulin antibody (DAKO, #A0564; 1:1000), mouse anti-glucagon (Sigma, # G2654; 1:1000), rabbit anti-CD63 (SY21-02; 1:100) (Novus, nbp2-67425), rabbit anti-Mafa (Bethyl, #IHC-00352; 1:100), rabbit anti-ALDH1A3 (Novus, #NBP2-15339; 1:100), mouse anti-human CD31 (Abcam, ab9498; 1:100), mouse Anti-GAPDH (Invitrogen, MA5-15738; 1:100), biotinylated goat anti-guinea pig, (Vector Laboratories, BA-7000-1.5; 1:250), DyLight 488 Goat anti-rabbit (Vector Laboratories, #DI-1488; 1:250), Alexa Fluor 647 goat anti-guinea pig (Invitrogen, #A21450; 1:250), Alexa Fluor Plus 405 Donkey anti-mouse (Invitrogen, A48257; 1:250), Alexa Fluor Plus 488 Donkey anti-Mouse (Invitrogen, A32766; 1:250), The antibodies used for flow cytometry were CD63 Monoclonal Antibody (NVG-2)-APC (eBioscience, 17-0631-82; 2.5 μg/ml), Anti-IgG Isotype Control Monoclonal Antibody (RTK2758)-APC (Biolegend, 400512; 2.5 μg/ml).
Histological studies.
Pancreata were dissected, fixed in 10% neutral-buffered formalin (VWR) overnight at 4°C. Then, tissues were transferred to 70% ethanol and subsequently embedded in paraffin and sectioned at 5 μm thickness. For immunohistochemistry/immunofluorescence, sections were dewaxed, and antigen retrieval was performed by boiling samples in 10 mM sodium citrate buffer (pH=6.0). For immunofluorescence staining, primary antibodies were incubated overnight at 4°C, followed by incubation with corresponding Alexa-Fluor conjugated secondary antibodies (Invitrogen). Nuclei were counterstained with DAPI. Images were obtained with a NIKON Eclipse Ti microscope (NIKON NIS-Elements AR 4.3 software) or a Zeiss Axio Observer.Z1 confocal microscope (ZEN 2.3 imaging software). CD63hi β cells were marked using an arbitrary threshold and the number of DAPI positive cells was counted in areas where Insulin and CD63 staining overlapped. Then the percentage of CD63hi β cells was calculated (CD63hi β cells/ total β cells). Quantification of CD63hi β cells was performed using with Fiji/ImageJ (NIH, Bethesda, MD) by two independent investigators and at least 10 islets per animal were scored. For MAFA staining, Tyramide Signal Amplification (TSA; Thermo) was used. HRP-conjugated secondary antisera were applied for 1 h at room temperature and posteriorly a Tyramide working solution was applied for 10 min at room temperature. Nuclei were counterstained with DAPI. To determine β cell mass, insulin immunohistochemistry was performed in at least 4 non-consecutive pancreatic sections for each mouse as previously described3. Briefly, guinea pig anti-insulin antibody was incubated overnight at 4°C, followed by incubation with biotinylated secondary antibody (goat anti- guinea pig, Vector Laboratories). Then avidin–biotin peroxidase complex (Vectastain ABC kit, PK-7200, Vector Laboratories) was added for 30 min. Sections were developed using 3,3’-diaminobenzidine or 3-amino-9-ethyl carbazole (Sigma) and counterstained with hematoxylin. Whole sections were scanned using a Leica SCN400 F whole-slide scanner. The fraction of insulin-positive areas compared to total pancreatic tissue area (hematoxylin) was determined with Fiji/ImageJ (NIH, Bethesda, MD). β cell mass was then determined by multiplying the obtained fraction by the initial pancreatic wet weight.
Flow cytometry.
For the isolation of CD63hi and CD63lo β cells, isolated islets were dissociated using Trypsin-EDTA (0.05%) (Corning) for 10 minutes at 37°C. Dead cells were first removed using DAPI staining (1:5000, Thermo Fisher). β cells were gated based on granularity and FAD/FMN (FITC) autofluorescence. CD63-APC monoclonal antibody (eBioscience) was used for the isolation of CD63hi and CD63lo β cells. This sorting strategy was used in pancreatic islets from C57BL/6J wild-type mice, MIP-GFP mice and human. FAD and NAD(P)H levels were assessed by measuring autofluorescence at and excitation wavelength of 488 and 350 nm respectively using a BD LSRFortessa flow cytometer as previously reported81. To assess mitochondrial mass, dissociated islet cells were incubated with MitoTracker Green FM (20 nM) (M7514, Invitrogen) according to the manufacturer’s instructions and analyzed by flow cytometry. MitoTracker Green FM accumulates in mitochondria regardless of the mitochondrial membrane potential (Δψm). To evaluate mitochondrial function, we determined Δψm using MitoTracker Red CMXRos (M7512, Invitrogen), a red-fluorescent dye that accumulates in mitochondria from live cells upon membrane potential. Briefly, dissociated islets cells were starved in 2.8 mM Glucose media for 1 hour. Then, islet cells were subjected to high glucose (20 mM) for 1 hour and stained with MitoTracker Red CMXRos (25 nM) for 15 minutes at 37°C. The data were acquired using DIVA software (Version 9.0) and analyzed with FlowJo™ v10.7 software (BD Biosciences, San Diego, CA).
Transmission electron microscopy (TEM)
Murine FAC-sorted β cells were placed in EM Fix solution containing 4% paraformaldehyde, 2.5% glutaraldehyde, and 0.02% picric acid, in 0.1M sodium cacodylate buffer. After fixation overnight at 4°C, cells were post-fixed for 1h in 1% OsO4 −1.5% K-ferricyanide. Then, cells were dehydrated, infiltrated with Embed 812 epoxy resin, and transferred to resin models at 50°C for 36 hours. Samples were viewed using a JEOL JSM1400 transmission electron microscope operating at 100 KV (JEOL USA, Peabody, MA) and images were captured on a Veleta 2K x 2K CCD camera (EMSIS, GbhM, Miuenster, Germany).
Oxygen consumption and extracellular acidification rates.
Real-time measurements of oxygen consumption and extracellular acidification rates were performed in both CD63hi and CD63lo pseudo-islets using a Seahorse XFe96 (Agilent Technologies, Santa Clara, CA, USA). In brief, 15 pseudo-islets were pre-cultured in 2.8 mM Glucose, 1% FBS, 1 mM sodium pyruvate XF DMEM (pH 7.4) for 1 h at 37°C without CO2 before transfer to XFe96 Spheroid microplates. Four baseline measurements were recorded before compound were injected in order to establish a stable basal OCR. Glucose-stimulated respiration was obtained after the addition of 20 mM glucose and mitochondrial efficiency was determined following injection of oligomycin (5 μM), FCCP (1 μM), and rotenone/antimycin A (5 μM).
In Vitro Glucose Stimulated Insulin Secretion.
For static incubation assays, 20 pseudo-islets were handpicked and transferred into basal Krebs buffer containing 2.8 mM Glucose. After 1 hour of starvation, pseudo-islets were transferred into Krebs solution containing 20 mM for 1 hour at 37°C. Supernatants were centrifuged and collected. The pellet was used to determine intracellular insulin content. For dynamic insulin secretion assays, 20 pseudo-islets were pre-incubated into basal Krebs buffer containing 2.8 mM Glucose. Then, pseudo-islets were transferred into perifusion chambers (BioRep) for GSIS: 2.8 mM glucose (15 min), 20 mM glucose (45 min), 2.8 mM glucose (20min) and 50mM KCl (20min). The perifusate was collected at one-minute intervals (100 μl). Insulin secretion was measured with Insulin ELISA kit (Mercodia).
Statistics and reproducibility
Data are represented as mean ± standard error of the mean (S.E.M.). Statistical tests were performed with GraphPad Prism V9.0. Welch’s unpaired two-tailed t test or paired two-tailed t test are used for comparison and are dented in the corresponding figure legend. Pearson analysis was used for correlations with bars indicating the 95% confidence intervals. In bulk RNA-Seq data, the significance of DEGs between pairwise comparisons were identified by Wald test using DESeq2 v1.26.0 and Benjamini–Hochberg corrected two-tiled p-values < 0.05 were considered statistically significant. In figures corresponding to pathway analyses, p-value was calculated using two-tailed Benjamini-Hochberg. Statistical details (sample size and specific test performed) for each experiment are denoted in the corresponding figure or figure legend. No statistical method was used to pre-determine sample size. Sample sizes were determined on the basis of previous experimental experience. No data were excluded from the analyses. In animal experiments, mice were allocated to groups ensuring same average body weight and glucose. Investigators were blinded during manual cell counting and imaging analysis. Collection of raw data (mouse weight, glucose) and tissue harvesting for analyses was not blinded because it was necessary to preserve treatment information of each animal. In experiments in vitro or ex vivo, blinding was not possible because investigators who performed the experiments also analyze the data.
Extended Data
Extended Data Figure 1. Characteristics of diet-induced obesity model used for scRNA-Seq.
