Abstract
Feline immunodeficiency virus (FIV) is a lentivirus causing immune suppression and neurological disease in cats. Like primate lentiviruses, FIV utilizes the chemokine receptor CXCR4 for infection. In addition, FIV gene expression has been demonstrated in immortalized human cell lines. To investigate the extent and mechanism by which FIV infected primary and immortalized human cell lines, we compared the infectivity of two FIV strains, V1CSF and Petaluma, after cell-free infection. FIV genome was detected in infected human peripheral blood mononuclear cells (PBMC) and macrophages at 21 and 14 days postinfection, respectively. Flow cytometry analysis of FIV-infected human PBMC indicated that antibodies to FIV p24 recognized 12% of the cells. Antibodies binding the CCR3 chemokine receptor maximally inhibited infection of human PBMC by both FIV strains compared to antibodies to CXCR4 or CCR5. Reverse transcriptase levels increased in FIV-infected human PBMC, with detection of viral titers of 101.3 to 102.1 50% tissue culture infective doses/106 cells depending on the FIV strain examined. Cell death in human PBMC infected with either FIV strain was significantly elevated relative to uninfected control cultures. These findings indicate that FIV can productively infect primary human cell lines and that viral strain specificity should be considered in the development of an FIV vector for gene therapy.
Feline immunodeficiency virus (FIV) is a lentivirus associated with immunological and neurological disease in cats and is similar to human immunodeficiency virus (HIV) in humans. Although antigenically and genetically distinct (32), FIV is believed to employ a similar mechanism of infection involving chemokine receptors as the primate lentiviruses (46). No evidence has been reported to indicate infection of humans by FIV, despite many opportunities for transmission of the virus through the bite of an infected cat. Like most retroviruses, FIV is considered highly species adapted, but the virus possesses a broad cell tropism within its natural host (32). In vivo, FIV primarily infects blood-derived cells, including T cells, B cells, and macrophages (11), but it also shares the neurotropism common to other lentiviruses, such as HIV-1 (10). The ability to infect terminally differentiated cells, such as macrophages, and the potential for greater safety implied by the failure of FIV to cause human disease, has made the virus a candidate for use as a nonprimate lentiviral vector in human gene therapy (35).
The tropism exhibited by retroviruses is determined by the ability to gain entry into target cells and viral gene expression following infection (24). A direct correlation between viral-envelope-mediated infection and FIV host cell range has been reported (15, 33), a correlation that is supported by reports showing that cell tropism was altered by changes in FIV envelope glycoproteins involving a single amino acid substitution (43, 44). Extensive sequence variation has been reported within the env gene of FIV isolates possessing different host cell ranges (14, 21, 30, 40), which is an effect associated with virus entry being restricted by changes in the env gene sequence of these strains (31). Like all retroviruses, FIV gene expression is regulated by sequence elements in the long terminal repeats (LTR) of the viral genome (41). The basal promoter activity of the LTR from different FIV strains varies with the cell line infected (24, 25) and correlates with the extent of virus production (19). As reported for the env gene, sequence diversity in the LTR also varies among different FIV strains (33), although a role for this diversity in infection remains uncertain at present.
The capacity of FIV to infect human cells in vitro is not clearly defined, but it may be influenced by the same factors that determine tropism in the feline host. Infection of primary human cells by cell-free FIV has not been observed previously; however, infection of human MOLT-4 lymphoblastoid (17, 42), U373 astrocyte (10), and HeLa epithelioid (24) cells was reported following transfection or cocultivation with FIV-infected feline cells. Productive replication of FIV in HeLa and U373 cells was suggested, but a latent state was observed in FIV-infected MOLT-4 cells in which neither infectious virus nor viral mRNA was detectable (17). Provirus integration was assumed to occur in these cell lines, since FIV DNA was detectable in high-molecular-weight DNA for extended periods of time (17, 42). Failure to detect virus-specific mRNA implies that transcription was inhibited in MOLT-4 cells, an assumption supported by studies showing that the promoter activity of the FIV LTR varies in different human cell types in a manner analogous to that observed in feline cells (24). Recently, transcriptional limitations associated with the U3 element of the FIV LTR were implicated as the sole restriction to productive infection of human cells by the virus (35), suggesting a role for transcriptional efficiency in influencing FIV tropism in human cells.
The CXCR4, CCR3, and CCR5 chemokine receptors, expressed on many cell types, are believed to be important primate lentivirus coreceptors (47). CCR5 and CXCR4 are associated with HIV infection of macrophages and T-cell lines, respectively, while the function of CCR3 is less clearly understood (2). Although the primary receptor for FIV has not been identified (45), FIV strains have been shown to interact with the human CXCR4 chemokine receptor, leading to cell fusion and infection of both human and rodent cell lines (34, 46). In addition, human stromal cell-derived factor (SDF-1) was shown to bind feline CXCR4 molecules and inhibit infection of CrFK cells by FIV (16). These observations suggest a common mechanism of infection for HIV and FIV involving the chemokine receptor family, indicating that the evolutionary link between the two viruses may be closer than previously assumed (46). Furthermore, differences in receptor distribution and utilization by different strains of lentiviruses may indicate a potential mechanism for strain-dependent infection of human cells.
In this study, we investigated the tropism of two primary isolates of FIV, blood-derived Petaluma and cerebrospinal fluid-derived V1CSF, in primary and immortalized human cell lines after cell-free infection. Our results indicate that Petaluma has a broader tropism for human cells than V1CSF but that both strains infected primary human peripheral blood mononuclear cells (PBMC) and macrophages. Infection of human PBMC by either strain could be inhibited to various degrees by antibodies that recognized the chemokine receptors CXCR4, CCR3, and CCR5. Furthermore, FIV gene expression and productive infection, as determined by the detection of virus-specific proteins and infectious FIV, was also observed in human PBMC. These results suggest that some FIV strains can productively infect human cells, possibly employing a mechanism involving the same chemokine receptors implicated as coreceptors for primate lentiviruses, and indicate that caution must be exercised in choosing strains of FIV on which to base a lentiviral vector system.
MATERIALS AND METHODS
Cell culture and primary cell purifications.
The murine IC-21 macrophage and human U373 astrocytoma and THP-1 monocyte cell lines were obtained from the American Type Culture Collection (Rockville, Md.). Primary human fetal astrocyte cultures were prepared from 12- to 17-week-old human fetal brain specimens as described previously (29). Human PBMC, from which macrophage cultures were prepared, were obtained from healthy donors as previously described (37). Feline PBMC cultures were obtained from specific-pathogen-free adult cats and prepared in the same manner as the human blood-derived cells. PBMC cultures were initially stimulated with 5 μg of concanavalin A (ConA) per ml for 3 days and maintained on 5 μg of interleukin-2 (IL-2) per ml. U373 cells and human fetal astrocyte cultures were maintained in minimum essential medium with 10% (vol/vol) heat-inactivated fetal bovine serum (FBS). Both human and feline PBMC and macrophage cultures were maintained in RPMI 1640 medium with 15% FBS. Murine macrophages and THP-1 monocytes were cultured in RPMI medium with 10% FBS and either 10 mM HEPES or 5.5 μM β-mercaptoethanol, respectively. All cultures were supplemented with 100 μg of streptomycin and 0.25 μg of amphotericin per ml. PBMC and THP-1 cultures were seeded at a density of 2 × 106 and 1 × 106 cells/ml, respectively, in 24-well plates. Murine and human macrophages were cultured at a density of 5 × 105 cells/ml. Both U373 and fetal astrocyte cells were seeded at 106 cells/ml.
