Key Points
CX3CR1+BIRC5+ immune cells expand with S. mansoni and S. aureus infection.
Leukocidin-deficient S. aureus does not induce CX3CR1+BIRC5+ myeloid cells.
Deletion of BIRC5 in CX3CR1+ cells improves survival with S. aureus infection.
Abstract
Our previous studies identified a population of stem cell–like proliferating myeloid cells within inflamed tissues that could serve as a reservoir for tissue macrophages to adopt different activation states depending on the microenvironment. By lineage-tracing cells derived from CX3CR1+ precursors in mice during infection and profiling by single-cell RNA sequencing, in this study, we identify a cluster of BIRC5+ myeloid cells that expanded in the liver during chronic infection with either the parasite Schistosoma mansoni or the bacterial pathogen Staphylococcus aureus. In the absence of tissue-damaging toxins, S. aureus infection does not elicit these BIRC5+ cells. Moreover, deletion of BIRC5 from CX3CR1-expressing cells results in improved survival during S. aureus infection. Hence the combination of single-cell RNA sequencing and genetic fate-mapping CX3CR1+ cells revealed a toxin-dependent pathogenic role for BIRC5 in myeloid cells during S. aureus infection.
Introduction
During inflammation, CX3CR1+ monocytes are recruited into inflamed sites and can respond to signals to adopt different activation states when exposed to different environmental stimuli in the tissues (1, 2). We had previously used a combination of single-cell RNA sequencing (scRNA-seq) and genetic fate mapping to profile cells derived from CX3CR1+ precursors in mice during atherosclerosis (3). The results revealed a spectrum of macrophage activation states more complex than the traditional M1 and M2 categories in the inflamed plaques (3). In addition, we identified an unexpected cluster of proliferating myeloid cells with a stem cell–like signature that we hypothesize may persist in a proliferating self-renewal state in inflamed tissue instead of differentiating immediately into macrophages after entering the tissue. We speculated that these proliferative cells may serve as a reservoir for tissue macrophages that adopt different activation states during an inflammatory response. However, it was unclear whether this population was unique to atherosclerosis or could be observed in other inflammatory conditions, such as during parasite and bacterial infections.
During infection with the helminth parasite Schistosoma mansoni, macrophages derived from Ly6ChighCX3CR1+ monocytes are required to protect the liver hepatocytes from tissue damage as part of the granulomas that are formed around the parasite eggs (4, 5). This chronic inflammatory response in the liver has been well characterized as a model of monocyte recruitment and differentiation into activated macrophages important for the prevention of tissue damage (6). Staphylococcus aureus is a Gram-positive bacterium that produces a collection of pore-forming leukocidins as a strategy for immune evasion and tissue damage (7, 8). Leukocidins are important virulence factors that target and kill a variety of immune cells, and S. aureus mutants that lack leukocidins are attenuated (9–12). However, the exact mechanisms by which leukocidins promote pathogenesis during S. aureus infection are still incompletely understood, and there is a possibility that increased inflammatory responses driven by these toxins as immune subversion molecules may be damaging instead of being protective to the host. In this study, we investigated whether the proliferative myeloid cells that we first observed in the context of atherosclerosis are also present in the liver during S. mansoni and S. aureus infections.
BIRC5, also termed “Survivin,” is an important protein for cell division and inhibition of apoptosis (13, 14). It is most well characterized in the context of cancer cells, where it has received considerable attention as a potential drug target (15–17). This small adaptor protein interacts with its cellular partners during mitosis as part of the chromosomal passenger complex, as well as with members of the inhibitors of apoptosis protein family to interfere with apoptosis (14). The BIRC5 inhibitor YM155 has been shown to have anticancer effects in cell culture and in mouse tumor models but has yet to be used as an effective clinical drug (17–19). A potential role in autoimmune diseases, such as rheumatoid arthritis (20) and multiple sclerosis (21, 22), as well as in CD4+ cells during HIV-1 infection (23), has also been described.
Although the role of BIRC5 has been well characterized at a cellular level in cancer cells, the role played by BIRC5 in myeloid cells during infection is not well understood. By using scRNA-seq with genetic fate mapping to characterize immune cells derived from CX3CR1+ precursors in the liver during S. mansoni and S. aureus infection, we identified and further investigated a population of BIRC5+ myeloid proliferating cells with stem cell–like signatures, which may play a pathogenic role during infection with S. aureus. Hence small molecules designed to inhibit the activity of BIRC5 may be beneficial as host-directed therapy for S. aureus infection.
Materials and Methods
Mice
B6.Cx3cr1CreERT2-IRES-EYFP/+ mice were generously provided by D. Littman (Department of Pathology, Department of Microbiology, NYU Langone Health, New York, NY). B6.Rosa26stop-tdTomato mice (JAX: 007914) were from Jackson Laboratories (Bar Harbor, ME). B6.Cx3cr1CreERT2-IRES-EYFP/+ and B6.Rosa26stop-tdTomato mice were crossed to generate Cx3cr1CreERT2-IRES-EYFP/+Rosa26tdTomato/+ mice as previously described. B6.129P2-Birc5tm1Mak/J mice were purchased from Jackson Laboratories (strain #031830) and crossed with B6.Cx3cr1CreERT2-IRES-EYFP/+ to generate Cx3cr1CreERT2-IRES-EYFP/+Birc5Flox/Flox conditional knockout mice (24). To induce labeling, we dissolved tamoxifen (Sigma-Aldrich) in corn oil and administered by oral gavage at a dose of 500 mg/kg body weight. Alternatively, mice were placed on a tamoxifen diet (Teklad Global, 250, 2016, Red) with diet code TD130856, to induce conditional deletion. A mix of male and female mice was used for experiments. All mice were age matched within experiments and used at 8–12 wk of age. All animal procedures were approved by the NYU School of Medicine Institutional Animal Care and Use Committee.
Models of infection
For experiments with S. mansoni infection, mice were infected percutaneously with 75 cercariae, given an oral gavage of tamoxifen at 7 wk postinfection, and analyzed at 8 wk postinfection. The reagent was provided by the National Institute of Allergy and Infectious Diseases Schistosomiasis Resource Center of the Biomedical Research Institute (Rockville, MD) through National Institutes of Health, National Institute of Allergy and Infectious Diseases Contract HHSN272201700014I. National Institutes of Health: Biomphalaria glabrata were exposed to S. mansoni (Puerto Rican strain NMRI).
