Abstract
DNA-dependent protein kinase (DNA-PK) plays a critical role in non-homologous end joining (NHEJ), the predominant pathway that repairs DNA double-strand breaks (DSB) in response to ionizing radiation (IR) to govern genome integrity. The interaction of the catalytic subunit of DNA-PK (DNA-PKcs) with the Ku70/Ku80 heterodimer on DSBs leads to DNA-PK activation; however, it is not known if upstream signaling events govern this activation. Here, we reveal a regulatory step governing DNA-PK activation by SIRT2 deacetylation, which facilitates DNA-PKcs localization to DSBs and interaction with Ku, thereby promoting DSB repair by NHEJ. SIRT2 deacetylase activity governs cellular resistance to DSB-inducing agents and promotes NHEJ. SIRT2 furthermore interacts with and deacetylates DNA-PKcs in response to IR. SIRT2 deacetylase activity facilitates DNA-PKcs interaction with Ku and localization to DSBs and promotes DNA-PK activation and phosphorylation of downstream NHEJ substrates. Moreover, targeting SIRT2 with AGK2, a SIRT2-specific inhibitor, augments the efficacy of IR in cancer cells and tumors. Our findings define a regulatory step for DNA-PK activation by SIRT2-mediated deacetylation, elucidating a critical upstream signaling event initiating the repair of DSBs by NHEJ. Furthermore, our data suggest that SIRT2 inhibition may be a promising rationale-driven therapeutic strategy for increasing the effectiveness of radiation therapy.
Graphical Abstract
Graphical Abstract.

INTRODUCTION
DNA double-strand breaks (DSB) are highly cytotoxic lesions, which must be repaired in order to preserve genome integrity. DSBs are induced by multiple sources, including ionizing radiation (IR), chemical exposures, reactive oxygen species (ROS), collapse of stalled replication forks, and immunoglobulin variable (diversity) joining [V (D)J] and class switch recombination (CSR). The predominant pathway for the repair of DSBs is non-homologous end joining (NHEJ), which involves direct ligation of broken DNA ends and is active in all phases of the cell cycle (1,2). NHEJ dysfunction can lead to chromosomal translocations and telomere fusions that contribute to cancer as well as immune deficiency and multiple human syndromes (2,3).
DNA-dependent protein kinase (DNA-PK), which consists of the DNA-PK catalytic subunit (DNA-PKcs) and the Ku70/Ku80 heterodimer, functions at the apex of the NHEJ repair pathway (4,5). In response to DSBs, the Ku70/Ku80 heterodimer recognizes and assembles onto DSB ends and recruits DNA-PKcs to form the DNA-PK holoenzyme (6,7), mediated by the carboxyl-terminus of Ku80 and the amino-terminus of DNA-PKcs (8–11). Interactions between the DNA-PK holoenzymes promote synapsis of the broken DNA ends (11). DNA-PK then recruits and activates the Artemis endonuclease for DNA end processing (12). Finally, DNA ligase IV in complex with XRCC4 and XLF further bind and catalyze ligation of the broken DNA ends (13,14). NHEJ is also facilitated by paralog of XRCC4 and XLF (PAXX), which stabilizes the core NHEJ components on chromatin (15–17).
The interaction of DNA-PKcs with the Ku70/80 heterodimer on DNA leads to its activation, likely through a conformational change (11,18). Once activated, DNA-PKcs phosphorylates itself (19–21) as well as a number of downstream substrates, including Ku70/Ku80 (22), XRCC4 (23), XLF (24), DNA ligase IV (25), Artemis (26), PNKP (27) and H2AX (28,29). DNA-PKcs is regulated by phosphorylation on a number of sites, including a serine 2056 (S2056) cluster [2023–2056] (30,31), a threonine 2609 (T2609) cluster [2609–2647] (19,32), and several other sites (4,5). S2056 is a specific DNA-PKcs autophosphorylation site (30,31), which limits DNA end processing (31) and promotes DNA end ligation by relieving a physical block by DNA-PKcs itself (33). The T2609 cluster is also a DNA-PKcs autophosphorylation site but is primarily phosphorylated by the related phosphatidylinositol 3-kinase (PI3K)-kinase-related protein kinases (PIKK) ATM and ATR (19,32,34,35). Phosphorylation at this site promotes DNA-PKcs dissociation from Ku70/Ku80 and release from DSB ends (20,36,37), DNA end processing (31,38) through recruitment (33) and activation of Artemis endonuclease activity (39), and DNA ligation (19,38,40). DNA-PKcs is also regulated by epidermal growth factor receptor (EGFR) (41), Akt (42), heterochromatin protein 1β (HP1β) (43), and CK2 (44). The interaction of DNA-PKcs with the Ku70/80 heterodimer is critical for DNA-PK activation; however, it is unknown if upstream signaling events govern this activation.
Sirtuin 2 (SIRT2) is a sirtuin family NAD+ dependent deacetylase, which regulates multiple biological processes, including genome maintenance, aging, tumorigenesis, and metabolism (45–47). Significantly, mice deficient in Sirt2 develop breast, liver, and other cancers (48,49), suggesting that SIRT2 functions in tumor suppression. Moreover, somatic cancer-associated SIRT2 mutations impair the activity of SIRT2 in maintaining genome integrity (50), suggesting that SIRT2’s role in tumor suppression may be attributed, at least in part, to its role in genome maintenance. In this regard, SIRT2 directs the replication stress response (RSR), a subset of the DNA damage response (DDR), through the acetylation status of ATRIP and CDK9 (47,51,52). SIRT2 also promotes homologous recombination by facilitating BRCA1-BARD1 heterodimerization through BARD1 deacetylation (53) and RPA70 and RAD51 recruitment to DSBs (54) and promotes nucleotide excision repair (NER) (55). However, a role for SIRT2 in directing NHEJ has not been established. Furthermore, DNA-PKcs acetylation at lysine (K) 3241 and K3260 promotes genome stability and radioresistance but not DNA-PK activation (56), and SIRT6 stabilizes DNA-PKcs on chromatin and promotes its activity in NHEJ, likely through a scaffold function (57,58); however, the role of SIRT2 deacetylation, and more generally of sirtuin deacetylation, in directing DNA-PK activation is not known. Moreover, SIRT2 inhibitors have been reported to have anti-proliferative activity against a number of cancer types (59); however, the specific cellular context, including synthetic lethal relationships with specific types of cancer treatments for which SIRT2 inhibitors may be most effective for cancer therapy are not clear.
In this study, we reveal a regulatory step governing DNA-PK activation by SIRT2 deacetylation, which facilitates DNA-PKcs localization to DSBs and interaction with Ku, thereby promoting DSB repair by NHEJ. Our findings define a mechanism for DNA-PK activation by SIRT2-mediated deacetylation, elucidating a critical upstream signaling event initiating the repair of DSBs by NHEJ. Furthermore, SIRT2 inhibition augments the efficacy of IR in cancer cells and tumors and may be a promising rationale-driven therapeutic strategy for increasing the effectiveness of radiation therapy.
MATERIALS AND METHODS
Transfections
siRNAs were obtained from Thermo Scientific or Qiagen. Transfections were performed using Lipofectamine 2000 (Invitrogen) or RNAi Max (Invitrogen) following the manufacturer's instructions. Individual siRNAs sequences include:
1. siSIRT2-10 UTR:(TGGGCAGAAGACATTGCTTAT);
2. siSIRT2-5:(GGAGAAAGCTGGCCAGTCG);
4. Nontargeting siRNA:(ATGAACGTGAATTGCTCAATT)
Generation of CRISPR/cas9 SIRT2 knockout cells
U2OS and HCT116 cells were transfected with a plasmid encoding Cas9-GFP construct and a single guide RNA (sgRNA), targeting an SIRT2 exon 6 (5′- CGGGCTCAAGTTCCGCTTCGGG-3′) (Sigma) (50). Cells were maintained for 72 h post transfection, and then harvested for fluorescence activated cell sorting based on GFP expression (50). Single cells were assayed in 96-well plates. Resulting cloned cell lines were tested for loss of SIRT2 protein by western blot analysis (50).
Clonogenic assays
For IR and CPT sensitivity assays, wildtype or SIRT2 knockout HCT116 cells were transfected with SIRT2-FLAG or SIRT2-FLAG-H187Y expression vectors, or were non-transfected. After 48 h to allow for protein expression, cells were seeded into 6-well plates (400 cells per well). Some groups were treated with DNA-PKcs inhibitor NU7441 as indicated in the figures. Three biological replicates were analyzed for each experimental group. After 24 h, cells were exposed to 0, 0.5, 1, 2 or 4 Gy of 320 kVp X-rays for IR sensitivity or grown in varying doses of CPT for 72 h. A portion of the cells from the 0 Gy groups were reserved for analysis of SIRT2 expression levels by western blotting. Remaining cells were used for clonogenic survival assays. Cells were incubated for a total of 10 days with media changed every 3 days. Cells were stained with crystal violet and colonies of ≥ 50 cells were counted.
EJ5 assay
In EJ5 U2OS reporter cells, genomically integrated GFP is separated from its promoter by a puromycin gene (60). The puromycin gene is flanked by two I-SceI sites (60). Therefore, expression of the endonuclease I-SceI generates a DSB that can only be repaired by NHEJ (60). Successful repair of the induced DSB results in genomic loss of the puromycin gene and GFP expression (60). If NHEJ repair efficiency is reduced due to the loss of an essential regulatory protein, GFP expression remains low following I-SceI expression (60). EJ5 U2OS reporter cells were transfected with SIRT2-FLAG-WT, SIRT2-FLAG-H187Y, Empty Vector, or were left untransfected. After 24 h, cells were transfected with non-targeting siRNA or SIRT2-10 UTR siRNA. After 48 h, cells were transfected with an I-SceI expression vector. At 72 h post I-SceI transfection, a portion of cells from each group were used for analysis of SIRT2 expression levels by western blotting. The remainder of cells were fixed in 1% PFA/PBS and analyzed for GFP expression via flow cytometry. Each group was performed in three biological replicates.