Viruses and infection.
Two primary isolates of FIV, blood-derived Petaluma (a gift from N. C. Pedersen) and cerebrospinal fluid-derived V1CSF (38), were used for these experiments because they were isolated from different tissue compartments and restriction enzyme analysis and sequencing revealed differences in the env gene between isolates (19). Both isolates were cultured in primary feline PBMC and had not undergone more than 10 passages in vitro to minimize the effects of culture adaptation on viral tropism.
Culture supernatant from feline PBMC infected with either Petaluma or V1CSF was used as a source of infectious virus. Supernatants were cleared of cellular debris by high-speed centrifugation, and the viral titers were determined to be 103.5 to 104.5 50% tissue culture infectious doses (TCID50)/106 cells. Human cells were incubated with 200 μl of culture supernatant at 37°C for 2 h, washed twice with serum-free medium, and seeded as described above. Infection of astrocyte cultures was preceded by incubation for 30 min with 5 μg of hexadimethrine bromide (Sigma, St. Louis, Mo.) per ml. For all cell lines, uninfected cultures served as negative controls. Culture supernatants and infected cells were harvested at various days postinfection (p.i.). To assess productive viral infection, supernatants from human cell cultures infected with either the V1CSF or Petaluma strains that were found to be PCR-positive for FIV DNA sequences were used to infect feline PBMC cultures. Cells and supernatants from feline cultures were harvested and analyzed in the same manner as the human cell cultures.
Isolation of high-molecular-weight DNA.
Cells were pelleted at 13,000 rpm for 5 min, washed once, and resuspended in 50 μl of H2O and 50 μl of 2% Triton X-100/TE buffer. Samples were boiled for 15 min, cooled on ice, and incubated with 14.4 μg of proteinase K for 2 h at 56°C. Samples were again boiled for 15 min, incubated on ice for 5 min, and centrifuged at high speed for 5 min to pellet the cellular debris. High-molecular-weight DNA was precipitated from the resulting supernatants, purified by using a Wizard DNA Clean-up System Kit (Promega, Madison, Wis.), and stored at 4°C in TE buffer. The concentration of DNA in each sample was assessed by measuring the absorbance at 260 nm.
Detection of FIV DNA sequences.
FIV DNA was detected by PCR amplification of a conserved region of either the FIV pol (38) or gag (33) genes. PCR reagents were prepared in three separate rooms and included 0.2 mM deoxynucleotide triphosphates, 2.5 mM MgCl2, 0.2 μM concentrations of each primer, 5 mM KCl, 1 mM Tris-HCl, 0.25 U of Taq DNA polymerase, and 2 μl of sample in a total volume of 25 μl. High-molecular-weight DNA isolated from feline PBMC cultures infected with either Petaluma or V1CSF served as a positive control. A 770-bp fragment corresponding to the region from 3361 to 4131 of the pol gene was generated with primers 3361 (5′-GAAGGATCCAGAAAAGATACTATGG-3′) and 4131C (5′-GGCAACATTAGCTTTACCCCTGTTGG-3′) following 30 cycles of 1 min–95°C of denaturation, 1 min–45°C of annealing, and 2 min–72°C of extension, which permitted a linear amplification of template. Nested PCR with primers 3860 (5′-CCAGATATGATGGAGGGAATCT-3′) and 4052 (5′-CATATCCTGCATCTTCTGAACT-3′) generated a 192-bp fragment corresponding to region 3860 to 4052 of the pol gene and was performed by using 2 μl of product amplified in the first PCR reaction with the same reagents at an annealing temperature of 50°C. The presence of FIV DNA was confirmed by Southern blot with an [α-32P]dCTP-labeled oligonucleotide probe (5′-TGTCAAACAATGATGATAATAGAAGG-3′) that detected both the 770- and 192-bp fragments. Amplification and detection of a 293-bp fragment of the gag gene was performed as previously detailed (7).
Flow cytometry.
Analysis of FIV p24 expression in human PBMC infected with Petaluma or V1CSF was performed on a Becton Dickinson FacScan fluorescence activated cell sorter with a 488-nm laser for excitation. Duplicate cell cultures (2 × 106) were harvested on day 4 p.i., washed twice with phosphate-buffered saline (PBS) by centrifugation, and fixed with a solution of 2% formaldehyde in PBS by incubation for 30 min at room temperature. Fixed PBMC were washed three times in PBS containing 0.5% Tween-20 (Sigma, St. Louis, Mo.) and 5% FCS to permeabilize the cells and enable detection of both surface-associated and intracellular p24. Permeabilized cells were incubated for 30 min at 4°C with a 1:100 dilution of one of two monoclonal antibodies specific to FIV p24 (clones 43-1B9 or 43-1E2; a gift of N. C. Pedersen) and washed twice with PBS containing 0.5% Tween 20. Washed PBMC were incubated for 30 min at 4°C with fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin G (0.25 μg/μl; Becton Dickinson, San Jose, Calif.), washed twice, and resuspended in 0.5 ml of PBS. Data were collected from approximately 15,000 events for each experimental condition, and results are expressed as a single-parameter log fluorescence histogram. FIV-infected feline PBMC and uninfected human PBMC served as positive and negative controls, respectively. Appropriate isotype-matched antibody controls were conducted with mouse IgG1 (1 μg/μl; Becton Dickinson).
Chemokine receptor blocking.
IL-2–ConA-stimulated human PBMC (approximately 2 × 106 cells/ml) were incubated at 37°C for 1.5 h with monoclonal antibodies (1 μg/ml) recognizing the chemokine receptors CCR3, CCR5, or CXCR4 (a gift from A. Nath). Cells were centrifuged for 10 min at 1,000 rpm, and the pellet was resuspended in 150 μl of antibody-containing RPMI medium and 150 μl of culture supernatant containing 104.0 TCID50/106 cells of either Petaluma or V1CSF. Cells were incubated at 37°C for 2 h, washed twice with medium, and seeded in 24-well plates. Uninfected PBMC and PBMC infected in the absence of antibodies to any of the chemokine receptors served as negative and positive controls, respectively. To investigate the effect of a nonspecific protein on FIV infection, human PBMC were incubated with 1 μg of either bovine serum albumin (BSA) or anti-rabbit polyclonal antibody (ARPA) per ml, and viral DNA levels were assessed in the same manner as with the chemokine receptor antibodies. Cultures were harvested at 24 h p.i., and high-molecular-weight DNA was isolated. FIV DNA levels were assessed by semiquantitative PCR amplification of the pol gene for 30 cycles and Southern blot by the methods described above. The abundance of FIV genome in each culture was determined by densitometry by using the public domain program NIH Image. Amplification of equal amounts of template DNA from all samples was ensured by PCR amplification of the housekeeping gene, HLA-DQα (22). Levels of FIV DNA in each sample were expressed relative to the amount of template DNA amplified and compared to the positive controls for each strain.
