Abstract
Previous studies have demonstrated the tumor-suppressive function of microRNA-22-3p (miR-22-3p) in several cancers, whereas the significance of miR-22-3p in non-small cell lung cancer (NSCLC) remains unclear. In this study, we explored the biological function and molecular mechanism of miR-22-3p in NSCLC cells. First, we assessed the expression of miR-22-3p in NSCLC tissues and cells based on RT-qPCR and TCGA database. Compared with normal lung tissues and cells, miR-22-3p expression was dramatically decreased in lung cancer tissues and cells. miR-22-3p expression was also correlated with lymph node metastasis and tumor size, but not TNM stages. We further explored the in vitro function of miR-22-3p on the migration and epithelial–mesenchymal transition (EMT) of NSCLC cells. The results showed that overexpression of miR-22-3p suppressed the migration and EMT of NSCLC cells, whereas silencing miR-22-3p showed the opposite effect. Luciferase assay demonstrated that RAS-related C3 botulinum toxin substrate 1 (RAC1) was the target gene for miR-22-3p. Mechanistically, we demonstrated that miR-22-3p suppressed the cell migration and EMT via downregulation of RAC1 because the inhibitory effect of miR-22-3p on cell migration and EMT of NSCLC cells was reversed by RAC1 overexpression. Based on these novel data, the miR-22-3p/RAC1 axis may be an alternative target in the therapeutic intervention of NSCLC.
Keywords: miR-22-3p, Lung cancer, RAC1, Migration, EMT, Luciferase assay
Introduction
Lung cancer is the second most commonly diagnosed cancer worldwide and was the leading cause of cancer-related deaths in 2020, with approximately 2.2 million new cancer cases (11.4%) and 1.8 million deaths (18%) (Sung et al. 2021). The main cause of lung carcinogenesis is tobacco consumption, followed by other etiological factors like occupational exposures, air pollution, poor diet, and genetic susceptibility; these may be individual risk factors or combined with cigarette smoking in shaping the descriptive epidemiology of lung cancer (Malhotra et al. 2016). Approximately 80% of lung cancer cases are non-small cell lung carcinomas (NSCLCs), while the remaining ones are small cell lung cancers (SCLCs). Recently, many revolutionary advances have been made in managing lung cancer in screening, diagnosis, and therapy using low-dose CT screening, immunotherapy, and molecular-targeted therapy (Alexander et al. 2020). The estimated 5-year survival rate of lung cancer patients diagnosed in 2010–2014 ranged between 10 and 30% in 34 countries, and the survival increased by more than 10% in China (Allemani et al. 2018).
miRNAs are small non-coding RNAs that regulate the expression of thousands of genes post-transcriptionally in normal physiological or disease contexts (Pritchard et al. 2012). miRNA expression profiling has been found to be related to tumor progression, development, and therapeutic reaction, which suggests their probable application as predictive, diagnostic, and prognostic biomarkers (Iorio and Croce 2012). In lung cancer, increasing research indicates that miRNAs are critical in modifying the microenvironment, enhancing tumor progression, angiogenesis, metastasis, invasion, and drug resistance (Wu et al. 2019; Hu et al. 2020). For example, microRNA-1246 was down-regulated and suppressed the invasion of NSCLC cells through negative regulation of CXCR4 (Xu et al. 2018). Another study identified that microRNA-218 down-regulation was associated with the worse prognosis of NSCLC patients. Silencing of microRNA-218 contributed to the enhanced the progression of NSCLC cells in vitro and in vivo by repressing IL-6 receptor (Yang et al. 2017). In contrast to the function of microRNA-1246 and microRNA-218, miR-21 functions as a tumor-promoting microRNA in NSCLC. miR-21-5p/SMAD7 axis promoted the progression of NSCLC (Tang et al. 2021). These evidences suggest that dysregulation of microRNAs play an essential role in NSCLC development.
According to various reports, miR-22-3p serves as a tumor suppressor in several cancers including breast (Wang et al. 2021), glioblastoma (Ma et al. 2021a), bladder (Tian et al. 2021), colorectal (Wang and Lin 2021), gastric (Zhang et al. 2020), hepatocellular (Chen et al. 2016), cervical (Lv et al. 2018), and osteosarcoma (Xue et al. 2022). In lung cancer, Ma et al. (Ma et al. 2021b) reported decreased miR-22-3p expression, correlating with patients’ prognosis of lung adenocarcinoma and clinicopathological characteristics. Yang et al. (Yang et al. 2021) found that miR-22-3p suppressed lung cancer cell growth through MET/STAT3 signaling. In addition, Jiang et al. (Jiang et al. 2019) revealed that miR-22-3p strengthened the radiosensitivity of small cell lung cancers via targeting WRNIP1. Furthermore, Zhang et al. (Zhang et al. 2017) showed that miR-22 inhibited lung cancer cell EMT and invasion through targeting snail. Targets of miR-22-3p involved in lung cancer progression included yes-associated protein (YAP1) (He et al. 2020), MET (Yang et al. 2021), ATP citrate lyase (ACLY) (Xin et al. 2016), enolase1 (ENO1) (Zhou et al. 2019), and MALAT1 (Li et al. 2020). These studies demonstrate the crucial role of miR-22-3p during malignant tumor progression. Targeting the substrates of miR-22-3p might be helpful for the treatment of cancer patients with lowly expressed miR-22-3p. Although numbers of targets of miR-22-3p have been identified by various studies, none of the drugs or inhibitors against these targets is under consideration in clinical studies. Thus, discovering novel substrates of miR-22-3p in malignant cancer, such as NSCLC, might help the pharmaceuticals companies developing effective drugs to treat the malignant cancers triggered by down-regulation of miR-22-3p.