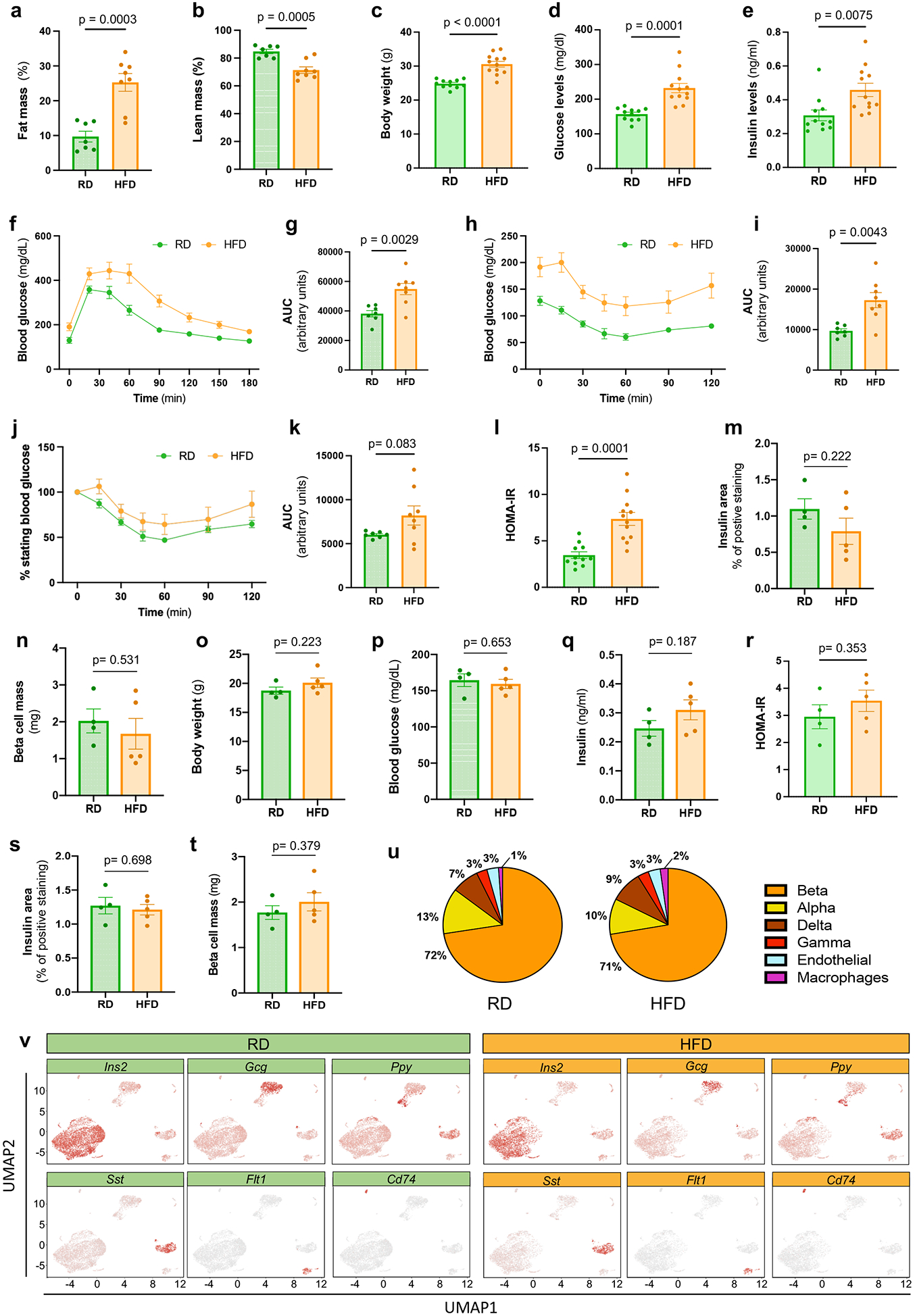
a-b, Percentages of fat mass (a), lean mass (b) in male mice fed with regular diet (RD, N=7) or high fat diet (HFD, N=8) for 6 weeks after weaning. c-e, Body weights (c), 6 hours fasting blood glucose (d), 6 hours fasting insulin (e) levels in male mice fed with RD (N=11) or HFD (N=12) for 6 weeks after weaning. f-g, Glucose tolerance test (GTT) was performed on RD (N=7) and HFD (N=8) fed male mice for 6 weeks with measurement of blood glucose concentrations (f). Area under the curve (AUC) of blood glucose levels (g). h-k, Insulin tolerance test (ITT) was performed on RD (N=7) and HFD (N=8) fed male mice for 7 weeks (h). AUC of blood glucose levels in h (i). ITT plotted as a % of starting glucose value (j). AUC of values plotted in j after t0 (k). l, Homeostasis model assessment for insulin resistance (HOMA-IR) index of male mice fed with RD (N=11) or HFD (N=12) for 6 weeks after weaning (insulin [mU/L]* glucose [mg/dL] )/405. m-n, Quantification of β cell area (m) and mass (n) in RD (n=4) and HFD (n=5) fed male mice. o-r, Body weights (o), 6 hours fasting blood glucose (p), 6 hours fasting insulin levels (q), HOMA-IR (r) of female mice fed with regular diet (RD, N=4) or high fat diet (HFD, N=5) for 6 weeks after weaning. s-t, Quantification of β cell area (s) and mass (t) in RD (n=4) and HFD (n=5) fed female mice. u, Distribution of pancreatic islet cell clusters from RD (left chart) and HFD (right chart) fed male mice. v, UMAPs representing expression of known marker genes across the cell clusters Ins1 (beta cells), Gcg (alpha cells), Sst (delta cells), Ftl1 (endothelial cells), Ppt (gamma cells), and Cd74 (immune cells) in male mice fed with RD (left panel) and HFD (right panel). All data are presented as mean ± S.E.M. and Welch’s unpaired two-tailed t test is used for comparison. Source numerical data are available in source data.
Extended Data Figure 2. Molecular analysis of β cell clusters.
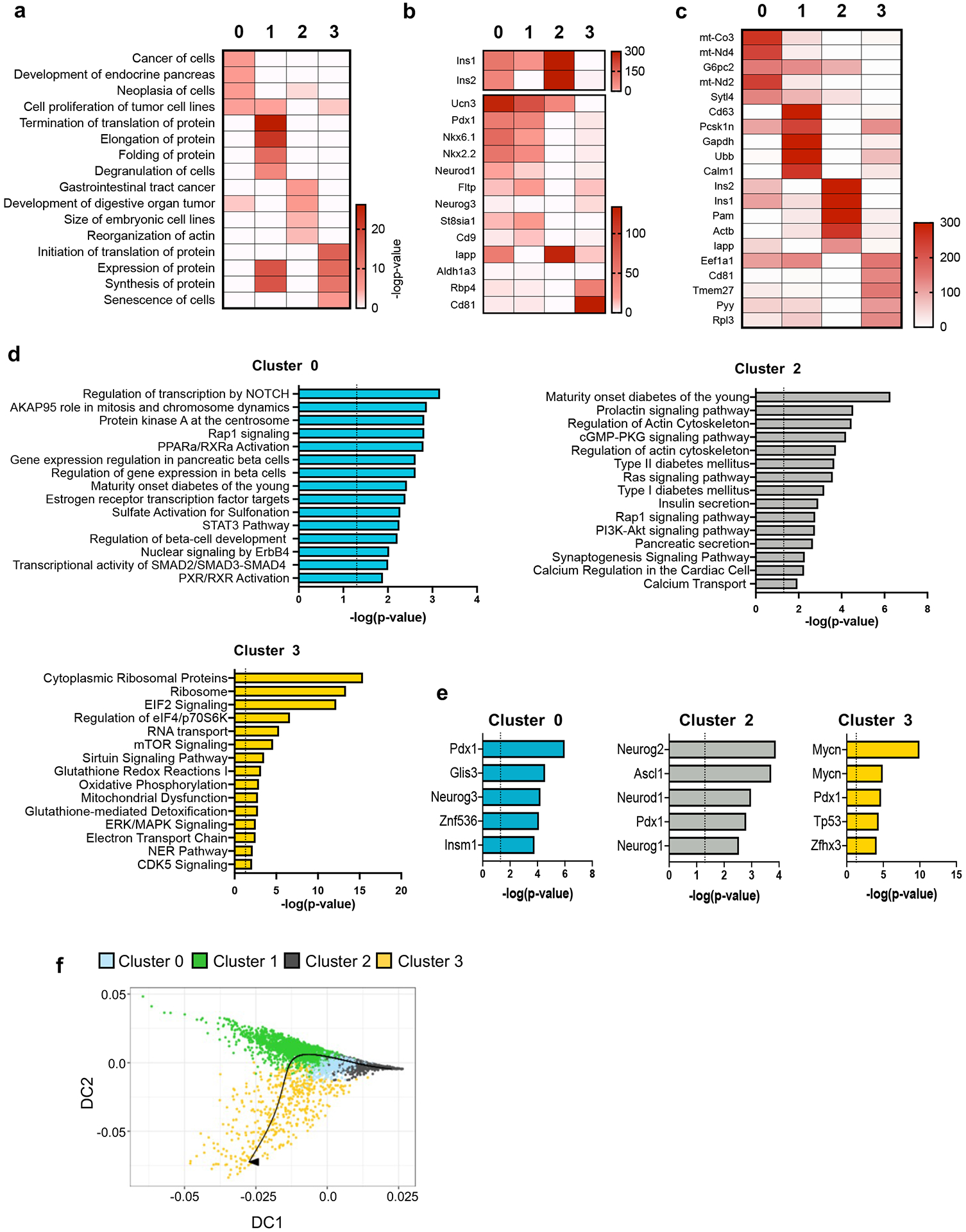
a, Top 4 enriched biological processes for the indicated β cell clusters (p-value was calculated using two-tailed Benjamini-Hochberg). b, Heatmap showing select genes previously identified to be heterogeneously expressed in β cells in each of the indicated β cell clusters. c, Top 5 differentially expressed genes (DEGs) for the indicated β cell clusters. d, Pathway enrichment analysis of DEGs in Cluster 0, Cluster 2, and Cluster 3 β cells ordered by statistical significance (p-value was calculated using two-tailed Benjamini-Hochberg). e, Top 5 transcription factors predicted in Cluster 0, Cluster 2, and Cluster 3 β cells ordered by statistical significance (p-value was calculated using two-tailed Benjamini-Hochberg). f, Trajectory of the different β cell clusters predicted by Slingshot analysis. Source numerical data are available in source data.