Reverse transcriptase (RTase) assay.
Mg2+-dependent RTase activity in culture supernatants was measured by limiting dilution using a protocol described previously (6). Briefly, 10 μl of culture supernatant was cleared of cellular debris by high-speed centrifugation and incubated with 10 μl of reaction cocktail containing [α-32P]TTP for 2 h at 37°C in a 96-well plate. Samples were blotted on DE81 ion-exchange chromatography paper (Whatman International Ltd., Maidstone, England) and washed three times for 5 min in 0.35 M Na2HPO4 and twice for 5 min in 95% ethanol. RTase levels were measured by liquid scintillation counting and expressed as the counts per minute per milliliter of culture supernatant. All assays were performed in triplicate and repeated a minimum of two times.
FIV-induced cell death.
Triplicate cultures of IL-2–ConA-stimulated human PBMC were seeded at 2 × 106 cells/ml in a 24-well plate and infected with either V1CSF or Petaluma strains of FIV at a titer of 104.0 TCID50/106 cells as described above. Uninfected PBMC served as a negative control. Cells were collected at 3, 7, 10, and 14 days p.i. and stained with trypan blue. Cell death in these cultures was expressed as the number of PBMC with trypan blue-stained nuclei relative to the total number of PBMC.
Statistical analysis.
The statistical significance of the differences between uninfected and FIV-infected cell cultures was determined by Student’s unpaired t test. P values of less than 0.05 were considered significant.
RESULTS
Detection of FIV DNA sequences in human cells.
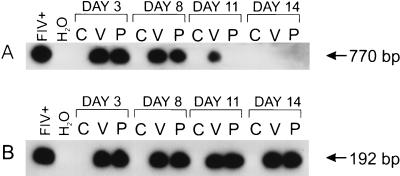
FIV DNA was detected in several primary and immortalized human cell lines after infection with either the Petaluma or V1CSF strains. Viral DNA was detected after a single round of PCR in V1CSF- and Petaluma-infected PBMC for 11 and 8 days p.i., respectively (Fig. 1A), and for 14 days p.i. following nested PCR amplification of these samples (Fig. 1B). Similar results were observed with FIV-infected macrophage cultures (data not shown), but viral DNA could not be detected beyond 5 days in any other human cell line tested (Table 1). In long-term studies, viral DNA was detected consistently in PBMC infected with either strain of FIV for a minimum of 21 days p.i. after nested PCR. Following semiquantitative PCR analysis of serial dilutions of high-molecular-weight DNA from FIV-infected human and feline PBMC, FIV genome was approximately 10 times more abundant in feline cultures than in human cultures (data not shown). These results suggest that FIV infects human cells to a lesser extent than feline cells and in a cell-type restricted manner.
FIG. 1.
Representative Southern blots showing detection of FIV pol sequences. (A) After 30 cycles of PCR, FIV DNA could be detected for a minimum of 8 days in human PBMC infected with Petaluma (P) and up to 11 days p.i. in V1CSF-infected cells (V) but not in uninfected controls (C). Petaluma-infected feline PBMC served as a positive control (FIV+). (B) FIV DNA was detected by nested PCR in human PBMC infected with either strain of the virus at days 3 to 14 p.i.
TABLE 1.
Detection of FIV DNA and RTase activity in infected human cell cultures
| Human cell line | FIV DNA detection (days p.i.)a
|
RTase activity (± SD)b with:
|
||||
|---|---|---|---|---|---|---|
| Petaluma at:
|
V1CSF at:
|
|||||
| Day 3 | Day 7 | Day 3 | Day 7 | Day 3 | Day 7 | |
| PBMC | 21 | 21 | 5.5 ± 0.5* | 7.6 ± 0.6* | 6.3 ± 0.3* | 8.2 ± 0.6* |
| Macrophages | 14 | 14 | 6.9 ± 0.9* | 6.1 ± 0.7* | 6.4 ± 0.9* | 9.2 ± 1.0* |
| THP-1 monocytes | 3 | ND | 2.7 ± 0.4* | 1.2 ± 0.2 | 1.6 ± 0.3 | 0.9 ± 0.1 |
| Fetal astrocytes | 5 | ND | 1.5 ± 0.3 | 1.5 ± 0.2 | 1.2 ± 0.2 | 1.2 ± 0.2 |
| U373 astrocytes | 5 | ND | 1.8 ± 0.2 | 0.9 ± 0.1 | 1.5 ± 0.2 | 0.9 ± 0.1 |
High-molecular-weight DNA from human PBMC and macrophages were amplified by nested PCR; FIV DNA was detected in the remaining cell lines by a single PCR round. ND, not detectable.
RTase activity was assessed as the mean fold increase over uninfected cells, for which RTase levels averaged 4,500 ± 400 cpm/ml. Significant differences between infected and uninfected cultures were determined by using a two-tailed Student’s t test (∗, P < 0.01).
Viral gene expression in FIV-infected human PBMC.
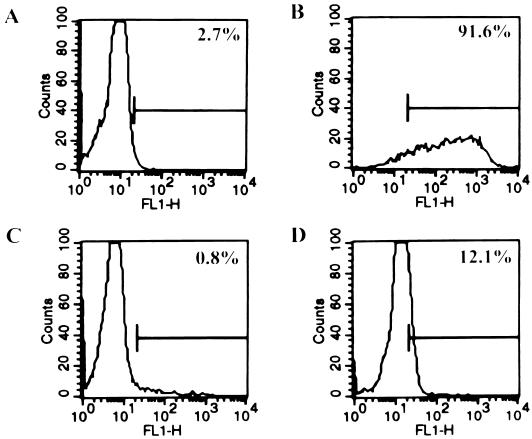
To determine whether the presence of FIV genome in human cells was accompanied by viral gene expression, human PBMC cultures were analyzed for the presence of FIV p24 reactivity by flow cytometry 4 days after infection with either Petaluma or V1CSF. Approximately 12% of cells in Petaluma (Fig. 2D)- and V1CSF-infected human cultures were recognized by monoclonal antibodies specific to FIV p24 compared to 0.8% in uninfected controls (Fig. 2C). In FIV-infected feline PBMC cultures, 91.6% of the cells expressed viral protein (Fig. 2B), supporting PCR evidence that FIV infection of human cells is inefficient compared to infection in its natural host.
FIG. 2.