S. aureus strains USA300 LAC clone AH1263 (wild type [WT]) (25) and AH-LAC ΔlukAB, hlg::tet, lukED::kan, pvl::epc, hla::erm (ΔTOX) were used in this study (26). For in vivo infection studies, WT S. aureus was grown on tryptic soy agar or tryptic soy broth. S. aureus was grown overnight, and a 1:100 dilution of overnight cultures was subcultured into fresh tryptic soy broth. S. aureus grown to early stationary phase (3 h) was collected and normalized by OD600 for further experimental analysis. Mice were anesthetized with Avertin (2,2,2-tribromoethanol) dissolved in tert-Amyl alcohol and diluted to a final concentration of 2.5% v/v in 0.9% sterile saline) by i.p. injection. For the bloodstream infection model, mice were challenged with 1 × 107 CFUs by retro-orbital injection, after being placed on tamoxifen diet 2 d prior. Survival was assessed by continuous monitoring and a 30% weight loss cutoff for humane euthanasia. To evaluate bacterial burden in organs, we sacrificed infected mice, collected the indicated organs in 1 ml PBS, and homogenized them to enumerate bacterial burden. Blood was collected by cardiac puncture and anticoagulated using heparin-coated tubes.
For sorting and scRNA-seq, mice were infected by retro-orbital injection with either 5 × 106 CFU WT or ΔTOX S. aureus and given an oral gavage of tamoxifen just before infection. Infected mice were analyzed at 7 d postinfection.
Liver immune cell isolation
Liver tissues were processed as described. Livers were chopped and incubated in RPMI + 10% FBS with collagenase VIII (100 U/ml; Sigma Aldrich) and DNase I (150 μg/ml; Sigma Aldrich) for 45 min at 37°C and then passed through a 70-μm cell strainer (Fisher Scientific). Leukocytes were enriched for by density-gradient centrifugation over a 40/80% Percoll (GE Healthcare) gradient, and remaining RBCs were lysed with ACK lysis buffer (Lonza) and washed in PBS and used for cell sorting or flow cytometry analysis.
Flow cytometry and cell sorting
Single-cell suspensions were stained with fluorescently conjugated Abs in a 1:100 dilution unless otherwise noted. For live/dead discrimination, cells were stained with either LIVE/DEAD Blue Reactive Dye (Invitrogen) (20 min, on ice, before primary) or Alexa Fluor NHS Ester (ThermoFisher) (with primary stain) and 4 μg/ml anti-CD16/32 (2.4G2; BioXCell) to block Fc receptors. The following anti-mouse Abs were used, with clone and source company listed: CD45 PerCP-Cyanine5.5 (30-F11; BioLegend), CD45 BV510 (30-F11; BioLegend), CX3CR1 BV605 (SA011F11; BioLegend), F4/80 BV711 (BM8; BioLegend), Ly6G PerCP-Cyanine5.5 (1A8; BioLegend), Ly6C PE/Cyanine7 (HK1.4; BioLegend), CD11b BUV395 (M1/70; BD Horizon), CD11c BV650 (N418; BioLegend), and Ki67 PE/Dazzle (16A8; BioLegend). Intracellular Ki67 was stained using the eBioscience Transcription Factor Staining Kit, following the included protocol. The BD FACSAria II was used for cell sorting, and the Bio-Rad ZE5 Yeti was used for flow cytometry analysis. Data analysis was performed using FlowJo v10 (FlowJo).
scRNA-seq
Single-cell suspensions were obtained from livers as described earlier. For each dataset, cells isolated from two to four mice were pooled together, with equal numbers of cells pooled from each animal used. A total of 12,000 cells from each group were loaded on a 10× Genomics Next GEM chip, and single-cell GEMs were generated on a 10× Chromium Controller. Subsequent steps to generate cDNA and sequencing libraries were performed following 10× Genomics’ protocol. Libraries were pooled and sequenced using Illumina NovaSeq SP 100 cycle as per 10× sequencing recommendations.
The sequenced data were processed using Cell Ranger (version 6.0) to demultiplex the libraries. The reads were aligned to Mus musculus mm10 and SCV2 (MN985325.1) genomes to generate count tables that were further analyzed using Seurat (version 4.1.2). Cell types were assigned manually based on expression of canonical markers and guided by SingleR (single-cell recognition) analysis (27). Data are displayed as Uniform Manifold Approximation and Projection (UMAP).
Cytokine analysis
Cytokine quantification was determined using the MILLIPLEX MAP Mouse Cytokine/Chemokine Magnetic Kit (MCYTMAG-70K-PX32; Millipore). The assay was conducted as per the manufacturer’s instructions, and plates were run on a MAGPIX instrument with xPONENT software. Statistical analyses were performed in Prism for each individual cytokine.
Results
scRNA-seq analysis of immune cells derived from CX3CR1+ precursors during S. mansoni infection
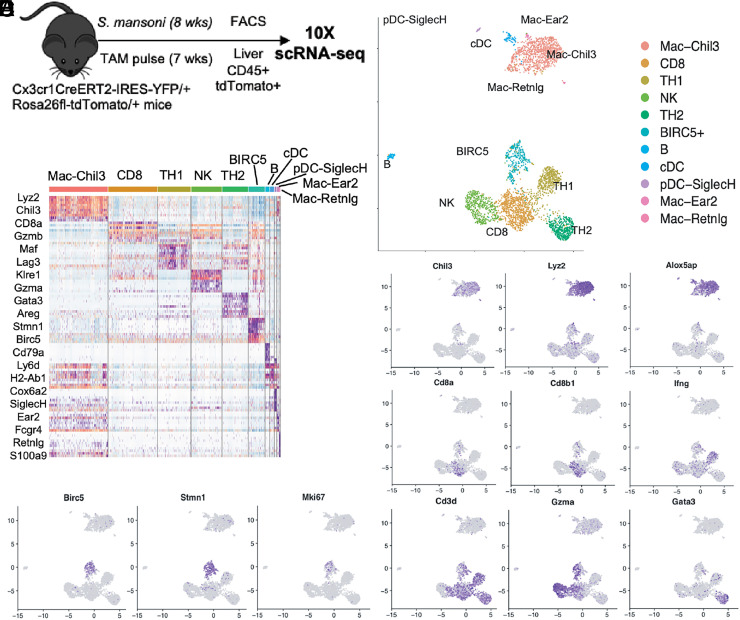
During infection with S. mansoni, extracellular parasite eggs lodged in the liver induce the recruitment of CX3CR1+ monocytes to form a granuloma as part of the host response in limiting tissue damage and hepatotoxicity (4, 6). To perform scRNA-seq on immune cells derived from CX3CR1+ precursors, we made use of a previously described fate-mapping mouse model in which Cx3cr1CreERT2-IRES-EYFP mice were crossed with Rosa26stop-tdTomato reporter mice (hereinafter referred to as Cx3cr1CreERT2-EYFP/+R26tdTomato/+ mice) (5). Tamoxifen treatment of these mice results in the irreversible labeling of all CX3CR1+ cells with tdTomato. We infected Cx3cr1CreERT2-EYFP/+R26tdTomato/+ mice with 75 cercariae of S. mansoni, and 7 d before analysis at 8 wk postinfection, mice were administered tamoxifen via oral gavage (Fig. 1A). At 8 wk postinfection, we FACS-purified tdTomato+CD45+ cells from the livers of infected mice, combined all samples, and submitted 12,000 cells for scRNA-seq. Sequencing was performed on the 10× Genomics platform, and cell types were assigned manually based on expression of canonical markers and guided by SingleR analysis (27). Cells were visualized by UMAP, a machine learning algorithm for dimensionality reduction (Fig. 1B). UMAP clustering revealed CX3CR1+-derived cells to be a heterogeneous population consisting of both a major myeloid and lymphoid cluster. The myeloid cluster consisted mostly of macrophages characterized by expression of Chil3, Lyz2, and Alox5, whereas the lymphoid cluster consisted of multiple cell types, including CD8+ T, NK, TH1, and TH2 CD4+ T cells (Fig. 1B–D). Interestingly, our analysis showed a separate cluster of cells defined by the gene Birc5 that consisted of predominantly lymphoid cells but also contains some Lyz2-expressing myeloid cells (Fig. 1B–D). This cluster was further defined by the expression of cell-cycle and proliferation genes Stmn1 and Ki67 (Fig. 1E), similar to the BIRC5+ population of monocytes we identified in a previous study (3). These data suggest that a population of BIRC5+ lymphocytes, which may be derived from a small subset of T cells that can also express CX3CR1 (28, 29), are present in the liver during chronic infection with S. mansoni. Hence, in addition to proliferating BIRC5+ myeloid cells, BIRC5+ lymphocytes could be a general feature of inflammation and not unique to atherosclerosis (3).