Extended Data Figure 3. CD63 expression in pancreatic islet cells.
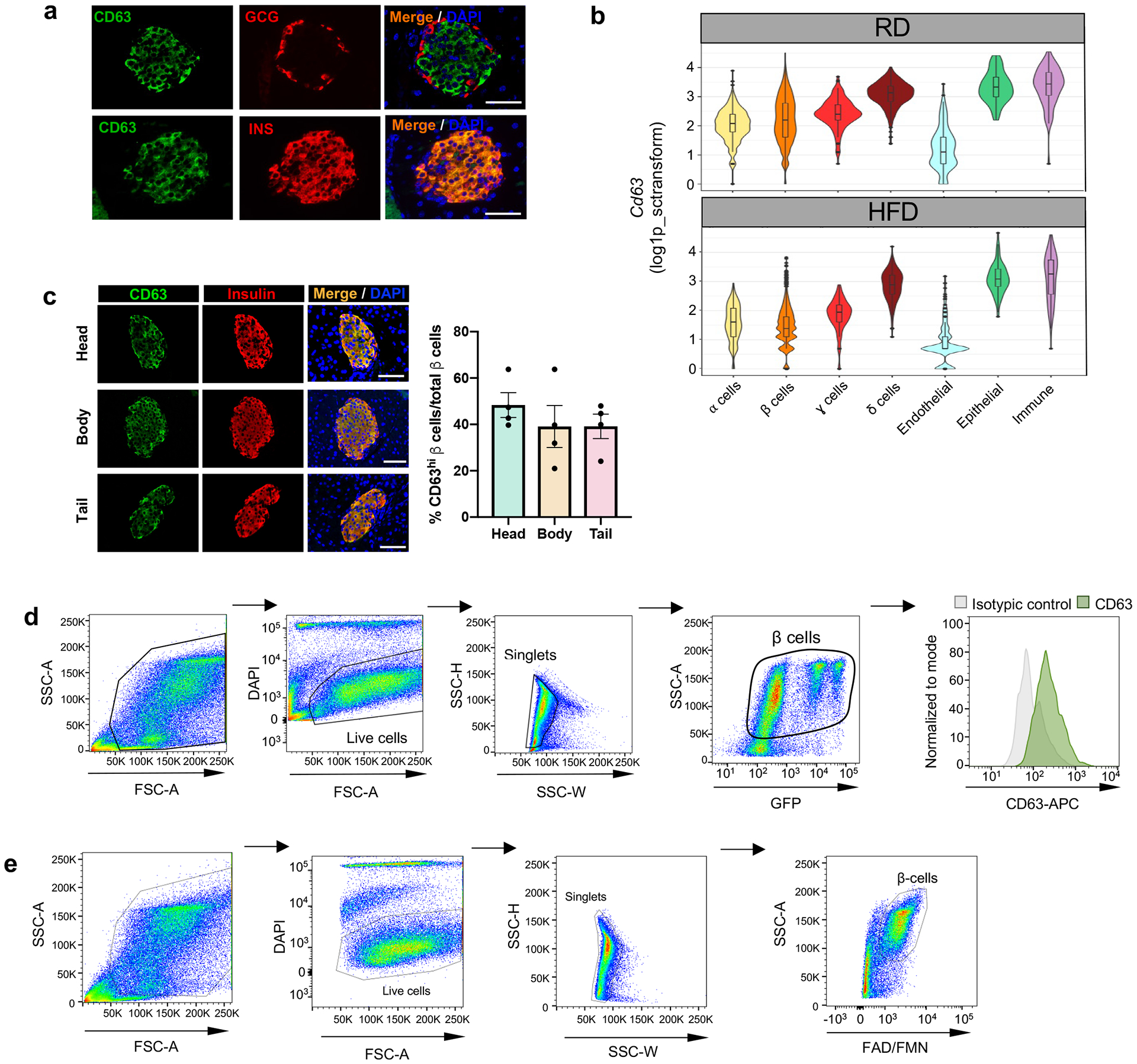
a, Representative immunofluorescence images of CD63 (green) and insulin (red) (upper panel), and CD63 (green) and glucagon (red) (lower panel) in pancreatic islets (Scale bars, 50 μm). Data are presented as mean±S.E.M. Paired two-tailed t test is used for comparison. (N=3 independent experiments). b, Violin plots representing expression of Cd63 across different pancreatic islet cell types from mice fed with RD and HFD. Minimum to maximum values are shown by the whiskers, the bounds of boxes represent the first and third quartiles and the center line indicates the median. α cells [RD= 802 cells, HFD=430 cells]; β cells [RD=4514 cells, HFD= 3050 cells]; γ cells [RD= 182 cells, HFD= 130 cells]; δ cells [RD= 461 cells, HFD= 377 cells]; Endothelial cells [RD= 196 cells, HFD 140 cells]; Epithelial cells [RD= 60 cells, HFD=79 cells]; Immune cells [RD= 69 cells, HFD= 92 cells]. c, Representative immunofluorescence images showing CD63 (green) and insulin (red) in the different portions of the pancreas (head, body and tail) from 10-week old WT male mice (left panel) (Scale bars, 50 μm). Quantification of the percentage of β cells that are CD63hi in the different parts of the pancreas (right panel) (N = 4 mice per group). Data are presented as mean ± S.E.M. Two-way ANOVA is used for comparison of multiple groups. d, Scatterplots showing the sequential gating scheme to determine CD63 protein expression by flow cytometry in β cells from mouse insulin promoter-GFP transgenic mice. Results are representative of 3 independent experiments. e, Scatterplots showing the sequential gating protocol to isolate CD63hi and CD63lo β cells from WT mice by FACS (in total 60057 CD63hi and 60064 CD63lo β cells). Data are presented as mean ± S.E.M. Source numerical data are available in source data.
Extended Data Figure 4. Molecular characterization of CD63hi β cells.
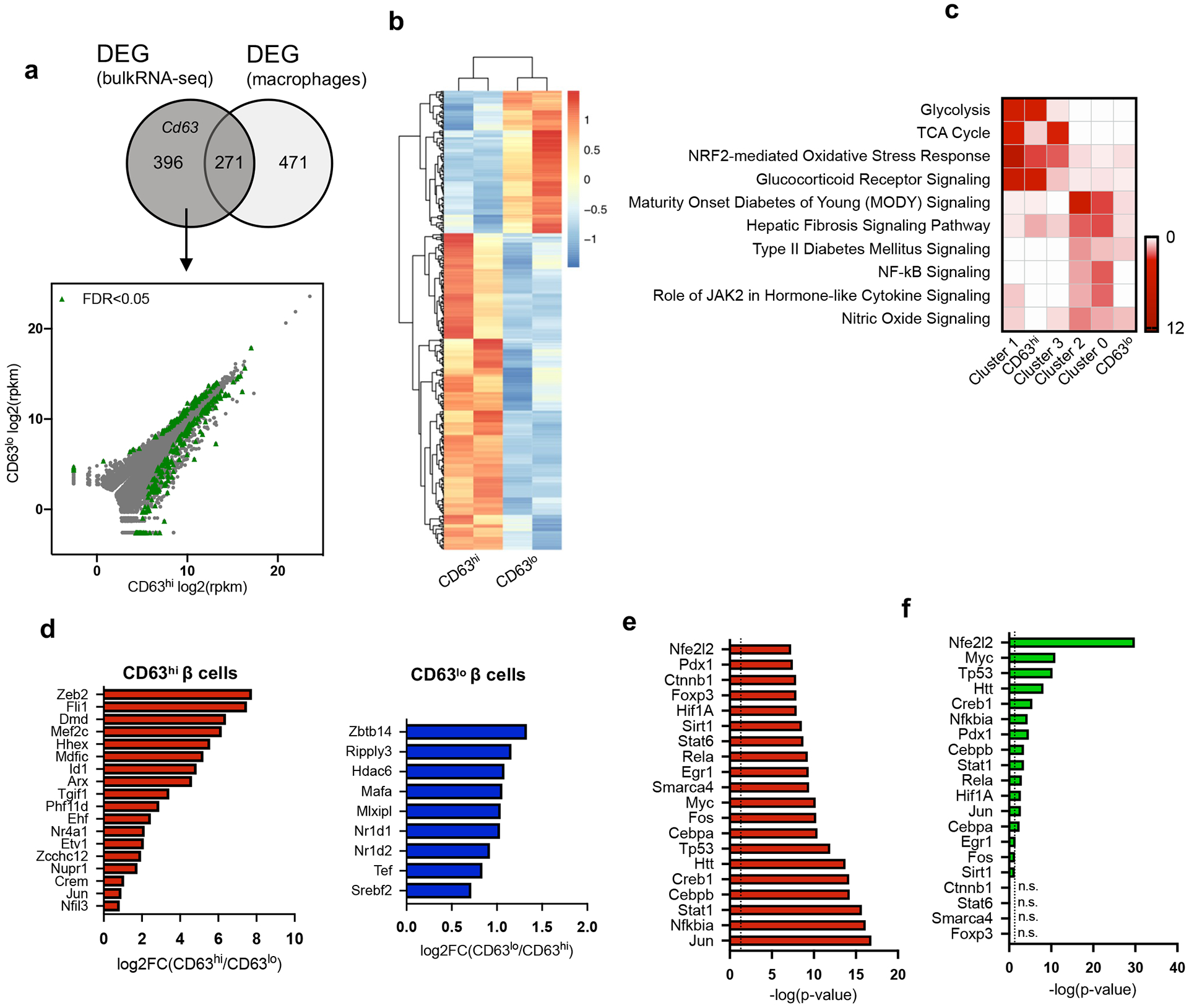
a, Venn diagram illustrating the overlap of differentially expressed genes (DEGs) obtained by bulk RNA-Seq and genes enriched in macrophages (left panel). Scatterplot of RNA-Seq analysis from FAC-sorted CD63hi and CD63lo β cells. DEGs with a false discovery rate (FDR) < 0.05 are in green and those that are not differentially expressed are in grey (right panel). b, Heatmap of 396 DEGs from FAC-sorted CD63hi and CD63lo β cells obtained through bulk RNA-Seq. (N = 2, each replicate is pooled from 5 mice). c, Heatmap showing the enriched pathways in β cell clusters 0-3 and CD63hi and CD63lo β cells. d, Differentially expressed transcription factors (DETs) in both CD63hi and CD63lo β cells (p-value was calculated using two-tailed Benjamini-Hochberg). e, Top 20 transcription factors predicted by upstream pathway analysis in CD63hi β cells ordered by statistical significance (p-value was calculated using two-tailed Benjamini-Hochberg). f, Statistical significance of the predicted transcription factors from CD63hi β cells (e) in Cluster 1 β cells (p-value was calculated using two-tailed Benjamini-Hochberg). Source numerical data are available in source data.