In this study, we aimed to explore the clinical relevance, biology function, and the downstream targets of miR-22-3p in NSCLC. Our findings highlight a new axis of miR-22-3p/RAS-related C3 botulinum toxin substrate 1 (RAC1) involved in lung cancer cell migration and EMT.
Materials and methods
Samples from The Genome Cancer Atlas (TCGA) and clinical patients
We collected a total of 23 pairs of NSCLC tissue samples and matched normal tissues from the Department of Thoracic Surgery in the Second Affiliated Hospital, the Air Force Medical University, which was authorized by the Ethics Committee of the Second Affiliated Hospital, the Air Force Medical University (ethical approval number was TDLL-202304–03). We analyzed clinical data of the 23 pairs of NSCLC samples to survey the relationship between miR-22-3p expression level and clinicopathological characteristics (including smoking history, age, tumor differentiation, gender, tumor size, lymphatic metastasis, and TNM classification). To further explore the clinical significance of miR-22-3p in NSCLC from TCGA data, we analyzed the expression miR-22-3p in NSCLC and normal samples according to the dataset of ENCORI (https://starbase.sysu.edu.cn/panCancer.php). A total of 1085 cancer samples and 104 normal tissues derived from NSCLC patients were included in this study.
Cell culture and transfection
We acquired the human NSCLC cell lines A549 and Calu-1, and the bronchial epithelial cell line BEAS-2B, from the American Type Culture Collection (ATCC, Manassas, VA, USA), with cells cultured in DMEM/F-12 medium (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, USA) and 100 U/mL penicillin/streptomycin (Sigma, USA) in a 5% CO2 incubator at 37 °C.
We synthesized mimics-NC (5′-UUUGUACUACACAAAAGUACUG-3′), miR-22-3p inhibitor (5′-ACAGUUCUUCAACUGGCAGCUU-3′), miR‐22-3p mimics (5′-AAGCUGCCAGUUGAAGAACUGU-3′), inhibitor-NC (5′-CAGUACUUUUGUGUAGUACAAA-3’) using HippoBiotec (Huzhou, China), and cloned ORF of RAC1 (NM_006908.5) into pcDNA3.1 vector (Promega) for overexpression of RAC1. Cell transfection was done using Lipofectamine 2000 (Invitrogen) as per the manufacturer’s protocol. Two days later, the cells were subjected to other experiments.
Real-time quantitative polymerase chain reaction (RT-qPCR)
Total RNA from tissues samples or transfected cells was extracted using RNAiso Plus Kit (TaKaRa, Japan) as per the manufacturer’s guidelines. The concentration of total RNA was determined by using the NanoDrop. We adopted SuperScript III® (Invitrogen) for cDNA amplification. qPCR analysis was carried out using an AceQ RT-qPCR Kit (Vazyme) on a BioRad CFX96 Sequence Detection System (BioRad company, Berkeley, CA, USA). The cycling condition was described as follows: step 1, holding stage, 95 ℃ for 3 min; step 2, cycling stage, 95 ℃ for 15 s; step 3, cycling stage, 60 ℃ for 30 s; step 4, cycling stage, 72 ℃ for 30 s; step 2 to step 4, 40 cycles. Primer of miR-22-3p and U6 was purchased from Ruibo (Guangzhou). Primer sequences of RAC1 and β-actin were as follows: RAC1, forward, 5′-ATGTCCGTGCAAAGTGGTATC-3′, reverse, 5′-CTCGGATCGCTTCGTCAAACA-3′; β-actin, forward, 5′-CATGTACGTTGCTATCCAGGC-3′, reverse, 5′-CTCCTTAATGTCACGCACGAT-3′. We normalized mRNA levels using U6 and β-actin. The relative expression of miR-22-3p or RAC1 was analyzed using the 2 − ΔΔCt method.
Wound healing assay
We tested the migratory capacity of cells using wound healing assays. Transfected cells were cultured in six-well plates for 1 day. Then, a 10-μL pipette was used to scratch a wound into the middle part of each well, followed by a change of medium to 1% FBS-containing DMEM medium. Photos of the wound at 0 and 24 h were taken. The migration percentage was then calculated using Image Pro Plus software (IPP; produced by Media Cybernetics Corporation, USA).
Transwell assay
Transwell cell migration assays were conducted using 24-well transwell chambers (Costar, San Diego, CA, USA). Briefly, transfected cells in non-serum medium with a density of 1 × 105 cells per 500 µL were placed in the upper chambers. In the lower chambers, 500 µL of absolute DMEM medium (containing 10% FBS) was added. After 2 days of incubation, the cells on the upper surface of the chambers were removed by cotton swab and migrated cells were fixed using 4% paraformaldehyde, then stained for 30 min using 1% crystal violet at normal atmospheric temperature. Invaded cells were counted using a microscope (IM 1200, Countstar).