Chromatin immunoprecipitation (ChIP)
ChIP assay was performed using ChromaFlash One-Step ChIP Kit (Epigentek, P-2025) according to the manufacturer's instruction. Briefly, Fok1-expressing U2OS-265 Fok1 cells were fixed with 1% formaldehyde for 15 mins at room temperature, followed by treatment with 0.125 M glycine. Cells were then washed three times with ice-cold PBS and the lysate was prepared using CHAPS lysis buffer as described below for immunoprecipitation assay. Lysate was sonicated to an average DNA size below 500 base pairs and subsequently immunoprecipitated with anti-Ligase IV (Genetex, GTX55592) antibody. Immunoprecipitation with IgG served as a negative control. DNA from the immunoprecipitated samples was extracted and subjected to quantitative real-time PCR (PowerTrack™ SYBR Green Master Mix, A46012; and PCR 7500 Fast Real-Time PCR system, ThermoFisher) using the following primers at the Fok1-induced DSB site:
Forward: 5′-GGAAGATGTCCCTTGTATCACCAT-3′
Reverse: 5′-TGGTTGTCAACAGAGTAGAAAGTGAA-3′
Immunoprecipitation
Harvested cells were lysed for 40 min on ice in IP lysis buffer (0.75% CHAPS, 10% (vol/vol) glycerol, 150 mM NaCl, 50mM Tris pH 7.5) freshly supplemented with protease inhibitors. Supernatants were diluted to adjust the CHAPS concentration to 0.375%. Whole cell lysates (3 to 5 mg total protein) were incubated with anti-IgG rabbit (Millipore, N101), anti-IgG mouse (Millipore, N103), anti-DNA-PKcs (Thermo Fisher, PIMA513244 or Invitrogen, MA5-132238), anti-SIRT2 (Abcam, ab67299) or anti-GFP antibody (Abcam; Ab6556). Protein G beads, A agarose beads (Invitrogen), or FLAG conjugated beads (Sigma A2095) were used to immunoprecipitate antibody bound protein. Complexes were washed 4 times with IP washing buffer (0.375% CHAPS, 10% glycerol, 150 mM NaCl, 50 mM Tris pH 7.5) supplemented with protease inhibitors.
Mass spectrometry
HCT116 protein lysates were used in immunoprecipitation experiments with IgG or SIRT2 antibody. IP’ed material was eluted from beads and subjected to LC–MS/MS analysis as described previously (52). SIRT2 IP and mass spectrometry were conducted in duplicates.
Cell cycle analysis
HCT116 cells were treated with 0.5 mM minosine for 24 h to block cells in G1. Cells were then washed 2× in PBS and released into DMEM media. Seven hours after release, a portion of the cells were harvested and collected for S phase population and 24 hours after release, the remaining cells were collected for the G2 phase population. Where required, cells were irradiated with 10 Gy IR for 4 hours prior to harvest. Harvested cells were subjected to immunoprecipitation studies.
Deacetylation experiments
For in vitro analysis of DNA-PKcs deacetylation (61), HeLa cells were treated with 0.5 μM trichostatin A (TSA) and 20 mM nicotinamide for 12 h. TSA inhibits class I and II HDACs but not class III HDACs while nicotinamide inhibits class III HDACs. Cells were lysed with IP buffer (20 mM HEPES pH 7.4, 180 mM KCl, 0.2 mM EGTA, 1.5 mM MgCl2, 20% glycerol, 1.0% Nonidet P-40) supplemented with 1 μM TSA and fresh protease inhibitors. Acetyl-DNA-PKcs was immunoprecipitated using anti-DNA-PKcs (Thermo Fisher, PIMA513244) and protein G agarose beads (Invitrogen). Samples of immunoprecipitated DNA-PKcs bound to G agarose beads were washed 4 times with IP buffer containing 1μM TSA followed by an additional 2 washes with deacetylation buffer (50 mM Tris pH 7.5, 150mM NaCl, and 1mM MgCl2) to remove nicotinamide. SIRT2-FLAG WT and SIRT2–FLAG H187Y expression vectors were transfected into 293T cells. After 48 h to allow for protein expression, SIRT2-FLAG WT and SIRT2-FLAG H187Y proteins were immunoprecipitated using anti-FLAG M2 agarose beads (Sigma) and washed with IP buffer and TBS (50 mM Tris pH 7.5 and 150 mM NaCl). SIRT2-FLAG WT and SIRT2-FLAG H187Y proteins were eluted from beads with 0.15 mg/ml 3× FLAG Peptide (Sigma) for 30 min at 4°C. SIRT2 protein concentrations were determined by SDS-PAGE with Coomassie stain. Purified acetyl-DNA-PKcs was incubated with 1 μg of SIRT2-FLAG WT with NAD+, SIRT2-FLAG WT with nicotinamide and NAD+, SIRT2-FLAG H187Y and NAD+, or with no SIRT2-FLAG in deacetylation reaction buffer (1 μM TSA, 50 mM Tris·HCl, pH 7.5, 150 mM NaCl, 1 mM MgCl2 and 1 mM NAD+) at 30 °C for 3 h. The reactions were stopped by the addition of 5x SDS loading buffer and were incubated for 5 min at 100 °C. Samples were analyzed for acetylation by western blot using an anti-acetyl lysine antibody (Immunechem, ICP0380). For cellular deacetylation analysis (62), HeLa cells were transiently transfected with SIRT2-FLAG or SIRT2-FLAG-H187Y expression vectors, or were non-transfected. Cells were cultured with 0.5 μM TSA and with or without 20 mM nicotinamide for 12 h prior to being lysed with IP buffer containing 1 μM TSA. Cells were also treated with or without irradiation. Protein lysates were immunoprecipitated using anti-DNA-PKcs (Thermo Fisher, PIMA513244) and protein G agarose beads. The immunocaptured protein was analyzed for deacetylation by immunoblotting with anti-acetyl lysine antibody (Immunechem, ICP0380), anti-DNA-PKcs (Thermo Fisher, PIMA513244) and FLAG (Sigma F4042).
Immunoblot
Harvested cells were lysed for 30 min on ice in Nonidet P-40 buffer (200 mM NaCl, 1% Nonidet P-40,50 mM Tris·HCl pH 8.0) or IP lysis buffer (0.75% CHAPS, 10% (vol/vol) glycerol, 150 mM NaCl, 50mM Tris pH 7.5) freshly supplemented with protease inhibitors. Protein samples were resolved by SDS-PAGE and probed with indicated antibodies: anti-SIRT2 (Abcam ab67299 or Santa Cruz sc-20966), GAPDH (Santa Cruz sc-25778 or sc-47724), FLAG (Sigma F4042), GFP (Abcam Ab6556), anti-DNA-PKcs (Thermo Fisher, PIMA513244 or Invitrogen, MA5-132238), Artemis (Abcam, ab3834), anti-Artemis phospho Ser516 (Genetex, GTX32292), XRCC4 (Thermo Fisher Scientific, MA5-24383), XRCC4 phospho Ser260 (Thermo Fisher Scientific, PA5-64731), Cyclin A (BD BioScience, 611268) and α-Tubulin (Sigma, T6074). Detection was performed with the Odyssey system.
Laser microirradiation assay
For SIRT2 knockdown experiments, U2OS cells stably transfected with DNA-PKcs-GFP were transected transiently with non-targeting siRNA or SIRT2 siRNA. After 48 h, cells were plated into 35-mm glass bottom dishes (MatTek Corporation). Laser microirradiation (UV laser with a wavelength of 365nm) was performed on a Zeiss Observer Z1 microscope equipped with a Micropoint® Laser Illumination and Ablation System (Photonic Instruments). The laser output was set to 75%, which can reproducibly give focused DNA-PKcs-GFP stripes. Images were taken every minute for 5 min following damage or cells were fixed 2 min following damage induction for immunofluorescence assays. Quantitation of DNA-PKcs localization (signal of DNA-PKcs-GFP at bands of microirradiation) was performed using Image Studio lite software. For each condition, 15 biological replicates were performed. For SIRT2 inhibition experiments, Chinese hamster ovarian (CHO) DNA-PKcs deficient cells (V3) were also used. These cells were also stably transfected with human DNA-PKcs-GFP. Cells were plated into 35-mm glass bottom dishes (MatTek Corporation) and after 16 h to allow or cell adhesion, treated with DMSO or SIRT2 specific inhibitor (SirReal2 (63)) at 50 uM for 6 h. Laser microirradiation was performed on a Zeiss Observer Z1 microscope equipped with a Micropoint® Laser Illumination and Ablation System (Photonic Instruments). Following 2 min irradiation, samples were fixed in fixed in 4% PFA and stained for ɣH2AX and DAPI. 15 biological replicates were performed for each condition (DMSO or SirReal2).
Immunofluorescence
Immunofluorescence studies were performed in WT U2OS, SIRT2 U2OS KO or U2OS-265 mCherry-LacI-Fok1 (64) (induced for Fok1 expression with 1 μM Shield-1 and 2 μM 4-OHT for 4 hours) cells. Cells were transfected with SIRT2-FLAG or SIRT2-FLAG-H187Y expression vectors, or SIRT2 siRNA where indicated. After 72 h (for protein expression) or 48 h (for siRNA silencing), cells were exposed to 10 Gy IR for various time periods as required. Cells were fixed on coverslips with 4% PFA for 10 min, and permeabilized in 0.5% triton X-100 for 10 min. Cells were blocked in 5% BSA and immunostained with anti-γH2AX (Millipore 05–636), DNA-PKcs phosphoserine 2056 (Abcam, ab18192), anti-FLAG (Cell Signaling: 2368P), Artemis pS516 (Abcam, ab138411) or XRCC4 pS260 (Sigma, HPA006801) primary antibodies, followed by Alexa Fluor 555/488 secondary antibodies and DAPI.