Extended Data Figure 5. CD63hi β cell maturation.
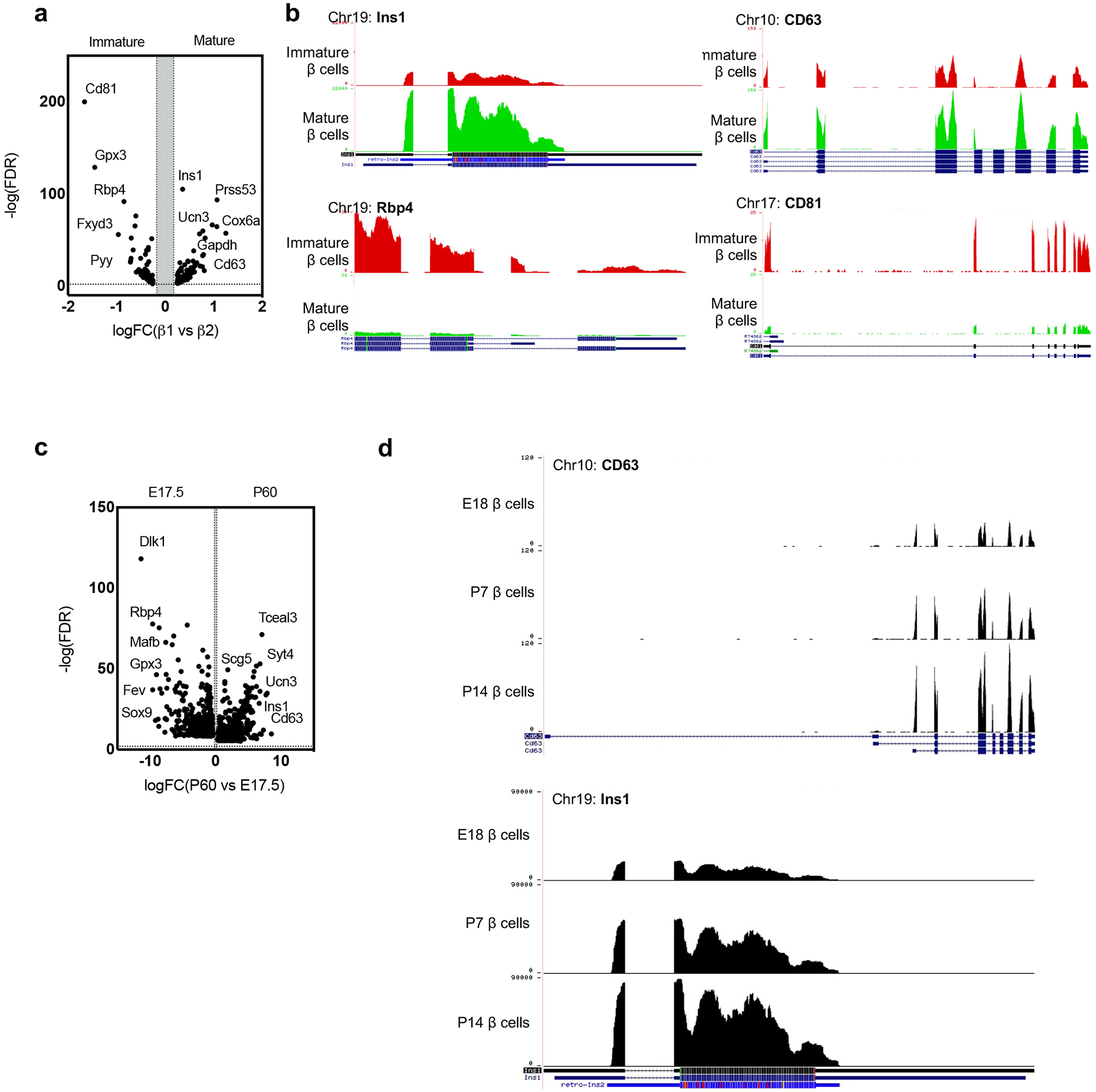
a, Volcano plot showing differentially expressed genes (DEGs) in immature and mature β cells52. b, Gene expression of Cd63, Ins1, Cd81 and Rbp4 by RNA-Seq on FAC-sorted β cells during maturation. Gene structure and chromosome number are indicated for each panel53. Data are publicly available at Huising Lab website https://www.huisinglab.com/). c, Volcano plot showing differentially expressed genes (DEGs) in embryonic and mature β cells54. d, Gene expression of Cd63 and Ins1 by RNA-seq on FAC-sorted β cells during perinatal maturation. Gene structure and chromosome number are indicated for each panel53. Data are publicly available at Huising Lab website (https://www.huisinglab.com/).
Extended Data Figure 6. Functional analysis of CD63hi β cells.
