Abstract
Background
Adjuvants are key for effective vaccination against cancer and chronic infectious diseases. Saponin-based adjuvants (SBAs) are unique among adjuvants in their ability to induce robust cell-mediated immune responses in addition to antibody responses. Recent preclinical studies revealed that SBAs induced cross-presentation and lipid bodies in otherwise poorly cross-presenting CD11b+ murine dendritic cells (DCs).
Method
Here, we investigated the response of human DC subsets to SBAs with RNA sequencing and pathway analyses, lipid body induction visualized by laser scanning microscopy, antigen translocation to the cytosol, and antigen cross-presentation to CD8+ T cells.
Results
RNA sequencing of SBA-treated conventional type 1 DC (cDC1) and type 2 DC (cDC2) subsets uncovered that SBAs upregulated lipid-related pathways in CD11c+ CD1c+ cDC2s, especially in the CD5− CD163+ CD14+ cDC2 subset. Moreover, SBAs induced lipid bodies and enhanced endosomal antigen translocation into the cytosol in this particular cDC2 subset. Finally, SBAs enhanced cross-presentation only in cDC2s, which requires the CD163+ CD14+ cDC2 subset.
Conclusions
These data thus identify the CD163+ CD14+ cDC2 subset as the main SBA-responsive DC subset in humans and imply new strategies to optimize the application of saponin-based adjuvants in a potent cancer vaccine.
Keywords: Antigen Presentation; Adaptive Immunity; Adjuvants, Immunologic; CD8-Positive T-Lymphocytes; Dendritic Cells
WHAT IS ALREADY KNOWN ON THIS TOPIC
Saponin-based adjuvants (SBAs) have been used in the clinic against infectious diseases. They have the advantage to induce both CD4+ T helper and CD8+ T cells in addition to antibody responses. We have shown previously that only the CD11b+ dendritic cell (DC) subset in the mouse responds to SBAs by cross-presentation of exogenous antigens to CD8+ T cells in a lipid-body dependent manner. Here, we expand our research to human DC subsets, including the recently discovered subpopulations within the cDC2s.
WHAT THIS STUDY ADDS
We defined that only the cDC2 subset is sensitive to SBAs, and not the cDC1 subset. Especially the recently identified subpopulation of the cDC2 subset, CD5− CD163+ CD14+ cDC2s, showed enhanced antigen translocation and upregulation of several lipid-related pathways, which correlated with increased lipid body presence in the cells. Most importantly, our study revealed that SBAs can specifically enhance antigen cross-presentation in CDC2s, which requires the CD5− CD163+ CD14+ cDC2s.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Understanding the DC subset-specificity and mechanisms of SBAs in humans is important for further development and application of SBA based vaccines against infectious diseases and cancer.
Introduction
The development of therapeutic vaccines against cancer and persistent infections has received much attention in recent decades. These vaccines are usually accompanied by an adjuvant to enhance the magnitude and durability of the immune response to the vaccine antigen.1 Saponin-based adjuvants (SBAs) belong to the group of new-generation of adjuvants. They consist of triterpene glycosides found in a variety of plants and trees including the South American soapbark tree Quillaja saponaria. Specific saponin fractions derived from this particular tree have been shown to exhibit potent immunostimulatory effects and are used for SBAs.2 The first approved human vaccines containing SBAs are against the herpes zoster virus (RZV, Shingrix, GlaxoSmithKline) and malaria (RTS, S/AS01, Mosquirix, GlaxoSmithKline). The SBA-containing COVID-19 vaccine (NVX-COV2373, Novavax) has also successfully been used in the clinic. SBAs are extensively investigated as an adjuvant since they can induce robust cell-mediated immune responses activating both CD4+ T helper and CD8+ T cells in addition to antibody responses.3–6 Especially for cancer, a strong CD8+ T- cell response is necessary to eradicate the tumor.7
Dendritic cells (DCs) are well known for their ability to capture exogenous (tumor) antigens, and present them on major histocompatibility complex class I (MHCI) molecules to CD8+ T cells, a process called antigen cross-presentation.8 Different subsets can be distinguished within the murine DCs, namely the CD8α+ CD103+ DCs (conventional type 1 DCs (cDC1s)) and the CD8α− CD11b+ DCs (conventional type 2 DCs (cDC2s)). The human BDCA-3+ Clec9a+ DCs (cDC1s) from human peripheral blood resemble murine CD8α+ CD103+ DCs, whereas the BDCA-1+ DCs (cDC2s) display a gene expression profile resembling that of murine CD8α− CD11b+ DCs.9 10 The general consensus for murine DCs is that cDC1s are superior in cross-presenting exogenous antigens to CD8+ T cells compared with cDC2s, whereas cDC2s mainly present antigens to CD4+ T cells.11–13 However, this distinction between cDC1s and cDC2s in humans is less pronounced.14–18
We and others have shown that an important characteristic of SBAs is their potency to enhance cross-presentation in murine DC and thereby induce efficient cytotoxic CD8+ T- cell immunity.19–21 Importantly, we have demonstrated using an in situ tumor ablation model that SBA injection induced tumor-specific CD8+ T- cell responses, resulting in superior synergistic antitumor immunity compared with other adjuvants.22 Interestingly, we also demonstrated that in vitro generated murine MHCIIlo CD11bhi DCs and the corresponding in vivo CD11b+ DCs from lymph nodes were most responsive to SBAs and exhibited high cross-presentation capacity.20 We further reported that SBAs enhance endosomal antigen translocation to the cytosol, and that the endoplasmic reticulum (ER) stress-related PKR-like ER kinase (PERK) pathway, and proteasomal antigen degradation are crucial for SBA-induced cross-presentation.20 21 Moreover, only the CD11b+ DCs showed induction of lipid bodies (LBs) after treatment with SBAs. LBs, also called lipid droplets, are cytoplasmic organelles that play a major role in the storage of neutral lipids and energy metabolism, but also play a role in immune regulation.23 24 Importantly, the presence of LBs is essential for SBA-induced increase in antigen cross-presentation and subsequent CD8+ T- cell activation.20
The SBA-responsiveness of human DC subsets is still largely unknown, especially concerning their capacity to cross-present exogenous antigens. Recent RNA sequencing studies uncovered new members in the human cDC2 subset family.25–27 Initially, it had been reported that cDC2s comprised two subsets, one is named DC2s which is genetically and functionally closer related to cDC1s, and the other is called CD14+ DC3s that are more related to monocytes.25 26 Now, cDC2s are proposed to be delineated and independent of monocytes and macrophages, and comprise four subsets where the CD5+ subset corresponds to DC2s, and CD5− subsets to DC3s.27 28 The CD5− cDC2s could further be refined with the additional expression of CD163+ or CD14+. The CD163+ CD14+ cDC2 subset has been described as being inflammatory.27 There is not much known yet about their function, although a recent study showed that CD1c+ CD163+ cDC2 can prime CD8+ CD103+ T cells.28
In the current study, we investigated SBA-mediated cross-presentation in human cDC1s, the total cDC2 fraction, and the four cDC2 subsets using SBA, formulated with immunoactive saponin Fraction C in combination with cholesterol and phospholipid to generate so-called immune stimulatory complexes (ISCOMs) Matrix C.29 We show that ISCOMs Matrix C can specifically induce cross-presentation in cDC2s, not cDC1s, and that this depends on the CD163+ CD14+ cDC2 subset. RNA sequencing analysis revealed that mainly lipid-related pathways were upregulated in the CD163+ CD14+ cDC2 subset. Additional data demonstrate enhanced antigen translocation and LB formation in this particular subset after treatment with ISCOMs. The discovery that specifically the human CD163+ CD14+ cDC2 subset can benefit from ISCOMs is an important step toward the design and development of new vaccines for diseases that require T cell-mediated immunity, including a potent cancer vaccine.
Results
ISCOMs induce lipid-related pathways specifically in cDC2s
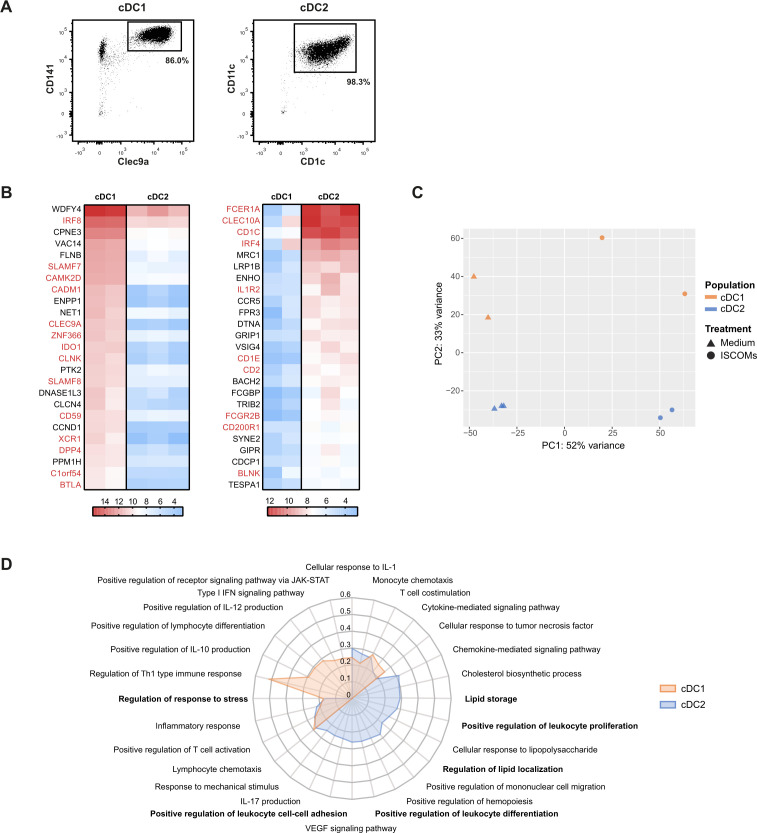
In order to identify the human DC subset that is responsive to ISCOMs, CD141+ Clec9a+ cDC1s and CD11c+ CD1c+ cDC2s were isolated from blood (figure 1A), stimulated with ISCOMs or mock-treated, and subjected to RNA sequencing. The top 25 expressed genes in the cDC1 (figure 1B, left) and cDC2 subsets (figure 1B, right) confirmed the purity of the isolated subsets, as most of these genes correspond with genes that were previously found to be associated with these subsets (gene names in red, figure 1B).26 27 30 Principal component analysis (PCA) indicated that cDC1s and cDC2s were separated along the PC2 axis, accounting for 33% variances, confirming substantial difference in gene expression between the subsets (figure 1C). Stimulation with ISCOMs showed 52% variances along the PC1 axis, indicating that ISCOMs treatment led to a substantial change in gene expression in both subsets (figure 1C). Analysis of differentially expressed genes (DEGs) indicated that in cDC1s 1327 and 1047 genes were significantly upregulated and downregulated upon ISCOMs treatment, respectively, whereas 1379 and 1174 genes were significantly upregulated and downregulated in cDC2s. Gene ontology (GO) analysis of upregulated genes was performed to indicate enrichment of Biological Processes upon ISCOMs treatment. Immunologically relevant GO pathways were selected and displayed as a radar plot showing the gene ratio. Upon ISCOMs treatment, mostly T-cell activation-related pathways were upregulated in cDC1s (figure 1D, orange), whereas lipid-related pathways, and leukocyte proliferation and differentiation pathways were upregulated in cDC2s (figure 1D, blue). Previously, we showed that induction of PERK, activated upon ER stress, is crucial for ISCOMs-induced cross-presentation in murine DCs.21 Here, both DC subsets induced “Regulation of response to stress” upon ISCOMs treatment, indicating that stress-related pathways could also play a role in human DC subsets. Interestingly, “Lipid storage” is specifically upregulated in the cDC2s upon ISCOMs treatment. Since LBs are the main lipid storage organelles in the cell, this data encouraged us to further investigate LB induction upon ISCOMs treatment in the cDC2 subset. Moreover, “Cholesterol biosynthetic process” and “Regulation of lipid localization” further point towards involvement of LBs. Overall, ISCOMs induce a variety of immune signaling and stress pathways in both human DC subsets, and specifically lipid-related pathways in the cDC2s.