Representative flow cytometric analysis of uninfected human PBMC (A) and FIV-infected feline (B) and human (C and D) PBMC. By using a monoclonal antibody specific to FIV p24 (clone 43-1B9), viral capsid protein was detected in 12% of Petaluma-infected human PBMC by flow cytometry (D) compared to 0.8 and 91.6% of cells in uninfected human PBMC (C) and FIV-infected feline PBMC (B) cultures, respectively. By using an isotype-matched control antibody, 2.4% of human PBMC were recognized (A).
RTase activity in FIV-infected human cells.
RTase activity was measured to assess viral production in FIV-infected cell cultures, revealing significantly elevated levels in supernatants from human PBMC and macrophage cultures infected with either strain of FIV compared to uninfected control cultures. RTase levels increased in V1CSF-infected PBMC from (6.3 ± 0.3)- to (8.2 ± 0.6)-fold greater than uninfected controls at day 3 and 7 p.i., respectively (Table 1). A similar increase in RTase activity was observed in macrophage cultures infected with the same strain of the virus. RTase levels were also higher in Petaluma-infected human PBMC at day 7 p.i. than day 3, increasing from (5.5 ± 0.5)- to (7.6 ± 0.6)-fold greater than uninfected cells (Table 1). Activity did not vary significantly in macrophages infected with this strain over the same period. A (2.7 ± 0.4)-fold increase in RTase activity was also detected in the THP-1 monocyte cultures infected with the Petaluma strain of FIV, but this activity was not detectable at subsequent time points (Table 1). No significant RTase activity was detected in the other FIV-infected human cell lines studied.
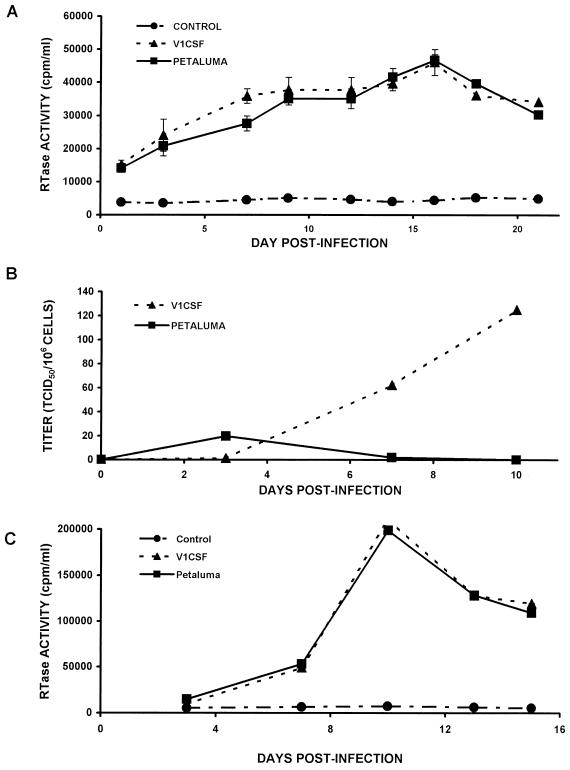
Since RTase levels increased from day 3 to day 7 p.i. and the FIV genome could be detected for at least 21 days p.i. in human PBMC infected with either FIV strain, these cells were chosen as the most promising cell line in which to investigate the infection of human cells beyond 2 weeks p.i. RTase activity was consistently greater in culture supernatants from Petaluma- and V1CSF-infected human PBMC than in supernatants harvested from uninfected controls over a 3-week period (Fig. 3A). RTase levels increased over the time course, reaching a peak activity of approximately 5 × 104 cpm/ml at day 16 p.i. for cells infected with either strain of the virus. In comparison, RTase activity reached maximal levels of 2 × 106 cpm/ml at day 10 p.i. in feline PBMC infected with either FIV strain (Fig. 3C), a finding further supporting the observation that FIV replication is decreased in human cells.
FIG. 3.
RTase activity and viral titer in culture supernatants from human and feline PBMC infected with either Petaluma or V1CSF strains of FIV. (A) RTase activity in human PBMC infected with either strain increased over the time course, reaching a maximum value of approximately 5 × 104 cpm/ml at day 16 p.i. for both viruses. Significant differences between uninfected cells (CONTROL) and PBMC infected by either virus were observed by using a two-tailed Student’s t test (P < 0.01 at all days). (B) Viral titers were assessed in feline PBMC by limiting dilution, and values are expressed as the number of TCID50/106 cells. Virus titers in duplicate cultures of human PBMC infected with V1CSF increased over the time course, reaching a maximum titer of 125 TCID50/106 at day 10 p.i. In contrast, the viral titer in Petaluma-infected human PBMC was maximal at day 3 p.i. and decreased to undetectable levels by day 10. (C) RTase activity in feline PBMC infected with Petaluma or V1CSF peaked at day 10 p.i., reaching maximal values of 2 × 105 cpm/ml.
Viral titer in feline cells.
To determine whether productive infection had occurred, the virus titers of supernatants from human cell cultures that were PCR-positive for FIV DNA sequences were determined in feline PBMC. Infectious virus was detected in supernatants harvested from human PBMC infected with both strains of FIV (Fig. 3B) and in Petaluma-infected THP-1 monocyte cultures. A limited viral titer was detectable in supernatants from V1CSF-infected human PBMC at day 3 p.i., but the virus titer increased to 62.5 × 101.8 and 125 × 102.1 TCID50/106 cells at days 7 and 10, respectively. A viral titer of 19.8 × 101.3 TCID50/106 cells was also detected at 3 days p.i. in culture supernatant from human PBMC infected with Petaluma, but it decreased to undetectable levels by day 10 p.i. A similar pattern was observed in Petaluma-infected THP-1 cultures, with a viral titer of 68.8 × 101.8 TCID50/106 cells detected in culture supernatant harvested on day 3 p.i. (data not shown).
The inability to detect infectious virus in human cultures immediately after inoculation with FIV-containing supernatant indicated that infectious virus from the original inoculum did not persist in these cultures. To further ensure that any residual FIV did not remain infectious and was not responsible for the viral titers detected in the above experiments, culture supernatants containing either Petaluma or V1CSF were incubated in the presence of nonpermissive IC-21 murine macrophages for 3 days prior to infection of human cells. FIV viral DNA, the virus titers that could be determined, and elevated RTase activity were not detected in feline PBMC cultures inoculated with supernatant that had been incubated as described above (data not shown), indicating that the RTase activity and detection of FIV genome resulted from infectious virus after productive infection of human PBMC.
Chemokine receptor utilization by FIV.