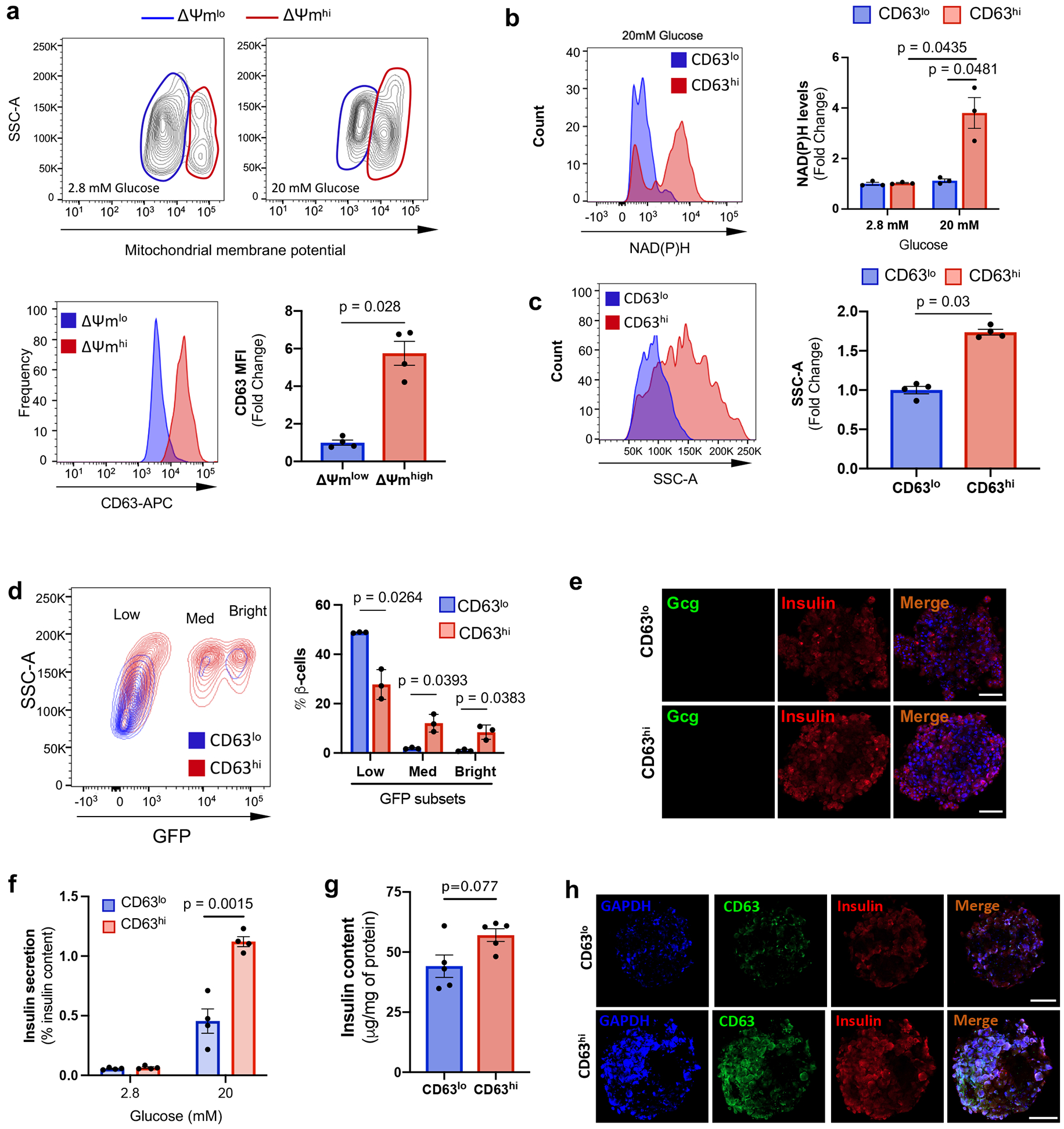
a, Contour plots of mitochondrial membrane potential in β cells incubated with 2.8 or 20 mM glucose. Histogram and quantification of CD63 expression in β cells with low and high mitochondrial membrane potential after incubation with 20 mM glucose. Results are representative of 4 independent experiments. Data are presented as mean ± S.E.M. Paired two-tailed t test is used for comparison. b, NAD(P)H fluorescent patterns of CD63hi and CD63lo β cells after incubation with 20 mM glucose. Quantification of NAD(P)H levels after incubation with 2.8 or 20 mM glucose (N = 3 independent experiments per group). Paired two-tailed t test is used for comparison. c, Analysis and quantification of cellular granularity in CD63hi and CD63lo β cells assessed by side scatter (SSC). (N = 4 independent experiments per group). Paired two-tailed t test is used for comparison. d, Scatterplots and quantification showing the distribution of CD63hi and CD63lo β cells in the different subpopulations of mouse insulin promoter-GFP transgenic β cells (N = 3 independent experiments per group; 5 mice per experiment). Welch’s unpaired two-tailed t test is used for comparison. e, Representative immunofluorescence images of pseudo-islets from mice showing β cells (Insulin, Red) and α cells (Glucagon, Green) (Scale bars, 50 μm). N = 5 pseudo-islets per group.. f, Static glucose-stimulated insulin secretion (GSIS) assay normalized by insulin content in CD63hi and CD63lo murine pseudo-islets. Data are pooled from 4 independent experiments. Paired two-tailed t test is used for comparison. g, Insulin content of CD63hi and CD63lo murine pseudo-islets normalized by intracellular protein (N = 5 independent experiments per group). Paired two-tailed t test is used for comparison. h, Representative immunofluorescence images of murine pseudo-islets showing Insulin (Red), CD63 (Green) and GADPH (Blue) (Scale bars, 50 μm). N = 5 pseudo-islets per group. Data are presented as mean ± S.E.M. Source numerical data are available in source data.
Extended Data Figure 7. Insulin secretion of CD63hi and CD63lo β cells in response to GLP-1 agonist.
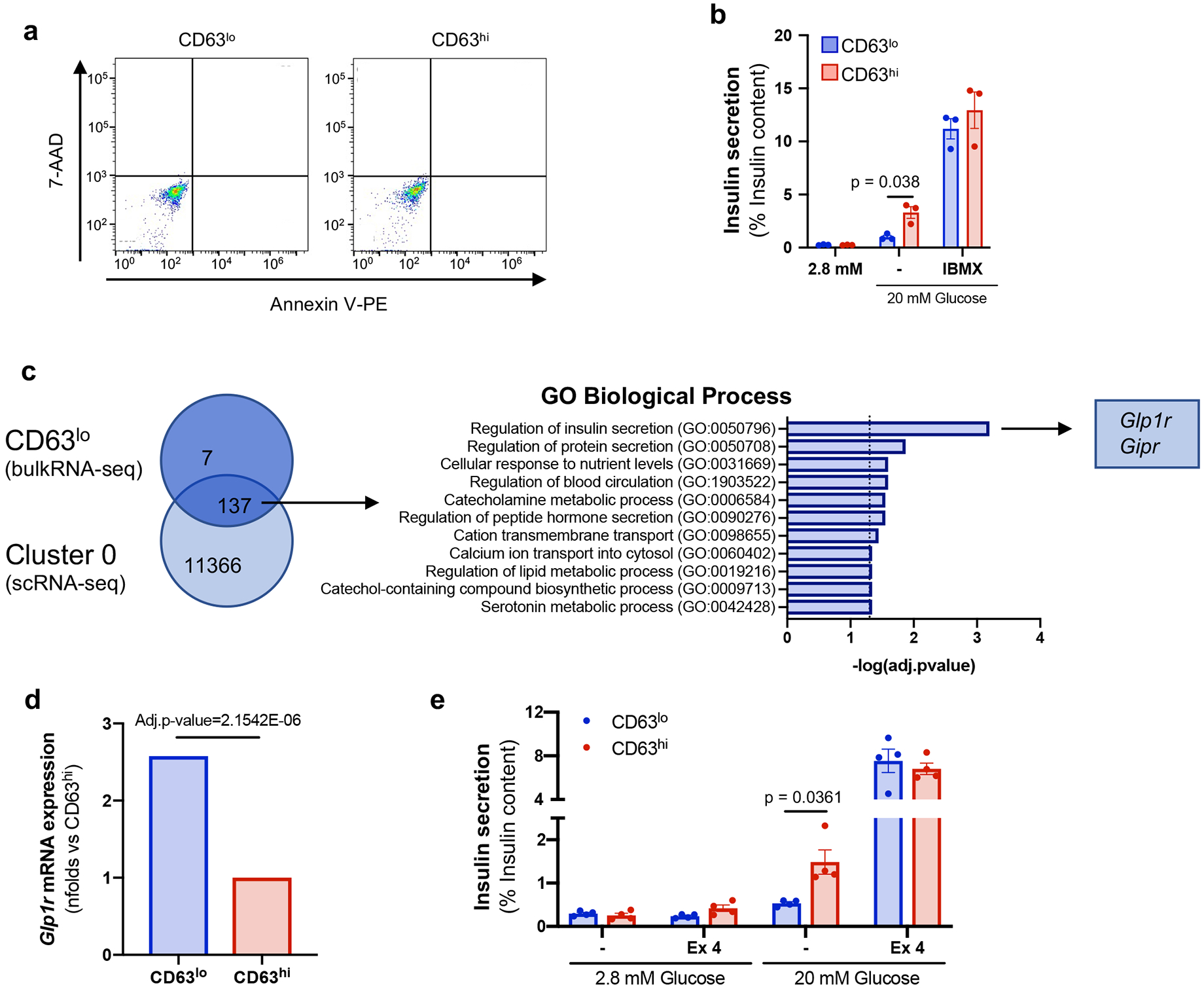
a, Flow cytometry analysis for annexin V and 7-AAD in isolated CD63hi and CD63lo β cells from WT mice. Data are representative of 3 independent experiments (cells pooled from at least 5 mice per experiment). b, Static glucose-stimulated insulin secretion (GSIS) assay in the absence or presence of 100 μM 3-isobutyl-1-methylxanthine (IBMX). Results are normalized by insulin content in CD63hi and CD63lo murine pseudo-islets (N = 3 per group). Welch’s unpaired two-tailed t test is used for comparison. c, Venn diagram illustrating the overlap of differentially expressed genes (FDR<0.01) between Cluster 0 β cells obtained by scRNA-seq and sorted CD63lo β cells determined through bulk RNA-Seq. Biological process enrichment analysis of differentially expressed genes in both Cluster 0 and CD63lo β cells. d, Glp1r mRNA expression in sorted CD63hi and CD63lo mouse β cells determined by bulk RNA-Seq. Comparisons were performed by Wald test. Benjamini–Hochberg corrected two-tiled p-value. e, Static GSIS assay in the absence or presence of 5 nM Exendin-4 (Ex4). Results are normalized by insulin content in CD63hi and CD63lo murine pseudo-islets (N = 4 replicates per group). Results are representative of 3 independent experiments. Data are presented as mean ± S.E.M. Paired two-tailed t test is used for comparison. Source numerical data are available in source data.
Extended Data Figure 8. Molecular analysis of human CD63hi β cells.