Figure 1.
ISCOMs upregulate distinct pathways in cDC2 and cDC1 subsets. (A) Purity of isolated cDC1s and cDC2s after MACS isolation analyzed by flow cytometry. (B) Top 25 highest expressed genes for cDC1s (left heatmap) and cDC2s (right heatmap) without ISCOMs stimulation based on transformed normalized counts from RNA sequencing. Known genes that are specific for each subset according to literature are depicted in red. (C) Principal component analysis (PCA) for bulk-sequenced cDC1s and cDC2s before and after 8 hours of ISCOMs stimulation. (D) Gene ontology (GO) enrichment for Biological Processes based on DEGs with p<0.05 and fold change ≥2 after stimulation with ISCOMs for 8 hours. Immunologically relevant GO pathways were selected and displayed as a radar plot showing the gene ratio. RNA sequencing for bulk subsets was performed on two to three healthy donors. cDC1, conventional type 1 DC; cDC2, conventional type 2 DC; DEGs, differentially expressed genes; IFN, interferon; IL, interleukin; ISCOMs, immune stimulatory complexes; JAK-STAT, Janus kinases-signal transducer and activator of transcription proteins; MACS, magnetic-activated cell sorting; Th, T helper; VEGF, vascular endothelial growth factor.
ISCOMs induce lipid bodies in cDC2s
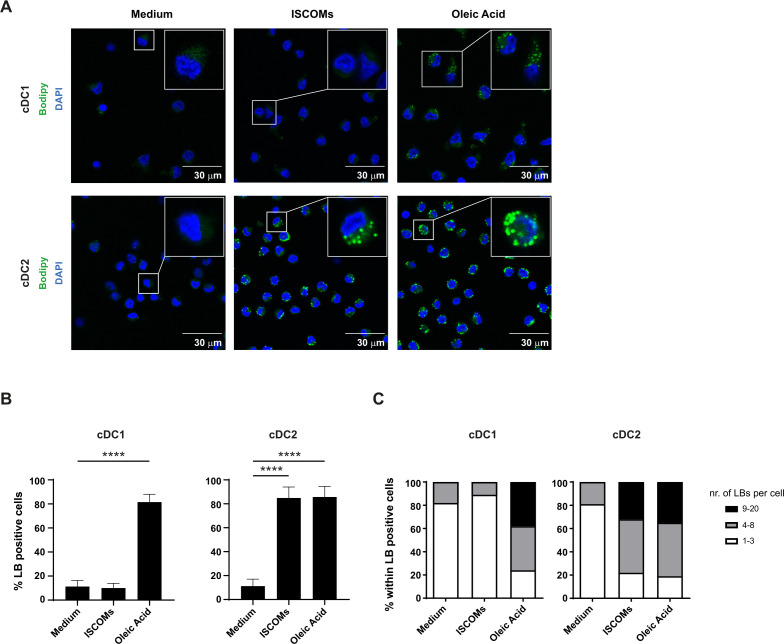
Since lipid-related pathways were upregulated in human cDC2s after ISCOMs stimulation, we investigated its impact on LB induction via staining with the neutral lipid dye Bodipy 493/503. Feeding the fatty acid oleic acid (OA) was used as a positive control for the overall capacity of the DC subsets to induce LBs. As expected, treatment with OA was able to induce LBs in both DC subsets (figure 2A,B). In contrast, treatment with ISCOMs for 18 hours showed more than 80% LB-positive cDC2s, with no detectable increase in cDC1s (figure 2A,B). Quantification of the number of ISCOMs-induced LBs within the LB-positive cells showed that the majority of cDC2s contained 4–8 LBs per cell, whereas ~40% of the cDC2 cells had even 9–20 LBs per cell after stimulation with ISCOMs (figure 2C). In the cDC1s, a very low number of LBs was observed upon ISCOMs treatment, and the number of LBs per cell was similar to untreated cells. Shorter stimulations of cDC2s with ISCOMs showed that LBs were still mostly absent after 5 hours, appearing in ~50% of the cells after 10 hours, reaching ~90% after 18 hours (figure 2 and online supplemental extended data figure 1A,B). OA addition induced LB formation in ~30% of the cells already after 5 hours incubation, ~70% LB positive cells were present after 10 hours incubation, reaching ~90% after 18 hours (figure 2 and online supplemental extended data figure 1A,B). The cDC2s showed a similar increase in the number of LBs per cell in time following ISCOMs or OA stimulation (online supplemental extended data figure 1C). These data indicate that only cDC2s are capable of inducing LBs after ISCOMs stimulation, consistent with the upregulation of genes in lipid-related pathways.
Figure 2.
ISCOMs induce lipid bodies in cDC2s. (A) LB staining in cDC1s and cDC2s with Bodipy 493/503 (green) after 18 hours stimulation with ISCOMs or OA, analyzed by laser scanning microscopy. A magnification of a selected cell in the overview picture is shown in the upper right corner. Nucleus staining is done with DAPI (blue). (B) Quantification of the frequency of LB-positive cells was done with Fiji. Statistical analyses were done using one-way ANOVA using post hoc Tukey. Data are shown here from two healthy donors. cDC1 ****p<0.0001 (DF=2; F (2, 24) = 560.3), cDC2 ****p<0.0001 (DF=2; F (2, 37) = 366.2). (C) The number of LBs within the LB-positive cells was quantified with Fiji and divided into three categories, 1–3 LBs, 4–8 LBs, and 9–20 LBs per cell, and shown as frequency per subset. At least five overview pictures per sample were taken for quantification. ANOVA, analysis of variance; cDC1, conventional type 1 DC; cDC2, conventional type 2 DC; ISCOMs, immune stimulatory complexes; LBs, lipid bodies; OA, oleic acid.
jitc-2023-007082supp001.pdf (2.3MB, pdf)
Enhanced antigen cross-presentation in cDC2s upon ISCOMs stimulation
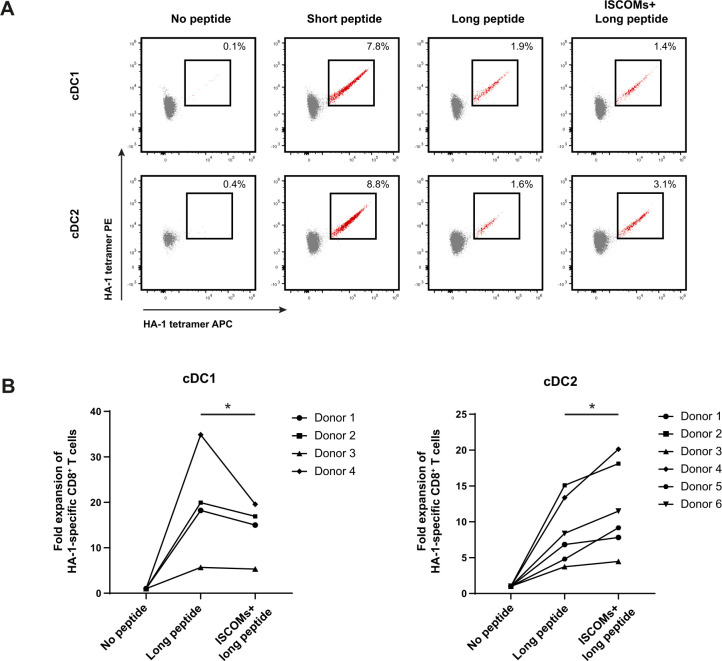
Next, we investigated the ability of ISCOMs to enhance antigen cross-presentation in human cDC1s and cDC2s. Here, purified cDC1s and cDC2s were either loaded with minor histocompatibility antigen (MiHA) HA-1 long peptide (VARFAEGLEKLKECVLHDDLLEARRPRAHEZL) in the presence or absence of ISCOMs or with the short nominal peptide epitope (VLHDDLLEA) and co-cultured with peripheral blood mononuclear cells (PBMCs) from patients containing HA-1-specific CD8+ T cells. Both cDC1s and cDC2s induced a comparable increase in frequencies of HA-1-specific CD8+ T cells after stimulation with the positive control HA-1 short peptide as detected by dual-color HA-1 tetramer staining (figure 3A). Loading with the HA-1 long peptide also led to HA-1-specific CD8+ T-cells expansion by both subsets, although to a lesser extent as compared with the nominal peptide, which is consistent with the requirement of prior antigen processing and cross-presentation. As expected, long peptide pulsed cDC1 (mean 19.7±12 SD) were much more effective than cDC2 (mean 8.7±4.6 SD) in cross-presenting antigen without adjuvant treatment (figure 3B). Strikingly, ISCOMs-treated long peptide loaded cDC2s increased frequencies and expansion of HA-1-specific CD8+ T cells in six independent donors consistent with an enhanced antigen cross-presentation capacity, while ISCOMs-treated cDC1s did not show any enhancement (figure 3A,B). Taken together, these data demonstrate that ISCOMs specifically enhance antigen cross-presentation in cDC2s.
Figure 3.
Enhanced antigen cross-presentation in cDC2s upon ISCOMs. (A) HA-1- specific CD8+ T-cell expansion indicated by double-positive HA-1 tetramer staining (in red) for cDC1s and cDC2s after stimulation with short peptide, long peptide, or ISCOMS+ long peptide. DCs (100,000 cells) were incubated with PBMCs (750,000 cells) in a 1:7.5 ratio. Representative flow cytometry data are shown from one healthy DC donor. (B) Fold expansion of HA-1-specific CD8+ T cells in cDC2s and cDC1s. The numbers of different healthy DC donors used are depicted in the graphs. Statistical analyses were done using paired non-parametric t-test. *p<0.05. cDC1, conventional type 1 DC; cDC2, conventional type 2 DC; ISCOMs, immune stimulatory complexes; PBMCs, peripheral blood mononuclear cells.