Western blot
Transfected cells were cultured for 2 days, then collected and lysed by vortexing with acid‐washed glass balls for 2 min, then placed on ice for 2 min; this was repeated 10 times. The supernatant was gathered after 30 min of centrifugation at 12,000 rpm, and protein concentration measured with NanoDrop (Thermo-Fisher Scientific). Protein (20 μg) was loaded onto 10% SDS-PAGE gels for 1.5 h, then transferred onto polyvinylidene difluoride membranes for 1 h, followed by blocking for 2 h in non-fat milk (5%) in Tris-buffered saline solution containing 0.1% Tween‐20, at normal atmospheric temperature. We then probed the membranes overnight at 4 °C with primary antibodies for Vimentin (1:1000, ab137321, Abcam), GAPDH (1:1000, ab9485, Abcam), N-cadherin (1:1000, ab76057, Abcam), E-cadherin (1:1000, ab231303, Abcam), β-actin (1:1000; sc-47778, Santa Cruz), and RAC1 (1:1000, ab155938, Abcam). We added horseradish peroxidase‐conjugated secondary antibody (anti‐mouse, 1:2000, Santa Cruz) for 2 h at normal atmospheric temperature and visualized protein bands using ECL chemiluminescence detection kit (Thermo-Fisher Scientific).
Luciferase assay
TargetScan (http://www.targetscan.org), a website widely used for prediction of microRNA targets, was applied to predict the targets of miR-22-3p. Based on the results, numbers of genes were the potential targets of miR-22-3p, including ENO1, SNAIL1, and RAC1. We focused on RAC1 gene in subsequent experiments. As shown in the website, the combination point of miR‐22-3p and RAC1 3′-UTR regions is GCAGCUC and GCAGCUU. The mutation sequence of RAC1‐3′-UTR is CGUCGAC and CGTCGAA, respectively. The 3′-UTR region (WT and Mut) of the RAC1 gene were sub-cloned into pGL 3.1 luciferase reporter vector (Promega). We cultured transfected 293 T cells into 96-well plates at a density of 5 × 104 cells per well, and measured luciferase activity with the Dual-Luciferase Reporter Assay System kit (Promega) on EnSpire Multimode Plate Reader (PerkinElmer, Waltham, MA, USA) according to manufacturer’s instruction, with Renilla luciferase activity as normalized control.
Statistical analysis
All data are presented as mean ± SEM with three independent repeats. Student’ t-test was used to compare the difference between two groups. One-way analysis of variance (ANOVA) followed by a Tukey’s post hoc test was applied to analyze the difference among more than two groups. P values < 0.05 were considered statistically significant.
Results
miR-22-3p expression is decreased in NSCLC tissue and cells
We assessed miR-22-3p expression in NSCLC tissues and cells with real-time quantitative polymerase chain reaction (RT-qPCR). We found that miR-22-3p expression in NSCLC tissues was significantly lower than that of neighboring normal tissues (Fig. 1A). miR-22-3p expression in NSCLC samples (n = 1085) was dramatically decreased as compared to normal samples (n = 104) from TCGA database (Fig. 1B). The patients’ clinical data showed that the expression of miR-22-3p influenced the patients’ lymphatic metastasis and tumor size (P < 0.05), but not in gender, smoking history, age, or TNM classification (Table 1). Additionally, miR-22-3p expression in lung cancer cell lines A549 and Calu-1 was detected with RT‐qPCR. The level of miR-22-3p was decreased in lung cancer cells when compared with human lung epithelial cells BEAS‐2B (Fig. 1C). These results are consistent with previous studies (Ma et al. 2021b; Zhou et al. 2019), demonstrating miR-22-3p as a potential suppressor in NSCLC.
Fig. 1.
Decreased miR-22-3p expression in NSCLC tissues and cells. A miR-22-3p expression measured in 23 paired NSCLC and neighboring normal tissues using RT‐qPCR. B miR-22-3p expression measured for NSCLC and normal samples in TCGA database. C miR-22-3p expression measured in two NSCLC cell lines (Calu-1 and A549) and normal lung epithelial cell line (BEAS‐2B) using real-time quantitative polymerase chain reaction (RT‐qPCR). ** P < 0.01, *** P < 0.001
Table 1.
Correlation of miR-22-3p expression with clinicopathological characteristics in 23 patients of lung cancer
| Variables | N | miR-22-3p expression | P Value | |
|---|---|---|---|---|
| High expression | Low expression | |||
| Gender | ||||
| Male | 12 | 6 | 4 | |
| Female | 11 | 5 | 6 | 0.505 |
| Age | ||||
| <60 | 9 | 7 | 2 | |
| >60 | 14 | 8 | 6 | 0.311 |
| Smoking history | ||||
| Yes | 10 | 4 | 6 | |
| No | 13 | 8 | 5 | 0.305 |
| Tumor differentiation | ||||
| High-Middle | 7 | 2 | 5 | |
| Low | 16 | 10 | 6 | 0.134 |
| Tumor size | ||||
| >3 | 13 | 4 | 9 | |
| <3 | 10 | 8 | 2 | 0.019 |
| Lymphatic matastasis | ||||
| Yes | 8 | 2 | 6 | |
| No | 15 | 11 | 4 | 0.026 |
| TNM calassification | ||||
| I+II | 9 | 4 | 5 | |
| III+IV | 14 | 9 | 5 | 0.349 |
miR-22-3p negatively regulates the migration of lung cancer cells
Both A549 and Calu-1 were NSCLC cell lines, and they had moderate level of miR-22-3p. The cell lines were suitable for overexpression and inhibition of miR-22-3p experiments. Thus, gain- and loss-of-function studies were carried out to investigate the effect of miR-22-3p on cell migration. RT-qPCR results confirmed the efficacy of miR-22-3p overexpression or knockdown in A549 and Calu-1 cells (Fig. 2A, 2B). Scratch wound assay results demonstrated that the migration of A549 and Calu-1cells was suppressed by miR-22-3p ectopic expression, whereas it was promoted by miR-22-3p silencing (Fig. 2C). The migrated cell numbers were statistically analyzed in Fig. 2D. This suggests the negative regulation of cell migration by miR-22-3p in NSCLC cells.