In vivo growth inhibition study
Female Balb/cnu/nu (athymic) mice (Charles River Laboratories, Wilmington, MA) were housed in a pathogen-free location, and all in vivo studies were conducted under an IACUC-approved protocol at Emory University. IRR-A549 cell lines (1.5 × 106) were injected subcutaneously into bilateral flanks of mice. When tumors reached 100–200 mm3 (day 3), mice were randomized into four groups with 5 mice per group. Group 1 served as the control group, and received vehicle (0.25% DMSO) via Intraperitoneal (i.p.) injection, twice weekly; Group 2 received single agent AGK2 (82 mg/kg body weight), Group 3 received 4Gy and Group 4 received AGK2 + 4 Gy twice a week, for 2 total weeks of treatment. Tumor growth was measured as described previously using the modified ellipsoid formula 1/2 (length × width2) with measurements taken every 4th or 3rd day for a total of two weeks. Following the last measurement, mice were anesthetized with ketamine and sacrificed by cervical dislocation per IACUC approved institutional protocol. The mean and standard deviation of the outcomes of different treatment groups at different measurement time points are calculated. For proliferation at different time point, One-way ANOVA and T-tests were conducted to test whether there were any differences amongst the treatment groups. For tumor volume of animal study, the linear mixed models are performed to test whether there are any significant differences of tumor volume among different treatments. The significance level is set at 0.05.
RESULTS
SIRT2 deacetylase activity promotes DNA double-strand break repair by NHEJ
To determine the role of SIRT2 in DSB repair, we examined HCT116 SIRT2 wildtype (WT) and knockout (KO) cells, generated by CRISPR/Cas9 (50), for sensitivity to ionizing radiation (IR) and camptothecin (CPT). Similar to cells treated with a DNA-PK inhibitor (NU7441) (65), HCT116 SIRT2 KO cells were hypersensitive to IR and CPT compared to WT cells (Figure 1A, B; Supplementary Figure S1). The IR and CPT hypersensitivity of HCT116 SIRT2 KO cells was rescued by expression of SIRT2-FLAG WT but not catalytically-inactive SIRT2-FLAG H187Y (66), suggesting that SIRT2 deacetylase activity is important for responding to DSBs. Moreover, treatment of HCT116 SIRT2 KO cells with NU7441 did not cause significantly greater sensitivity to IR and CPT compared to SIRT2 KO or NU7441 treatment alone, implying that SIRT2 and DNA-PKcs function in a common pathway in responding to DSBs.
Figure 1.
SIRT2 deacetylase activity promotes DNA double-strand break repair by NHEJ. (A) Wildtype (WT) and SIRT2 KO HCT116 cells were transfected with or without SIRT2-FLAG WT or H187Y and treated with or without NU7441. Cells were seeded for colony formation, treated with indicated doses of ionizing radiation (IR), and assayed for surviving colonies 10 days later. Total colony number for each condition was normalized to non-damaged controls. Experiments were performed in biological replicates of 3. Error bars represent standard deviation: NS indicates P ≥ 0.05, *P< 0.05, **P< 0.01. (B) Western blot analysis of cells used in clonogenic assays in (A). (C) Diagram of EJ5 NHEJ reporter assay. (D) EJ5 U2OS reporter cells were transfected with SIRT2-FLAG WT, H187Y or empty vector (EV). After 24 h, cells were transfected with non-targeting (NT) siRNA or SIRT2-10 UTR siRNA (siSIRT2). After 48 h, cells were transfected with or without I-SceI and later analyzed for GFP expression using flow cytometry. The percentage of GFP positive cells in each sample was normalized to the percentage of GFP positive cells in NT siRNA + I-SceI control samples. Experiments were performed in biological replicates of 3 and the means were plotted. Error bars represent standard deviation: NS indicates P ≥ 0.05, *P< 0.05, **P< 0.01. (E) Representative western blot analysis of cells used in (D). (F) Western blot of U2OS-265 Fok1 cells used in (G), showing SIRT2 downregulation and overexpression of SIRT2-FLAG plasmids. (G) ChIP-qPCR analysis showing enrichment of Ligase IV at the Fok1-induced DSB site under the conditions indicated. Mean and SEM of three biological replicates is shown. ***P < 0.001.
To directly determine if SIRT2 functions in DSB repair by NHEJ, similar to DNA-PKcs, we examined SIRT2 depletion in U2OS cells integrated with a EJ5 NHEJ GFP reporter substrate, in which expression of the I-SceI endonuclease generates a DSB that when repaired by NHEJ restores GFP expression (60) (Figure 1C). SIRT2 depletion in these cells significantly impaired NHEJ compared to a non-targeting (NT) siRNA control (Figure 1D, E). In addition, expression of siRNA-resistant SIRT2-FLAG WT but not SIRT2-FLAG H187Y restored NHEJ to control levels, suggesting that SIRT2 deacetylase activity facilitates NHEJ repair. Consistently, SIRT2 depletion impaired the binding of endogenous DNA ligase IV to mCherry-LacI-FokI endonuclease-induced DSBs in U2OS reporter cells integrated with lac operator repeats (64), which was more efficiently rescued with expression of SIRT2-FLAG WT but not H187Y (Figure 1F, G). Moreover, SIRT2 depletion impaired the localization of Artemis and XRCC4 (phosphorylated at S516 and S260, respectively) to IR-induced foci (Supplementary Figure S2A-B) and to Fok1-induced DSBs (Supplementary Figure S2C–G). Expression of SIRT2-FLAG WT but not H187Y efficiently rescued the localization of phospho-Artemis S516 and phospho-XRCC4 S260 to Fok1-induced DSBs (Supplementary Figure S2C–G). Collectively, these data strongly suggest that SIRT2 deaetylase activity promotes NHEJ.
SIRT2 interacts in a complex with DNA-PKcs
To identify potential SIRT2 substrates in NHEJ repair, SIRT2 was immunopurified from HCT116 cells and subjected to mass spectrometry analysis, which showed enrichment of the NHEJ repair kinase DNA-PKcs (Figure 2A). The endogenous interaction of SIRT2 and DNA-PKcs in HCT116 and HeLa cells was validated by reciprocal co-immunoprecipitation (co-IP) (Figure 2B, C; Supplementary Figure S3). This interaction did not change appreciably in the absence and presence of IR (Figure 2C and Supplementary Figure S3) or across asynchronized, S-phase, and G2 phases of the cell cycle (Supplementary Figure S3). Furthermore, this interaction was preserved in the presence of ethidium bromide (EtBr), which intercalates with DNA and disrupts protein-protein interactions that are dependent on the presence of DNA (Figure 2C). In addition, although SIRT2 co-IPs with DNA-PKcs, it does not co-IP with Ku80-GFP expressed in HeLa cells either before or after IR treatment (Figure 2D). Collectively, these results indicate that SIRT2 specifically interacts with DNA-PKcs and that this interaction is not dependent on DNA damage, DNA binding, cell cycle, or DNA-PKcs complexed with Ku at sites of DNA damage.
Figure 2.
SIRT2 interacts in a complex with DNA-PKcs. (A) SIRT2 was IP’ed from HCT116 cells and IP’ed lysates were subjected to mass spectrometry analysis to identify SIRT2-interacting proteins. Total (T) and unique (U) number of peptides pulled-down for each IP’ed protein was measured and DNA-PKcs was identified as one of the top SIRT2-interactors. IP and mass spectrometry were conducted in duplicate (SIRT2 IP-1 and SIRT2 IP-2) with comparable results. IP with IgG (Control) was included. (B) Immunoprecipitation (IP) of endogenous DNA-PKcs or IgG was carried out on HeLa whole cell lysates, run on SDS-PAGE and immunoblotted for DNA-PKcs and SIRT2. The adjoining input is seen directly adjacent to the IP. (C) IP of endogenous SIRT2 or IgG was carried out on HeLa whole cell lysates with or without ethidium bromide (EtBr 50 μg/ml) or IR treatment (0 or 10 Gy IR), run on SDS-PAGE and immunoblotted for DNA-PKcs and SIRT2. The adjoining input is seen directly below the IP. (D) IP of overexpressed Ku80-GFP or IgG was carried out on HeLa whole cell lysates, run on SDS-PAGE and immunoblotted for DNA-PKcs, SIRT2, and GFP. The adjoining input is seen directly below the IP.
SIRT2 deacetylates DNA-PKcs in response to DNA damage
To determine if SIRT2 deacetylates DNA-PKcs, we performed an in vitro deacetylation assay with purified acetylated DNA-PKcs, SIRT2-FLAG WT and H187Y, and NAD + with or without nicotinamide, an inhibitor of sirtuin activity. SIRT2-FLAG WT but not H187Y deacetylated DNA-PKcs, and deacetylation was inhibited by nicotinamide (Figure 3A). To determine whether SIRT2 deacetylates endogenous DNA-PKcs in cells, we transfected HeLa cells with SIRT2-FLAG WT or H187Y and treated cells with or without nicotinamide. SIRT2-FLAG WT but not H187Y deacetylated endogenous DNA-PKcs in cells in a manner that was inhibited by nicotinamide (Figure 3B, C), confirming that SIRT2 deacetylates DNA-PKcs in cells. Moreover, treatment of the cells with IR resulted in similar level of deacetylation of DNA-PKcs (Figure 3B, C), indicating that DNA-PKcs is deacetylated in response to DNA damage potentially by SIRT2. To determine unequivocally if SIRT2 deacetylates endogenous DNA-PKcs in response to DNA damage, we examined DNA-PKcs acetylation in HeLa cells transfected with SIRT2 or NT siRNA and treated with or without IR. Endogenous DNA-PKcs was acetylated under basal conditions and deacetylated in response to IR, and the IR-regulated deacetylation of DNA-PKcs was rescued by SIRT2 depletion (Figure 3D, E), demonstrating that DNA-PKcs is acetylated in cells and subsequently deacetylated by SIRT2 in response to DNA damage. SIRT6 has been reported to stabilize DNA-PKcs at chromatin and promote its activity in NHEJ, likely through a scaffold function (57,58). To determine if SIRT6 has a role in deacetylating DNA-PKcs, we examined DNA-PKcs acetylation in HeLa cells overexpressing SIRT6-FLAG WT. In contrast with SIRT2, endogenous DNA-PKcs acetylation did not decrease following SIRT6-FLAG overexpression (Figure 3F), suggesting that DNA-PKcs deacetylation is mediated specifically by SIRT2 and not SIRT6.
Figure 3.