FIGURE 1.
scRNA-seq of fate-mapped CD45+ cells derived from CX3CR1+ precursors during S. mansoni infection reveals BIRC5+ immune cells. (A) Experimental design for scRNA-seq of CD45+ cells from Cx3cr1CreERT2-EYFP/+R26tdTomato/+ mice infected cutaneously with 75 S. mansoni and sorted 8 wk postinfection. Gating was on singlets, live cells, CD45+ cells, and tdTomato+ cells. (B) UMAP representation and unbiased clustering of major immune cell populations from the CD45+tdTomato+ cells sequenced, with each dot representing individual cells. (C) Expression of canonical cellular markers for the major clusters of immune cells. (D) Heatmap showing the markers of the main populations of immune cell clusters. (E) Identification of BIRC5+ proliferative cluster.
scRNA-seq analysis reveals leukotoxin-dependent BIRC5+ cells during S. aureus infection in the liver
We next investigated whether the population of BIRC5+ myeloid cells derived from CX3CR1+ precursors are also observed in an unrelated infection model characterized by a type 1 immune response in the liver. For this we infected mice with the Gram-positive bacteria S. aureus, which is known to infect the liver and would allow us to compare immune cell infiltrates in the same tissue affected by S. mansoni infection (30).
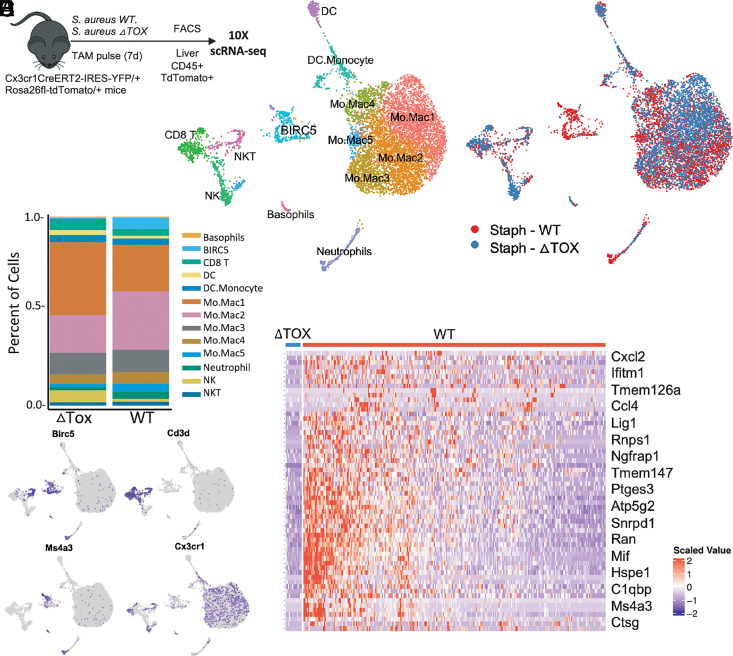
S. aureus has evolved to produce virulence factors including leukocidins that target leukocytes and are required for infection (8, 12, 31). We hypothesized that leukocidins may affect CX3CR1+ immune cells during S. aureus infection; thus, we infected Cx3cr1CreERT2-EYFP/+R26tdTomato/+ mice via retro-orbital injection with 5 × 106 CFUs of either WT S. aureus or a strain in which the genes hlgABC, lukSF-PVL, lukAB, lukED, and hla have been deleted, referred to as ΔTOX (Fig. 2A). All mice were given an oral gavage of tamoxifen at the time of infection to label cells that express CX3CR1 at that time. At 7 d postinfection, we FACS-purified tdTomato+CD45+ cells from the livers of both WT and ΔTOX-infected mice and submitted 12,000 cells each from both groups, separately, for scRNA-seq. Cell types were assigned manually based on expression of canonical markers and guided by SingleR analysis (27). All cells were visualized together in two-dimensional space using UMAP, with distinct sequencing libraries allowing us to distinguish between cells derived from either WT- or ΔTOX-infected mice (Fig. 2B, 2C). Similar to our results from S. mansoni–infected mice, we identified distinct myeloid and lymphoid clusters derived from CX3CR1+ precursors in the livers of S. aureus–infected mice. The myeloid cluster consisted mostly of monocyte and macrophage cell types; however, uniquely in this dataset, a neutrophil cluster was also identified. The lymphoid cluster consisted of three main cell types: CD8 T cells, NK cells, and NKT cells. Notably, CD4 T cells were absent from this cluster, suggesting the expression of CX3CR1 on CD4 T cells depends on the type of infection and can occur during helminth infection, but not S. aureus infection.