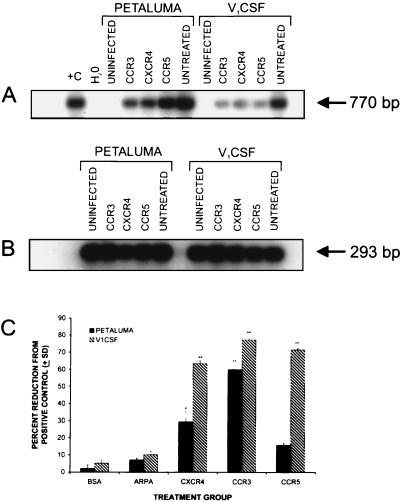
The role of the chemokine receptors in FIV infection of human cells was investigated by treating human PBMC with antibodies that recognized the CCR3, CCR5, or CXCR4 chemokine receptors prior to infection with either Petaluma or V1CSF. The levels of viral genome in FIV-infected human PBMC was determined by semiquantitative PCR and densitometric analysis and compared to PBMC infected in the absence of chemokine receptor antibodies (Fig. 4A). FIV DNA levels were decreased in both Petaluma- and V1CSF-infected cultures after treatment with each of the antibodies, but differences were observed in the extent of inhibition depending on the infecting strain. Levels of FIV genome in Petaluma-infected human PBMC were decreased by 59.8 ± 2.4 and 29.3 ± 5.7% with antibodies to either the CCR3 or CXCR4 receptors compared to untreated controls (Fig. 4C). A decrease in the amount of viral DNA was observed in Petaluma-infected cultures treated with antibodies to the CCR5 receptor, but this effect was not statistically significant. The amount of FIV DNA in V1CSF-infected human PBMC decreased equally after treatment with antibodies to either the CCR3 or the CCR5 receptors, declining by 77.1 ± 1.1 and 71.2 ± 0.7%, respectively, of that detected in untreated control cultures (Fig. 4C). A similar decrease of 63.3 ± 1.6% was observed after treatment with antibodies to the CXCR4 receptor. These findings indicate that antibodies to all of the chemokine receptors investigated reduced FIV infection, but the extent of inhibition varied depending on the viral strain and the specific chemokine receptor, implying a different pattern of usage by each strain.
FIG. 4.
(A) FIV viral DNA detection after incubation with antibodies recognizing the CCR3, CXCR4, or CCR5 chemokine receptors. Uninfected PBMC and PBMC infected with either viral strain but without pretreatment with antibody served as negative and positive controls, respectively. (B) FIV DNA levels were measured relative to the amount of template DNA, as determined by amplification of the HLA-DQα gene. (C) The greatest decrease in the viral DNA levels compared to positive control cultures was observed after treatment with antibodies to the CCR3 chemokine receptor for both strains (59.8 ± 2.4% for Petaluma, 77.1 ± 1.1% for V1CSF). Antibodies to the CCR5 receptor significantly inhibited infection with V1CSF, decreasing detectable FIV DNA levels by 71.2 ± 0.7% of the control value, but not with Petaluma. For both viruses, infection was decreased in the presence of antibodies recognizing the CXCR4 receptor (29.3 ± 5.7% for Petaluma, 63.3 ± 1.6% for V1CSF). Viral DNA levels were not affected by preincubation with nonspecific ARPA or BSA. ∗∗, P < 0.01; ∗, P < 0.05.
FIV-mediated cytotoxicity.
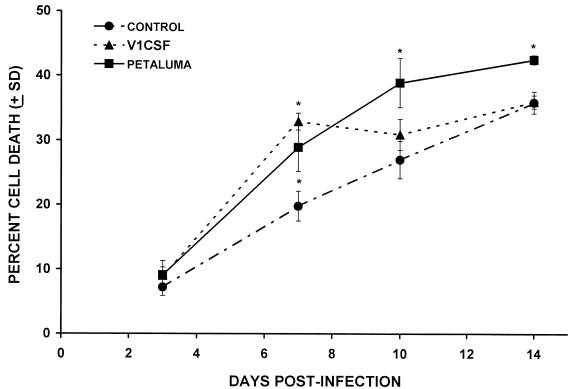
The cell death induced by FIV infection of human PBMC cultures was compared to uninfected cultures by trypan blue staining at sequential days p.i. Although cell death in uninfected PBMC cultures increased with time, a significantly higher level of cell death was observed in Petaluma-infected human PBMC compared to control cultures at days 7, 10, and 14 p.i. (Fig. 5). The greatest difference occurred at day 10 when the nuclei of 38.9 ± 2.4% cells in cultures infected with Petaluma were stained with trypan blue, compared to 27.0 ± 2.9% in uninfected controls. Cell death in PBMC cultures infected with V1CSF was significantly greater than the controls only at day 7 p.i., when a 13.1 ± 5.2% increase in cell death was observed (32.8 ± 3.7% for V1CSF versus 19.8 ± 2.3% for controls), implying that strain-dependent cytotoxicity of PBMC occurred.
FIG. 5.
FIV-induced cytotoxicity in human PBMC cultures infected with either Petaluma or V1CSF strains. Results are expressed as the percentage of cell death and are assessed relative to uninfected (CONTROL) cultures. Petaluma-infected cultures exhibited increased cytotoxicity at days 7, 10, and 14 p.i., with the greatest difference occurring at day 10 when cell death was 38.9 ± 2.4% (P < 0.05) in infected PBMC compared to 27.0 ± 2.9% in control cultures. In comparison, significant cell death of 32.9 ± 3.7% (P < 0.05) was observed only at day 7 p.i. in V1CSF-infected PBMC compared to 19.8 ± 2.3% in control cultures. Significant increases in cell death were determined using a two-tailed Student’s t-test (∗, P < 0.05).
DISCUSSION
In this study, we have demonstrated that cell-free isolates of FIV Petaluma and V1CSF infect several human cell lines in vitro, although prolonged infection is restricted to primary PBMC and monocyte-derived macrophages. In addition to the viral genome, expression of FIV-specific proteins was observed in human PBMC cultures infected with either viral strain. Furthermore, the presence of elevated RTase activity and FIV titers in supernatants from human PBMC infected with V1CSF and Petaluma indicated that productive infection of human cells could occur. The mechanism by which FIV infects human cells is not known, but antibodies recognizing the CXCR4, CCR3, or CCR5 chemokine receptors inhibited the infection of human PBMC by Petaluma and V1CSF to varying degrees. It is also of note that cell death was greater in FIV-infected PBMC relative to uninfected control cultures. In general, FIV infection of human cultures was characterized by viral-strain-dependent properties, most notably, the ability of Petaluma to infect a broader range of human cells than V1CSF.
Recently, Poeschla et al. have proposed that FIV provides a safer alternative to HIV-based viral vectors because wild-type FIV is unable to productively infect human cells (35). However, other reports have shown sustained, productive infection of human cell lines by FIV (10, 26) after transfection or coinfection with FIV-infected cell lines. The current studies pertain to the use of FIV as a viral vector in human gene therapy and indicate that some caution should be exercised in developing lentivirus-based vectors for therapeutic use because of the potential pathogenicity of these vectors. Differences in viral strain pathogenicity and tropism have been reported for several lentiviruses (1, 23, 28), and the present work demonstrates similar differences between V1CSF and Petaluma, in particular, the ability of V1CSF to infect primary human PBMC productively. Therefore, the strains from which FIV vectors are derived are important in terms of safety.