SIRT2 deacetylates DNA-PKcs in response to DNA damage. (A) In vitro deacetylation assay of DNA-PKcs by SIRT2. Acetylated endogenous DNA-PKcs was immunopurified from HeLa cells treated overnight with TSA and nicotinamide. SIRT2-FLAG WT and SIRT2-FLAG H187Y proteins were purified from 293T cells and eluted from beads. Acetylated DNA-PKcs was incubated in an in vitro deacetylation reaction with SIRT2-FLAG WT or SIRT2-FLAG H187Y with or without NAD+. Nicotinamide was also added, where indicated, as an inhibitor of SIRT2. (B, C) Cellular deacetylation assay. HeLa cells were transfected with SIRT2-FLAG WT, H187Y, or mock transfected. Cells received overnight TSA treatment with or without nicotinamide as indicated. Cells were also treated with or without 10 Gy IR. Endogenous DNA-PKcs or IgG was IP’d from whole cell lysate. The resulting western blots for IP and input were immunoblotted with DNA-PKcs, pan-acetyl, FLAG, and GAPDH antibodies. Shown are a representative western blot (B) and quantification of average and SD of co-IP’ed acetylated DNA-PKcs from three independent experiments (C). Acetylated DNA-PKcs was quantified using the Image Studio software and is expressed relative to the acetylated DNA-PKcs in the no treatment control sample, after normalization to total DNA-PKcs IP’ed. ** P< 0.01, *** P< 0.001. (D, E) Endogenous DNA-PKcs acetylation before and after damage and before and after SIRT2 knockdown was assessed through IP of DNA-PKcs or IgG from HeLa cells. HeLa cells were transfected with or without NT siRNA or SIRT2 siRNA. The resulting blots were stained for pan-acetyl, DNA-PKcs, SIRT2, and/or GAPDH. A representative western blot (D) and quantification of average and SD of co-IP’ed acetylated DNA-PKcs from three independent experiments (E) are shown. Acetylated DNA-PKcs was calculated as described in (C). *** P< 0.001. (F) HeLa cells were transfected with or without SIRT6-FLAG WT. Endogenous DNA-PKcs or IgG was IP’d from whole cell lysate and run on SDS-PAGE gel. The resulting western blots for IP and input were immunoblotted for DNA-PKcs, pan-acetyl, FLAG and GAPDH.
SIRT2 deacetylase activity facilitates DNA-PKcs localization to DNA damage sites and interaction with ku
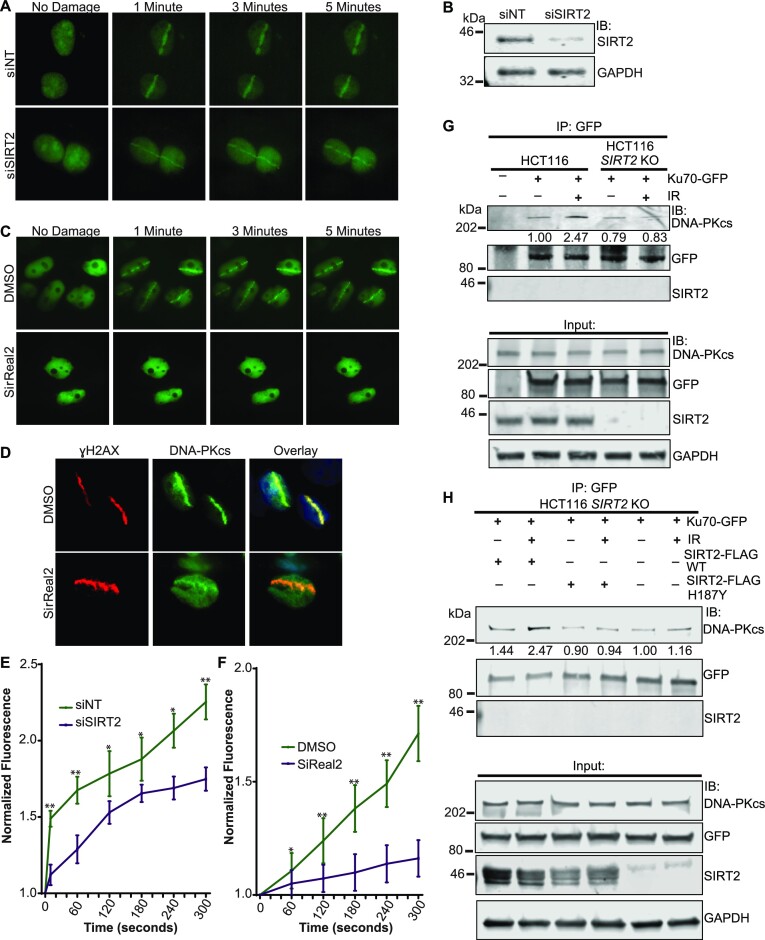
To determine if SIRT2 facilitates DNA-PKcs localization to DNA damage sites, we performed laser microirradiation experiments in U2OS cells stably transfected with DNA-PKcs-GFP (37). SIRT2 depletion delayed the early recruitment of DNA-PKcs-GFP to DNA damage sites induced by laser microirradiation (Figure 4A-B and 4E). Moreover, SIRT2 inhibition with SirReal2 (63) in CHO V3 cells stably transfected with human DNA-PKcs-GFP (37) also impaired the recruitment of DNA-PKcs-GFP to DNA damage sites induced by laser microirradiation (Figure 4C and F), indicating that SIRT2 deacetylase activity facilitates DNA-PKcs recruitment to DNA damage sites. Significantly, SIRT2 inhibition with SirReal2 did not impair the induction of DNA damage as determined by γH2AX staining following laser microirradiation (Figure 4D).
Figure 4.
SIRT2 deacetylase activity promotes DNA-PKcs localization to DNA damage sites and interaction with Ku. (A) DNA-PKcs-GFP stably transfected U2OS cells were subjected to laser microirradiation following transfection with SIRT2 or NT siRNA. The laser output was set to 75%, which can reproducibly give focused DNA-PKcs-GFP stripes. (B) Western blot demonstrating SIRT2 knockdown generated from cells used in (A). (C) DNA-PKcs-GFP stably transfected CHO V3 cells were subjected to laser microirradiation with and without SIRT2 inhibition by SirReal2 (50 uM) or DMSO. (D) CHO V3 cells treated with or without SirReal2 or DMSO were fixed 2 min post microirradiation and stained for γH2AX in red, DNA-PKcs-GFP in green, and DAPI stain in the overlay. (E) Fluorescence of the GFP stripes was measured for part A, recorded at indicated time points, and analyzed for quantitation using Image Studio Lite Software. The average of 15 biological replicates at each time point per condition was plotted. Error bars represent standard deviation: NS indicates P ≥ 0.05, *P< 0.05, **P< 0.01. (F) Quantitation of DNA-PKcs-GFP localization to sites of damage was performed for 15 biological replicates per condition. Fluorescence of the GFP stripes were measured for part C, recorded at indicated time points, and analyzed for quantitation using Image Studio Lite Software. The average of each time point's replicates was plotted. Error bars represent standard deviation. NS indicates P ≥ 0.05, *P< 0.05, **P< 0.01. (G) WT and SIRT2 KO HCT116 cells were transfected with and without Ku70-GFP and treated with or without 10 Gy IR. Ku70-GFP was IP’d, run on SDS-PAGE gels, and immunoblotted for DNA-PKcs, GFP, SIRT2, and GAPDH. DNA-PKcs pulled down with Ku70-GFP was quantified (values shown under the DNA-PKcs IP blot) using the Image Studio software and is shown as a fraction of DNA-PKcs co-IP’ed under control conditions, after normalization to IP’ed Ku70-GFP. (H) SIRT2 KO HCT116 cells were transfected with and without Ku70-GFP, SIRT2-FLAG WT, and H187Y and treated with or without 10 Gy IR. Ku70-GFP was IP’d, run on SDS-PAGE gel, and, immunoblotted for DNA-PKcs, GFP, SIRT2, and GAPDH. DNA-PKcs co-IP’ed with Ku70-GFP was quantified as described in (G).
It has previously been shown that DNA-PKcs forms a complex with Ku at DNA damage sites (6,7). To determine if SIRT2 facilitates the interaction between DNA-PKcs and Ku, we performed co-IP experiments in HCT116 SIRT2 WT and KO cells. As expected, we observed an increase in co-IP of DNA-PKcs with Ku70-GFP following IR treatment in HCT116 SIRT2 WT cells (Figure 4G). However, the IR-regulated increase in interaction of DNA-PKcs with Ku70-GFP was impaired in HCT-116 SIRT2 KO cells (Figure 4G, H). Expression of SIRT2-FLAG WT but not SIRT2-FLAG H187Y rescued the impairment in IR-induced interaction of DNA-PKcs with Ku70-GFP (Figure 4H), suggesting that SIRT2 deacetylase activity facilitates the interaction of DNA-PKcs with Ku70 in response to DNA damage.