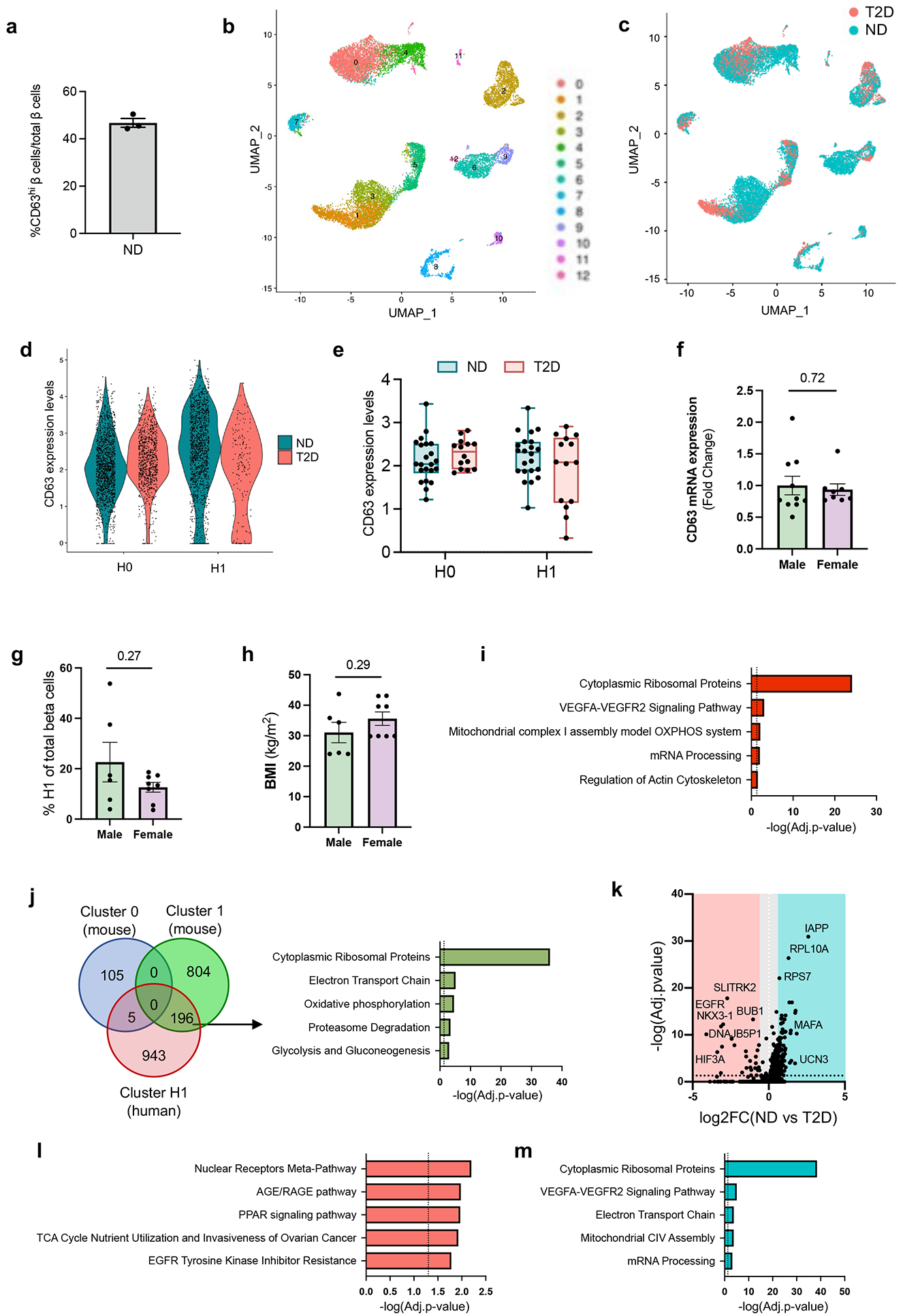
a, Quantification of the percentage of human β cells that are CD63hi (Fig. 3a). N = 3 non-diabetic human donors. Data are presented as mean ± S.E.M. b, UMAP showing the clustering of cells by constructing a Shared Nearest Neighbor (SNN) graph with resolution of 0.5 integrating cells from non-diabetic (ND) and donors with type 2 diabetes (T2D). c, UMAP indicating cells from ND and T2D donors in Extended Data Fig. 8a. d, Violin plots showing CD63 expression in H0 and H1 β cell subclusters in ND and T2D. e, Box plots showing the pseudo bulked median expression levels of CD63 in H0 and H1 β cell subclusters from ND (N=22) and T2D (N=14) donors. Each dot represents the median expression of one donor. Boxes represent the first and third quartiles, center line denotes the median and whiskers showed the maximum and minimum value. f, CD63 mRNA expression in whole islets from male (N=10) and female (N=8) ND donors determined by RT-qPCR. g-h, Frequency of Cluster H1 cells (g) and body mass index (BMI) (h) in T2D donors by sex. Donors with at least 13 β cells are included in the analysis. Male (N=6), Female (N=8). i, Pathway enrichment analysis of differentially expressed genes (DEGs) in human Cluster H1 β cells. j, Venn diagram illustrating the overlap of DEGs (FDR<0.01) among Cluster 0 and 1 mouse β cells obtained by scRNA-Seq and human H1 subcluster. Pathway enrichment analysis of DEGs by scRNA-Seq in Cluster 1 mouse and human H1 β cells. k, Volcano plot showing DEGs in H1 β cells between T2D and ND donors. l, Pathway enrichment analysis of DEGs in H1 β cells from T2D donors. m, Pathway enrichment analysis of DEGs in H1 β cells from ND donors. Data are presented as mean ± S.E.M. and Welch’s unpaired two-tailed t test is used for comparison (f, g, and h), and p-values were calculated using two-tailed Benjamini-Hochberg in I, j, l and m. Source numerical data are available in source data.
Extended Data Figure 9. Effects of obesity on functional and transcriptional profiles of β cells.
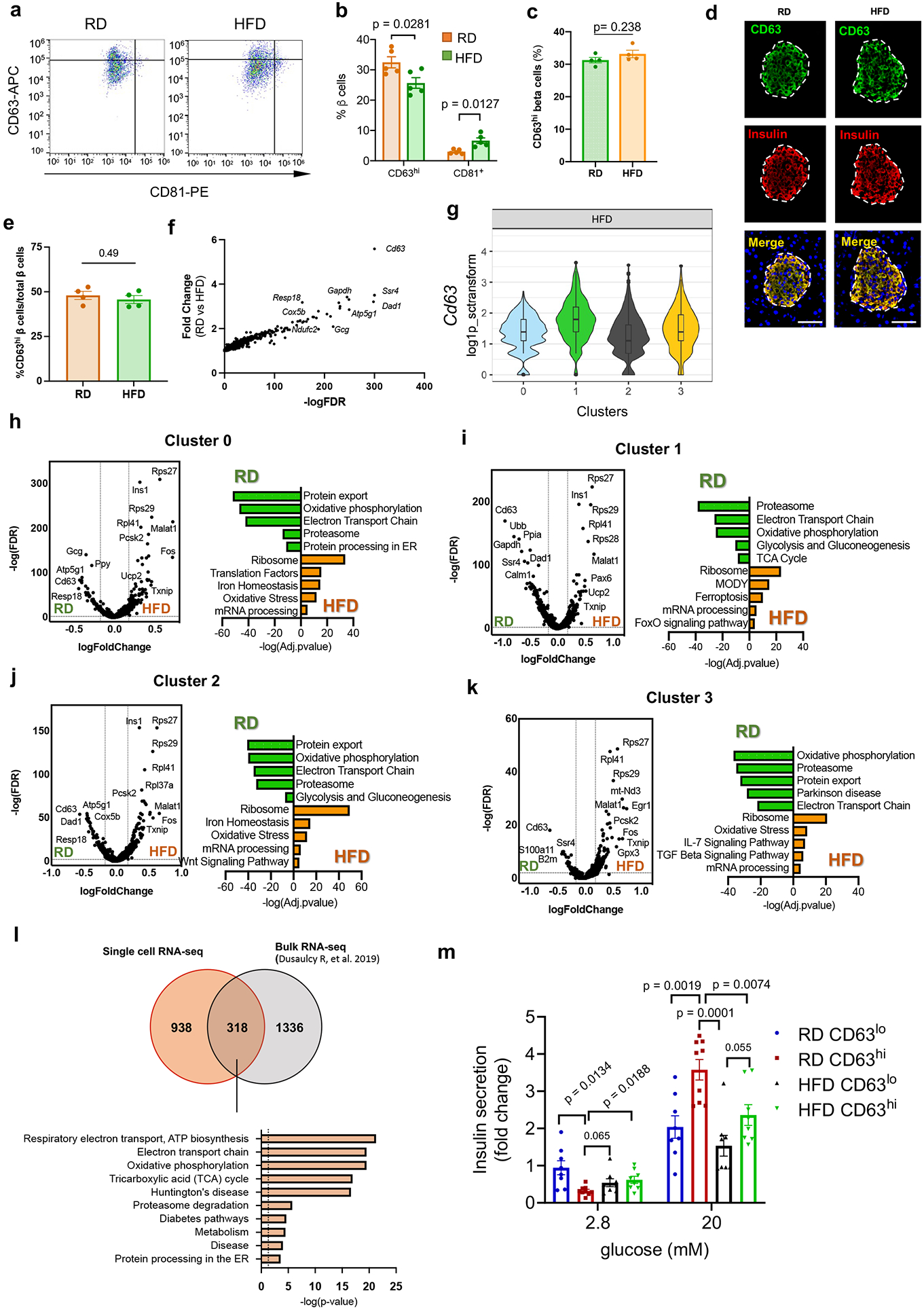
a, Representative flow cytometry plot for CD63 and CD81 staining on β cells from regular diet (RD) and high fat diet (HFD) fed male mice. b, Percentage of CD63hi and CD81+ β cells from RD and 6-weeks HFD-fed male mice. N = 5 mice per group. c, Percentage of CD63hi β cells from RD and 6-weeks HFD-fed female mice using flow cytometry. N = 4 mice per group. d, Representative immunofluorescence images showing CD63 (green) and insulin (red) in pancreas from WT female mice fed with RD or HFD for 6 weeks. (Scale bars, 50 μm). e, Quantification of the percentage of β cells that are CD63hi in female mice fed RD or HFD for 67 weeks (N = 4 mice per group). f, Plot showing differentially expressed genes (DEG) between β cells from RD and HFD-fed male mice. g, Violin plots representing expression of Cd63 across different β cell clusters from male mice fed with HFD. Whiskers show the minimum to maximum values, bounds of boxes represent first and third quartiles and the center line indicates the median. Cluster 0 (N= 1281 cells), Cluster 1 (N= 758), Cluster 2 (N= 767), Cluster 3 (N= 244 cells). h-k, Volcano plot showing DEGs between RD- and HFD-fed mice and pathways enriched in Cluster 0 (h), Cluster 1 (i), Cluster 2 (j) and Cluster 3 (k) β cells. l, Venn diagram illustrating the overlap of DEGs (FDR<0.01) between the bulk β cell population (all β cells from this scRNA-Seq study) from mice fed with HFD and β cell population from mice fed with HFD determined through bulk RNA-Seq56. Pathway analysis of shared DEGs. m, Glucose stimulated insulin secretion assay from pseudo-islets derived from CD63hi and CD63lo β cells from RD and HFD-fed mice. CD63lo (RD [N = 8 Samples]; HFD [N = 8 Samples) and CD63hi β cells (RD [N=9 Samples]; HFD (N= 8 Samples). Data are presented as mean ± S.E.M. and P values were determined by Welch’s unpaired two-tailed t test (b, c, e, and m). Source numerical data are available in source data.