ISCOMs upregulate lipid-related pathways in CD163+ CD14+ cDC2s
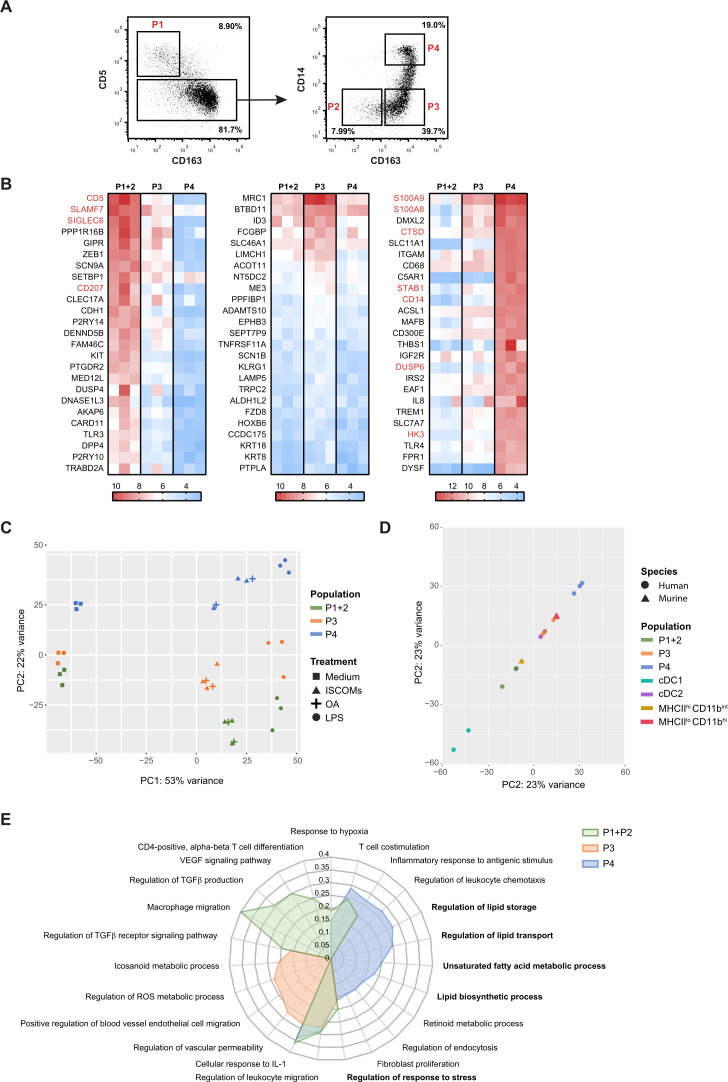
Since our data so far indicated that especially the cDC2s were responsive to ISCOMs and that ISCOMs induced higher numbers of LBs (9–20 per cell) in ~40% of the total cDC2 fraction, it was imperative to further investigate the impact of SBAs on the cDC2 subsets. Dutertre et al proposed the presence of at least four subsets within the cDC2s based on the expression of CD5, CD163, and CD14.27 We have isolated these four cDC2 subsets according to the following markers: subset 1 (P1, CD11c+ CD1c+ CD5+), subset 2 (P2, CD11c+ CD1c+ CD5− CD163− CD14−), subset 3 (P3, CD11c+ CD1c+ CD5− CD163+ CD14−), and subset 4 (P4, CD11c+ CD1c+ CD5− CD163+ CD14+) (figure 4A). Since P1 and P2 were described to be closely related28 and due to the low amounts of P1 and P2 that could be isolated per donor, we combined these two subsets for subsequent RNA sequencing analysis.
Figure 4.
ISCOMs upregulate lipid-metabolism-mediated pathways in CD163+ CD14+ cDC2s. (A) Gating strategy of the CD11c+ CD1c+ cDC2 subsets: P1 (CD5+), P2 (CD5− CD163−), P3 (CD5− CD163+ CD14−), and P4 (CD5− CD163+ CD14+) by flow cytometry. (B) Top 25 highest expressed genes for cDC2 subsets P1+P2 (left heatmap), P3 (middle heatmap), and P4 (right heatmap) without ISCOMs stimulation based on transformed normalized counts from RNA sequencing. Known genes for each subset according to literature are depicted in red. (C) PCA for bulk-sequenced P1+2, P3, and P4 cDC2s before and after 8 hours of ISCOMs, LPS, or OA stimulation. (D) PCA of bulk-sequenced human cDC1, cDC2, P1+2, P3, P4 after 8 hour ISCOM stimulation and murine MHCIIhi CD11bint and MHCIIlo CD11bhi DCs after 5 hours of ISCOMs stimulation. (E) GO enrichment for Biological Processes based on DEGs with p<0.05 and fold change ≥2 after stimulation with ISCOMs for 8 hours. Immunologically relevant GO pathways were selected and displayed as a radar plot showing the gene ratio. RNA sequencing for bulk cDC1s and cDC2s was performed on two healthy donors, for cDC2 subsets on three healthy donors, and for the murine subsets from two mice. cDC1, conventional type 1 DC; cDC2, conventional type 2 DC; DEGs, differentially expressed genes; GO, gene ontology; IL, interleukin; ISCOMs, immune stimulatory complexes; LPS, lipopolysaccharide; MHC, major histocompatibility complex; OA, oleic acid; PCA, principal component analysis; ROS, reactive oxygen species; TGF, transforming growth factor; VEGF, vascular endothelial growth factor.
The top 25 expressed genes were identified in untreated cDC2 subsets and compared with the genes found by Dutertre et al (figure 4B. P1+2, left; P4 right). Similar genes (gene names in red) were identified for P1+2 and P4. Despite the fact that none of the genes identified by Dutertre et al were expressed in the top 25 of P3, no P1+2 nor P4 genes were found in P3 either (figure 4B. middle). This confirms that all cDC2 subsets exhibit distinct marker and gene expression profiles. Interestingly, one of the highest expressed genes within P4 is ITGAM, which encodes CD11b, the marker that distinguishes the SBA-responsive DC subset in mice.
PCA indicated that untreated P1+2 and P3 were clustered more closely together, and that P4 was clearly separated from P1+2 and P3 based on gene expression on the PC2 axis (figure 4C). After ISCOMs stimulation, all cDC2 subsets responded with a change in gene expression on the PC1 axis. P4 was still the most separated from the other subsets on the PC2 axis (figure 4C). The shift induced by OA stimulation in the subsets appears closely related to the shift observed for ISCOMs treatment, while their activation using the Toll-like receptor 4 (TLR4) ligand lipopolysaccharide (LPS) induced a gene expression profile distinct from the other treatments (figure 4C). Interestingly, PCA of the different human and murine DC subset after ISCOMs stimulation showed a closer similarity between the human total cDC2s, P3 cDC2s, P4 cDC2s, and the SBA-responsive murine MHCIIlo CD11bhi DC subset based on the PC2 axis, whereas human P1+2 cDC2s were more closely related to the non-responsive murine MHCIIhi CD11bint DC subset (figure 4D). Analysis of DEGs in the cDC2 subsets induced upon ISCOMs treatment indicated that in P1+2 cDC2s 1721 and 1529 genes were significantly upregulated and downregulated, respectively, whereas 1346 and 1472 genes were significantly upregulated and downregulated in P3 cDC2s, respectively. In P4 cDC2s 1288 were significantly upregulated and 1468 genes were downregulated upon ISCOMs treatment. GO enrichment analysis and subsequent selection of immunologically relevant pathways of upregulated genes upon ISCOMs treatment revealed distinctive pathways, but also some overlap in upregulated pathways in the different cDC2 subsets (figure 4E). One of the pathways upregulated in all subsets is “Regulation of response to stress”, which is in line with what was described earlier for the total cDC2s. While different immune pathways were induced in P1+2 and P3, the pathways induced in P4 were mostly lipid-related. Interestingly, lipid-related pathways were also observed earlier in total cDC2s upon ISCOMs stimulation. The “Regulation of lipid storage”, “Regulation of lipid transport”, “Unsaturated fatty acid metabolic process” and “Lipid biosynthetic process” pathways all strongly suggest the induction of LBs in this subset. Upon LPS treatment, a different set of induced pathways was observed in P4 cDC2s showing upregulation of inflammatory response pathways including “Positive regulation of acute inflammatory response” and “Inflammatory response to antigenic stimulus” (online supplemental extended data figure 2A). This is in line with the description of P4 as the inflammatory subset in literature.27 By using a combinatorial approach filtering out ISCOMs-induced genes in the other subsets and OA/ LPS-induced genes in P4, we identified 65 genes that are specifically upregulated by ISCOMs in P4, but not upregulated upon ISCOMs treatment in cDC1s, P1+2, P3, nor upon OA or LPS treatment in P4 (online supplemental extended data figure 2B and table 1). Strikingly, this included genes such as TRIB3 (~4-fold induction), which is an ER stress-inducible gene, downstream of the PERK pathway. We have published before that TRIB3 and other genes downstream of PERK are upregulated in the murine MHCIIlo CD11bhi DC subset upon ISCOMs stimulation and that PERK activation is crucial for ISCOMs-induced cross-presentation in this DC subset.21 Moreover, lipid-related genes APOE (~5-fold induction), CDS1 (~5-fold induction), GBA (~4-fold induction), and HSD17B14 (~14-fold induction) are specifically upregulated in P4. APOE is best known for its role in lipoprotein metabolism,31 whereas CDS1, also known as CPG-diacylglycerol synthase 1, is crucial for the regulation of lipid droplet growth.32 Mutations in the GBA gene have been linked to lipid alterations that are associated with Parkinson’s disease. HSD17B14 is a 17-beta-hydroxysteroid dehydrogenase that is involved in peroxisomal β-oxidation of fatty acids, and the breakdown of fatty acid chain elongation in the ER.33 34 In conclusion, these data indicate that ISCOMs upregulate lipid-related pathways specifically in the P4 (CD163+ CD14+) cDC2 subset.
Table 1.