Deacetylation by SIRT2 directs DNA-PK activation and signaling
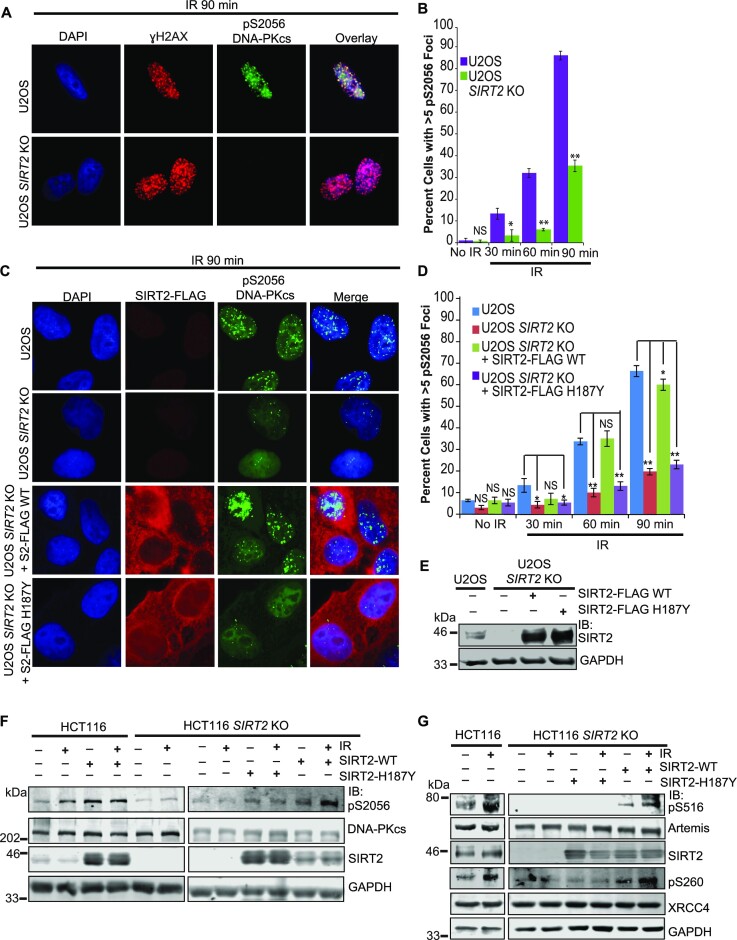
To determine whether SIRT2 directs DNA-PK activation in response to DNA damage, we examined DNA-PKcs autophosphorylation at serine 2056 (pS2056) following IR treatment in U2OS SIRT2 WT and KO cells. A significant impairment in DNA-PKcs pS2056 but not γH2AX foci induction in response to IR was observed in U2OS SIRT2 KO cells when compared with WT cells (Figure 5A, B), which was rescued by expression of SIRT2-FLAG WT but not H187Y (Figure 5C–E; Supplementary Figure S4A–C), suggesting that SIRT2 deacetylase activity promotes DNA-PK activation. Consistent with these findings, IR-induced DNA-PKcs autophosphorylation at S2056 but not total DNA-PKcs levels was also reduced in HCT116 SIRT2 KO cells compared with WT cells as determined by western blot analysis (Figure 5F; Supplementary Figure S4D and Figure S5A, B). This impairment was more fully rescued by expression of SIRT2-FLAG WT but not H187Y (Figure 5F; Supplementary Figure S4E). Furthermore, treatment of HCT116 SIRT2 KO cells expressing SIRT2-FLAG WT with DNA-PK inhibitor NU7441 resulted in inhibition of the rescue (Supplementary Figure S5A, B). Overall, these results imply that SIRT2 deacetylase activity directs DNA-PK activation in response to IR in a manner that requires DNA-PK catalytic activity.
Figure 5.
Deacetylation by SIRT2 directs DNA-PK activation and signaling. (A) WT and SIRT2 KO U2OS cells were treated with or without 10 Gy IR. Cells were fixed at 30, 60 or 90 min post IR and stained with anti-γH2AX (red), anti-DNA-PKcs pS2056 (green) and DAPI (blue). A representative 90 min post IR time point is shown. (B) Quantitation of the percentage of cells with >5 DNA-PKcs pS2056 foci is shown for (A). The mean from each group was calculated from three biological replicates (100 cells per replicate) and error bars represent S.D. *P< 0.05; **P< 0.01. (C) WT and SIRT2 KO U2OS cells were transfected with SIRT2-FLAG WT, H187Y or mock transfected and treated with or without 10 Gy IR. Cells were fixed at 30, 60 or 90 min post IR and stained with anti-FLAG (red), anti-DNA-PKcs pS2056 (green) and DAPI (blue). A representative 90 min post IR time point is shown. (D) Quantitation of the percentage of cells with >5 DNA-PKcs pS2056 foci is shown for (C) and Figure S3. The mean for each group was calculated from three biological replicates (100 cells per replicate) and error bars represent S.D. *P< 0.05; **P< 0.01. (E) Western blot analysis for (C) and (D). (F) Western blot analysis of DNA-PKcs autophosphorylation at S2056 in response to IR in WT and SIRT2 KO HCT116 cells transfected with or without SIRT2-FLAG WT and H187Y. Quantification and statistical analysis is shown in Supplementary Figure S4D-E. (G) Western blot analysis of Artemis and XRCC4 phosphorylation at S516 and S260 respectively in response to IR in WT and SIRT2 KO HCT116 cells transfected with or without SIRT2-FLAG WT and H187Y.
To determine if SIRT2 functions in DNA-PK-dependent signaling, we examined HCT116 SIRT2 WT and KO cells for phosphorylation of a panel of DNA-PK substrates in response to IR treatment. Phosphorylation of Artemis at serine 516 (pS516) and XRCC4 at serine 260 (pS260) following IR treatment was impaired in HCT116 SIRT2 KO cells compared with WT cells (Figure 5G). As observed with DNA-PKcs autophosphorylation at serine 2056, DNA-PK-dependent phosphorylation of Artemis at S516 and XRCC4 at S260 was rescued in HCT116 SIRT2 KO cells by expression of SIRT2-FLAG but not SIRT2-FLAG-H187Y (Figure 5G). Collectively, combined with our data indicating that SIRT2 interacts with and deacetylates DNA-PKcs, these results support a model in which DNA-PKcs deacetylation by SIRT2 directs DNA-PK activation and signaling.
SIRT2 inhibitor augments the efficacy of IR in cancer cells and tumors
Our observation that SIRT2 deficiency in cancer cells causes IR hypersensitivity and can be rescued by SIRT2-FLAG WT but not H187Y suggest that SIRT2 may be a promising therapeutic target and that SIRT2 activity could be targeted to sensitize cancer cells to IR. SIRT2 inhibitors have been reported to have anti-proliferative activity against a number of cancer types (59); however, the specific cellular context, including synthetic lethal relationships with specific types of cancer treatments for which SIRT2 inhibitors may be most effective for cancer therapy are not clear. Indeed, similar to SIRT2 deficiency, HCT116 cells treated with AGK2, a selective SIRT2 inhibitor (67), were sensitized to IR (Figure 6A). Treatment of the cells with NU7441, a DNA-PK inhibitor, caused a similar level of IR hypersensitivity, and combined treatment of cells with AGK2 and NU7441 did not further sensitize cells to IR (Figure 6A), confirming that SIRT2 and DNA-PKcs function in a common pathway in governing resistance to IR.
Figure 6.
SIRT2 inhibitor augments the efficacy of IR in cancer cells and tumors. (A) Clonogenic assay of HCT116 cells using SIRT2 inhibitor AGK2. WT HCT116 cells were treated with NU7441 or with AGK2. Cells were then seeded for colony formation, treated with indicated doses of IR, and assayed for surviving colonies 10 days later. Total colony number for each condition was normalized to non-damaged controls. Experiments were performed in biological replicates of three. Error bars represent standard deviation: NS indicates P ≥ 0.05, *P< 0.05, **P< 0.01, ***P< 0.001. (B) In vivo growth inhibition study of IRR-A549 xenographs in female Balb/cnu/nu (athymic) mice. Mice had IRR-A549 cells implanted subcutaneously as described in the Materials and Methods section. Animals received IR and/or intraperitoneal (IP) injection of AGK2. Tumor volume was measured once every three to four days. Tumors were resected from animals at the completion of treatment. There was no significance seen between tumor volumes between control IRR-A549 and IRR-A549 + IR groups. A significant decrease in the average tumor volume was observed in animals treated with AGK2 or AGK2 + IR as compared to controls (NS indicates P ≥ 0.05, *P< 0.05, **P< 0.01, ***P< 0.001). A significant decrease was observed in tumors from animals treated with AGK2 + IR as compared to AGK2 alone (NS indicates P ≥ 0.05, #P< 0.05, ##P< 0.01, ###P< 0.001). Each group included 5 mice. (C) Weight of mice during course of treatment did not significantly differ between treatment groups.
To determine if SIRT2 inhibitor can be used to sensitize resistant tumors to IR treatment, we generated tumor xenografts using IR resistant A549 cells (IRR-A549) (68) in female Balb/cnu/nu (athymic) mice. As expected, IR treatment did not significantly delay growth of the radioresistant tumors (Figure 6B). However, treatment of the mice with AGK2 and IR significantly delayed tumor growth compared with treatment with IR alone (Figure 6B), suggesting that SIRT2 inhibitor can cause resensitization of the resistant tumors to IR. Consistent with findings in other tumor types (59), treatment of the mice with AGK2 alone also significantly delayed tumor growth (Figure 6B) but to a lesser extent than with treatment with both AGK2 and IR together. No significant difference in body weight was observed in mice treated with AGK2 compared to controls (Figure 6C).
DISCUSSION
Our findings reveal a regulatory step governing DNA-PK activation by SIRT2 deacetylation, which facilitates DNA-PKcs localization to DSBs and interaction with Ku, thereby promoting DSB repair by NHEJ. These findings elucidate a critical upstream signaling event directing the interaction of DNA-PKcs and Ku that activates DNA-PK, identify DNA-PKcs as a unique interacting partner and substrate for SIRT2, and establish SIRT2 as a positive regulator of DSB repair by NHEJ that broadens its role in DNA repair. This work also provides important insights into how Sirt2 deficiency results in genomic instability and carcinogenesis and provides mechanistic rationale and preclinical evidence for SIRT2 inhibition as a potential therapeutic strategy for radiation sensitization. In this regard, we found that SIRT2 deacetylase activity governs cellular resistance to IR and CPT and promotes NHEJ through a direct reporter assay, defining a role for SIRT2 in DSB repair by NHEJ. We further found that SIRT2 interacts with DNA-PKcs but not Ku and deacetylates DNA-PKcs in response to IR, providing evidence that DNA-PKcs is regulated by SIRT2 deacetylation in a damage regulated manner and that DNA-PKcs is an interacting partner and substrate for SIRT2. Moreover, we found that SIRT2 deacetylation facilitates DNA-PKcs interaction with Ku and early recruitment to DNA damage sites, thereby leading to DNA-PKcs autophosphorylation and DNA-PK signaling to downstream NHEJ substrates, providing a mechanistic model for how DNA-PKcs deacetylation by SIRT2 promotes DNA-PK activation in the NHEJ pathway. Finally, we found that SIRT2 inhibitor augments the efficacy of IR in cancer cells and resistant tumors, providing rationale-driven preclinical evidence for the use of SIRT2 inhibition in sensitizing cancer to radiation therapy.