Supplementary Material
Acknowledgments
This work was supported by the following grants: NIH R01 DK121140 (J.C.L.), R01 DK121844 (J.C.L.), P30 DK063608 pilot feasibility grant (J.C.L.), R01 DK056626 (D.E.C.), R37 DK048873 (D.E.C.), R01 DK103046 (D.E.C.), and Human Pancreas Analysis Program for Type 1 Diabetes 2U01DK112217-02A1 (A.N). Human pancreatic islets were provided by the NIDDK-funded Integrated Islet Distribution Program (IIDP) at City of Hope, NIH Grant # U24DK098085 and the JDRF-funded IIDP Islet Award Initiative (J.C.L.). A.R.-N. was supported by a Maria Zambrano Research Fellowship (Spanish Ministry of Universities and European Next Generation Funds). N.G.-B. (1-18-PMF-032) and A.G. (9-22-PDFPM-01) were supported by American Diabetes Association postdoctoral fellowships. L.S. was supported by an American Heart Association (AHA) postdoctoral fellowship (908952). N.I. was supported by an AHA Postdoctoral Fellowship (19POST34380692) and an American Liver Foundation Irwin M. Arias, MD Postdoctoral Research Fellowship Award American. L.M. was supported by a Chinese Scholarship Council award. The views expressed in this manuscript are those of the authors and do not necessarily represent the official views of the National Institute of Diabetes and Digestive and Kidney Diseases or the National Institutes of Health.
Footnotes
Reporting Summary. Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Declaration of Interests
No competing interests from the authors.
Data availability
Single-cell RNA-seq and Bulk RNA-seq data have been deposited at the Gene Expression Omnibus (GEO) database under accession number GSE203151 and are publicly available. Bulk RNA-seq data have been deposited at the GEO database under accession number GSE205023 and are publicly available. The previously published human single cell RNA-sequencing data that were re-analyzed are obtained from GSE84133, GSE86469, GSE85241, GSE81608 and E-MTAB-5061. Meta-analysis is available from the Huising Lab Website (https://www.huisinglab.com/). Data from Extended Data Figure 5a were obtained from GSE128565. Data from Extended Data Figure 5b were obtained from GSE88778. Data from Extended Data Figure 5c were obtained from GSE87375. Data from Extended Data Figure 5d were obtained from GSE88779. Data from Extended Figure 9l are obtained from European Nucleotide Archive (ENA). Accession number PRJEB30761 (https://www.ebi.ac.uk/ena/data/view/PRJEB30761). Source data have been provided in Source Data. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Code availability
All bioinformatics analyses code has been deposited at Gitub and is publicly available as of the date of publication. https://github.com/abcwcm/RubioNavarro_Pancreas.
References
- 1.Festa A, Williams K, D'Agostino R Jr., Wagenknecht LE & Haffner SM The natural course of beta-cell function in nondiabetic and diabetic individuals: the Insulin Resistance Atherosclerosis Study. Diabetes 55, 1114–1120 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Ahlqvist E et al. Novel subgroups of adult-onset diabetes and their association with outcomes: a data-driven cluster analysis of six variables. Lancet Diabetes Endocrinol 6, 361–369 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Gomez-Banoy N et al. Adipsin preserves beta cells in diabetic mice and associates with protection from type 2 diabetes in humans. Nat Med 25, 1739–1747 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tabak AG et al. Trajectories of glycaemia, insulin sensitivity, and insulin secretion before diagnosis of type 2 diabetes: an analysis from the Whitehall II study. Lancet 373, 2215–2221 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wagner R et al. Pathophysiology-based subphenotyping of individuals at elevated risk for type 2 diabetes. Nat Med 27, 49–57 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Haythorne E et al. Diabetes causes marked inhibition of mitochondrial metabolism in pancreatic beta-cells. Nat Commun 10, 2474 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma RB, Landa-Galvan HV & Alonso LC Living Dangerously: Protective and Harmful ER Stress Responses in Pancreatic beta-Cells. Diabetes 70, 2431–2443 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Accili D et al. When beta-cells fail: lessons from dedifferentiation. Diabetes Obes Metab 18 Suppl 1, 117–122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hunter CS & Stein RW Evidence for Loss in Identity, De-Differentiation, and Trans-Differentiation of Islet beta-Cells in Type 2 Diabetes. Front Genet 8, 35 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knudsen JG & Rorsman P beta Cell Dysfunction in Type 2 Diabetes: Drained of Energy? Cell Metab 29, 1–2 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Gutierrez GD, Gromada J & Sussel L Heterogeneity of the Pancreatic Beta Cell. Front Genet 8, 22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pipeleers DG Heterogeneity in pancreatic beta-cell population. Diabetes 41, 777–781 (1992). [DOI] [PubMed] [Google Scholar]
- 13.Muller A et al. 3D FIB-SEM reconstruction of microtubule-organelle interaction in whole primary mouse beta cells. J Cell Biol 220 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katsuta H et al. Subpopulations of GFP-marked mouse pancreatic beta-cells differ in size, granularity, and insulin secretion. Endocrinology 153, 5180–5187 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salomon D & Meda P Heterogeneity and contact-dependent regulation of hormone secretion by individual B cells. Exp Cell Res 162, 507–520 (1986). [DOI] [PubMed] [Google Scholar]
- 16.Schuit FC, In't Veld PA & Pipeleers DG Glucose stimulates proinsulin biosynthesis by a dose-dependent recruitment of pancreatic beta cells. Proc Natl Acad Sci U S A 85, 3865–3869 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jetton TL & Magnuson MA Heterogeneous expression of glucokinase among pancreatic beta cells. Proc Natl Acad Sci U S A 89, 2619–2623 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiekens R et al. Differences in glucose recognition by individual rat pancreatic B cells are associated with intercellular differences in glucose-induced biosynthetic activity. J Clin Invest 89, 117–125 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giordano E, Bosco D, Cirulli V & Meda P Repeated glucose stimulation reveals distinct and lasting secretion patterns of individual rat pancreatic B cells. J Clin Invest 87, 2178–2185 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Schravendijk CF, Kiekens R & Pipeleers DG Pancreatic beta cell heterogeneity in glucose-induced insulin secretion. J Biol Chem 267, 21344–21348 (1992). [PubMed] [Google Scholar]
- 21.Heimberg H et al. Heterogeneity in glucose sensitivity among pancreatic beta-cells is correlated to differences in glucose phosphorylation rather than glucose transport. EMBO J 12, 2873–2879 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soria B, Chanson M, Giordano E, Bosco D & Meda P Ion channels of glucose-responsive and -unresponsive beta-cells. Diabetes 40, 1069–1078 (1991). [DOI] [PubMed] [Google Scholar]
- 23.Holz G.G.t., Kuhtreiber WM & Habener JF Pancreatic beta-cells are rendered glucose-competent by the insulinotropic hormone glucagon-like peptide-1(7-37). Nature 361, 362–365 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baron M et al. A Single-Cell Transcriptomic Map of the Human and Mouse Pancreas Reveals Inter- and Intra-cell Population Structure. Cell Syst 3, 346–360 e344 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muraro MJ et al. A Single-Cell Transcriptome Atlas of the Human Pancreas. Cell Syst 3, 385–394 e383 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Segerstolpe A et al. Single-Cell Transcriptome Profiling of Human Pancreatic Islets in Health and Type 2 Diabetes. Cell Metab 24, 593–607 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xin Y et al. Use of the Fluidigm C1 platform for RNA sequencing of single mouse pancreatic islet cells. Proc Natl Acad Sci U S A 113, 3293–3298 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lawlor N et al. Single-cell transcriptomes identify human islet cell signatures and reveal cell-type-specific expression changes in type 2 diabetes. Genome Res 27, 208–222 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xin Y et al. Pseudotime Ordering of Single Human beta-Cells Reveals States of Insulin Production and Unfolded Protein Response. Diabetes 67, 1783–1794 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Fang Z et al. Single-Cell Heterogeneity Analysis and CRISPR Screen Identify Key beta-Cell-Specific Disease Genes. Cell Rep 26, 3132–3144 e3137 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xin Y et al. Single-Cell RNAseq Reveals That Pancreatic beta-Cells From Very Old Male Mice Have a Young Gene Signature. Endocrinology 157, 3431–3438 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Dominguez-Gutierrez G, Xin Y & Gromada J Heterogeneity of human pancreatic beta-cells. Mol Metab 27S, S7–S14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mawla AM & Huising MO Navigating the Depths and Avoiding the Shallows of Pancreatic Islet Cell Transcriptomes. Diabetes 68, 1380–1393 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Camunas-Soler J et al. Patch-Seq Links Single-Cell Transcriptomes to Human Islet Dysfunction in Diabetes. Cell Metab 31, 1017–1031 e1014 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bar Y & Efrat S The NOTCH pathway in beta-cell