Fig. 2.
miR-22-3p negatively regulates the migration of NSCLC cells. A, B mRNA expression of miR-22-3p in Calu-1 and A549 cells transfected with control, mimics negative control (NC), miR-22-3p mimics, inhibitor NC, and miR-22-3p inhibitor. C, D Representative images and statistical analysis of cell migration of A549 and Calu-1 cells transfected with control, mimics NC, miR-22-3p mimics, inhibitor NC, and miR-22-3p inhibitor measured using wound healing assay. ** P < 0.01, *** P < 0.001
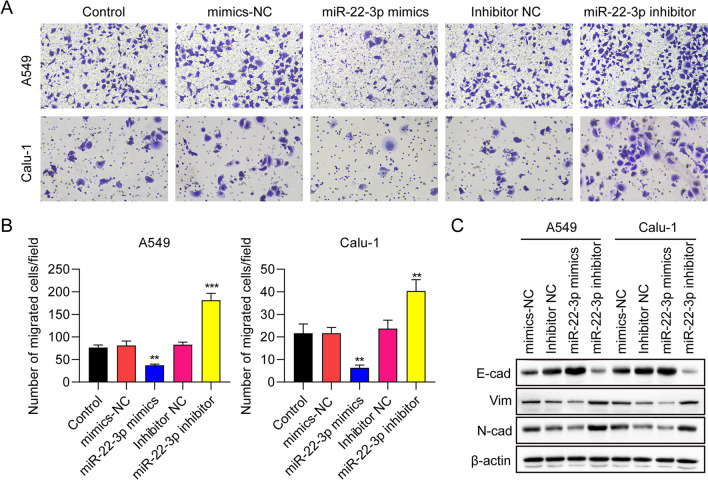
miR-22-3p negatively regulated epithelial-mesenchymal transition (EMT) of NSCLC cells
In order to explore the effects of miR-22-3p on EMT in NSCLC cells, we used the transwell migration assay. We found that overexpression of miR-22-3p reduced the migration of A549 and Calu-1 cells, while silencing of miR-22-3p enhanced the migration of both cell lines (Fig. 3A). The migrated cell numbers were statistically analyzed in Fig. 3B. Western blot data demonstrated increased expression of the epithelial marker E-cadherin and decreased expression of the mesenchymal markers N-cadherin and Vimentin, after miR-22-3p overexpression. In contrast, miR-22-3p knockdown showed the opposite trend (Fig. 3C). Collectively, these results indicate that miR-22-3p negatively regulates EMT in NSCLC cells.
Fig. 3.
miR-22-3p suppresses epithelial-mesenchymal transition (EMT) of NSCLC cells. A, B Representative images and statistical analysis of EMT of A549 and Calu-1 cells transfected with control, mimics NC, miR-22-3p mimics, inhibitor NC, and miR-22-3p inhibitor determined by transwell assay. C Protein expression of E-cadherin, N-cadherin, and vimentin and in transfected A549 and Calu-1 cells measured by Western blot. ** P < 0.01, *** P < 0.001
miR-22-3p regulated RAS-related C3 botulinum toxin substrate 1 (RAC1) expression
To investigate possible downstream gene targets of miR-22-3p, the bioinformatics tool TargetScan was adopted to predict putative targets, discovering the 3′-UTR of RAC1 gene (Fig. 4A). Based on this, the relationship between RAC1 and miR-22-3p expression in lung cancer cell lines was studied. Seen from RT-qPCR results, the mRNA levels of RAC1 were dramatically decreased following miR-22-3p overexpression in A549 and Calu-1 cells, whereas they were greatly increased following miR-22-3p knockdown (Fig. 4B and 4C). Moreover, RAC1 protein expression in the lung cancer cell lines was measured with Western blot. The results showed downregulated RAC1 levels in miR-22-3p overexpressed cells, and increased RAC1 levels in miR-22-3p silenced cells (Fig. 4D). Then, to generate mutant plasmids, we cloned the wild-type 3′-UTR of human RAC1 mRNA into a luciferase reporter and conducted site-directed mutagenesis. Luciferase activity of the wild-type reporter was downregulated by miR-22-3p overexpression, while the mutant reporter reversed the suppressive effect (Fig. 4E). These findings indicate that RAC1 serves as a direct substrate of miR-22-3p in NSCLC cells.
Fig. 4.
miR-22-3p inhibits RAS-related C3 botulinum toxin substrate 1 (RAC1) expression. A Putative miR-22-3p connecting point in 3′-UTR of RAC1 mRNA. B–D RAC1 mRNA and protein levels were detected in A549 cells and Calu-1 cell transfected using mimics NC, miR-22-3p mimics, inhibitor NC and miR-22-3p inhibitor. E Relative luciferase activities of reporter plasmids of 293 T cells co-transfected with miR-22-3p mimics, 3′-UTR NC, RAC1-3′-UTR-WT, or RAC1-3′-UTR-Mut. ** P < 0.01
RAC1 overexpression recovers the inhibitory effect of miR-22-3p on RAC1
To further explore the relationship between miR-22-3p and RAC1, we overexpressed RAC1 in A549 and Calu-1 cells transfected with miR-22-3p mimics. Western blot results showed that comparing with the miR-22-3p mimics group, protein expression of RAC1 in the miR-22-3p mimics + RAC1-overexpressed vector group was significantly increased in A549 and Calu1 cells (Fig. 5).