Our findings are consistent with a model in which in response to IR and other DSB-inducing agents, SIRT2 deacetylates DNA-PKcs, which facilitates its interaction with Ku and early recruitment to DSBs, leading to DNA-PK activation in which DNA-PKcs phosphorylates itself and a number of downstream substrates, including Artemis and XRCC4 to promote NHEJ (Figure 7). Dysregulation of this pathway leads to genomic instability and ultimately carcinogenesis. Our observation that SIRT2 complexes with DNA-PKcs but not Ku suggests that SIRT2 interacts with DNA-PKcs specifically and regulates its function prior to its recruitment and interaction with Ku at DSBs. In this regard, while our data indicate that SIRT2 deacetylates DNA-PKcs in response to DNA damage, the interaction of SIRT2 with DNA-PKcs does not appear to change appreciably with DNA damage. Thus, it is possible that the IR-regulated deacetylation of DNA-PKcs by SIRT2 results from an increase in SIRT2 deacetylase activity triggered by IR. Indeed, we have previously found that SIRT2 deacetylase activity increases in response to DNA damage (51). Furthermore, while SIRT2 is predominantly cytoplasmic, we found no evidence of a significant change in nuclear localization after IR. Thus, we favor a model in which IR triggers the activation of a sufficient amount of nuclear SIRT2 that then deacetylates DNA-PKcs, although our model does not exclude the possibility of a role for cytoplasmic SIRT2.
Figure 7.
Model of SIRT2 regulation of DNA-PKcs in NHEJ repair. DNA damage results in a DSB (1) which leads to the recruitment of Ku70 and Ku80 heterodimers to both sides of the DSB (2). SIRT2 deacetylates DNA-PKcs in response to damage on its N terminus (light brown), which is important for DNA-PKcs localization to the sites of DNA damage, its interaction with Ku, and the formation of DNA-PK holoenzyme dimers (3). Once dimerized, DNA-PK phosphorylates downstream NHEJ repair factor Artemis, and Artemis resects DNA ends (4,5). DNA-PKcs autophosphorylates at serine 2056 (5) and phosphorylates XRCC4 (6). XRCC4 with DNA ligase 4 and DNA-PKcs promotes end ligation (6). DNA-PKcs releases the repaired DNA (7).
How might DNA-PKcs deacetylation by SIRT2 promote its activation via its interaction with Ku? DNA-PKcs interfaces with Ku at multiple sites within its amino-terminal N-HEAT (aa 1–872) and circular cradle M-HEAT domains (aa 890–2580) (8–11,69–74). Indeed, a large number of acetylation sites have been identified in these regions as reported in PhosphoSitePlus (75). It is possible that SIRT2 deacetylates critical acetyl-lysine residues in these regions that may mediate electrostatic interactions with Ku and/or Ku bound to DNA. These interactions likely contribute to a conformational change in DNA-PKcs (11,18), which leads to DNA-PK activation. While our findings indicate that SIRT2 deacetylation facilitates the damage-regulated interaction of DNA-PKcs with Ku, some interaction between DNA-PKcs and Ku is observed in SIRT2 KO cells, albeit this interaction is not regulated by DNA damage. Thus, while our data strongly suggest that SIRT2 deacetylation of DNA-PKcs plays a critical role in the damage-regulated activation of DNA-PK by facilitating its interaction with Ku, our model does not rule out the possibility that other mechanisms may also contribute to the DNA-PKcs-Ku interaction such as a scaffold function for SIRT6 (57). In addition, it was recently reported that Sirt2 KO mouse embryonic fibroblasts (MEFs) are paradoxically resistant to IR and hypersensitive to etoposide, a topoisomerase II inhibitor, which also induces DSBs (76). In contrast, we found that both SIRT2 deficiency and inhibition in several human cancer cell types causes hypersensitivity to IR, which can be rescued by expression of exogenous SIRT2 WT but not H187Y. It is possible that SIRT2’s role in responding to DSBs could be species dependent, perhaps dependent on a dominant downstream substrate that governs response to DSBs or compensatory mechanisms resulting from Sirt2 deficiency in the context of dysregulation of other DNA damage response genes. It has also been reported that SIRT2 and SIRT3 depletion in H1299 non-small cell lung cancer cells impair HR but promote NHEJ (54). This discrepancy with our findings may be due to differences in cell type as similar to our finding that SIRT2 depletion in U2OS cells impairs NHEJ, it has also been reported that SIRT3 depletion in U2OS cells impairs NHEJ (77).
SIRT2 has previously been shown to direct the RSR through deacetylation of ATRIP and CDK9 (51,52) and HR through deacetylation of BARD1 (53). We now define a role for SIRT2 deacetylation in activating DNA-PK to promote DSB repair by NHEJ. Furthermore, the identification of DNA-PKcs as a binding partner and substrate of SIRT2 adds to the growing number of SIRT2 substrates that function in promoting genome integrity, providing support for SIRT2 in regulating a network of proteins involved in the DDR (47). Our finding that SIRT2 directs DNA-PK activation in NHEJ provides an additional layer of insight into how SIRT2 dysregulation leads to genomic instability and carcinogenesis.
SIRT2 inhibitors have been reported to have anti-proliferative activity against a number of cancer types (59); however, the specific cellular context for which SIRT2 inhibitors may be most effective for cancer therapy have not been clearly defined. We now show that SIRT2 inhibition sensitizes cancer cells and resistant tumors to IR, providing preclinical evidence for the use of SIRT2 inhibition as a rationale-driven therapeutic strategy for increasing the effectiveness of radiation therapy. Our observation that SIRT2 deficiency and inhibition is epistatic with DNA-PKcs inhibition in mediating IR sensitivity, suggests that SIRT2 governs IR sensitivity largely by activating DNA-PK to direct NHEJ despite its role in both NHEJ and HR. This is consistent with NHEJ’s role as the predominant pathway for the repair of DSBs induced by IR. However, we acknowledge that in the in vivo context, our model does not rule out the possibility that SIRT2 inhibition may sensitize tumors to IR via other DSB repair pathways such as HR. In summary, our findings define a mechanism for DNA-PK activation by SIRT2-mediated deacetylation, elucidating a critical upstream signaling event initiating the repair of DSBs by NHEJ to promote genome integrity and furthermore provide mechanistic rationale and preclinical evidence for the rational targeting of SIRT2 as an adjunct to radiation therapy.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Yu lab for helpful discussion. We kindly thank Manfred Jung for SirReal2 inhibitor, Eric Verdin for the SIRT2-FLAG plasmid, Anthony Davis for DNA-PKcs antibody, DNA-PKcs and Ku70/80 plasmids, and DNA-PKcs-GFP stable cell lines (both U2OS and CHO V3), and Roger Greenberg for mCherry-LacI-FokI U2OS reporter cells integrated with lac operator repeats.
Author contributions: P.E.H. and D.S.Y. conceived and designed the study. P.E.H., P.K., N.P.G., S.K.R., N.C.L., R.H.S.J., H.Z., F.S., S.W., D.M.D., W.D., E.V.M., B.S., D.D., X.L. and D.M.S. performed and analyzed the experiments. P.E.H., P.K., N.P.G., S.K.R., N.C.L., H.Z., F.S., S.W., D.M.D., W.D., E.V.M., B.S., D.D., X.L., S.L., E.A.O., N.T.S., D.M.S., Y.W., X.D., W.S.D., B.E., A.J.D. and D.S.Y. interpreted the findings. P.E.H., P.K. and D.S.Y. wrote the manuscript with input from all authors.
Contributor Information
PamelaSara E Head, Department of Radiation Oncology and Winship Cancer Institute, Emory University School of Medicine, Atlanta, GA 30322, USA.
Priya Kapoor-Vazirani, Department of Radiation Oncology and Winship Cancer Institute, Emory University School of Medicine, Atlanta, GA 30322, USA.
Ganji P Nagaraju, School of Medicine, Division of Hematology and Medical Oncology, University of Alabama, Birmingham, AL 35233, USA.
Hui Zhang, Department of Radiation Oncology and Winship Cancer Institute, Emory University School of Medicine, Atlanta, GA 30322, USA.
Sandip K Rath, Department of Radiation Oncology and Winship Cancer Institute, Emory University School of Medicine, Atlanta, GA 30322, USA.
Nho C Luong, Department of Radiation Oncology and Winship Cancer Institute, Emory University School of Medicine, Atlanta, GA 30322, USA.
Ramona Haji-Seyed-Javadi, Department of Radiation Oncology and Winship Cancer Institute, Emory University School of Medicine, Atlanta, GA 30322, USA.
Fatmata Sesay, Department of Radiation Oncology and Winship Cancer Institute, Emory University School of Medicine, Atlanta, GA 30322, USA.
Shi-Ya Wang, Department of Radiation Oncology, UT Southwestern Medical School, Dallas, TX 75390, USA.
Duc M Duong, Department of Biochemistry, Emory University School of Medicine, Atlanta, GA 30322, USA.
Waaqo Daddacha, Department of Biochemistry and Molecular Biology, Augusta University, Augusta, GA 30912, USA.
Elizabeth V Minten, Department of Radiation Oncology and Winship Cancer Institute, Emory University School of Medicine, Atlanta, GA 30322, USA.
Boying Song, Department of Radiation Oncology and Winship Cancer Institute, Emory University School of Medicine, Atlanta, GA 30322, USA.
Diana Danelia, Department of Radiation Oncology and Winship Cancer Institute, Emory University School of Medicine, Atlanta, GA 30322, USA.
Xu Liu, Department of Biochemistry, Emory University School of Medicine, Atlanta, GA 30322, USA.
Shuyi Li, Department of Radiation Oncology and Winship Cancer Institute, Emory University School of Medicine, Atlanta, GA 30322, USA; Department of Biochemistry, Emory University School of Medicine, Atlanta, GA 30322, USA.
Eric A Ortlund, Department of Biochemistry, Emory University School of Medicine, Atlanta, GA 30322, USA.
Nicholas T Seyfried, Department of Biochemistry, Emory University School of Medicine, Atlanta, GA 30322, USA.
David M Smalley, Parker H. Petit Institute for Bioengineering and Bioscience, Georgia Institute of Technology, Atlanta, GA 30332, USA.
Ya Wang, Department of Radiation Oncology and Winship Cancer Institute, Emory University School of Medicine, Atlanta, GA 30322, USA.
Xingming Deng, Department of Radiation Oncology and Winship Cancer Institute, Emory University School of Medicine, Atlanta, GA 30322, USA.
William S Dynan, Department of Radiation Oncology and Winship Cancer Institute, Emory University School of Medicine, Atlanta, GA 30322, USA; Department of Biochemistry, Emory University School of Medicine, Atlanta, GA 30322, USA.