growth and differentiation. Vitam Horm 95, 391–405 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Bartolome A, Zhu C, Sussel L & Pajvani UB Notch signaling dynamically regulates adult beta cell proliferation and maturity. J Clin Invest 129, 268–280 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelly P et al. Rap1 promotes multiple pancreatic islet cell functions and signals through mammalian target of rapamycin complex 1 to enhance proliferation. J Biol Chem 285, 15777–15785 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kramer A, Green J, Pollard J Jr. & Tugendreich S Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30, 523–530 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang HS et al. Transcription factor Glis3, a novel critical player in the regulation of pancreatic beta-cell development and insulin gene expression. Mol Cell Biol 29, 6366–6379 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osipovich AB et al. Insm1 promotes endocrine cell differentiation by modulating the expression of a network of genes that includes Neurog3 and Ripply3. Development 141, 2939–2949 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Newsholme P, Keane KN, Carlessi R & Cruzat V Oxidative stress pathways in pancreatic beta-cells and insulin-sensitive cells and tissues: importance to cell metabolism, function, and dysfunction. Am J Physiol Cell Physiol 317, C420–C433 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Lee JH et al. Overexpression of SIRT1 protects pancreatic beta-cells against cytokine toxicity by suppressing the nuclear factor-kappaB signaling pathway. Diabetes 58, 344–351 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh CK et al. The Role of Sirtuins in Antioxidant and Redox Signaling. Antioxid Redox Signal 28, 643–661 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baumel-Alterzon S, Katz LS, Brill G, Garcia-Ocana A & Scott DK Nrf2: The Master and Captain of Beta Cell Fate. Trends Endocrinol Metab 32, 7–19 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gupta RK et al. The MODY1 gene HNF-4alpha regulates selected genes involved in insulin secretion. J Clin Invest 115, 1006–1015 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miura A et al. Hepatocyte nuclear factor-4alpha is essential for glucose-stimulated insulin secretion by pancreatic beta-cells. J Biol Chem 281, 5246–5257 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Flannick J, Johansson S & Njolstad PR Common and rare forms of diabetes mellitus: towards a continuum of diabetes subtypes. Nat Rev Endocrinol 12, 394–406 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Lee AH, Heidtman K, Hotamisligil GS & Glimcher LH Dual and opposing roles of the unfolded protein response regulated by IRE1alpha and XBP1 in proinsulin processing and insulin secretion. Proc Natl Acad Sci U S A 108, 8885–8890 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Juliana CA et al. A PDX1-ATF transcriptional complex governs beta cell survival during stress. Mol Metab 17, 39–48 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salinno C et al. CD81 marks immature and dedifferentiated pancreatic beta-cells. Mol Metab 49, 101188 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xie Z et al. Gene Set Knowledge Discovery with Enrichr. Curr Protoc 1, e90 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sachs S et al. Targeted pharmacological therapy restores beta-cell function for diabetes remission. Nat Metab 2, 192–209 (2020). [DOI] [PubMed] [Google Scholar]
- 53.van der Meulen T et al. Virgin Beta Cells Persist throughout Life at a Neogenic Niche within Pancreatic Islets. Cell Metab 25, 911–926 e916 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qiu WL et al. Deciphering Pancreatic Islet beta Cell and alpha Cell Maturation Pathways and Characteristic Features at the Single-Cell Level. Cell Metab 25, 1194–1205 e1194 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Farack L et al. Transcriptional Heterogeneity of Beta Cells in the Intact Pancreas. Dev Cell 48, 115–125 e114 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Dusaulcy R et al. High-fat diet impacts more changes in beta-cell compared to alpha-cell transcriptome. PLoS One 14, e0213299 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Backe MB, Moen IW, Ellervik C, Hansen JB & Mandrup-Poulsen T Iron Regulation of Pancreatic Beta-Cell Functions and Oxidative Stress. Annu Rev Nutr 36, 241–273 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Talchai C, Xuan S, Lin HV, Sussel L & Accili D Pancreatic beta cell dedifferentiation as a mechanism of diabetic beta cell failure. Cell 150, 1223–1234 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim-Muller JY et al. Metabolic inflexibility impairs insulin secretion and results in MODY-like diabetes in triple FoxO-deficient mice. Cell Metab 20, 593–602 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim-Muller JY et al. Aldehyde dehydrogenase 1a3 defines a subset of failing pancreatic beta cells in diabetic mice. Nat Commun 7, 12631 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pasquier A et al. Lysosomal degradation of newly formed insulin granules contributes to beta cell failure in diabetes. Nat Commun 10, 3312 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dorrell C et al. Human islets contain four distinct subtypes of beta cells. Nat Commun 7, 11756 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ellenbroek JH et al. Topologically heterogeneous beta cell adaptation in response to high-fat diet in mice. PLoS One 8, e56922 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tersey SA et al. Episodic beta-cell death and dedifferentiation during diet-induced obesity and dysglycemia in male mice. FASEB J, fj201800150RR (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Benninger RKP & Kravets V The physiological role of beta-cell heterogeneity in pancreatic islet function. Nat Rev Endocrinol (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bader E et al. Identification of proliferative and mature beta-cells in the islets of Langerhans. Nature 535, 430–434 (2016). [DOI] [PubMed] [Google Scholar]
- 67.Karaca M et al. Exploring functional beta-cell heterogeneity in vivo using PSA-NCAM as a specific marker. PLoS One 4, e5555 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang YJ et al. Single-Cell Transcriptomics of the Human Endocrine Pancreas. Diabetes 65, 3028–3038 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li J et al. Single-cell transcriptomes reveal characteristic features of human pancreatic islet cell types. EMBO Rep 17, 178–187 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang YJ & Kaestner KH Single-Cell RNA-Seq of the Pancreatic Islets--a Promise Not yet Fulfilled? Cell Metab 29, 539–544 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huotari MA et al. ErbB signaling regulates lineage determination of developing pancreatic islet cells in embryonic organ culture. Endocrinology 143, 4437–4446 (2002). [DOI] [PubMed] [Google Scholar]
- 72.Doyle ME & Egan JM Mechanisms of action of glucagon-like peptide 1 in the pancreas. Pharmacol Ther 113, 546–593 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lun AT, McCarthy DJ & Marioni JC A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res 5, 2122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Amezquita RA et al. Orchestrating single-cell analysis with Bioconductor. Nat Methods 17, 137–145 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hafemeister C & Satija R Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol 20, 296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stuart T et al. Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902 e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Becht E et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol (2018). [DOI] [PubMed] [Google Scholar]
- 78.Waltman L & van Eck NJ A smart local moving algorithm for large-scale modularity-based community detection. The European Physical Journal B 86, 471 (2013). [Google Scholar]
- 79.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Smelt MJ, Faas MM, de Haan BJ & de Vos P Pancreatic beta-cell purification by altering FAD and NAD(P)H metabolism. Exp Diabetes Res 2008, 165360 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell RNA-seq and Bulk RNA-seq data have been deposited at the Gene Expression Omnibus (GEO) database under accession number GSE203151 and are publicly available. Bulk RNA-seq data have been deposited at the GEO database under accession number GSE205023 and are publicly available. The previously published human single cell RNA-sequencing data that were re-analyzed are obtained from GSE84133, GSE86469, GSE85241, GSE81608 and E-MTAB-5061. Meta-analysis is available from the Huising Lab Website (https://www.huisinglab.com/). Data from Extended Data Figure 5a were obtained from GSE128565. Data from Extended Data Figure 5b were obtained from GSE88778. Data from Extended Data Figure 5c were obtained from GSE87375. Data from Extended Data Figure 5d were obtained from GSE88779. Data from Extended Figure 9l are obtained from European Nucleotide Archive (ENA). Accession number PRJEB30761 (https://www.ebi.ac.uk/ena/data/view/PRJEB30761). Source data have been provided in Source Data. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
All bioinformatics analyses code has been deposited at Gitub and is publicly available as of the date of publication. https://github.com/abcwcm/RubioNavarro_Pancreas.