Fig. 5.

RAC1 overexpression recovers the inhibitory effect of miR-22-3p on RAC1. The immunoblotting results of RAC1 in A549 and Calu-1 cells transfected with mimics NC, miR-22-3p mimics, mimics + Ctrl and miR-22-3p mimics + RAC1-overexpressed vectors
miR-22-3p regulates cell migration by inhibiting RAC1 expression
Rescue assays were carried out with EMT and cell migration experiments. A549 and Calu-1 cells were transfected with miR-22-3p mimics + RAC1-overexpressed vector, corresponding controls and mimics NC. Based on wood healing assay, miR-22-3p ectopic expression significantly repressed the invasion of A549 and Calu-1 cells, which were restored by RAC1 overexpression (Fig. 6A). Additionally, transwell assays demonstrated that overexpression of miR-22-3p caused a significant decrease in cell migration numbers in Calu-1 and A549 cells, while RAC1 ectopic expression rescued the migration ability (Fig. 6B). These results suggest that miR-22-3p inhibits NSCLC cell migration and invasion through targeting RAC1.
Fig. 6.
miR-22-3p suppresses the migration of NSCLC cells by inhibiting RAC1 expression. A Representative images and statistical analysis of cell migration of A549 and Calu-1 cells transfected with mimics NC, miR-22-3p mimics, mimics + Ctrl, and miR-22-3p mimics + RAC1-overexpressed vector measured using wound healing assay. B Representative images and statistical analysis of migration of A549 and Calu-1 cells measured using transwell assay. ** P < 0.01, *** P < 0.001
Discussion
miRNAs are small, non-coding RNA molecules that regulate gene expression through direct interaction with the 3′-UTR of related target mRNAs (Kim 2005). miR-22-3p has been found to serve as a tumor suppressor in various cancers. For example, down-regulation of miR-22-3p was associated with the progression and poor prognosis of cervical cancer (Kwon et al. 2022). In breast cancer, miR-22-3p exhibited a tumor-suppressive function by targeting the expression of PLAGL2 (Fan et al. 2021). miR-22-3p was also reduced in colorectal cancer patients and inhibited the cancer malignancy through inhibition of KDM3A (Jin et al. 2022). Despite that several evidences have reported the function and potential mechanisms of miR-22-3p in lung cancer (Yang et al. 2021; He et al. 2020), the clinical significance, biology function, and especially the downstream target of miR-22-3p in NSCLC cell invasion need to be further illustrated. Here, we discovered a dramatic decrease in miR-22-3p expression in NSCLC tissues from clinical patients, lung cancer samples from TCGA dataset, and NSCLC cells compared with controls. These results are consistent with previous studies (Ma et al. 2021b). We also found that miR-22-3p expression was correlated with lymph node metastasis and tumor size, but not TNM stages. This result was consistent with the report of Ma et al. (Ma et al. 2021b) and differed from the study of Yang et al. (Yang et al. 2021), which showed that the TNM stages was also significant. And miR-22-3p overexpression also significantly suppressed the migration and epithelial-mesenchymal transition (EMT) of Calu-1 and A549 cells, regulating the expression of EMT-related proteins (vimentin, E-cadherin and N-cadherin). However, silencing of miR-22-3p exhibited the opposite effect, confirming the tumor suppressive role of miR-22-3p in NSCLC.
EMT is the biological process of changing non-motile epithelial cells into mesenchymal phenotypes with invasive capacities. The hallmark of EMT is the loss of epithelial surface markers, such as E-cadherin, and the gain of mesenchymal markers, including vimentin and N-cadherin (Na et al. 2020). Clinical studies from Grigoraş et al. indicated that reduced E-cadherin expression was related to tumoral differentiation and was associated with unfavorable diagnosis and lymph node metastasis of NSCLC patients (Grigoras et al. 2017). N-cadherin, encoded by the CDH2 gene, is a transmembrane protein critical to cell adhesion (Reid and Hemperly 1990). Sher et al. indicated that CDH2 was the target gene of miR-218 for lung adenocarcinoma (Sher et al. 2014). The expression of vimentin, an intermediate filament protein, is associated with higher metastatic disease, and poor prognosis and survival of patients with multiple tumor types (Havel et al. 2015), including NSCLC patients (Soltermann et al. 2008; Dauphin et al. 2013). In addition, Havel et al. reported that vimentin affected lung cancer cell adhesion via a VAV2-Rac1 pathway to promote focal adhesion kinase stability (Havel et al. 2015). Our study demonstrated that miR-22-3p suppressed the EMT and invasion of NSCLC cells through the alteration of critical EMT markers, including E-cadherin, N-cadherin, and vimentin. These in vitro findings might explain why down-regulation of miR-22-3p was significantly correlated with NSCLC patients’ lymphatic metastasis status in clinic.