FIGURE 2.
scRNA-seq reveals BIRC5+ immune cells to be specifically elicited by S. aureus toxins during infection. (A) Experimental design for scRNA-seq of CD45+ from Cx3cr1CreERT2-EYFP/+R26tdTomato/+ mice infected intravascularly with 5 × 106 CFUs of S. aureus and sorted 1 wk postinfection. Gating was on singlets, live cells, CD45+ cells, and tdTomato+ cells. (B) UMAP clustering of CX3CR1-derived CD45+ cells with phenotypic classification. (C) UMAP clustering of isolated cells differentiating between cells from WT versus ΔTOX-infected mice. (D) Percentage of total cells defined as each immune subset from either WT or ΔTOX-infected mice. (E) Expression of Birc5 and other lineage-defining markers across both datasets. (F) Heatmap showing differential gene expression between identified BIRC5+ immune clusters in mice infected with either WT or ΔTOX S. aureus.
Consistent with our data from S. mansoni infection, we saw a distinct BIRC5+ cluster, separate from both the myeloid and the lymphoid clusters (Fig. 2B). However, distinguishing cells derived from either WT- or ΔTOX-infected mice revealed that it was only in mice infected with the WT strain of S. aureus that this BIRC5+ population was elicited (Fig. 2C, 2D). There were other minor differences, with slightly different proportions of the monocyte/macrophage clusters and a greater percentage of cells characterized as NK cells in the ΔTOX-infected mice (Fig. 2D). The BIRC5 cluster can be identified as myeloid cells based on expression of Ms4a3 (Fig. 2E). Some NKT cells that express Cd3d also express Birc5, but they fall clearly into the lymphoid clusters (Fig. 2E). Within the BIRC5+ cluster, differential analysis of gene expression between cells from WT and ΔTOX-infected mice showed that the few cells in this cluster from ΔTOX-infected mice express lower levels of inflammation-related genes, such as Ccl4, Mif, and Ctsg, as well as Ms4a3 (Fig. 2F). These data suggest that the elicitation of a population of BIRC5+ myeloid cells in the liver during S. aureus infection is dependent on the production of leukotoxins by the bacteria.
BIRC5+ immune cells share a common transcriptional program across a range of inflammatory states
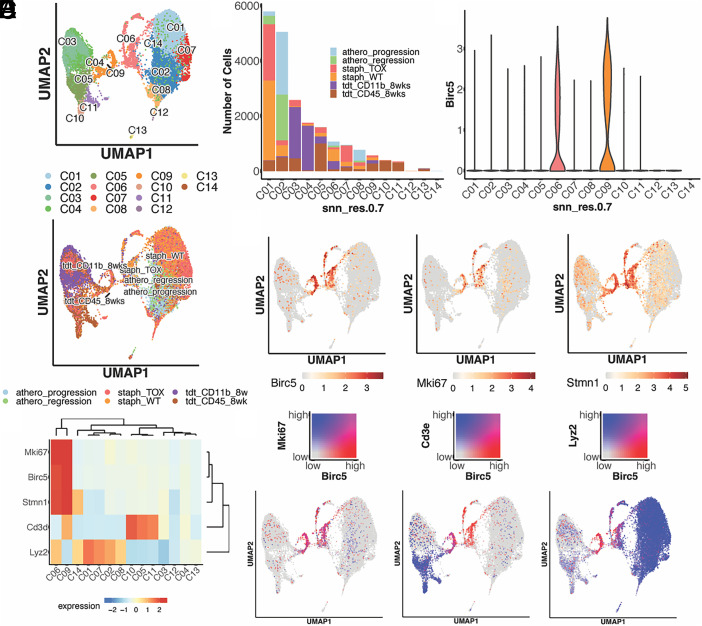
To determine whether BIRC5+ myeloid cells derived from CX3CR1+ precursors are present across a variety of inflammatory states, we integrated scRNA-seq datasets from several fate-mapping experiments that we have performed. The combined dataset consisted of data from CD45+tdTomato+ cells isolated from the liver of S. mansoni– and S. aureus–infected mice as described earlier (Figs. 1, 2), a subset of liver CD11b+ tdTomato+ cells isolated during S. mansoni infection, and a previously published dataset in which BIRC5+ monocytes were initially identified in the aorta from a model of atherosclerosis (3). After integration and dimension reduction, we visualized the cells according to their transcriptional profile and the dataset of origin (Fig. 3A). Many of the clusters corresponded to the original datasets, for example, cluster 1 consisted mostly of cells from mice infected with S. aureus, although other datasets are represented in this cluster as well (Fig. 3B). Cells derived from S. mansoni–infected mice appeared to be the most transcriptionally distinct based on the clustering. The expression of Birc5 was concentrated in clusters 6 and 9, each of which consisted of cells drawn from multiple datasets, suggesting that the BIRC5+ population has a common transcriptional profile across multiple inflammatory conditions (Fig. 3C–E). These same clusters had higher expression of Ki67 and Stmn1, indicating a shared capacity for proliferation.
FIGURE 3.
Combined UMAP clustering of scRNA-seq datasets reveals common transcriptional profile of BIRC5+ immune cells across multiple inflammatory states. (A) UMAP clustering of CX3CR1+-derived cells from multiple datasets based on transcriptional profile and experimental origin. (B) Number of cells present in each defined cluster, demonstrating common transcriptional profiles across multiple inflammatory settings. (C) Violin plot of Birc5 expression. (D) Gene expression levels of Birc5 coinciding with expression of the proliferation markers Mki67 and Stmn1. (E) Heatmap of relevant gene expression across all clusters. (F) Coexpression of Birc5 with Mki67, Cd3e, and Lyz2, showing Birc5 expression in both a lymphoid and a myeloid population. Figures were generated using SHINY app (https://ruggleslab.shinyapps.io).
Using the Shiny application, a computational tool for analysis of integrated scRNA-seq datasets, we generated gene-pair heat plots to visualize overlaps in expression. Comparing Birc5 with Ki67 expression, we see near-complete overlap, indicating the BIRC5+ population as the major proliferating population across all datasets (Fig. 3F). Similar to what was observed in the individual analyses, we found two clusters of BIRC5+ cells, one lymphoid and the other myeloid, as indicated by the overlap between the genes CD3e and Lyz2, respectively, and Birc5 expression (Fig. 3F). Cluster 6 corresponded to the BIRC5+ lymphoid cluster, whereas cluster 9 corresponded to the BIRC5+ myeloid cluster. The BIRC5+ myeloid cluster is also marked by expression of Ms4a3 and other myeloid markers. Overall, combined analysis revealed BIRC5+ immune cells share a transcriptional profile across multiple inflammatory states, with a clear distinction between BIRC5+ myeloid cells and BIRC5+ lymphocytes.