Genes specific for P4 ISCOMs
| Genes | Fold change |
| ABCB1 | 7.713929243 |
| APOE | 4.975581838 |
| CAPNS2 | 9.979767603 |
| CCAT1 | 28.39131712 |
| CDKN3 | 6.914625195 |
| CDS1 | 5.098935519 |
| CHST1 | 27.72334149 |
| CLEC4A | 4.008313234 |
| COL22A1 | 33.00824066 |
| COL4A2-AS1 | 14.13743764 |
| FBXO10 | 4.190391801 |
| FBXO2 | 26.89625517 |
| FIGF | 33.40963375 |
| GAL | 44.38457189 |
| GBA | 4.231682052 |
| GIN1 | 4.366585996 |
| GNAO1 | 36.76779046 |
| HEBP1 | 4.068944088 |
| HSD17B14 | 14.44046381 |
| KHDRBS3 | 4.952637264 |
| LANCL3 | 25.65677356 |
| LINC00659 | 15.24906197 |
| LOC101929371 | 26.25785865 |
| LOC102723524 | 6.926784041 |
| LOC102723569 | 11.42477861 |
| LOC102724153 | 4.150846501 |
| LOC102724316 | 6.632749517 |
| LOC102724369 | 4.265184456 |
| LOC102724571 | 20.1669791 |
| LOC285638 | 4.432657248 |
| MAS1 | 117.7526355 |
| MICA | 6.128474378 |
| MIR4257 | 4.978114229 |
| MIR499B | 31.61367763 |
| MIR591 | 6.12076253 |
| MKNK1 | 4.082959086 |
| MPP4 | 9.526311288 |
| NDUFAF4P1 | 18.4677358 |
| NRBF2 | 4.098259785 |
| OR10P1 | 50.69324162 |
| PCDHGA11 | 21.87064488 |
| PCDHGA12 | 26.06845056 |
| PCDHGB5 | 25.65677356 |
| PHF23 | 4.04618254 |
| PITX1 | 25.15984089 |
| POM121L9P | 34.77758985 |
| RBM47 | 4.153873052 |
| RBP1 | 18.80462484 |
| RP11-265P11.2 | 28.33723066 |
| RP11-290F20.2 | 47.3134694 |
| SCARNA20 | 7.661874874 |
| SDS | 5.593669543 |
| SLC4A2 | 4.11043159 |
| SPOCD1 | 92.05931005 |
| TACSTD2 | 34.8083394 |
| TMEM52B | 5.153274705 |
| TNFSF18 | 43.01283957 |
| TRIB3 | 4.091024102 |
| TRIM47 | 12.67510751 |
| TRNAR22 | 9.36268282 |
| TRNQ | 6.691803218 |
| UNC5B | 4.492870072 |
| YPEL4 | 6.67719103 |
| ZBTB7C | 27.06881449 |
| ZNF827 | 6.877323214 |
ISCOMs, immune stimulatory complexes.
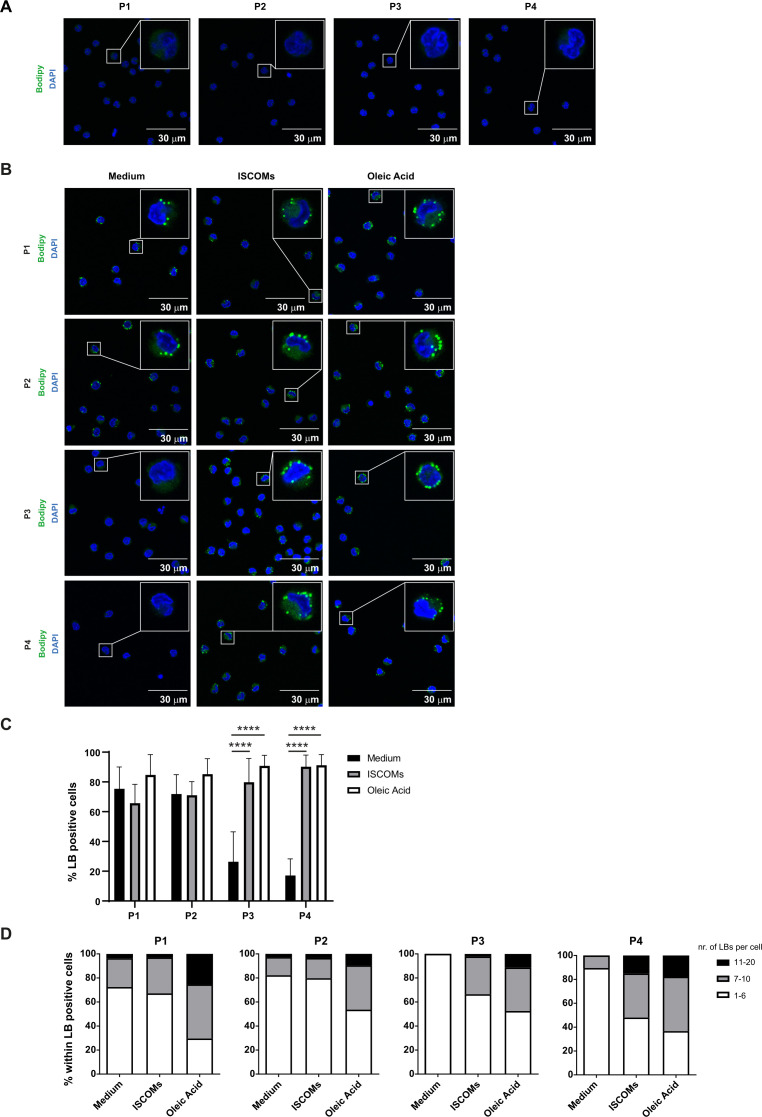
ISCOMs induce lipid bodies mainly in CD163+ CD14+ cDC2s
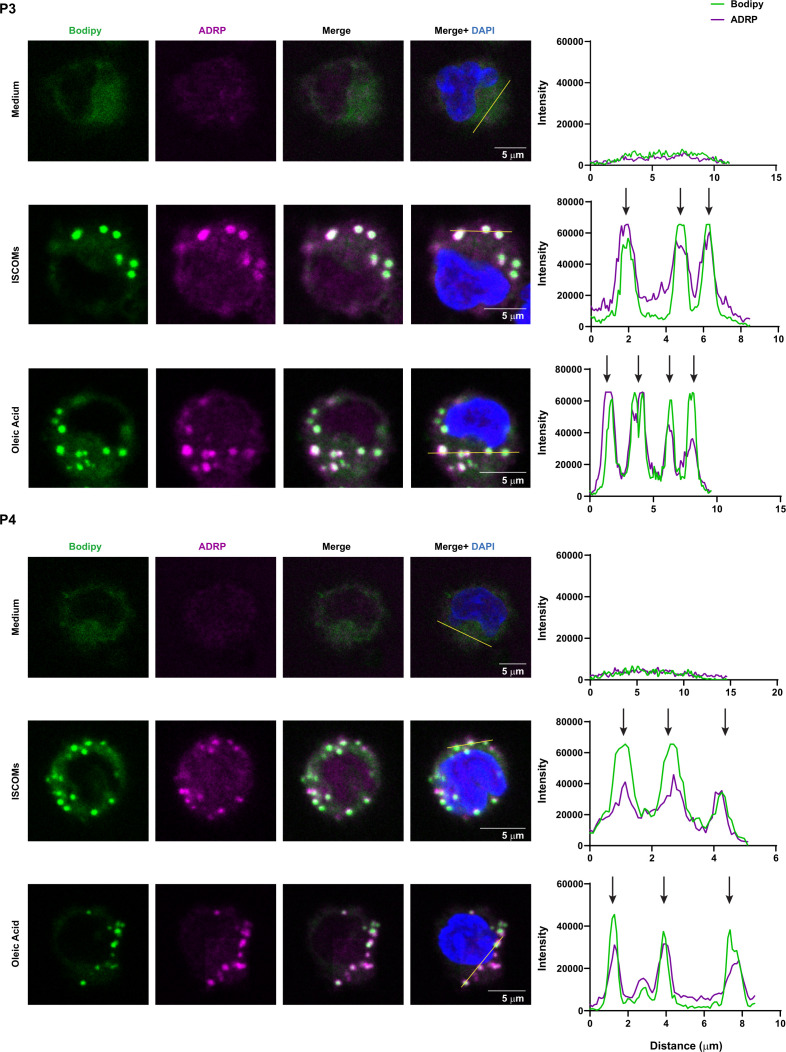
Since ISCOMs upregulate lipid-related pathways in the CD163+ CD14+ (P4) cDC2 subset, we further investigated their ability to induce LBs. No LBs were detected in all isolated cDC2 subsets directly after cell sorting (figure 5A). Analysis of the P1 and P2 subsets revealed that there were already LBs present in ~70% of the cells after 18 hours in medium (figure 5B,C). However, this was not further enhanced by ISCOMs stimulation. LBs were also present in populations P3 and P4 (~30% and 20%, respectively) when cultured with medium (figure 5B,C). Interestingly, the frequency of LB-containing cells in P3 and P4 was significantly increased after ISCOMs stimulation to ~80% and 90%, respectively. Further quantification of the number of LBs per cell revealed that ~15% of P4 contained ~11–20 LBs per cell compared with ~3% in the P1, P2, and P3 subsets when treated with ISCOMs (figure 5D). Shorter stimulation with ISCOMs showed comparable results, where significant induction of LBs was observed in P3 and P4 (online supplemental extended data figure 3A–C). To further confirm the identity of the lipid aggregates, we co-stained the cells with Bodipy and the LB marker ADRP (also known as Perilipin 2) after stimulation with ISCOMs or OA. By performing a fluorescence intensity profile analysis, we observed that the Bodipy signals are highly correlated with the localization of ADRP in all four cDC2 subsets, indicating that the lipid aggregates were indeed LBs (figure 6, online supplemental extended data figure 4). Our results indicate that ISCOMs induce LBs mostly in the P4 (CD163+ CD14+) cDC2s, both with respect to the frequency of LB positive cells and the number of LBs per cell.
Figure 5.
ISCOMs induce lipid bodies mostly in CD163+ CD14+ cDC2 s. (A) LB staining with Bodipy 493/503 in P1, P2, P3, and P4 cDC2 subsets immediately after cell sort was analyzed by laser scanning microscopy. A magnification of a selected cell in the overview picture is shown in the upper right corner. Nucleus staining is done with DAPI (blue). (B) LB (green) and nucleus (blue) staining in the four cDC2 subsets after 18 hours stimulation with ISCOMs or OA. (C) Quantification of the frequency of LB-positive cells was done with Fiji. (D) The number of LBs within LB-positive cells was quantified with Fiji and divided into three categories, 1–6 LBs, 7–10 LBs, and 11–20 LBs per cell, and shown as frequency per cDC2 subset. At least five overview pictures per sample were taken for quantification. Statistical analyses were done using one-way ANOVA using post hoc Tukey. Data are shown here from two healthy donors. ****p<0.0001 (DF=2; F (2, 172) = 167.2). ANOVA, analysis of variance; ISCOMs, immune stimulatory complexes; LBs, lipid bodies; OA, oleic acid.
Figure 6.
Co-localization between separate markers for LBs in cDC2 subsets. LB staining with Bodipy 493/503 (green) and ADRP (magenta) in P3 and P4 cDC2 subsets after 18 hours stimulation with ISCOMs or OA. Overlay pictures between Bodipy and ADRP are shown as Merge. Co-localization histograms for each fluorophore were created for a selected area in Fiji (indicated by a line on the Merge+DAPI picture). Co-localizations were depicted by arrows in the histograms. At least five pictures were taken per sample, and representative data are shown here for two healthy donors. ISCOMs, immune stimulatory complexes; LBs, lipid bodies; OA, oleic acid.