Bassel El-Rayes, School of Medicine, Division of Hematology and Medical Oncology, University of Alabama, Birmingham, AL 35233, USA.
Anthony J Davis, Department of Radiation Oncology, UT Southwestern Medical School, Dallas, TX 75390, USA.
David S Yu, Department of Radiation Oncology and Winship Cancer Institute, Emory University School of Medicine, Atlanta, GA 30322, USA.
Data Availability
The data underlying this article will be shared on reasonable request to the corresponding author.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH)/National Cancer Institute (NCI) [R01CA178999, R01CA254403]; Department of Defense (DOD) Ovarian Cancer Research Program (OCRP) [OC160540]; DOD Breast Cancer Research Program (BCRP) [BC180883]; Basser Center for BRCA External Research Grant Program Innovation Award 32356; Winship Cancer Institute (WCI) Brenda Nease Breast Cancer Research Fund Pilot Award [53237 to D.S.Y.]; NIH/National Institutes of General Medical Sciences (NIGMS) [T32GM008490 to P.E.H.]; NIH/NCI [CA92584, CA162804]; NIH/NIGMS [GM047251 to A.D.]. Funding for open access charge: Startup Funding.
Conflict of interest statement. None declared.
REFERENCES
- 1. Davis A.J., Chen D.J.. DNA double strand break repair via non-homologous end-joining. Transl. Cancer Res. 2013; 2:130–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lieber M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010; 79:181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Woodbine L., Gennery A.R., Jeggo P.A.. The clinical impact of deficiency in DNA non-homologous end-joining. DNA Repair (Amst.). 2014; 16:84–96. [DOI] [PubMed] [Google Scholar]
- 4. Davis A.J., Chen B.P., Chen D.J.. DNA-PK: a dynamic enzyme in a versatile DSB repair pathway. DNA Repair (Amst.). 2014; 17:21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jette N., Lees-Miller S.P.. The DNA-dependent protein kinase: a multifunctional protein kinase with roles in DNA double strand break repair and mitosis. Prog. Biophys. Mol. Biol. 2015; 117:194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dvir A., Peterson S.R., Knuth M.W., Lu H., Dynan W.S.. Ku autoantigen is the regulatory component of a template-associated protein kinase that phosphorylates RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 1992; 89:11920–11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gottlieb T.M., Jackson S.P.. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993; 72:131–142. [DOI] [PubMed] [Google Scholar]
- 8. Davis A.J., Lee K.J., Chen D.J.. The N-terminal region of the DNA-dependent protein kinase catalytic subunit is required for its DNA double-stranded break-mediated activation. J. Biol. Chem. 2013; 288:7037–7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sibanda B.L., Chirgadze D.Y., Blundell T.L.. Crystal structure of DNA-PKcs reveals a large open-ring cradle comprised of HEAT repeats. Nature. 2010; 463:118–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Singleton B.K., Torres-Arzayus M.I., Rottinghaus S.T., Taccioli G.E., Jeggo P.A.. The C terminus of Ku80 activates the DNA-dependent protein kinase catalytic subunit. Mol. Cell. Biol. 1999; 19:3267–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Spagnolo L., Rivera-Calzada A., Pearl L.H., Llorca O.. Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol. Cell. 2006; 22:511–519. [DOI] [PubMed] [Google Scholar]
- 12. Ma Y., Pannicke U., Schwarz K., Lieber M.R.. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V (D)J recombination. Cell. 2002; 108:781–794. [DOI] [PubMed] [Google Scholar]
- 13. Ahnesorg P., Smith P., Jackson S.P.. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006; 124:301–313. [DOI] [PubMed] [Google Scholar]
- 14. Andres S.N., Vergnes A., Ristic D., Wyman C., Modesti M., Junop M.. A human XRCC4-XLF complex bridges DNA. Nucleic Acids Res. 2012; 40:1868–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ochi T., Blackford A.N., Coates J., Jhujh S., Mehmood S., Tamura N., Travers J., Wu Q., Draviam V.M., Robinson C.V.et al.. DNA repair. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair. Science. 2015; 347:185–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tadi S.K., Tellier-Lebegue C., Nemoz C., Drevet P., Audebert S., Roy S., Meek K., Charbonnier J.B., Modesti M.. PAXX Is an Accessory c-NHEJ Factor that Associates with Ku70 and Has Overlapping Functions with XLF. Cell Rep. 2016; 17:541–555. [DOI] [PubMed] [Google Scholar]
- 17. Xing M., Yang M., Huo W., Feng F., Wei L., Jiang W., Ning S., Yan Z., Li W., Wang Q.et al.. Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway. Nat. Commun. 2015; 6:6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dobbs T.A., Tainer J.A., Lees-Miller S.P.. A structural model for regulation of NHEJ by DNA-PKcs autophosphorylation. DNA Repair (Amst.). 2010; 9:1307–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chan D.W., Chen B.P., Prithivirajsingh S., Kurimasa A., Story M.D., Qin J., Chen D.J.. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 2002; 16:2333–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chan D.W., Lees-Miller S.P.. The DNA-dependent protein kinase is inactivated by autophosphorylation of the catalytic subunit. J. Biol. Chem. 1996; 271:8936–8941. [DOI] [PubMed] [Google Scholar]
- 21. Meek K., Douglas P., Cui X., Ding Q., Lees-Miller S.P.. trans Autophosphorylation at DNA-dependent protein kinase's two major autophosphorylation site clusters facilitates end processing but not end joining. Mol. Cell. Biol. 2007; 27:3881–3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chan D.W., Ye R., Veillette C.J., Lees-Miller S.P.. DNA-dependent protein kinase phosphorylation sites in Ku 70/80 heterodimer. Biochemistry. 1999; 38:1819–1828. [DOI] [PubMed] [Google Scholar]
- 23. Leber R., Wise T.W., Mizuta R., Meek K.. The XRCC4 gene product is a target for and interacts with the DNA-dependent protein kinase. J. Biol. Chem. 1998; 273:1794–1801. [DOI] [PubMed] [Google Scholar]
- 24. Yu Y., Mahaney B.L., Yano K., Ye R., Fang S., Douglas P., Chen D.J., Lees-Miller S.P.. DNA-PK and ATM phosphorylation sites in XLF/Cernunnos are not required for repair of DNA double strand breaks. DNA Repair (Amst.). 2008; 7:1680–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang Y.G., Nnakwe C., Lane W.S., Modesti M., Frank K.M.. Phosphorylation and regulation of DNA ligase IV stability by DNA-dependent protein kinase. J. Biol. Chem. 2004; 279:37282–37290. [DOI] [PubMed] [Google Scholar]
- 26. Ma Y., Pannicke U., Lu H., Niewolik D., Schwarz K., Lieber M.R.. The DNA-dependent protein kinase catalytic subunit phosphorylation sites in human Artemis. J. Biol. Chem. 2005; 280:33839–33846. [DOI] [PubMed] [Google Scholar]
- 27. Zolner A.E., Abdou I., Ye R., Mani R.S., Fanta M., Yu Y., Douglas P., Tahbaz N., Fang S., Dobbs T.et al.. Phosphorylation of polynucleotide kinase/phosphatase by DNA-dependent protein kinase and ataxia-telangiectasia mutated regulates its association with sites of DNA damage. Nucleic Acids Res. 2011; 39:9224–9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lu H., Saha J., Beckmann P.J., Hendrickson E.A., Davis A.J.. DNA-PKcs promotes chromatin decondensation to facilitate initiation of the DNA damage response. Nucleic Acids Res. 2019; 47:9467–9479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stiff T., O’Driscoll M., Rief N., Iwabuchi K., Lobrich M., Jeggo P.A.. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 2004; 64:2390–2396. [DOI] [PubMed] [Google Scholar]
- 30. Chen B.P., Chan D.W., Kobayashi J., Burma S., Asaithamby A., Morotomi-Yano K., Botvinick E., Qin J., Chen D.J.. Cell cycle dependence of DNA-dependent protein kinase phosphorylation in response to DNA double strand breaks. J. Biol. Chem. 2005; 280:14709–14715. [DOI] [PubMed] [Google Scholar]
- 31. Cui X., Yu Y., Gupta S., Cho Y.M., Lees-Miller S.P., Meek K.. Autophosphorylation of DNA-dependent protein kinase regulates DNA end processing and may also alter double-strand break repair pathway choice. Mol. Cell. Biol. 2005; 25:10842–10852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Douglas P., Sapkota G.P., Morrice N., Yu Y., Goodarzi A.A., Merkle D., Meek K., Alessi D.R., Lees-Miller S.P.. Identification of in vitro and in vivo phosphorylation sites in the catalytic subunit of the DNA-dependent protein kinase. Biochem. J. 2002; 368:243–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jiang W., Crowe J.L., Liu X., Nakajima S., Wang Y., Li C., Lee B.J., Dubois R.L., Liu C., Yu X.et al.. Differential phosphorylation of DNA-PKcs regulates the interplay between end-processing and end-ligation during nonhomologous end-joining. Mol. Cell. 2015; 58:172–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen B.P., Uematsu N., Kobayashi J., Lerenthal Y., Krempler A., Yajima H., Lobrich M., Shiloh Y., Chen D.J.. Ataxia telangiectasia mutated (ATM) is essential for DNA-PKcs phosphorylations at the Thr-2609 cluster upon DNA double strand break. J. Biol. Chem. 2007; 282:6582–6587. [DOI] [PubMed] [Google Scholar]