Conditional deletion of BIRC5 in CX3CR1+ cells improves survival during S. aureus infection
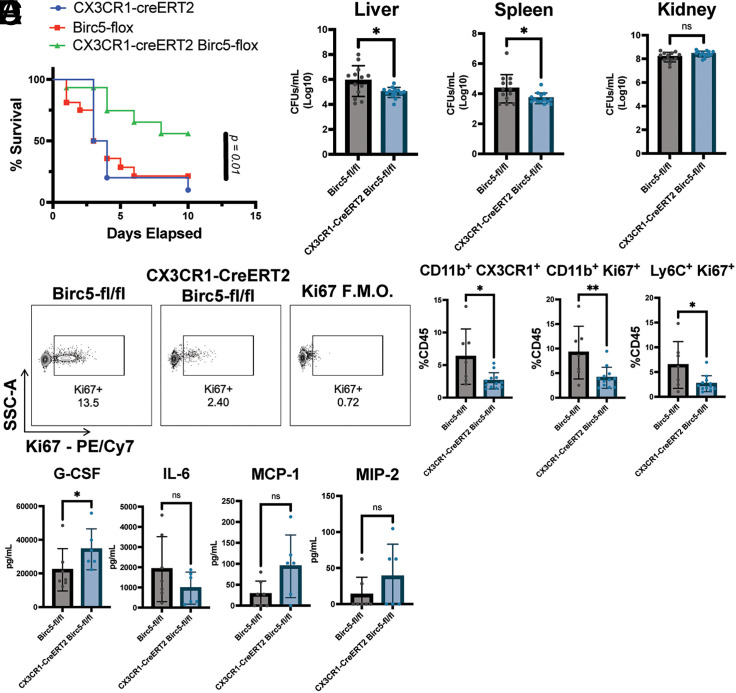
To further assess the role of BIRC5+ immune cells in vivo, we crossed mice in which the BIRC5 gene was flanked by two loxP sequences to the CX3CR1CreERT2-IRES-eYFP mice, so that when tamoxifen is administered, the Birc5 gene is conditionally deleted. Progeny were then crossed to generate mice homozygous for both alleles. We infected double-homozygous mice, along with mice homozygous for either the CX3CR1CreERT2-IRES-eYFP or the BIRC5-Flox alone, with 1 × 107 CFUs of S. aureus. To induce deletion, we fed mice chow containing tamoxifen, starting 2 d before infection, up until the end of monitoring at day 10 postinfection. Mice were then monitored for weight loss and survival. We observed significantly less mortality in the double-homozygous group as compared with other groups, suggesting that deletion of Birc5 in CX3CR1+ cells is protective against lethality (Fig. 4A). Approximately 50% of mice in the double-homozygous group survived to day 10, whereas only 5–15% of mice in the control group survived until day 10. Furthermore, mice in the control group started to die sooner during the course of infection, mostly between days 2 and 4. Observing this difference in survival, we assessed bacterial burden in both CX3CR1+/CreERT2-IRES-eYFP BIRC5-Flox and BIRC5-Flox–alone mice postinfection with 1 × 107 CFUs of S. aureus. All mice were placed on tamoxifen diet 2 d before infection, and at day 2 postinfection, liver, spleen, heart, kidney, and lung were harvested to quantify bacterial burdens. There was a lower bacterial burden in the livers and spleens of mice in which Birc5 was conditionally deleted in CX3CR1-expressing cells, with no significant difference in the kidney, heart, and lung (Fig. 4B, data not shown). The differences are likely to be more significant than shown because three mice in the BIRC5-Flox–alone group died before bacterial burden could be measured. These results show that the deletion of Birc5 in CX3CR1+ cells protects mice from acute lethality from S. aureus infection, with reduced bacterial burden in the liver and spleen. Because this difference occurs within the first 3 d of infection, the results implicate CX3CR1+ myeloid cells as playing a pathogenic role by promoting an increased bacterial burden during acute S. aureus infection.
FIGURE 4.
Conditional deletion of Birc5 in CX3CR1-expressing cells is protective during infection with S. aureus. (A) Survival curve of mice infected with 1 × 107 CFUs of S. aureus (showing combination of two different experiments, n = 13–14 mice per genotype). (B) Bacterial burden across tissues at day 2 postinfection, with values log transformed for standardization (showing combination of two different experiments, n = 12–14 mice per genotype; note that three mice in the Birc5-Flox group died before bacterial burden could be assessed). (C) Representative flow cytometry plots of Ki67 expression in CD11b+ leukocytes in the liver at day 2 postinfection, gated on live cells, CD45+ cells, and CD11b+ cells. (D) Percentage of monocyte populations as part of total CD45+ at day 2 postinfection in the liver (showing combination of two different experiments, n = 7–12 mice per genotype). (E) Concentration of cytokines/chemokines per milliliter of blood was quantified by the magnetic bead assay for blood infected from either Birc5-Flox or CX3CR1-creERT2 Birc5-Flox after sera were harvested at day 2 postinfection (n = 6–7 per genotype). Error bars represent SEM. Student t tests were performed to determine significance. *p < 0.05, **p < 0.01.
To assess the immune response in these mice, we performed flow cytometry on cells isolated from the liver. After gating on CD45+ CD11b+ cells, we observed a lower incidence of Ki67 expression in mice in which BIRC5 was conditionally deleted, suggesting that BIRC5 is needed for proliferation of the myeloid cells in the liver (Fig. 4C). Quantifying cells as a percentage of total CD45 cells showed there to be a significant reduction in proliferating monocytes upon deletion of BIRC5 (Fig. 4D). We next determined whether deletion of BIRC5 from CX3CR1+ cells resulted in altered cytokine and chemokine profiles at a systemic level. We noticed very little difference between the two genotypes, with the most notable difference being a modest increase in the levels of G-CSF in mice in which BIRC5 was conditionally deleted (Fig. 4E). Although not statistically significant, there was a trend toward higher levels of MCP-1 (CCL2) and MIP-2 in the conditional knockout mice (Fig. 4E). Inflammatory cytokines showed little difference, although there was a trend toward lower levels of IL-6 in the conditional knockout (Fig. 4E). Overall, these data show that the deletion of BIRC5 in CX3CR1+ cells results in reduced proliferation within the monocyte population, as well as lower numbers of total monocytes in the liver, but with minor effects on other immune cell populations and inflammatory parameters.