Enhanced antigen translocation by ISCOMs in CD163+ CD14+ cDC2s
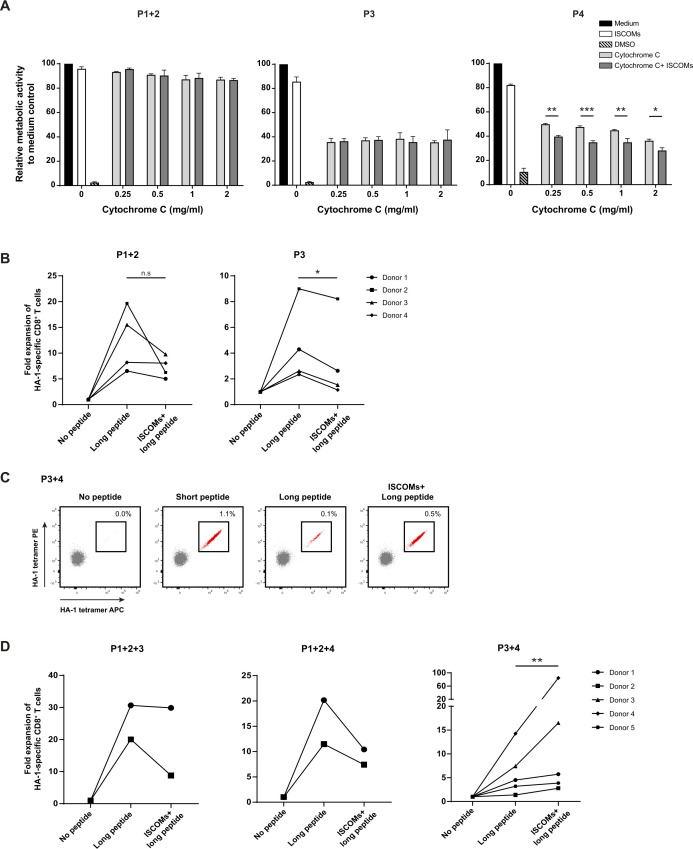
We and others have proposed that enhanced endosomal antigen translocation to the cytosol, thereby allowing more antigens to be available for processing, could play a role in SBA-induced cross-presentation.20 35 To investigate antigen translocation, the cDC2 subsets received mitochondrial protein cytochrome c in addition to ISCOMs. The translocation and subsequent exposure of cytochrome c from endosomes to the cytosol induces cell apoptosis, which can be measured by reduced cell metabolic activity and viability. The translocation of cytochrome c in non-stimulated conditions and subsequent decrease in cell metabolic activity is higher in P3 and P4 (~60%) compared with P1+2 (~10–15%) (figure 7A). This baseline difference might be caused by differences in the uptake of cytochrome c or by the differential capacity of translocation between the subsets. Strikingly, P4 was the only cDC2 subset that showed significantly reduced cell metabolism activity in the presence of ISCOMs in combination with different concentrations of cytochrome c (figure 7A). Antigen translocation of cytochrome c was observed in cDC1s, however, this was not further enhanced by ISCOMs (online supplemental extended data figure 5A). These data demonstrate that ISCOMs specifically enhance antigen translocation in the CD163+ CD14+ (P4) cDC2s subset.
Figure 7.
Enhanced antigen translocation and cross-presentation by CD14+ CD163+ cDC2 s upon ISCOMs. (A) Antigen translocation measured by CCK8 assay after stimulation with ISCOMs and titration of cytochrome c, indicated as relative metabolic activity and viability to medium control. Statistical analyses were done comparing using one-way ANOVA using post hoc Tukey. Representative data from one donor with two technical replicates are shown for two healthy donors. *p<0.05, **p<0.01, ***p<0.001 (DF=10; F (10, 11) = 444.0). (B) Fold expansion of HA-1-specific CD8+ T cells in P1+2 and P3. P1+2 (25,000+25,000 cells) and P3 (50,000 cells) were incubated with PBMCs (500,000 cells) in a 1:10 ratio. Data are shown for four healthy DC donors. (C) HA-1-specific CD8+ T-cell expansion indicated by double-positive HA-1 tetramer staining (in red) for P3+4 (87,000+12,500 cells) after stimulation with short peptide, long peptide, or ISCOMS+long peptide. DCs (100,000 cells) were incubated with PBMCs (1×106 cells) in a 1:10 ratio. Representative flow cytometry data are shown from one healthy DC donor. (D) Fold expansion of HA-1-specific CD8+ T cells in different combinations of cDC2 subsets for the number of donors indicated per graph. P1+2 (12,500+12,500 cells, respectively), P3 (25,000 cells), and P4 (25,000 cells) were combined into P1+2+3, and P1+2+4 (50,000 cells total). For P3+4 the following amounts were used: Donor 1 and 2 (P3 25,000+P4 25,000), donor 3 (P3 75,000+P4 25,000 cells), donor 4 (P3 87,000+P4 12,500 cells), and donor 5 (P3 93,750+P4 6,250 cells). The DCs were incubated with PBMCs containing HA-1-specific CD8+ T cells in a 1:10 ratio. Statistical analyses were done using paired non-parametric t-test. N.s. non significant, *p<0.05, **p<0.01. ANOVA, analysis of variance; DCs, dendritic cells; DMSO, dimethyl sulfoxide; ISCOMs, immune stimulatory complexes; PBMCs, peripheral blood mononuclear cells.
Enhanced antigen cross-presentation by ISCOMs requires CD163+ CD14+ cDC2s
Since we demonstrated earlier that ISCOMs could only augment antigen cross-presentation to HA-1+ T cells in cDC2s, we further zoomed in on the cDC2 subsets. The subsets P1+2, P3, and P4 were individually pulsed with HA-1 long peptide in the presence or absence of ISCOMs stimulation and then incubated with PBMCs containing HA-1 specific CD8+ T cells. HA-1 long peptide loading onto P1+2 and P3 cDC2 subsets resulted in an increase in absolute numbers of HA-1 specific CD8+ T cells, however, there was no further enhancement by addition of ISCOMs (figure 7B). Unexpectedly, co-culture of subset P4 and PBMCs showed a remarkable decrease in cell viability of the PBMCs containing HA-1 specific CD8+ T cells regardless of the stimuli that were added (online supplemental extended data figure 5B). Upon co-culture with P4 there is a decrease in CD3+ T cells, especially in the amount of CD4+ T cells (online supplemental extended data figure 5B). Further experiments using PBMCs from healthy donors showed similar results, ruling out that this is a specific effect of the PBMCs from patients containing HA-1 CD8+ T cells (online supplemental extended data figure 5C). As this effect was not observed in the PBMCs co-cultured with the total cDC2 population, we combined P4 with either P1+2 or P3. Combining P4 with either of the other populations prevented the loss of CD3+ CD4+ T cells induced by population P4 only (online supplemental extended data figure 5C). Therefore, we loaded different subset combinations with HA-1 long peptide with or without ISCOMs. Combining the subsets P1+2 with P3 or P4 showed no enhanced expansion of HA-1 specific CD8+ T cells after ISCOMs stimulus (figure 7D). However, the combination of P3 with P4 showed a superior increase in the frequencies and amount of HA-1 specific CD8+ T cells in the presence of ISCOMs for multiple donors (figure 7C,D). Titrating the amounts of P4 when combined with P3 implies that there is a ratio optimum for the enhancement of ISCOM-mediated antigen cross-presentation, too high or too low amounts of P4 will cause diminished effects (online supplemental extended data figure 6A). Taken together, our results indicate that enhanced antigen cross-presentation is induced by ISCOMs in cultures consisting of P4 cDC2s supplemented with P3 cDC2s.
Discussion
SBAs are new-generation adjuvants that are recently implemented in vaccination strategies against diseases such as COVID-19. The most outstanding advantage of SBAs over older adjuvant types, for example, aluminum salts or oil-in-water emulsion, is their ability to induce both CD4+ T helper and CD8+ T-cell responses.3–6 CD8+ T cells play a prominent role in battling cancer and in infectious diseases such as HIV and COVID-19.7 36 37 Although the efficiency of SBA-containing vaccines has clearly been demonstrated, its effect on inducing cross-presentation in human DCs remains largely unknown. In the current study, we demonstrate that SBAs can enhance antigen cross-presentation in a specific human cDC2 subset, identified as CD11c+ CD1c+ CD5− CD163+ cDC2s, leading to antigen-specific CD8+ T-cell expansion.
It has been suggested that human cDC2s are the homologue of murine CD8α− CD11b+ DCs.9 10 Recent studies have reported the heterogenicity of the human cDC2s.25–27 One out of the four newly identified cDC2 subsets is the CD163+ CD14+ cDC2s.27 We have previously shown that SBAs could only induce cross-presentation in murine MHCIIlo CD11bhi DCs.20 With the current study, we discovered a similar specificity of SBAs for their human counterpart, most specifically in CD163+ CD14+ (P4) cDC2s. Interestingly, the CD163+ CD14+ (P4) cDC2s highly expressed ITGAM (CD11b), which corresponds to the murine SBA-responsive DC subset. The CD163+ CD14+ (P4) cDC2s are the cells most likely responsible for the enhanced cross-presentation ability upon SBA-treatment. However, as the addition of the CD163+ CD14− (P3) subset to the CD163+ CD14+ (P4) co-culture with PBMCs was required to sustain the viability of PBMCs, particularly the CD4+ T cells, we cannot formally rule out a contribution of P3. It has been suggested that splenic CD8α+ DCs from mice can kill CD4+ T cells through Fas/Fas-ligand interaction as part of regulating T-cell responses.38 However, we could not detect high expression of tumor necrosis factor superfamily ligands (such as TRAIL, FasL, or OX40L) or PD-L1 on the CD163+ CD14+ (P4) cDC2s based on our RNA sequencing data. Our preliminary data showed that overnight cDC2 stimulation with TLR ligands LPS, R848, or Poly(I:C) did not prevent the decrease in T-cell numbers after 7 days, but rather resulted in a further reduction in T-cell viability in case of P3 and P4. This suggests that in our co-culture system with purified DCs the T cells are sensitive for (over)stimulation and require further studies. Since these cDC2 subsets have been discovered only recently, there is still much unknown about their individual functions and their relation to each other. It has been suggested that the four cDC2 subsets reflect their differentiation stages starting from CD5+ cells (P1), to CD5− CD163− cells (P2), to CD5− CD163+ CD14− cells (P3), and finishing with CD163+ CD14+ (p4) cells, proposing a differentiation and/or activation continuum.27 An important note is that CD163+ CD14− (P3) cells are the largest subset of cDC2s followed by CD163+ CD14+ (P4) cells. We demonstrated that there was an optimum ratio between the two subsets for cross-presentation and T-cell expansion, where lower amounts of CD163+ CD14+ (P4) cells were required compared with CD163+ CD14− (P3) cells. This might be a reflection of how the subsets are naturally occurring in the body with yet unidentified mechanisms of interplay between the subsets. Interestingly, a recent study showed that CD163+ cDC2s, encompassing both CD14+ and CD14− cDC2s (comparable with our P3 and P4 combined), could prime CD8+ CD103+ tissue-resident memory T cells.28 They investigated whether CD163+ cDC2s could differentiate from the CD163− cDC2s (comparable with our P1 and P2 combined) or from monocytes in the presence of granulocyte-macrophage colony-stimulating factor (GM-CSF)-expressing stromal cells. They showed that after 2 days neither CD163− cDC2s nor monocytes differentiated into CD163+ cDC2s, whereas CD163+ cDC2s did upregulate CD14 expression. We observed similar results when we incubated the four different cDC2 subsets individually for 1 day in the presence of GM-CSF. Interestingly, the authors also showed that CD163+ cDC2s could only be differentiated from human cord blood-derived CD34+ hematopoietic stem and progenitor cells in the presence of GM-CSF but not FLT3L, while FLT3L was already sufficient to induce the differentiation of CD163− cDC2s.28 Taken together, these results indicate that CD163+ CD14− (P3) and CD163+ CD14+ (P4) cDC2s are probably more closely related compared with CD5+ (P1) and CD5− CD163− (P2) cDC2s. The reason why only cDC2s, and specifically the CD163+ CD14+ (P4) subset, are sensitive to SBAs remains not fully understood. There could be a difference in SBAs and antigen uptake between the different DC subsets. Another possibility is that there are distinct pathways activated in the different DC subsets on SBA uptake. We have recently published that the PERK pathway of the unfolded protein response is selectively upregulated in murine MHCIIlo CD11bhi DCs upon SBA stimulation.21 One of the genes that is related to the PERK pathway, TRIB3, was specifically upregulated in both the murine CD11b+ DCs and the human CD163+ CD14+ (P4) subset upon ISCOMs stimulation. It would be of interest to investigate whether PERK also plays a dominant role in SBA-induced cross-presentation in CD163+ CD14+ (P4) cDC2s.