The present study also illustrates the potential capacity of lentiviruses to infect a new host by a common coreceptor mechanism, perhaps providing insight into the evolution of lentiviral infections. Sequence diversity in the env gene of FIV has been shown to influence cell tropism by altering the ability of the envelope from different FIV strains to bind cell surface receptors and mediate viral penetration (43). The differences observed in the patterns of chemokine receptor usage and env gene sequences (19) of V1CSF and Petaluma in the present study may provide a mechanism for explaining the human cell tropism exhibited by these strains. Antibodies that recognized CCR3, the chemokine receptor implicated as a coreceptor for T-cell adapted and primary isolates of primate lentiviruses (2), inhibited Petaluma and V1CSF infection of human PBMC to a greater extent than antibodies to the CCR5 and CXCR4 receptors. Recent studies have demonstrated that human CCR5 does not mediate the infection of human cells by FIV 34TF10, a molecular clone of the FIV Petaluma strain (34). Similarly, antibodies to the CCR5 receptor did not inhibit the infection of human PBMC by a primary isolate of Petaluma in the current experiments, but infection by V1CSF was decreased significantly in the presence of these antibodies. Since CCR5 is most closely associated with infection of macrophages (2), this finding complements the higher levels of RTase activity and FIV DNA detected in V1CSF-infected human macrophages compared to those infected with Petaluma. Antibodies recognizing CXCR4 inhibited infection by either strain of FIV, supporting earlier reports that FIV can use this receptor (45, 34). It is conceivable that the chemokine receptor antibodies investigated nonspecifically blocked viral binding and entry, but this is unlikely given that the two viruses differed in their utilization of chemokine receptors despite similar input titers. The observation that antibodies to more than one chemokine receptor inhibited the infection of human PBMC by FIV is not surprising considering the range of human cells that could be infected by FIV and the fact that the viruses used in the current study were nonhomogeneous primary isolates. Previously, the ability of other lentiviruses to employ multiple chemokine receptors as cofactors for infection has been demonstrated (9). Since a single antibody did not completely block FIV infection of human cells, it is likely that other coreceptors for FIV infection of human cells exist.
Although retroviruses are considered to be species specific, cross-species infection by lentiviruses in vitro has been documented (13, 20). Previous studies report infection of immortalized human cell lines by molecular clones of feline retroviruses, such as FIV (10, 17, 24, 42) and feline leukemia virus (27). However, we have shown cell-free infection by FIV of both proliferating and terminally differentiated primary human cells. Of interest, long-term infection of human cells by FIV was observed only in primary blood-derived cell cultures, as evidenced by the consistent detection of increased RTase activity and viral DNA in FIV-infected human PBMC at consecutive time points. A corresponding increase in infectious virus in human PBMC cultures from day 3 to day 10 p.i. by V1CSF confirmed that productive infection of these cells occurred. Although PCR detection of virus was correlated with RTase activity, the decreasing viral titer in Petaluma-infected human PBMC despite increasing RTase levels indicated that elevated RTase activity did not correlate with the presence of infectious FIV. Since the expression of virus-specific proteins was detected in these cultures, this observation suggests that perhaps replication-defective particles were being produced in human cultures, similar to findings reported for env deletion mutants of FIV that have been adapted to replicate in human cells (35).
Limited viral load and replication has been shown in the early stages of lentiviral infection of the brain (3, 5). Viral DNA was not detectable in FIV-infected human astrocyte and THP-1 monocyte cultures beyond 5 days p.i., possibly because the more sensitive nested PCR was not used. Since limited viral genome was detected in these cultures (data not shown) and FIV replication was not observed, the percentage of cells containing FIV DNA may have fallen below the level of detection of a single round of PCR. Alternatively, the failure to detect FIV DNA in some human cell types may result from an inability of the virus to integrate in these cells. Viral integration into the host cell genome is a requisite feature of productive retroviral infection (12, 39); therefore, integration by FIV is also likely to be necessary for productive infection of human cells. Although integrated FIV provirus was not detectable by PCR analysis in human cells infected with Petaluma or V1CSF in the present study (data not shown), the presence of FIV genome in high-molecular-weight DNA from these cultures implies that integration may have occurred, as was suggested for Petaluma-infected human MOLT-4 cells (42).
The increased cell death that preceded a loss of infectious FIV in infected human PBMC supports previous findings that infection of human cells by FIV is cytopathic, which is probably due to the expression of FIV envelope glycoproteins (34). FIV infection of relatively few cells in culture has been associated with increased cytotoxicity in feline cultures due to the release of cytotoxic molecules (4, 18), which is similar to reports of other lentiviruses. Hence, it is conceivable that FIV-mediated cytotoxicity may limit the number of infected and potentially infectable cells leading to the loss of detectable FIV DNA in infected human cultures. Furthermore, the percentage of human PBMC expressing FIV p24 after infection with either Petaluma or V1CSF is consistent with the increased levels of cell death observed in FIV-infected human PBMC cultures.
FIV may be a suitable candidate for use as a vector in human gene therapy because viral gene expression occurs in human cells, but consideration should be given to the viral strain being used. Replacing the U3 region of the LTR of the molecular clone of Petaluma (34TF10) reveals that this domain is the principal limiting factor to productive infection of human cells by FIV (35). As the current study demonstrates, however, some primary isolates of FIV, such as V1CSF, can also replicate in human cells. Although the three-vector system reported by Poeschla et al. is unlikely to result in infectious FIV through recombination, greater safety can be attained in the development of vectors for clinical use by selecting viral strains that are unable to productively infect human cells.
ACKNOWLEDGMENTS
We wish to thank W. Maury, K. Coombs, P. Lee, A. Nath, and M. Mayne for helpful discussions and N. C. Pedersen for supplying reagents.
These studies were supported by NSERC (Canada) and the Hospital for Sick Children Research Foundation. J.J. is supported by an MHRC studentship. C.P. is supported by an NHRDP/MRC scholarship.