Discussion
In this study, we determined that a population of BIRC5+ cells derived from CX3CR1+ precursors is a general feature of both acute and chronic infections in the liver, during both bacterial and helminth infections. Although there are both BIRC5+ lymphocytes and BIRC5+ myeloid cells, the deletion of BIRC5 from CX3CR1-expressing cells results in improved survival during acute S. aureus infection. The 3-d time scale implicates BIRC5 in the myeloid cell compartment as playing a pathogenic role during S. aureus infection, by promoting bacterial burden in the livers of infected mice. In the absence of toxins, S. aureus infection does not elicit these BIRC5+ cells, implicating a toxin-dependent pathogenic role for BIRC5 in myeloid cells during S. aureus infection.
As part of this study, we have further defined the subsets of immune cells that express the chemokine receptor CX3CR1 using a combination of fate-mapping mouse models and scRNA-seq. We found this receptor to be expressed on multiple immune cell types, including members of both the myeloid and the lymphoid lineages, demonstrating a greater level of diversity in expression than previously realized. For our infection studies, we focused on a single tissue—the liver—but compared disparate infections to determine how immune cells respond to diseases with categorically different pathogen biology and etiologies. During infection with the trematode parasite S. mansoni, the liver is the main site of pathology, and the immune response is characterized by the infiltration of monocytes to form a granuloma structure around the parasite eggs (4–6). During infection with the Gram-positive bacteria S. aureus, the liver is also one of the primary tissues affected, because the bacteria is sequestered in the liver within hours of an intravascular infection (30, 32). By conducting our analysis on both a “type 2” and a “type 1” infection, respectively, and combining this with our analysis of a sterile inflammatory setting—atherosclerosis—we were able to identify transcriptional signatures, both common and distinct, of CX3CR1-expressing immune cells responding to inflammation. Most notable was our identification of a broadly occurring BIRC5+ population comprising both lymphoid and myeloid cells exhibiting stem cell–like properties. Numerous studies have demonstrated a role for BIRC5+ T cells in infection and autoimmunity; however, our study is unique in demonstrating the occurrence of a BIRC5+ myeloid population during multiple inflammatory settings, as well as showing this population to be pathogenic during S. aureus infection.
Due to its role in cell-cycle progression, BIRC5 has been hypothesized to be an important biomarker of many cancers. Indeed, multiple studies have shown a positive correlation between expression levels of BIRC5 in different tumors and a poorer prognostic outcome (15, 16, 33–35). Furthermore, although not true for all cancers, many tumors displayed a positive correlation between BIRC5 expression and immune cell infiltration, suggesting a potential immunogenic role for BIRC5 (15, 35). These data suggest BIRC5 could be a therapeutic target in certain tumor settings, although the efficacy of such a therapy remains to be tested. However, our own data suggest that many immune cells themselves would be targeted by such a therapy, and so it is crucial to take this into consideration when assessing the potential for targeting BIRC5.
One limitation of this study is that it is unclear why conditional deletion of BIRC5 in CX3CR1+ cells is protective during S. aureus infection. It is known that tissue-resident macrophages in the liver (Kupffer cells) will rapidly sequester the bacteria upon intravascular infection (30, 32). During the first 24 h of infection, many of these cells will manage to kill the bacterium; however, a significant portion will instead be overwhelmed by bacterial replication, subsequently lysed, and the bacteria allowed to disseminate. It is only after this initial phase of infection in the liver that the bacteria become systemic, spreading to other tissues and the blood. Recent work has shown that after the liver phase of the infection, the bacteria spread to the peritoneal cavity, where they are taken up by a population of GATA6+ macrophages before spreading widely (36). However, the same study showed multiple cell types take up the bacteria, including monocytes, both in the liver and the peritoneal cavity. In the context of our own study, it is possible that by deleting BIRC5 in CX3CR1+ cells we are reducing the cellular hosts for the bacteria, which prevents uncontrolled growth and in turn results in better survival. It has been shown that monocytes are able to engraft in the liver to replace depleted Kupffer cells, so deletion of BIRC5 in monocytes could prevent the replenishment of these macrophages and starve the bacteria of cellular hosts after the initial wave of lysis and dissemination (37). Furthermore, the reduction in monocytes upon conditional deletion could be what is causing increased systemic levels of G-CSF and CCL2 because there is an open niche for recruitment of new myeloid cells. Given the wide range of tissues affected by infection and the many different myeloid cells present in said tissues, it is possible that conditional deletion has a stronger effect in some tissues over others. We observed lowered bacterial burden in the liver and spleen, but not in the kidney, lung, or heart at 2 d postinfection, suggesting that we are not preventing spread to other tissues but arresting the growth of bacteria in the liver and spleen specifically. Remarkably, this was enough to improve survival.
Another possibility that we did not fully explore in this study is whether CX3CR1+BIRC5+ immune cells are regulatory in nature. It is known that S. aureus infection can actually suppress the host immune system in ways that are still being determined but include toxin-mediated killing of leukocytes (9, 10, 12). One interesting observation from our study was that infection with the ΔTOX strain of bacteria did not elicit the BIRC5+ population in the liver. Because the absence of this population in our genetic models improved survival, it could be mimicking an infection seen with a ΔTOX strain of the bacteria where lethality is reduced (11, 38). This phenotype could be mediated by toxin killing of immune cells or simply bacterial toxins inducing a dampened immune response, unable to strongly respond to infection (12). With the current lack of vaccines to protect against S. aureus infections, as well as the development of antimicrobial-resistant strains, it is imperative to pursue alternate treatment options. One such approach that has gained traction against infectious disease has been host-directed therapies, whereby the host cellular processes needed by the pathogen life cycle are disrupted (39). Other studies have shown this approach to be effective by, for example, disrupting the apoptosis pathway (40–42). Our study has revealed, to our knowledge, a new possible target for host-directed therapy in BIRC5, because conditional deletion of this gene in immune cells significantly improved survival. Further understanding of the role of BIRC5+ immune cells during infection with S. aureus is crucial for determining the possibility of targeting this protein therapeutically.
Acknowledgments
We acknowledge help from NYU Langone’s Genome Technology Center for performing all the RNA sequencing. Cell sorting/flow cytometry technologies were provided by NYU Langone’s Cytometry and Cell Sorting Laboratory. We thank Stephen T. Yeung for assistance with manuscript preparation.