In addition to SBA-mediated cross-presentation, we showed an increase in antigen translocation in CD163+ CD14+ (P4) cDC2s, indicating that SBAs facilitate antigen escape from endosomes. Others have shown comparable results in human monocyte-derived DCs, where SBAs could induce efficient cross-presentation and antigen translocation of the cancer testis antigen NY-ESO-1 based on interferon (IFN)-γ production by CD8+ T cells.35 We have published before that SBA-induced cross-presentation is proteasome-dependent, suggesting that antigens are processed and loaded on MHCI molecules through the cytosolic pathway.20 Therefore, antigen translocation is a critical step to transport antigens that are initially stored in endosomal compartments into the cytosol for proteasomal degradation and subsequent antigen cross-presentation. It is still not fully understood how SBAs facilitate antigen translocation. It has been proposed that the SBA QS-21 mediates pore formation in lysosomes, resulting in the release of macromolecules to the cytosol.39 However, others argue that antigen translocation is a highly regulated process that involves dislocation through the endoplasmic-reticulum-associated protein degradation (ERAD) machinery or a transmembrane pore complex instead of a simple lysosome leakage.40–42 Their main argument is that uncontrolled lysosome leakage would lead to the cytosolic release of cathepsins and activate the NLRP3 inflammasome resulting in cell death.43 44 Recent work by Gros et al showed that the repair of endosomes is a highly regulated process, which means that endosomal leakage can still be controlled within the cell.45 Since our previous studies in mice demonstrated that not all pore-forming saponins (which mostly interact with the cell membrane) showed adjuvant activity in DCs, SBA probably induce endosomal translocation in a more controlled and localized manner.20
One of the characteristics of SBA sensitivity in DCs is their ability to induce LBs. We have demonstrated before that LB induction was observed only in SBA-stimulated murine MHCIIlo CD11bhi DCs and that this is correlated with their enhanced cross-presentation ability.20 Importantly, we showed in the same study that blocking LB formation with inhibitors such as 5-tetradecyloxy-2-furoic acid, diacylglycerol acyltransferase 1 and 2, or by inhibiting acyl-CoA synthetase or acetyl-CoA carboxylase, was correlated with impaired antigen cross-presentation by DCs. Moreover, IGTP (an IFN-related GTPase associated with lipid bodies) and ADRP knockout mice showed reduced lipid body accumulation and levels of cross-presentation after ISCOM treatment.20 These results indicate the vital role of LB formation for SBA-induced cross-presentation by DCs. We now show that SBAs could only augment LBs in cDC2s, especially in the CD163+ CD14− (P3) and CD163+ CD14+ (P4) cDC2 subsets, and not in cDC1s. Interestingly, we already found many CD5+ (P1) and CD5− CD163− cDC2s (P2) containing LBs after incubation with medium for 18 hours, although the frequency or number of LBs were not further enhanced upon SBA stimulation. Importantly, we show with RNA sequencing that lipid-associated pathways and genes were specifically upregulated in the CD163+ CD14+ (P4) cDC2s by SBAs, the same subset where we found the highest induction of LBs and enhanced cross-presentation capacity. Although ISCOMs could also induce LBs in the CD163+CD14− (P3) subset, it was to a much lower extent compared with P4. Quantification of the data from confocal microscopy analysis showed that the amount of LBs per cell is much higher in P4 than P3. Moreover, RNA sequencing analysis showed that the lipid metabolic pathways that were upregulated in P3 were less dominant than in P4 after ISCOMs treatment, and therefore did not fall into the selection of top upregulated pathways in P3. It is known that different stimuli or metabolic states in cells can affect their protein and/or lipid compositions, and even different LB populations could be observed within the same cell.46–48 While some studies have pointed out the essential role of LBs in inducing DC cross-presentation,20 49–51 contradictory studies showed the opposite where LBs hamper cross-presentation in tumor-associated DCs.52–54 This suggests that the type of LBs and their content is crucial for the desired functional outcome. Further analysis of lipid content in the different DC subsets, for example, by lipidomics, would give better insight into the different types of LBs and their distinct functions.
A recent study demonstrated that the infiltration of CD163+ cDC2s in luminal breast cancer primary tumors, containing both CD14+ and CD14− cells, was associated with the abundance of CD8+ CD103+ T cells,28 which can contribute to improved breast cancer prognosis.55 56 In contrast, a study reported that CD1c+ CD14+ cells were expanded in patients with melanoma and that cDC1 vaccines with high amounts of CD1c+ CD14+ cells were less effective in inducing antigen-specific CD4+ T-cell responses in patients with melanoma.57 Another study described that CD14+ cDC2s have a higher capacity to prime naïve CD4+ T cells towards Th2 and Th17 cells, which might correspond to disease activity in patients with systemic lupus erythematosus.27 It would be interesting to investigate whether the use of SBAs could skew the immune responses of CD14+ cDC2s into a more favorable outcome in those patients. Moreover, it is important to further investigate the functions of CD14+ cDC2s in various tissues and tumor microenvironments besides blood circulation. Interestingly, Bosteels et al recently showed that the activity of the adjuvant AS01, a combination of saponin and monophosphoryl lipid A (MPL), is associated with the recruitment of cDC1s and cDC2, including the inflammatory cDC2s.58 They showed that these (inflammatory) cDC2s are equally efficient at priming both OVA-specific CD4+ and CD8+ T cells. Mice lacking CCR2-dependent and Flt3-dependent inflammatory cDC2s failed to raise proper antibody and T-cell responses. All in all, these and our data support the finding that the inflammatory cDC2 subset could be a key target for adjuvant systems like saponin to induce adaptive immunity in humans.
In conclusion, we show that SBAs enhance antigen cross-presentation in recently identified human CD11c+ CD1c+ CD5− CD163+ cDC2s. Since SBAs are currently gaining ground in vaccination strategies, it is important to understand the mechanisms of SBA adjuvant activity and their specificity for DC subsets. Further work would be essential to understand the roles of these subsets in SBA-mediated immune responses and contribute to improving cancer vaccination development.
Methods
Cell isolation
PBMCs were isolated from buffy coats or apheresis from healthy donors (Sanquin, Nijmegen, The Netherlands) using Lymphoprep (STEMCELL Technologies). Total CD141+ CLEC9A+ DCs (cDC1s) were MACS isolated from PBMCs using the human CD141 (BDCA-3) MicroBead kit (Miltenyi Biotec), followed by isolation of total CD1c+ DCs (cDC2s) using the human CD1c (BDCA-1)+ dendritic cell isolation kit (Miltenyi Biotec). A purity check of the subsets after isolation was performed by additional FACS staining and measured flow cytometry (BD FACSCanto II). The cDC1s were stained with anti-Clec9a (SONY Biotechnology) and anti-CD141 antibodies (BioLegend), cDC2s were stained with anti-CD11c (BD Biosciences) and anti-CD1c antibodies (Miltenyi Biotec). Further cDC2 subset isolation was performed by FACS sorting (BD FACS Aria II SORP) using anti-CD11c (BC Biosciences), anti-CD1c (Miltenyi Biotec), anti-CD5 (BioLegend), anti-CD163 (eBioscience), and anti-CD14 (BD Biosciences) antibodies. The four cDC2 subsets were gated according to the following markers: P1 (CD11c+ CD1c+ CD5+), P2 (CD11c+ CD1c+ CD5− CD163−), P3 (CD11c+ CD1c+ CD5− CD163+ CD14−), and P4 (CD11c+ CD1c+ CD5− CD163+ CD14+). Cells were cultured in X-VIVO 15 serum-free hematopoietic cell medium (Lonza) with an addition of 10% fetal bovine serum (Gibco), and 40 U/mL human granulocyte-macrophage colony-stimulating factor (hGM-CSF) (Immunotools).
RNA sequencing
The different cell populations were isolated, as described above, from healthy apheresis donors. Cells were stimulated with Matrix C ISCOMs (400 ng/mL, MSD Animal Health, Boxmeer, the Netherlands), LPS (1 µg/mL, Sigma-Aldrich), or OA (50 µM, Sigma-Aldrich) for 8 hours. RNA isolation for each subset was performed with TRIzol Reagent (Invitrogen) and DNAse treatment (DNA-free DNA Removal Kit, Invitrogen). RNA sequencing was performed and Fastq files were mapped to the reference human genome GRCh38 using the Seq2science pipeline (https://github.com/vanheeringen-lab/seq2science) (STAR as default aligner). Tag count normalization, dimension reduction, and differential expression analysis (twofold change, p value<0.05) were performed using DESeq2 in R.59 To investigate the subset purity, the top 25 highest expressed genes of the transformed normalized counts were selected per subset and compared with the other subsets and genes known from literature. Subsequently, enriched gene sets were determined by Generally Applicable Gene-set Enrichment.60 L2F change and go.bp.gs set were used to determine the enriched biological processes (GO-terms). Immunologically relevant GO pathways that were upregulated with p<0.05 were selected and displayed as a radar plot showing the gene ratio (observed gene count/ total gene count per pathway). Murine RNA sequencing data for MHCIIhi CD11bint and MHCIIlo CD11bhi were from our previous work.21
Lipid body staining
Isolated DC subsets were incubated with Matrix C ISCOMs (400 ng/mL) or OA (50 µM) for 5, 10, or 18 hours. Cells (100,000) were then washed and seeded on chamber slides (Nunc Lab-Tek II) precoated with Poly-L-Lysine (100 µg/mL, Sigma-Aldrich). After 1 hour attachment, cells were fixated with 4% formaldehyde (Merck) and stained with Bodipy 493/503 (7 μg/ml in phophate-buffered saline (PBS), Invitrogen) for 10 min at room temperature. For the co-staining studies, cells were first permeabilized with saponin (0.1% in PBS, Sigma) for 5 min, followed by 5% human blocking serum (30 min, 37°C). Cells were then incubated with anti-ADRP Alexa-647 (in 0.1% saponin in PBA, 30 min at 37°C, Abcam), washed, and incubated with Bodipy 493/503 as described earlier. DAPI (Santa Cruz Biotechnology) was used for nucleus staining and the chamber slides were mounted with Prolong Diamond Antifade Mountant (Invitrogen) and analyzed with laser scanning microscopy (Leica SP8 SMD, x63 water objective). At least five pictures were taken per sample for quantification. The LB frequency and number of LBs per cell were quantified with Fiji and a script developed by Paul Rijken (Department of Radiation Oncology, Radboud University Medical Center, Nijmegen, The Netherlands). Co-localization histograms for each fluorophore were created for a selected area (indicated by a line on the image) in Fiji.