REFERENCES
- 1.Andresdottir V, Tang X, Agnarsdottir G, Andresson O S, Georgsson G, Skraban R, Torsteinsdottir S, Rafnar B, Benediktsdottir E, Matthiasdottir S, Arnadottir S, Hognadottir S, Palsson P A, Petursson G. Biological and genetic differences between lung- and brain-derived isolates of maedi-visna virus. Virus Genes. 1998;16:281–293. doi: 10.1023/a:1008030706308. [DOI] [PubMed] [Google Scholar]
- 2.Bazan H A, Alkhatib G, Broder C C, Berger E A. Patterns of CCR5, CXCR4, and CCR3 usage by envelope glycoproteins from human immunodeficiency virus type 1 primary isolates. J Virol. 1998;72:4485–4491. doi: 10.1128/jvi.72.5.4485-4491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell J E, Busuttil A, Ironside J W, Rebus S, Donaldson Y K, Simmonds P, Peutherer J F. Human immunodeficiency virus and the brain: investigation of virus load and neuropathologic changes in pre-AIDS subjects. J Infect Dis. 1993;168:818–824. doi: 10.1093/infdis/168.4.818. [DOI] [PubMed] [Google Scholar]
- 4.Bishop S A, Williams N A, Gruffydd Jones T J, Harbour D A, Stokes C R. Impaired T-cell priming and proliferation in cats infected with feline immunodeficiency virus. AIDS. 1992;6:287–293. doi: 10.1097/00002030-199203000-00005. [DOI] [PubMed] [Google Scholar]
- 5.Boche D, Hurtrel M, Gray F, Claessens-Maire M A, Ganiere J P, Montagnier L, Hurtrel B. Virus load and neuropathology in the FIV model. J Neurovirol. 1996;2:377–387. doi: 10.3109/13550289609146903. [DOI] [PubMed] [Google Scholar]
- 6.Chesebro B, Wehrly K. Development of a sensitive quantitative focal assay for human immunodeficiency virus infectivity. J Virol. 1988;62:3779–3788. doi: 10.1128/jvi.62.10.3779-3788.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diehl L J, Mathiason DuBard C K, O’Neil L L, Hoover E A. Longitudinal assessment of feline immunodeficiency virus kinetics in plasma by use of a quantitative competitive reverse transcriptase PCR. J Virol. 1995;69:2328–2332. doi: 10.1128/jvi.69.4.2328-2332.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diehl L J, Mathiason Dubard C K, O’Neil L L, Obert L A, Hoover E A. Induction of accelerated feline immunodeficiency virus disease by acute-phase virus passage. J Virol. 1995;69:6149–6157. doi: 10.1128/jvi.69.10.6149-6157.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doranz B J, Rucker J, Yanjie Y, Smyth R, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptor CKR-5, CKR-3, and CKR-2b as fusion co-factors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 10.Dow S W, Dreitz M J, Hoover E A. Feline immunodeficiency virus neurotropism: evidence that astrocytes and microglia are the primary target cells. Vet Immunol Immunopathol. 1992;35:23–35. doi: 10.1016/0165-2427(92)90118-a. [DOI] [PubMed] [Google Scholar]
- 11.English R V, Johnson C M, Gebhard D H, Tompkins M B. In vivo lymphocyte tropism of feline immunodeficiency virus. J Virol. 1993;67:5175–5186. doi: 10.1128/jvi.67.9.5175-5186.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Englund G, Theodore T S, Freed E O, Engleman A, Martin M A. Integration is required for productive infection of monocyte-derived macrophages by human immunodeficiency virus type 1. J Virol. 1995;69:3216–3219. doi: 10.1128/jvi.69.5.3216-3219.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Georgiades J A, Billiau A, Vanderschueren B. Infection of human cell cultures with bovine visna virus. J Gen Virol. 1978;38:375–381. doi: 10.1099/0022-1317-38-2-375. [DOI] [PubMed] [Google Scholar]
- 14.Greene W K, Meers J, del Fierro G, Carnegie P R, Robinson W F. Extensive sequence variation of feline immunodeficiency virus env genes in isolates from naturally infected cats. Arch Virol. 1993;133:51–62. doi: 10.1007/BF01309743. [DOI] [PubMed] [Google Scholar]
- 15.Hohdatsu T, Hirabayashi H, Motokawa K, Koyama H. Comparative study of the cell tropism of feline immunodeficiency virus isolates of subtypes A, B and D classified on the basis of the env gene V3-V5 sequence. J Gen Virol. 1996;77:93–100. doi: 10.1099/0022-1317-77-1-93. [DOI] [PubMed] [Google Scholar]
- 16.Hosie M J, Broere N, Hesselgesser J, Turner J D, Hoxie J A, Neil J C, Willett B J. Modulation of feline immunodeficiency virus infection by stromal cell-derived factor. J Virol. 1998;72:2097–2104. doi: 10.1128/jvi.72.3.2097-2104.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ikeda Y, Tomonaga K, Kawaguchi Y, Kohmoto M, Inoshima Y, Tohya Y, Miyazawa T, Kai C, Mikami T. Feline immunodeficiency virus can infect a human cell line (MOLT-4) but establishes a state of latency in the cells. J Gen Virol. 1996;77:1623–1630. doi: 10.1099/0022-1317-77-8-1623. [DOI] [PubMed] [Google Scholar]
- 18.Johnson C M, Benson N A, Papadi G P. Apoptosis and CD4+ lymphocyte depletion following feline immunodeficiency virus infection of a T-lymphocyte cell line. Vet Pathol. 1996;33:195–203. doi: 10.1177/030098589603300209. [DOI] [PubMed] [Google Scholar]
- 19.Johnston J, Power C. The FASEB journal volume 12, experimental biology 98, abstracts part II. San Francisco, Calif: Federation of American Societies for Experimental Biology; 1998. Feline immunodeficiency virus tropism in human cells: evidence for strain-dependent infection, abstr. 4689; p. A809. [Google Scholar]
- 20.Koenig S, Hirsch V M, Olmsted R A, Powell D, Maury W, Rabson A, Fauci A S, Purcell R H, Johnson P R. Selective infection of human CD4+ cells by simian immunodeficiency virus: productive infection associated with envelope glycoprotein-induced fusion. Proc Natl Acad Sci USA. 1989;86:2443–2447. doi: 10.1073/pnas.86.7.2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maki N, Miyazawa T, Fukasawa M, Hasegawa A, Hayami M, Miki K, Mikami T. Molecular characterization and heterogeneity of feline immunodeficiency virus isolates. Arch Virol. 1992;123:29–45. doi: 10.1007/BF01317136. [DOI] [PubMed] [Google Scholar]
- 22.Mayne M, Krishnan J, Metz L, Nath A, Auty A, Sahai B, Power C. Infrequent detection of human herpesvirus 6 DNA in peripheral blood mononuclear cells from multiple sclerosis patients. Ann Neurol. 1998;44:391–394. doi: 10.1002/ana.410440317. [DOI] [PubMed] [Google Scholar]
- 23.McCarthy M, He J, Wood C. HIV-1 strain-associated variability in infection of primary neuroglia. J Neurovirol. 1998;4:80–89. doi: 10.3109/13550289809113484. [DOI] [PubMed] [Google Scholar]
- 24.Miyazawa T, Kawaguchi Y, Kohmoto M, Sakuragi J, Adachi A, Fukasawa M, Mikami T. Production of feline immunodeficiency virus in feline and non-feline non-lymphoid cell lines by transfection of an infectious molecular clone. J Gen Virol. 1992;73:1543–1546. doi: 10.1099/0022-1317-73-6-1543. [DOI] [PubMed] [Google Scholar]