This article is featured in Top Reads, p. 707
Footnotes
This work was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH) (Grants AI099394, AI105129, and AI133977 to V.J.T.; Grant AI143861 to K.M.K.; and Grant AI143861-S1 to K.M.K.). E.E.Z. was supported by National Institute of Allergy and Infectious Diseases (NIAID)-Supported Institutional Research Training Grant on Infectious Disease & Basic Microbiological Mechanisms T32 AI007180. J.C.D. was supported by NIAID-Supported Institutional Research Training Grant in Immunology and Inflammation Grant T32 AI100853. K.A.L. was supported by Cystic Fibrosis Postdoctoral Research Fellowship Award LACEY19FO. J.-D.L. was supported by Young Scholar Fellowship-Einstein Grant and Yushan Scholar Program from the National Science and Technology Council and the Ministry of Education, Taiwan. This work was also supported by the Intramural Research Program of the NIAID, NIH (P.L.). Research in the laboratory of V.J.T. was supported by a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Diseases award. The NYU Langone Health Genome Technology Center and the Cytometry and Cell Sorting Laboratory are shared resources partially supported by Laura and Isaac Perlmutter Cancer Center Support Grant P30CA016087 from the NIH, National Cancer Institute.
The sequences presented in this article have been submitted to the National Center for Biotechnology Information Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE232996) under accession number GSE232996.
- NIAID
- National Institute of Allergy and Infectious Diseases
- NIH
- National Institutes of Health
- scRNA-seq
- single-cell RNA sequencing
- SingleR
- single-cell recognition
- ΔTOX
- Staphylococcus aureus strain USA300 LAC clone AH-LAC ΔlukABhlg::tet
- lukED::kan
- pvl::epchla::erm
- UMAP
- Uniform Manifold Approximation and Projection
- WT
- wild type
Disclosures
The authors have no financial conflicts of interest.
References
- 1. Loke, P., Lin J. D.. 2022. Redefining inflammatory macrophage phenotypes across stages and tissues by single-cell transcriptomics. Sci. Immunol. 7: eabo4652. [DOI] [PubMed] [Google Scholar]
- 2. van de Laar, L., Saelens W., De Prijck S., Martens L., Scott C. L., Van Isterdael G., Hoffmann E., Beyaert R., Saeys Y., Lambrecht B. N., Guilliams M.. 2016. Yolk sac macrophages, fetal liver, and adult monocytes can colonize an empty niche and develop into functional tissue-resident macrophages. Immunity 44: 755–768. [DOI] [PubMed] [Google Scholar]
- 3. Lin, J. D., Nishi H., Poles J., Niu X., Mccauley C., Rahman K., Brown E. J., Yeung S. T., Vozhilla N., Weinstock A., et al. 2019. Single-cell analysis of fate-mapped macrophages reveals heterogeneity, including stem-like properties, during atherosclerosis progression and regression. JCI Insight 4: e124574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Girgis, N. M., Gundra U. M., Ward L. N., Cabrera M., Frevert U., Loke P.. 2014. Ly6C(high) monocytes become alternatively activated macrophages in schistosome granulomas with help from CD4+ cells. PLoS Pathog. 10: e1004080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gundra, U. M., Girgis N. M., Gonzalez M. A., San Tang M., Van Der Zande H. J. P., Lin J. D., Ouimet M., Ma L. J., Poles J., Vozhilla N., et al. 2017. Vitamin A mediates conversion of monocyte-derived macrophages into tissue-resident macrophages during alternative activation. Nat. Immunol. 18: 642–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Herbert, D. R., Hölscher C., Mohrs M., Arendse B., Schwegmann A., Radwanska M., Leeto M., Kirsch R., Hall P., Mossmann H., et al. 2004. Alternative macrophage activation is essential for survival during schistosomiasis and downmodulates T helper 1 responses and immunopathology. [Published erratum appears in 2004 Immunity 21: 455.] Immunity 20: 623–635. [DOI] [PubMed] [Google Scholar]
- 7. Thammavongsa, V., Kim H. K., Missiakas D., Schneewind O.. 2015. Staphylococcal manipulation of host immune responses. Nat. Rev. Microbiol. 13: 529–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Spaan, A. N., van Strijp J. A. G., Torres V. J.. 2017. Leukocidins: staphylococcal bi-component pore-forming toxins find their receptors. Nat. Rev. Microbiol. 15: 435–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. DuMont, A. L., Yoong P., Day C. J., Alonzo F. III, McDonald W. H., Jennings M. P., Torres V. J.. 2013. Staphylococcus aureus LukAB cytotoxin kills human neutrophils by targeting the CD11b subunit of the integrin Mac-1. Proc. Natl. Acad. Sci. USA 110: 10794–10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Berends, E. T. M., Zheng X., Zwack E. E., Ménager M. M., Cammer M., Shopsin B., Torres V. J.. 2019. Staphylococcus aureus impairs the function of and kills human dendritic cells via the LukAB toxin. MBio 10: e01918-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lubkin, A., Lee W. L., Alonzo F. III, Wang C., Aligo J., Keller M., Girgis N. M., Reyes-Robles T., Chan R., O’Malley A., et al. 2019. Staphylococcus aureus leukocidins target endothelial DARC to cause lethality in mice. Cell Host Microbe 25: 463–470.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zwack, E. E., Chen Z., Devlin J. C., Li Z., Zheng X., Weinstock A., Lacey K. A., Fisher E. A., Fenyö D., Ruggles K. V., et al. 2022. Staphylococcus aureus induces a muted host response in human blood that blunts the recruitment of neutrophils. Proc. Natl. Acad. Sci. USA 119: e2123017119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tamm, I., Wang Y., Sausville E., Scudiero D. A., Vigna N., Oltersdorf T., Reed J. C.. 1998. IAP-family protein survivin inhibits caspase activity and apoptosis induced by Fas (CD95), Bax, caspases, and anticancer drugs. Cancer Res. 58: 5315–5320. [PubMed] [Google Scholar]
- 14. Altieri, D. C. 2015. Survivin—the inconvenient IAP. Semin. Cell Dev. Biol. 39: 91–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xu, L., Yu W., Xiao H., Lin K.. 2021. BIRC5 is a prognostic biomarker associated with tumor immune cell infiltration. Sci. Rep. 11: 390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fäldt Beding, A., Larsson P., Helou K., Einbeigi Z., Parris T. Z.. 2022. Pan-cancer analysis identifies BIRC5 as a prognostic biomarker. BMC Cancer 22: 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sasaki, R., Ito S., Asahi M., Ishida Y.. 2015. YM155 suppresses cell proliferation and induces cell death in human adult T-cell leukemia/lymphoma cells. Leuk. Res. 39: 1473–1479. [DOI] [PubMed] [Google Scholar]