Antigen translocation
Isolated DC subsets (50,000 cells) were seeded in a flat-bottom 96-wells plate (Costar) and incubated with Matrix C ISCOMs (400 ng/mL) with or without cytochrome c (2.5 mg/mL unless indicated otherwise, Sigma-Aldrich) for 18 hours. As a control, 10% DMSO (Merck) was added to the cells to induce cell death. Cell metabolic activity and viability were measured by Cell Counting Kit-8 (CCK8, Sigma-Aldrich) following the manufacturer’s protocol. The absorbance was measured by a spectrophotometer at 450 nm. Relative metabolic activity was calculated as (treatment-blank)/(control-blank)×100%.
DC cross-presentation
For MiHA-specific T-cell expansion assays, cryopreserved PBMCs containing HA-1-specific CD8+ memory T cells from allogeneic stem cell transplantation patients were used.61 All patient material was obtained in accordance with the Declaration of Helsinki and institutional guidelines and regulations (CMO 2012/064). The different DC populations were isolated, as described above, from HLA-A*02:01+ buffy coats from healthy donors (Sanquin, Nijmegen, The Netherlands) and typed HA-1− with PCR to prevent endogenous expression of HA-1 by the DCs. The different DC subsets were incubated with Matrix C ISCOMs (400 ng/mL) in combination with or without HA-1 long peptide (5 µM, VARFAEGLEKLKECVLHDDLLEARRPRAHEZL) in Iscove's modified Dulbecco's medium (IMDM) (Gibco) containing 2% human serum, 40 U/mL hGM-CSF (Immunotools), and 1% penicillin-streptomycin (PS) (Gibco) for 3 hours. After 3 hours of pulse, additional human serum was added until 10% end concentration and cells were further incubated overnight. Positive control samples were incubated with HA-1 short peptide (5 µM, VLHDDLLEA) for 2 hours in the presence of 2% human serum. After washing the DCs, PBMCs containing HA-1 specific CD8+ T cells were added and incubated for 7 days in IMDM containing 10% human serum, 40 U/mL hGM-CSF, and 1% PS. On day 4, medium was refreshed with IMDM containing 10% human serum, 1% PS, 50 U/mL human interleukin (hIL)-2 (Immunotools), and 40 U/mL hIL-15 (Immunotools). Cells were harvested and counted after 7 days and HA-1-specific CD8+ T-cell expansion was measured by flow cytometry (Cytoflex LX 21-color, Beckman Coulter). Cells were stained with anti-CD3 (BioLegend), anti-CD8 (Invitrogen), HA-1 tetramer PE, and HA-1 tetramer APC. Fold expansions of tetramer+ HA-1 specific T cells were quantified by dividing the absolute numbers of double tetramer+ T cells after treatment by the absolute numbers of double tetramer+ T cells in the no peptide sample. The HA-1 tetramers, were kindly gifted from Mirjam Heemskerk and Frederik Falkenburg (Department of Hematology, Leiden University Medical Center, Leiden, The Netherlands).
PBMC viability assay
The isolated DC subsets (50,000 cells) were incubated with freshly isolated PBMCs (500,000 cells) from healthy donors in IMDM, 10% human serum, 40 U/mL hGM-CSF, and 1% PS. On day 4, medium was refreshed with IMDM with 10% human serum, 1% PS, 50 U/mL hIL-2, and 40 U/mL hIL-15. Cells were harvested and counted after 7 days, and cell viability was measured by flow cytometry (BD FACSCanto II). Cells were stained with anti-CD8 (BioLegend), anti-CD3 (BD Biosciences), anti-CD4 (BioLegend), and Fixable Viability Dye eFluor 780 (eBioscience).
Statistical analysis
One-way analysis of variance with post hoc Tukey, or paired non-parametric t-test was performed as indicated in the figure legends. All results are expressed as mean values with SD. The following indications are used in all figures: n.s., non significant; *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001.
Acknowledgments
We thank Lonneke van der Linden, Nicolette Scholtes, and Yvonne Biermann (Vaxxinova Netherlands B.V.) for their fruitful discussions. We thank Paul Rijken (Department of Radiation Oncology, Radboud University Medical Center, Nijmegen, The Netherlands) for developing a quantification script for Fiji. We thank Wendy de Jong, Henk Tijssen, and Arnold van der Meer (Department of Laboratory Medicine, Radboud University Medical Center, Nijmegen, The Netherlands) for performing HA-1 PCR typing of buffy coats from healthy donors. We thank Mirjam Heemskerk and Frederik Falkenburg (Department of Hematology, Leiden University Medical Center, Leiden, The Netherlands) for providing the HA-1 tetramers.
Footnotes
Contributors: NIH, LGMHi'tV, and GJA designed the study. NIH, LGMHi'tV, JvEvdS, MWGL, and EDK-R carried out research experiments. BMHH and JHAM performed and assisted in RNA sequencing and analysis. HD and WH assisted and interpreted the cross-presentation assays. KvdD assisted with lipid body co-localization analysis. NIH, LGMHi’tV, and GA wrote the manuscript. All authors have read and approved the final manuscript. GJA is the guarantor of the manuscript.
Funding: Research grant from Vaxxinova Netherlands B.V. to GJA.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available upon reasonable request. The data that support the findings of this study are available from the corresponding author upon request.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study involves human participants and was approved by Sanquin NVT0371.01 Participants gave informed consent to participate in the study before taking part.
References
- 1.Ho NI, Huis in ’t Veld LGM, Raaijmakers TK, et al. Adjuvants enhancing cross-presentation by Dendritic cells: the key to more effective vaccines Front Immunol 2018;9:2874. 10.3389/fimmu.2018.02874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kensil CR, Patel U, Lennick M, et al. Separation and characterization of saponins with adjuvant activity from Quillaja Saponaria Molina cortex. J Immunol 1991;146:431–7. [PubMed] [Google Scholar]
- 3.Sun H-X, Xie Y, Ye Y-P. Advances in Saponin-based Adjuvants. Vaccine 2009;27:1787–96. 10.1016/j.vaccine.2009.01.091 [DOI] [PubMed] [Google Scholar]
- 4.Pearse MJ, Drane D. ISCOMATRIXTM AdjuvaNt: A potent inducer of humoral and cellular immune responses. Vaccine 2004;22:2391–5. 10.1016/j.vaccine.2003.12.031 [DOI] [PubMed] [Google Scholar]
- 5.Maraskovsky E, Schnurr M, Wilson NS, et al. Development of prophylactic and therapeutic vaccines using the ISCOMATRIX adjuvant. Immunol Cell Biol 2009;87:371–6. 10.1038/icb.2009.21 [DOI] [PubMed] [Google Scholar]
- 6.Lövgren Bengtsson K, Morein B, Osterhaus AD. ISCOM technology-based matrix MTM Adjuva: success in future vaccines relies on formulation. Expert Rev Vaccines 2011;10:401–3. 10.1586/erv.11.25 [DOI] [PubMed] [Google Scholar]
- 7.Raskov H, Orhan A, Christensen JP, et al. Cytotoxic Cd8+ T cells in cancer and cancer Immunotherapy. Br J Cancer 2021;124:359–67. 10.1038/s41416-020-01048-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Embgenbroich M, Burgdorf S. Current concepts of antigen cross-presentation. Front Immunol 2018;9:1643. 10.3389/fimmu.2018.01643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guilliams M, Ginhoux F, Jakubzick C, et al. Dendritic cells, monocytes and Macrophages: a unified nomenclature based on Ontogeny. Nat Rev Immunol 2014;14:571–8. 10.1038/nri3712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Macri C, Pang ES, Patton T, et al. Dendritic cell Subsets. Semin Cell Dev Biol 2018;84:11–21. 10.1016/j.semcdb.2017.12.009 [DOI] [PubMed] [Google Scholar]
- 11.Vu Manh TP, Bertho N, Hosmalin A, et al. Investigating evolutionary conservation of Dendritic cell subset identity and functions. Front Immunol 2015;6:260. 10.3389/fimmu.2015.00260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutiérrez-Martínez E, Planès R, Anselmi G, et al. Cross-presentation of cell-associated antigens by MHC class I in Dendritic cell Subsets. Front Immunol 2015;6:363. 10.3389/fimmu.2015.00363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cruz FM, Colbert JD, Merino E, et al. The biology and underlying mechanisms of cross-presentation of exogenous antigens on MHC-I molecules. Annu Rev Immunol 2017;35:149–76. 10.1146/annurev-immunol-041015-055254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mittag D, Proietto AI, Loudovaris T, et al. Human Dendritic cell Subsets from spleen and blood are similar in phenotype and function but modified by donor health status. The Journal of Immunology 2011;186:6207–17. 10.4049/jimmunol.1002632 [DOI] [PubMed] [Google Scholar]
- 15.Segura E, Durand M, Amigorena S. Similar antigen cross-presentation capacity and Phagocytic functions in all freshly isolated human Lymphoid organ-resident Dendritic cells. J Exp Med 2013;210:1035–47. 10.1084/jem.20121103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nizzoli G, Krietsch J, Weick A, et al. Human Cd1C+ Dendritic cells Secrete high levels of IL-12 and potently prime cytotoxic T-cell responses. Blood 2013;122:932–42. 10.1182/blood-2013-04-495424 [DOI] [PubMed] [Google Scholar]
- 17.Cohn L, Chatterjee B, Esselborn F, et al. Antigen delivery to early Endosomes eliminates the superiority of human blood Bdca3+ Dendritic cells at cross presentation. J Exp Med 2013;210:1049–63. 10.1084/jem.20121251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sittig SP, Bakdash G, Weiden J, et al. A comparative study of the T cell stimulatory and polarizing capacity of human primary blood Dendritic cell Subsets. Mediators Inflamm 2016;2016:3605643. 10.1155/2016/3605643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson NS, Yang B, Morelli AB, et al. ISCOMATRIX vaccines mediate Cd8 T-cell cross-priming by a Myd88-dependent signaling pathway. Immunol Cell Biol 2012;90:540–52. 10.1038/icb.2011.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.den Brok MH, Büll C, Wassink M, et al. Saponin-based Adjuvants induce cross-presentation in Dendritic cells by intracellular lipid body formation. Nat Commun 2016;7. 10.1038/ncomms13324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huis in ’t Veld LGM, Ho NI, Wassink M, et al. Saponin-based adjuvant-induced Dendritic cell cross-presentation is dependent on PERK activation. Cell Mol Life Sci 2022;79:231. 10.1007/s00018-022-04253-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.den Brok MH, Nierkens S, Wagenaars JA, et al. Saponin-based Adjuvants create a highly effective anti-tumor vaccine when combined with in situ tumor destruction. Vaccine 2012;30:737–44. 10.1016/j.vaccine.2011.11.080 [DOI] [PubMed] [Google Scholar]