- 25.Miyazawa T, Kawaguchi Y, Kohmoto M, Tomonaga K, Mikami T. Comparative functional analysis of the various lentivirus long terminal repeats in human colon carcinoma cell line (SW480 cells) and feline renal cell line (CRFK cells) J Vet Med Sci. 1994;56:895–899. doi: 10.1292/jvms.56.895. [DOI] [PubMed] [Google Scholar]
- 26.Miyazawa T, Tomonaga K, Kawaguchi Y, Mikami T. The genome of feline immunodeficiency virus. Arch Virol. 1994;134:221–234. doi: 10.1007/BF01310563. [DOI] [PubMed] [Google Scholar]
- 27.Morgan R A, Dornsife R E, Anderson W F, Hoover E A. In vitro infection of human bone marrow by feline leukemia viruses. Virology. 1993;193:439–442. doi: 10.1006/viro.1993.1141. [DOI] [PubMed] [Google Scholar]
- 28.Naidu Y M, Kestler H W D, Li Y, Butler C V, Silva D P, Schmidt D K, Troup C D, Sehgal P K, Sonigo P, Daniel M D, et al. Characterization of infectious molecular clones of simian immunodeficiency virus (SIVmac) and human immunodeficiency virus type 2: persistent infection of rhesus monkeys with molecularly cloned SIVmac. J Virol. 1988;62:4691–4696. doi: 10.1128/jvi.62.12.4691-4696.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nath A, Hartloper V, Furer M, Fowke K R. Infection of human fetal astrocytes with HIV-1: viral tropism and the role of cell-to-cell contact in viral transmission. J Neuropathol Exp Neurol. 1995;54:320–330. doi: 10.1097/00005072-199505000-00005. [DOI] [PubMed] [Google Scholar]
- 30.Olmsted R A, Hirsch V M, Purcell R H, Johnson P R. Nucleotide sequence analysis of feline immunodeficiency virus: genome organization and relationship to other lentiviruses. Proc Natl Acad Sci USA. 1989;86:8088–8092. doi: 10.1073/pnas.86.20.8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pancino G, Castelot S, Sonigo P. Differences in feline immunodeficiency virus host cell range correlate with envelope fusogenic properties. Virology. 1995;206:796–806. doi: 10.1006/viro.1995.1002. [DOI] [PubMed] [Google Scholar]
- 32.Pedersen N C. The feline immunodeficiency virus. In: Levy J A, editor. The Retroviridae. New York, N.Y: Plenum Press, Inc.; 1993. pp. 181–228. [Google Scholar]
- 33.Phillips T R, Talbott R L, Lamont C, Muir S, Lovelace K, Elder J H. Comparison of two host cell range variants of feline immunodeficiency virus. J Virol. 1990;64:4605–4613. doi: 10.1128/jvi.64.10.4605-4613.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poeschla E M, Looney D J. CXCR4 is required by a nonprimate lentivirus: heterologous expression of feline immunodeficiency virus in human, rodent, and feline cells. J Virol. 1998;72:6858–6866. doi: 10.1128/jvi.72.8.6858-6866.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poeschla E M, Wong-Staal F, Looney D J. Efficient transduction of nondividing human cells by feline immunodeficiency virus lentiviral vectors. Nat Med. 1998;4:354–357. doi: 10.1038/nm0398-354. [DOI] [PubMed] [Google Scholar]
- 36.Poss M L, Dow S W, Hoover E A. Cell-specific envelope glycosylation distinguishes FIV glycoproteins produced in cytopathically and noncytopathically infected cells. Virology. 1992;188:25–32. doi: 10.1016/0042-6822(92)90731-4. [DOI] [PubMed] [Google Scholar]
- 37.Power C, McArthur J C, Johnson R T, Griffin D E, Glass J D, Dewey R, Chesebro B. Distinct HIV-1 env sequences are associated with neurotropism and neurovirulence. Curr Top Microbiol Immunol. 1995;202:89–104. doi: 10.1007/978-3-642-79657-9_7. [DOI] [PubMed] [Google Scholar]
- 38.Power C, Moench T, Peeling J, Kong P A, Langelier T. Feline immunodeficiency virus causes increased glutamate levels and neuronal loss in brain. Neuroscience. 1997;77:1175–1185. doi: 10.1016/s0306-4522(96)00531-3. [DOI] [PubMed] [Google Scholar]
- 39.Sakai H, Kawamura M, Sakuragi J, Sakuragi S, Shibata R, Ishimoto A, Ono N, Ueda S, Adachi A. Integration is essential for efficient gene expression of human immunodeficiency virus type 1. J Virol. 1993;67:1169–1174. doi: 10.1128/jvi.67.3.1169-1174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sodora D L, Shpaer E G, Kitchell B E, Dow S W, Hoover E A, Mullins J I. Identification of three feline immunodeficiency virus (FIV) env gene subtypes and comparison of the FIV and human immunodeficiency virus type 1 evolutionary patterns. J Virol. 1994;68:2230–2238. doi: 10.1128/jvi.68.4.2230-2238.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sparger E E, Shacklett B L, Renshaw Gegg L, Barry P A, Pedersen N C, Elder J H, Luciw P A. Regulation of gene expression directed by the long terminal repeat of the feline immunodeficiency virus. Virology. 1992;187:165–177. doi: 10.1016/0042-6822(92)90305-9. [DOI] [PubMed] [Google Scholar]
- 42.Tochikura T S, Tanabe Tochikura A, Hayes K A, Lazo A, Bailer R T, Blakeslee J R, Jr, Lafrado L J, Roy Burman P, Pandey R, Olsen R G, et al. Fusion activity dissociated from replication ability in feline immunodeficiency virus (FIV) in human cells. J Acquired Immune Defic Syndr. 1993;6:1301–1310. [PubMed] [Google Scholar]
- 43.Vahlenkamp T W, Verschoor E J, Schuurman N N, van Vliet A L, Horzinek M C, Egberink H F, de Ronde A. A single amino acid substitution in the transmembrane envelope glycoprotein of feline immunodeficiency virus alters cellular tropism. J Virol. 1997;71:7132–7135. doi: 10.1128/jvi.71.9.7132-7135.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verschoor E J, Boven L A, Blaak H, van Vliet A L, Horzinek M C, de Ronde A. A single mutation within the V3 envelope neutralization domain of feline immunodeficiency virus determines its tropism for CRFK cells. J Virol. 1995;69:4752–4757. doi: 10.1128/jvi.69.8.4752-4757.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Willett B J, Hosie M J, Jarrett O, Neil J C. Identification of a putative cellular receptor for feline immunodeficiency virus as the feline homologue of CD9. Immunology. 1994;81:228–233. [PMC free article] [PubMed] [Google Scholar]
- 46.Willett B J, Picard L, Hosie M J, Turner J D, Adema K, Clapham P R. Shared usage of the chemokine receptor CXCR4 by the feline and human immunodeficiency viruses. J Virol. 1997;71:6407–6415. doi: 10.1128/jvi.71.9.6407-6415.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang L, He T, Talal A, Wang G, Frankel S S, Ho D D. In vivo distribution of the human immunodeficiency virus/simian immunodeficiency virus coreceptors: CXCR4, CCR3, and CCR5. J Virol. 1998;72:5035–5045. doi: 10.1128/jvi.72.6.5035-5045.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]