RAC1 is listed as a significant member of the Rho GTPases, which are involved in the tumor cell cycle, invasion, proliferation, apoptosis, angiogenesis, and migration, and are viewed as promising targets for preventing and treating cancers (Liang et al. 2021). RAC1 is a target of many miRNAs in various cancers, such as miR-194-5p in osteoclasts (Ni et al. 2021), miR‑142‑3p in colorectal cancer (Xie et al. 2021), and miR-509-3p in cervical cancer (Xu et al. 2021), miR-4715-5p in lung cancer (Yang et al. 2019). In lung cancer tissues, RAC1 expression was dramatically increased, and knockdown of RAC1 remarkably decreased cell migration, invasion, and proliferation. Akunuru et al. found that RAC1 knockdown with shRNA restrained the tumorigenic activities of human non-small cell lung adenocarcinoma (NSCLA) cells (Akunuru et al. 2011). Kaneto et al. reported that RAC1 inhibition could serve as a therapeutic target for gefitinib-resistant NSCLC (Kaneto et al. 2014). Tan et al. discovered RAC1 strengthened radioresistance by enhancing EMT through PAK1-LIMK1-Cofilins signaling in lung cancer, indicating that RAC1 may serve as a potential treatment target in radioresistant lung cancer cells (Tan et al. 2020). In this study, a new downstream target gene of miR-22-3p was discovered based on TargetScan, dual-luciferase reporter, RT-qPCR, and immunoblotting assays. The results showed that miR-22-3p directly interacted with the 3′-UTR of RAC1 mRNA, which resulted in reduced mRNA and protein expression of RAC1 in NSCLC cells. Importantly, inhibition of RAC1 contributing to the tumor-suppressive function of miR-22-3p because overexpression of RAC1 restored the migration and invasion ability of NSCLC cells inhibited by miR-22-3p mimics. Therefore, we demonstrated that miR-22-3p retarded the migration and invasion NSCLC cells through inhibition of RAC1’s expression via directly binding its 3′-UTR region.
There were several limitations in this study. (1) The clinical significance and biology function of miR-22-3p in small cell lung cancers (SCLCs) were not clarified. (2) Whether miR-22-3p/RAC1 axis suppressed the development of SCLC needed to be determined. In the future, we will perform experiments to illustrate the role and mechanisms of miR-22-3p, as well as other microRNAs in SCLC.
In conclusion, we provided the first evidence that RAC1, a well-known oncogene, acted as the direct downstream target of miR-22-3p in NSCLC. We demonstrated that miR-22-3p was down-regulated in NSCLC tissues and cells and suppressed cell migration and EMT via targeting RAC1. The miR-22-3p/RAC1 axis offers a novel therapeutic target for NSCLC.
Author contributions
GY and NLX designed the study. XJW and XBW performed the experiments and drafted the manuscript. TH and ZPZ analyzed the data. XJW, GY, and XBW revised the manuscript.
Funding
This work was supported by grants from the Key Research and Development Plan of Shaanxi Province (2019SF-005).
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The study was performed in accordance with the ethical standards as laid down in the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards. All the experiments in this study were approved by the Ethics Committee of the Second Affiliated Hospital of the Air Force Medical University.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xuejiao Wang and Xiaobin Wang contributed equally.
Contributor Information
Nianlin Xie, Email: xienianlin@126.com.
Guang Yang, Email: guang83731yg@sina.com.
References
- Akunuru S, Palumbo J, Zhai QJ, Zheng Y. Rac1 targeting suppresses human non-small cell lung adenocarcinoma cancer stem cell activity. PLoS One. 2011;6(2):e16951. doi: 10.1371/journal.pone.0016951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander M, Kim SY, Cheng H. Update 2020: Management of non-small cell lung cancer. Lung. 2020;198(6):897–907. doi: 10.1007/s00408-020-00407-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allemani C, Matsuda T, Di Carlo V, Harewood R, Matz M, Niksic M, Bonaventure A, Valkov M, Johnson CJ, Esteve J, et al. Global surveillance of trends in cancer survival 2000–14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet. 2018;391(10125):1023–1075. doi: 10.1016/S0140-6736(17)33326-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Wu FX, Luo HL, Liu JJ, Luo T, Bai T, Li LQ, Fan XH. Berberine upregulates miR-22-3p to suppress hepatocellular carcinoma cell proliferation by targeting Sp1. Am J Transl Res. 2016;8(11):4932–4941. [PMC free article] [PubMed] [Google Scholar]
- Dauphin M, Barbe C, Lemaire S, Nawrocki-Raby B, Lagonotte E, Delepine G, Birembaut P, Gilles C, Polette M. Vimentin expression predicts the occurrence of metastases in non small cell lung carcinomas. Lung Cancer. 2013;81(1):117–122. doi: 10.1016/j.lungcan.2013.03.011. [DOI] [PubMed] [Google Scholar]