- 23.Welte MA, Gould AP. Lipid Droplet functions beyond energy storage. Biochim Biophys Acta Mol Cell Biol Lipids 2017;1862(10 Pt B):1260–72. 10.1016/j.bbalip.2017.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olzmann JA, Carvalho P. Dynamics and functions of lipid droplets. Nat Rev Mol Cell Biol 2019;20:137–55. 10.1038/s41580-018-0085-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yin X, Yu H, Jin X, et al. Human blood Cd1C+ Dendritic cells encompass Cd5High and Cd5Low Subsets that differ significantly in phenotype, gene expression, and functions. The Journal of Immunology 2017;198:1553–64. 10.4049/jimmunol.1600193 [DOI] [PubMed] [Google Scholar]
- 26.Villani A-C, Satija R, Reynolds G, et al. Single-cell RNA-Seq reveals new types of human blood Dendritic cells, monocytes, and progenitors. Science 2017;356. 10.1126/science.aah4573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dutertre C-A, Becht E, Irac SE, et al. Single-cell analysis of human mononuclear phagocytes reveals subset-defining markers and identifies circulating inflammatory Dendritic cells. Immunity 2019;51:573–89. 10.1016/j.immuni.2019.08.008 [DOI] [PubMed] [Google Scholar]
- 28.Bourdely P, Anselmi G, Vaivode K, et al. Transcriptional and functional analysis of Cd1C + human Dendritic cells identifies a Cd163 + subset priming Cd8 + Cd103 + T cells. Immunity 2020;53:335–352. 10.1016/j.immuni.2020.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morein B, Sundquist B, Höglund S, et al. Iscom, a novel structure for Antigenic presentation of membrane proteins from enveloped viruses. Nature 1984;308:457–60. 10.1038/308457a0 [DOI] [PubMed] [Google Scholar]
- 30.van der Aa E, Biesta PJ, Woltman AM, et al. Transcriptional patterns associated with Bdca3 expression on Bdca1 + myeloid Dendritic cells. Immunol Cell Biol 2018;96:330–6. 10.1111/imcb.12002 [DOI] [PubMed] [Google Scholar]
- 31.Kockx M, Traini M, Kritharides L. Cell-specific production, secretion, and function of apolipoprotein E. J Mol Med (Berl) 2018;96:361–71. 10.1007/s00109-018-1632-y [DOI] [PubMed] [Google Scholar]
- 32.Qi Y, Kapterian TS, Du X, et al. CDP-Diacylglycerol Synthases regulate the growth of lipid droplets and Adipocyte development. J Lipid Res 2016;57:767–80. 10.1194/jlr.M060574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lukacik P, Keller B, Bunkoczi G, et al. Structural and biochemical characterization of human orphan Dhrs10 reveals a novel cytosolic enzyme with steroid dehydrogenase activity. Biochem J 2007;402:419–27. 10.1042/BJ20061319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hiltunen JK, Kastaniotis AJ, Autio KJ, et al. 17B-Hydroxysteroid Dehydrogenases as Acyl Thioester Metabolizing enzymes. Mol Cell Endocrinol 2019;489:107–18. 10.1016/j.mce.2018.11.012 [DOI] [PubMed] [Google Scholar]
- 35.Schnurr M, Orban M, Robson NC, et al. ISCOMATRIX adjuvant induces efficient cross-presentation of tumor antigen by Dendritic cells via rapid cytosolic antigen delivery and processing via Tripeptidyl Peptidase II. The Journal of Immunology 2009;182:1253–9. 10.4049/jimmunol.182.3.1253 [DOI] [PubMed] [Google Scholar]
- 36.Collins DR, Gaiha GD, Walker BD. Cd8+ T cells in HIV control, cure and prevention. Nat Rev Immunol 2020;20:471–82. 10.1038/s41577-020-0274-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bange EM, Han NA, Wileyto P, et al. Cd8+ T cells contribute to survival in patients with COVID-19 and hematologic cancer. Nat Med 2021;27:1280–9. 10.1038/s41591-021-01386-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Süss G, Shortman K. A Subclass of Dendritic cells kills Cd4 T cells via Fas/Fas-ligand-induced apoptosis. J Exp Med 1996;183:1789–96. 10.1084/jem.183.4.1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Welsby I, Detienne S, N’Kuli F, et al. Lysosome-dependent activation of human Dendritic cells by the vaccine adjuvant QS-21. Front Immunol 2016;7:663. 10.3389/fimmu.2016.00663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koopmann JO, Albring J, Hüter E, et al. Export of Antigenic peptides from the Endoplasmic Reticulum INTERSECTS with retrograde protein translocation through the Sec61P channel. Immunity 2000;13:117–27. 10.1016/s1074-7613(00)00013-3 [DOI] [PubMed] [Google Scholar]
- 41.Ackerman AL, Giodini A, Cresswell P. A role for the Endoplasmic Reticulum protein Retrotranslocation machinery during Crosspresentation by Dendritic cells. Immunity 2006;25:607–17. 10.1016/j.immuni.2006.08.017 [DOI] [PubMed] [Google Scholar]
- 42.Zehner M, Marschall AL, Bos E, et al. The Translocon protein Sec61 mediates antigen transport from Endosomes in the Cytosol for cross-presentation to Cd8(+) T cells. Immunity 2015;42:850–63. 10.1016/j.immuni.2015.04.008 [DOI] [PubMed] [Google Scholar]
- 43.Hornung V, Bauernfeind F, Halle A, et al. Silica crystals and aluminum salts activate the Nalp3 Inflammasome through Phagosomal Destabilization. Nat Immunol 2008;9:847–56. 10.1038/ni.1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Próchnicki T, Mangan MS, Latz E. Recent insights into the molecular mechanisms of the Nlrp3 Inflammasome activation. F1000Res 2016;5:F1000 Faculty Rev-1469. 10.12688/f1000research.8614.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gros M, Segura E, Rookhuizen DC, et al. Endocytic membrane repair by ESCRT-III controls antigen export to the Cytosol during antigen cross-presentation. Cell Rep 2022;40:111205. 10.1016/j.celrep.2022.111205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng J, Fujita A, Ohsaki Y, et al. Quantitative electron microscopy shows uniform incorporation of triglycerides into existing lipid droplets. Histochem Cell Biol 2009;132:281–91. 10.1007/s00418-009-0615-z [DOI] [PubMed] [Google Scholar]
- 47.Soni KG, Mardones GA, Sougrat R, et al. Coatomer-dependent protein delivery to lipid droplets. J Cell Sci 2009;122(Pt 11):1834–41. 10.1242/jcs.045849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hodges BDM, Wu CC. Proteomic insights into an expanded cellular role for cytoplasmic lipid droplets. J Lipid Res 2010;51:262–73. 10.1194/jlr.R003582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bougnères L, Helft J, Tiwari S, et al. A role for lipid bodies in the cross-presentation of Phagocytosed antigens by MHC class I in Dendritic cells. Immunity 2009;31:232–44. 10.1016/j.immuni.2009.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ibrahim J, Nguyen AH, Rehman A, et al. Dendritic cell populations with different concentrations of lipid regulate tolerance and immunity in Mouse and human liver. Gastroenterology 2012;143:1061–72. 10.1053/j.gastro.2012.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee W, Kingstad-Bakke B, Paulson B, et al. Carbomer-based adjuvant elicits Cd8 T-cell immunity by inducing a distinct metabolic state in cross-presenting Dendritic cells. PLoS Pathog 2021;17:e1009168. 10.1371/journal.ppat.1009168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Herber DL, Cao W, Nefedova Y, et al. Lipid accumulation and Dendritic cell dysfunction in cancer. Nat Med 2010;16:880–6. 10.1038/nm.2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cao W, Ramakrishnan R, Tuyrin VA, et al. Oxidized lipids block antigen cross-presentation by Dendritic cells in cancer. The Journal of Immunology 2014;192:2920–31. 10.4049/jimmunol.1302801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Veglia F, Tyurin VA, Mohammadyani D, et al. Lipid bodies containing Oxidatively TRUNCATED lipids block antigen cross-presentation by Dendritic cells in cancer. Nat Commun 2017;8:2122. 10.1038/s41467-017-02186-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Z-Q, Milne K, Derocher H, et al. Cd103 and Intratumoral immune response in breast cancer. Clin Cancer Res 2016;22:6290–7. 10.1158/1078-0432.CCR-16-0732 [DOI] [PubMed] [Google Scholar]
- 56.Kathleen Cuningham Foundation Consortium for Research into Familial Breast Cancer (kConFab), Savas P, Virassamy B, et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat Med 2018;24:986–93. 10.1038/s41591-018-0078-7 [DOI] [PubMed] [Google Scholar]
- 57.Bakdash G, Buschow SI, Gorris MAJ, et al. Expansion of a Bdca1+Cd14+ myeloid cell population in Melanoma patients may attenuate the efficacy of Dendritic cell vaccines. Cancer Res 2016;76:4332–46. 10.1158/0008-5472.CAN-15-1695 [DOI] [PubMed] [Google Scholar]
- 58.Bosteels C, Fierens K, De Prijck S, et al. Ccr2- and Flt3-dependent inflammatory conventional type 2 Dendritic cells are necessary for the induction of adaptive immunity by the human vaccine adjuvant system As01. Front Immunol 2020;11:606805. 10.3389/fimmu.2020.606805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with Deseq2. Genome Biol 2014;15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luo W, Friedman MS, Shedden K, et al. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics 2009;10:161. 10.1186/1471-2105-10-161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Eck van der Sluijs J, van Ens D, Thordardottir S, et al. Clinically applicable Cd34+-Derived blood Dendritic cell Subsets exhibit key subset-specific features and potently boost anti-tumor T and NK cell responses. Cancer Immunol Immunother 2021;70:3167–81. 10.1007/s00262-021-02899-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2023-007082supp001.pdf (2.3MB, pdf)
Data Availability Statement
Data are available upon reasonable request. The data that support the findings of this study are available from the corresponding author upon request.