- 18. Cheng, X. J., Lin J. C., Ding Y. F., Zhu L., Ye J., Tu S. P.. 2016. Survivin inhibitor YM155 suppresses gastric cancer xenograft growth in mice without affecting normal tissues. Oncotarget 7: 7096–7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Véquaud, E., Séveno C., Loussouarn D., Engelhart L., Campone M., Juin P., Barillé-Nion S.. 2015. YM155 potently triggers cell death in breast cancer cells through an autophagy-NF-kB network. Oncotarget 6: 13476–13486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Andersson, K. M., Svensson M. N., Erlandsson M. C., Jonsson I. M., Bokarewa M. I.. 2015. Down-regulation of survivin alleviates experimental arthritis. J. Leukoc. Biol. 97: 135–145. [DOI] [PubMed] [Google Scholar]
- 21. Sharief, M. K., Semra Y. K.. 2001. Heightened expression of survivin in activated T lymphocytes from patients with multiple sclerosis. J. Neuroimmunol. 119: 358–364. [DOI] [PubMed] [Google Scholar]
- 22. Sharief, M. K., Noori M. A., Douglas M. R., Semra Y. K.. 2002. Upregulated survivin expression in activated T lymphocytes correlates with disease activity in multiple sclerosis. Eur. J. Neurol. 9: 503–510. [DOI] [PubMed] [Google Scholar]
- 23. Kuo, H. H., Ahmad R., Lee G. Q., Gao C., Chen H. R., Ouyang Z., Szucs M. J., Kim D., Tsibris A., Chun T. W., et al. 2018. Anti-apoptotic protein BIRC5 maintains survival of HIV-1-infected CD4+ T cells. Immunity 48: 1183–1194.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Okada, H., Bakal C., Shahinian A., Elia A., Wakeham A., Suh W. K., Duncan G. S., Ciofani M., Rottapel R., Zúñiga-Pflücker J. C., Mak T. W.. 2004. Survivin loss in thymocytes triggers p53-mediated growth arrest and p53-independent cell death. J. Exp. Med. 199: 399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Boles, B. R., Thoendel M., Roth A. J., Horswill A. R.. 2010. Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS One 5: e10146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Blake, K. J., Baral P., Voisin T., Lubkin A., Pinho-Ribeiro F. A., Adams K. L., Roberson D. P., Ma Y. C., Otto M., Woolf C. J., et al. 2018. Staphylococcus aureus produces pain through pore-forming toxins and neuronal TRPV1 that is silenced by QX-314. Nat. Commun. 9: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Aran, D., Looney A. P., Liu L., Wu E., Fong V., Hsu A., Chak S., Naikawadi R. P., Wolters P. J., Abate A. R., et al. 2019. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 20: 163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Batista, N. V., Chang Y. H., Chu K. L., Wang K. C., Girard M., Watts T. H.. 2020. T cell-intrinsic CX3CR1 marks the most differentiated effector CD4+ T cells, but is largely dispensable for CD4+ T cell responses during chronic viral infection. Immunohorizons 4: 701–712. [DOI] [PubMed] [Google Scholar]
- 29. Sun, H., He T., Wu Y., Yuan H., Ning J., Zhang Z., Deng X., Li B., Wu C.. 2022. Cytotoxin-associated gene A-negative Helicobacter pylori promotes gastric mucosal CX3CR1+CD4+ effector memory T cell recruitment in mice. Front. Microbiol. 13: 813774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Surewaard, B. G., Deniset J. F., Zemp F. J., Amrein M., Otto M., Conly J., Omri A., Yates R. M., Kubes P.. 2016. Identification and treatment of the Staphylococcus aureus reservoir in vivo. [Published erratum appears in 2016 J. Exp. Med. 213: 3087.] J. Exp. Med. 213: 1141–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reyes-Robles, T., Lubkin A., Alonzo F. III, Lacy D. B., Torres V. J.. 2016. Exploiting dominant-negative toxins to combat Staphylococcus aureus pathogenesis. EMBO Rep. 17: 428–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zeng, Z., Surewaard B. G., Wong C. H., Geoghegan J. A., Jenne C. N., Kubes P.. 2016. CRIg functions as a macrophage pattern recognition receptor to directly bind and capture blood-borne gram-positive bacteria. Cell Host Microbe 20: 99–106. [DOI] [PubMed] [Google Scholar]
- 33. Zhao, Y., Liu S., Li S., Zhang G., Tian A., Wan Y.. 2022. BIRC5 regulates inflammatory tumor microenvironment-induced aggravation of penile cancer development in vitro and in vivo. BMC Cancer 22: 448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ye, H. B., Ma B. J., Meng G. Q., Tao S., Wang Y., Chen Z., Zhao W., Ren B. Y., Ye Z.. 2022. Bioinformatics analysis of BIRC5 in human cancers. Ann. Transl. Med. 10: 888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ma, T., Gu J., Wen H., Xu F., Ge D.. 2022. BIRC5 modulates PD-L1 expression and immune infiltration in lung adenocarcinoma. J. Cancer 13: 3140–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jorch, S. K., Surewaard B. G., Hossain M., Peiseler M., Deppermann C., Deng J., Bogoslowski A., van der Wal F., Omri A., Hickey M. J., Kubes P.. 2019. Peritoneal GATA6+ macrophages function as a portal for Staphylococcus aureus dissemination. J. Clin. Invest. 129: 4643–4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Scott, C. L., Zheng F., De Baetselier P., Martens L., Saeys Y., De Prijck S., Lippens S., Abels C., Schoonooghe S., Raes G., et al. 2016. Bone marrow-derived monocytes give rise to self-renewing and fully differentiated Kupffer cells. Nat. Commun. 7: 10321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tam, K., Lacey K. A., Devlin J. C., Coffre M., Sommerfield A., Chan R., O’Malley A., Koralov S. B., Loke P., Torres V. J.. 2020. Targeting leukocidin-mediated immune evasion protects mice from Staphylococcus aureus bacteremia. J. Exp. Med. 217: e20190541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wallis, R. S., O’Garra A., Sher A., Wack A.. 2023. Host-directed immunotherapy of viral and bacterial infections: past, present and future. Nat. Rev. Immunol. 23: 121–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Parmanik, A., Das S., Kar B., Bose A., Dwivedi G. R., Pandey M. M.. 2022. Current treatment strategies against multidrug-resistant bacteria: a review. Curr. Microbiol. 79: 388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Alphonse, M. P., Rubens J. H., Ortines R. V., Orlando N. A., Patel A. M., Dikeman D., Wang Y., Vuong I., Joyce D. P., Zhang J., et al. 2021. Pan-caspase inhibition as a potential host-directed immunotherapy against MRSA and other bacterial skin infections. Sci. Transl. Med. 13: eabe9887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xu, H., Yu J., Cui J., Chen Z., Zhang X., Zou Y., Du Y., Li Y., Le S., Jiang L., et al. 2021. Ablation of survivin in T cells attenuates acute allograft rejection after murine heterotopic heart transplantation by inducing apoptosis. Front. Immunol. 12: 710904. [DOI] [PMC free article] [PubMed] [Google Scholar]