- Fan T, Wang CQ, Li XT, Yang H, Zhou J, Song YJ. MiR-22-3p Suppresses cell migration and invasion by targeting PLAGL2 in breast cancer. J Coll Phys Surg-Pak : JCPSP. 2021;31(8):937–940. doi: 10.29271/jcpsp.2021.08.937. [DOI] [PubMed] [Google Scholar]
- Grigoras ML, Arghirescu TS, Folescu R, Talpos IC, Gindac CM, Zamfir CL, Cornianu M, Anghel MD, Levai CM. Expression of E-cadherin in lung carcinoma, other than those with small cells (NSCLC) Rom J Morphol Embryol. 2017;58(4):1317–1325. [PubMed] [Google Scholar]
- Havel LS, Kline ER, Salgueiro AM, Marcus AI. Vimentin regulates lung cancer cell adhesion through a VAV2-Rac1 pathway to control focal adhesion kinase activity. Oncogene. 2015;34(15):1979–1990. doi: 10.1038/onc.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Zhang Y, Xia S. LncRNA NNT-AS1 promotes non-small cell lung cancer progression through regulating miR-22-3p/YAP1 axis. Thoracic Cancer. 2020;11(3):549–560. doi: 10.1111/1759-7714.13280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C, Meiners S, Lukas C, Stathopoulos GT, Chen J. Role of exosomal microRNAs in lung cancer biology and clinical applications. Cell Prolif. 2020;53(6):e12828. doi: 10.1111/cpr.12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4(3):143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Han X, Wang J, Wang L, Xu Z, Wei Q, Zhang W, Wang H. miR-22 enhances the radiosensitivity of small-cell lung cancer by targeting the WRNIP1. J Cell Biochem. 2019;120(10):17650–17661. doi: 10.1002/jcb.29032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin RR, Zeng C, Chen Y. MiR-22-3p regulates the proliferation, migration and invasion of colorectal cancer cells by directly targeting KDM3A through the Hippo pathway. Histol Histopathol. 2022;37(12):1241–1252. doi: 10.14670/HH-18-526. [DOI] [PubMed] [Google Scholar]
- Kaneto N, Yokoyama S, Hayakawa Y, Kato S, Sakurai H, Saiki I. RAC1 inhibition as a therapeutic target for gefitinib-resistant non-small-cell lung cancer. Cancer Sci. 2014;105(7):788–794. doi: 10.1111/cas.12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6(5):376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Kwon AY, Jeong JY, Park H, Hwang S, Kim G, Kang H, Heo JH, Lee HJ, Kim TH, An HJ (2022) miR-22–3p and miR-30e-5p Are associated with prognosis in cervical squamous cell carcinoma. Int J Mol Sci 23(10):5623 [DOI] [PMC free article] [PubMed]
- Li X, Zhao J, Zhang H, Cai J. Silencing of LncRNA metastasis-associated lung adenocarcinoma transcript 1 inhibits the proliferation and promotes the apoptosis of gastric cancer cells through regulating microRNA-22-3p-mediated ErbB3. Onco Targets Ther. 2020;13:559–571. doi: 10.2147/OTT.S222375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Oyang L, Rao S, Han Y, Luo X, Yi P, Lin J, Xia L, Hu J, Tan S, et al. Rac1, A Potential Target for Tumor Therapy. Front Oncol. 2021;11:674426. doi: 10.3389/fonc.2021.674426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv KT, Liu Z, Feng J, Zhao W, Hao T, Ding WY, Chu JP, Gao LJ. MiR-22-3p regulates cell proliferation and inhibits cell apoptosis through targeting the eIF4EBP3 gene in human cervical squamous carcinoma cells. Int J Med Sci. 2018;15(2):142–152. doi: 10.7150/ijms.21645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Wang H, Zong G, He J, Wang Y, Yang F, Yang Z, Bian E, Zhao B. EGR1 modulated LncRNA HNF1A-AS1 drives glioblastoma progression via miR-22-3p/ENO1 axis. Cell Death Discov. 2021;7(1):350. doi: 10.1038/s41420-021-00734-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DJ, Zhou XY, Qin YZ, Tian ZH, Liu HS, Li SQ. MiR-22-3p Expression is down-regulated in lung adenocarcinoma. Acta Biochim Pol. 2021;68(4):667–672. doi: 10.18388/abp.2020_5540. [DOI] [PubMed] [Google Scholar]
- Malhotra J, Malvezzi M, Negri E, La Vecchia C, Boffetta P. Risk factors for lung cancer worldwide. Eur Respir J. 2016;48(3):889–902. doi: 10.1183/13993003.00359-2016. [DOI] [PubMed] [Google Scholar]
- Na TY, Schecterson L, Mendonsa AM, Gumbiner BM. The functional activity of E-cadherin controls tumor cell metastasis at multiple steps. Proc Natl Acad Sci U S A. 2020;117(11):5931–5937. doi: 10.1073/pnas.1918167117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Zhang X, Li J, Zheng Z, Zhang J, Zhao W, Liu L. Tumour-derived exosomal lncRNA-SOX2OT promotes bone metastasis of non-small cell lung cancer by targeting the miRNA-194-5p/RAC1 signalling axis in osteoclasts. Cell Death Dis. 2021;12(7):662. doi: 10.1038/s41419-021-03928-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet. 2012;13(5):358–369. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid RA, Hemperly JJ. Human N-cadherin: nucleotide and deduced amino acid sequence. Nucleic Acids Res. 1990;18(19):5896. doi: 10.1093/nar/18.19.5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sher YP, Wang LJ, Chuang LL, Tsai MH, Kuo TT, Huang CC, Chuang EY, Lai LC. ADAM9 up-regulates N-cadherin via miR-218 suppression in lung adenocarcinoma cells. PLoS One. 2014;9(4):e94065. doi: 10.1371/journal.pone.0094065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltermann A, Tischler V, Arbogast S, Braun J, Probst-Hensch N, Weder W, Moch H, Kristiansen G. Prognostic significance of epithelial-mesenchymal and mesenchymal-epithelial transition protein expression in non-small cell lung cancer. Clin Cancer Res. 2008;14(22):7430–7437. doi: 10.1158/1078-0432.CCR-08-0935. [DOI] [PubMed] [Google Scholar]