- 35. Yajima H., Lee K.J., Chen B.P.. ATR-dependent phosphorylation of DNA-dependent protein kinase catalytic subunit in response to UV-induced replication stress. Mol. Cell. Biol. 2006; 26:7520–7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hammel M., Yu Y., Mahaney B.L., Cai B., Ye R., Phipps B.M., Rambo R.P., Hura G.L., Pelikan M., So S.et al.. Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. J. Biol. Chem. 2010; 285:1414–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Uematsu N., Weterings E., Yano K., Morotomi-Yano K., Jakob B., Taucher-Scholz G., Mari P.O., van Gent D.C., Chen B.P., Chen D.J.. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J. Cell Biol. 2007; 177:219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ding Q., Reddy Y.V., Wang W., Woods T., Douglas P., Ramsden D.A., Lees-Miller S.P., Meek K.. Autophosphorylation of the catalytic subunit of the DNA-dependent protein kinase is required for efficient end processing during DNA double-strand break repair. Mol. Cell. Biol. 2003; 23:5836–5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Goodarzi A.A., Yu Y., Riballo E., Douglas P., Walker S.A., Ye R., Harer C., Marchetti C., Morrice N., Jeggo P.A.et al.. DNA-PK autophosphorylation facilitates Artemis endonuclease activity. EMBO J. 2006; 25:3880–3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Block W.D., Yu Y., Merkle D., Gifford J.L., Ding Q., Meek K., Lees-Miller S.P.. Autophosphorylation-dependent remodeling of the DNA-dependent protein kinase catalytic subunit regulates ligation of DNA ends. Nucleic Acids Res. 2004; 32:4351–4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Das A.K., Chen B.P., Story M.D., Sato M., Minna J.D., Chen D.J., Nirodi C.S.. Somatic mutations in the tyrosine kinase domain of epidermal growth factor receptor (EGFR) abrogate EGFR-mediated radioprotection in non-small cell lung carcinoma. Cancer Res. 2007; 67:5267–5274. [DOI] [PubMed] [Google Scholar]
- 42. Toulany M., Kasten-Pisula U., Brammer I., Wang S., Chen J., Dittmann K., Baumann M., Dikomey E., Rodemann H.P.. Blockage of epidermal growth factor receptor-phosphatidylinositol 3-kinase-AKT signaling increases radiosensitivity of K-RAS mutated human tumor cells in vitro by affecting DNA repair. Clin. Cancer Res. 2006; 12:4119–4126. [DOI] [PubMed] [Google Scholar]
- 43. Liu H., Galka M., Mori E., Liu X., Lin Y.F., Wei R., Pittock P., Voss C., Dhami G., Li X.et al.. A method for systematic mapping of protein lysine methylation identifies functions for HP1beta in DNA damage response. Mol. Cell. 2013; 50:723–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Olsen B.B., Issinger O.G., Guerra B.. Regulation of DNA-dependent protein kinase by protein kinase CK2 in human glioblastoma cells. Oncogene. 2010; 29:6016–6026. [DOI] [PubMed] [Google Scholar]
- 45. Finkel T., Deng C.X., Mostoslavsky R.. Recent progress in the biology and physiology of sirtuins. Nature. 2009; 460:587–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Guarente L., Franklin H.. Epstein Lecture: sirtuins, aging, and medicine. N. Engl. J. Med. 2011; 364:2235–2244. [DOI] [PubMed] [Google Scholar]
- 47. Zhang H., Head P.E., Yu D.S.. SIRT2 orchestrates the DNA damage response. Cell Cycle. 2016; 15:2089–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim H.S., Vassilopoulos A., Wang R.H., Lahusen T., Xiao Z., Xu X., Li C., Veenstra T.D., Li B., Yu H.et al.. SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell. 2011; 20:487–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Serrano L., Martinez-Redondo P., Marazuela-Duque A., Vazquez B.N., Dooley S.J., Voigt P., Beck D.B., Kane-Goldsmith N., Tong Q., Rabanal R.M.et al.. The tumor suppressor SirT2 regulates cell cycle progression and genome stability by modulating the mitotic deposition of H4K20 methylation. Genes Dev. 2013; 27:639–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Head P.E., Zhang H., Bastien A.J., Koyen A.E., Withers A.E., Daddacha W.B., Cheng X., Yu D.S.. Sirtuin 2 mutations in human cancers impair its function in genome maintenance. J. Biol. Chem. 2017; 292:9919–9931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang H., Head P.E., Daddacha W., Park S.H., Li X., Pan Y., Madden M.Z., Duong D.M., Xie M., Yu B.et al.. ATRIP Deacetylation by SIRT2 Drives ATR Checkpoint Activation by Promoting Binding to RPA-ssDNA. Cell Rep. 2016; 14:1435–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang H., Park S.H., Pantazides B.G., Karpiuk O., Warren M.D., Hardy C.W., Duong D.M., Park S.J., Kim H.S., Vassilopoulos A.et al.. SIRT2 directs the replication stress response through CDK9 deacetylation. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:13546–13551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Minten E.V., Kapoor-Vazirani P., Li C., Zhang H., Balakrishnan K., Yu D.S.. SIRT2 promotes BRCA1-BARD1 heterodimerization through deacetylation. Cell Rep. 2021; 34:108921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yasuda T., Takizawa K., Ui A., Hama M., Kagawa W., Sugasawa K., Tajima K.. Human SIRT2 and SIRT3 deacetylases function in DNA homologous recombinational repair. Genes Cells. 2021; 26:328–335. [DOI] [PubMed] [Google Scholar]
- 55. Zhang M., Du W., Acklin S., Jin S., Xia F.. SIRT2 protects peripheral neurons from cisplatin-induced injury by enhancing nucleotide excision repair. J. Clin. Invest. 2020; 130:2953–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mori E., Davis A.J., Hasegawa M., Chen D.J.. Lysines 3241 and 3260 of DNA-PKcs are important for genomic stability and radioresistance. Biochem. Biophys. Res. Commun. 2016; 477:235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chen W., Liu N., Zhang H., Zhang H., Qiao J., Jia W., Zhu S., Mao Z., Kang J.. Sirt6 Promotes DNA End Joining in iPSCs Derived from Old Mice. Cell Rep. 2017; 18:2880–2892. [DOI] [PubMed] [Google Scholar]
- 58. McCord R.A., Michishita E., Hong T., Berber E., Boxer L.D., Kusumoto R., Guan S., Shi X., Gozani O., Burlingame A.L.et al.. SIRT6 stabilizes DNA-dependent protein kinase at chromatin for DNA double-strand break repair. Aging. 2009; 1:109–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jing H., Hu J., He B., Negron Abril Y.L., Stupinski J., Weiser K., Carbonaro M., Chiang Y.L., Southard T., Giannakakou P.et al.. A SIRT2-Selective Inhibitor Promotes c-Myc Oncoprotein Degradation and Exhibits Broad Anticancer Activity. Cancer Cell. 2016; 29:607. [DOI] [PubMed] [Google Scholar]
- 60. Pierce A.J., Johnson R.D., Thompson L.H., Jasin M.. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999; 13:2633–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Minten E.V., Yu D.S.. Protocol for in vitro lysine deacetylation to test putative substrates of class III deacetylases. STAR Protoc. 2022; 3:101313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kapoor-Vazirani P., Rath S.K., Liu X., Shu Z., Bowen N.E., Chen Y., Haji-Seyed-Javadi R., Daddacha W., Minten E.V., Danelia D.et al.. SAMHD1 deacetylation by SIRT1 promotes DNA end resection by facilitating DNA binding at double-strand breaks. Nat. Commun. 2022; 13:6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rumpf T., Schiedel M., Karaman B., Roessler C., North B.J., Lehotzky A., Olah J., Ladwein K.I., Schmidtkunz K., Gajer M.et al.. Selective Sirt2 inhibition by ligand-induced rearrangement of the active site. Nat. Commun. 2015; 6:6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shanbhag N.M., Rafalska-Metcalf I.U., Balane-Bolivar C., Janicki S.M., Greenberg R.A.. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010; 141:970–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Leahy J.J., Golding B.T., Griffin R.J., Hardcastle I.R., Richardson C., Rigoreau L., Smith G.C.. Identification of a highly potent and selective DNA-dependent protein kinase (DNA-PK) inhibitor (NU7441) by screening of chromenone libraries. Bioorg. Med. Chem. Lett. 2004; 14:6083–6087. [DOI] [PubMed] [Google Scholar]
- 66. North B.J., Marshall B.L., Borra M.T., Denu J.M., Verdin E.. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell. 2003; 11:437–444. [DOI] [PubMed] [Google Scholar]
- 67. Outeiro T.F., Kontopoulos E., Altmann S.M., Kufareva I., Strathearn K.E., Amore A.M., Volk C.B., Maxwell M.M., Rochet J.C., McLean P.J.et al.. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson's disease. Science. 2007; 317:516–519. [DOI] [PubMed] [Google Scholar]
- 68. Park D., Magis A.T., Li R., Owonikoko T.K., Sica G.L., Sun S.Y., Ramalingam S.S., Khuri F.R., Curran W.J., Deng X.. Novel small-molecule inhibitors of Bcl-XL to treat lung cancer. Cancer Res. 2013; 73:5485–5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chaplin A.K., Hardwick S.W., Liang S., Kefala Stavridi A., Hnizda A., Cooper L.R., De Oliveira T.M., Chirgadze D.Y., Blundell T.L.. Dimers of DNA-PK create a stage for DNA double-strand break repair. Nat. Struct. Mol. Biol. 2021; 28:13–19. [DOI] [PubMed] [Google Scholar]
- 70. Chen X., Xu X., Chen Y., Cheung J.C., Wang H., Jiang J., de Val N., Fox T., Gellert M., Yang W.. Structure of an activated DNA-PK and its implications for NHEJ. Mol. Cell. 2021; 81:801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gell D., Jackson S.P.. Mapping of protein-protein interactions within the DNA-dependent protein kinase complex. Nucleic Acids Res. 1999; 27:3494–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sharif H., Li Y., Dong Y., Dong L., Wang W.L., Mao Y., Wu H.. Cryo-EM structure of the DNA-PK holoenzyme. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:7367–7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sibanda B.L., Chirgadze D.Y., Ascher D.B., Blundell T.L.. DNA-PKcs structure suggests an allosteric mechanism modulating DNA double-strand break repair. Science. 2017; 355:520–524. [DOI] [PubMed] [Google Scholar]
- 74. Yin X., Liu M., Tian Y., Wang J., Xu Y.. Cryo-EM structure of human DNA-PK holoenzyme. Cell Res. 2017; 27:1341–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hornbeck P.V., Zhang B., Murray B., Kornhauser J.M., Latham V., Skrzypek E.. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015; 43:D512–D520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Nguyen P., Shukla S., Liu R., Abbineni G., Smart D.K.. Sirt2 Regulates Radiation-Induced Injury. Radiat. Res. 2019; 191:398–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sengupta A., Haldar D. Human sirtuin 3 (SIRT3) deacetylates histone H3 lysine 56 to promote nonhomologous end joining repair. DNA Repair (Amst.). 2018; 61:1–16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.