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- Tan S, Yi P, Wang H, Xia L, Han Y, Wang H, Zeng B, Tang L, Pan Q, Tian Y, et al. RAC1 involves in the radioresistance by mediating epithelial-mesenchymal transition in lung cancer. Front Oncol. 2020;10:649. doi: 10.3389/fonc.2020.00649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Li X, Cheng T, Wu J. miR-21-5p/SMAD7 axis promotes the progress of lung cancer. Thoracic Cancer. 2021;12(17):2307–2313. doi: 10.1111/1759-7714.14060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Guan Y, Su Y, Yang T, Yu H. TRPM2-AS promotes bladder cancer by targeting miR-22-3p and regulating GINS2 mRNA expression. Onco Targets Ther. 2021;14:1219–1237. doi: 10.2147/OTT.S282151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Lin C. Exosomes miR-22-3p Derived from Mesenchymal Stem Cells Suppress Colorectal Cancer Cell Proliferation and Invasion by Regulating RAP2B and PI3K/AKT Pathway. J Oncol. 2021;2021:3874478. doi: 10.1155/2021/3874478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Yao Z, Fang L. miR-22-3p/PGC1beta suppresses breast cancer cell tumorigenesis via PPARgamma. PPAR Res. 2021;2021:6661828. doi: 10.1155/2021/6661828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KL, Tsai YM, Lien CT, Kuo PL, Hung AJ (2019) The roles of microRNA in lung cancer. Int J Mol Sci 20(7):1611 [DOI] [PMC free article] [PubMed]
- Xie N, Meng Q, Zhang Y, Luo Z, Xue F, Liu S, Li Y, Huang Y (2021) MicroRNA1423p suppresses cell proliferation, invasion and epithelial to mesenchymal transition via RAC1ERK1/2 signaling in colorectal cancer. Mol Med Rep 24(2):568 [DOI] [PMC free article] [PubMed]
- Xin M, Qiao Z, Li J, Liu J, Song S, Zhao X, Miao P, Tang T, Wang L, Liu W, et al. miR-22 inhibits tumor growth and metastasis by targeting ATP citrate lyase: evidence in osteosarcoma, prostate cancer, cervical cancer and lung cancer. Oncotarget. 2016;7(28):44252–44265. doi: 10.18632/oncotarget.10020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Cao L, Zhang Y, Lian H, Sun Z, Cui Y. MicroRNA-1246 inhibits cell invasion and epithelial mesenchymal transition process by targeting CXCR4 in lung cancer cells. Cancer Biomarkers : Sect A Dis Markers. 2018;21(2):251–260. doi: 10.3233/CBM-170317. [DOI] [PubMed] [Google Scholar]
- Xu J, Ma X, Yang H, Zhang J, Cai G, Yao N. MiR-509-3p induces apoptosis and affects the chemosensitivity of cervical cancer cells by targeting the RAC1/PAK1/LIMK1/Cofilin pathway. Chem Pharm Bull (Tokyo) 2021;69(4):325–332. doi: 10.1248/cpb.c20-00600. [DOI] [PubMed] [Google Scholar]
- Xue Y, Guo Y, Liu N, Meng X. MicroRNA-22-3p targeted regulating transcription factor 7-like 2 (TCF7L2) constrains the Wnt/beta-catenin pathway and malignant behavior in osteosarcoma. Bioengineered. 2022;13(4):9135–9147. doi: 10.1080/21655979.2021.2003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Ding L, Hu Q, Xia J, Sun J, Wang X, Xiong H, Gurbani D, Li L, Liu Y, et al. MicroRNA-218 functions as a tumor suppressor in lung cancer by targeting IL-6/STAT3 and negatively correlates with poor prognosis. Mol Cancer. 2017;16(1):141. doi: 10.1186/s12943-017-0710-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Qiu Q, Qian X, Yi J, Jiao Y, Yu M, Li X, Li J, Mi C, Zhang J, et al. Long noncoding RNA LCAT1 functions as a ceRNA to regulate RAC1 function by sponging miR-4715-5p in lung cancer. Mol Cancer. 2019;18(1):171. doi: 10.1186/s12943-019-1107-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Su W, Li Y, Zhou Z, Zhou Y, Shan H, Han X, Zhang M, Zhang Q, Bai Y, et al. MiR-22-3p suppresses cell growth via MET/STAT3 signaling in lung cancer. Am J Transl Res. 2021;13(3):1221–1232. [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Li XY, Wang ZM, Han ZF, Zhao YH. MiR-22 inhibits lung cancer cell EMT and invasion through targeting Snail. Eur Rev Med Pharmacol Sci. 2017;21(16):3598–3604. [PubMed] [Google Scholar]
- Zhang Z, Li M, Zhang Z. lncRNA MALAT1 modulates oxaliplatin resistance of gastric cancer via sponging miR-22-3p. Onco Targets Ther. 2020;13:1343–1354. doi: 10.2147/OTT.S196619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Zhang S, Chen Z, He Z, Xu Y, Li Z. CircRNA-ENO1 promoted glycolysis and tumor progression in lung adenocarcinoma through upregulating its host gene ENO1. Cell Death Dis. 2019;10(12):885. doi: 10.1038/s41419-019-2127-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.