Significance
The neural retina, which provides visual input to the brain, is encased in a complex set of ocular tissues, generally grouped into the anterior and posterior segments. Defects in these extraretinal structures underlie a major fraction of vision loss. Recent studies used high-throughput single-cell RNA sequencing to generate cell atlases of the human retina and anterior segment. To complete a cell atlas of the eye, the current study analyzed ~151,000 single nuclei from components of the posterior segment: optic nerve, optic nerve head, peripapillary sclera, peripheral sclera, choroid, and retinal pigment epithelium. This atlas provides a strong foundation for investigating the pathogenesis of hundreds of ocular diseases, most of which remain incurable.
Keywords: astrocytes, optic nerve head, glaucoma, lamina cribrosa, choroid
Abstract
Although the visual system extends through the brain, most vision loss originates from defects in the eye. Its central element is the neural retina, which senses light, processes visual signals, and transmits them to the rest of the brain through the optic nerve (ON). Surrounding the retina are numerous other structures, conventionally divided into anterior and posterior segments. Here, we used high-throughput single-nucleus RNA sequencing (snRNA-seq) to classify and characterize cells in six extraretinal components of the posterior segment: ON, optic nerve head (ONH), peripheral sclera, peripapillary sclera (PPS), choroid, and retinal pigment epithelium (RPE). Defects in each of these tissues are associated with blinding diseases—for example, glaucoma (ONH and PPS), optic neuritis (ON), retinitis pigmentosa (RPE), and age-related macular degeneration (RPE and choroid). From ~151,000 single nuclei, we identified 37 transcriptomically distinct cell types, including multiple types of astrocytes, oligodendrocytes, fibroblasts, and vascular endothelial cells. Our analyses revealed a differential distribution of many cell types among distinct structures. Together with our previous analyses of the anterior segment and retina, the data presented here complete a “Version 1” cell atlas of the human eye. We used this atlas to map the expression of >180 genes associated with the risk of developing glaucoma, which is known to involve ocular tissues in both anterior and posterior segments as well as the neural retina. Similar methods can be used to investigate numerous additional ocular diseases, many of which are currently untreatable.
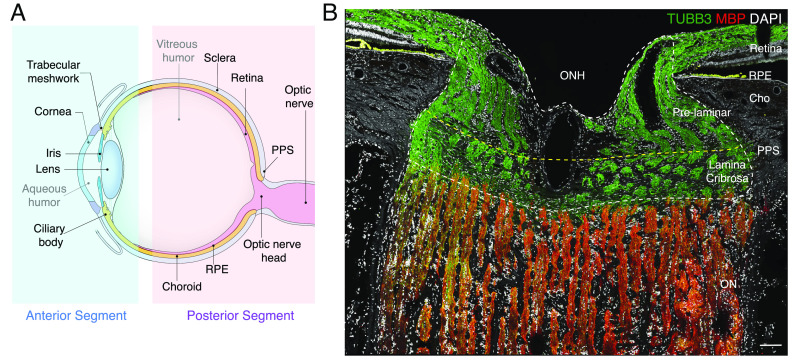
Cells of the neural retina sense light (photoreceptors), process visual signals (interneurons), and transmit them to retinal output neurons (retinal ganglion cells, RGCs), which pass the signals to the brain through the optic nerve (ON). The retina does not, however, act alone: It is surrounded by a complex set of ocular tissues, which are conventionally divided into those of the anterior and posterior segments (Fig. 1A). The anterior segment includes the cornea, iris, and lens, which ensure that focused light reaches the photoreceptors; and the ciliary body, ciliary muscle, trabecular meshwork, and Schlemm’s canal, which produce and drain aqueous humor that nourishes avascular structures, clears away waste, and maintains intraocular pressure (IOP) (1). The posterior segment includes, in addition to the neural retina, the ON and its specialized proximal region, the optic nerve head (ONH); the sclera and a specialized region, peripapillary sclera (PPS), that encircles the ONH; retinal pigment epithelium (RPE), which supports the health and function of photoreceptors; and the choroid/choriocapillaris, which is the vascular supply that supports photoreceptors and RPE (Fig. 1A) (2, 3).
Fig. 1.
Anterior and posterior tissues of the human eye. (A) Diagram of the human eye and the optic nerve, depicted in sagittal cross-section. Structures represented in the ocular cell atlas are labeled. (B) Section of the optic nerve head (ONH) and surrounding tissues immunostained for myelin basic protein (MBP, red) and beta-tubulin (TUBB3, green). TUBB3 highlights bundles of axons in the retina and ONH, and MBP highlights myelinating oligodendrocytes in the optic nerve (ON). The scale bar shows 100 µm. PPS, peripapillary sclera; RPE, retinal pigment epithelium; Cho, choroid.
All of these structures have been implicated in conditions that impact vision. In the anterior segment, they include cataracts (lens), corneal dystrophies (cornea), uncorrected refractive error (cornea and lens), and anterior uveitis (iris/ciliary body), trachoma and onchocerciasis (cornea and conjunctiva) (4). Diseases of the posterior segment include optic neuritis, a frequent manifestation of multiple sclerosis (ON), posterior uveitis and scleritis (choroid/sclera), and diabetic retinopathy (retina) (5). Moreover, many blinding diseases involve multiple tissues—for example, trabecular meshwork, ONH, PPS and RGCs for glaucoma; photoreceptors, and RPE for retinitis pigmentosa; and photoreceptors, RPE and choroid for age-related macular degeneration (6–8). Thus, elucidating the pathogenic mechanisms underlying ocular diseases requires a comprehensive understanding of all ocular cell types and their interactions.
In previous studies, we used high-throughput single-cell and single-nucleus RNA sequencing (scRNA-seq and snRNA-seq) to catalog and molecularly characterize the cell types of the human anterior segment (9, 10). We and others also used scRNA-seq and snRNA-seq to characterize the adult human neural retina and RPE (11–19). Here, to complete a cell atlas of the human eye, we used snRNA-seq to analyze the extraretinal tissues of the posterior segment. We profiled ~151,000 nuclei from the ON, ONH, PPS, RPE, choroid, and sclera, identified 37 cell clusters, and characterized the transcriptome of each. By dissecting tissues prior to profiling, we were able to better assess the tissue distribution of each cell type, identifying some types shared among tissues and others with striking tissue specificity.
Finally, to assess the expression of genes implicated in ocular diseases, we focused on glaucoma, which is the leading cause of irreversible blindness worldwide (20). As noted above, multiple tissues are involved in this disease. The major modifiable risk factor for glaucoma is increased IOP, which is usually ascribed to defects in the outflow pathways of the anterior segment. Increased pressure leads to stress on the PPS and ONH, resulting in reactions of astrocytes and other cells in the lamina cribrosa (LC), a sieve-like structure in the ONH through which RGC axons pass (Fig. 1B) (21–23). These reactions, along with remodeling of the extracellular matrix, cause or exacerbate compression of RGC axons. The compression then leads to dysfunction and eventual death of the RGCs which is the proximate cause of glaucomatous vision loss (7, 21, 24). To date, at least 13 genes have been associated with early onset or congenital forms of glaucoma (21, 25–29). In addition, large GWAS (genome-wide association study) meta-analyses have associated 127 genomic loci with primary open-angle glaucoma (POAG; the most common late-onset form of glaucoma) and 112 loci with IOP (30, 31). As an initial step in disentangling these interactions, we mapped the expression of >180 genes implicated in the pathogenesis of glaucoma or IOP regulation in the cells of the anterior and posterior segments.
Results
We profiled 36 tissue samples from 19 human donors with no history of ocular disease: 8 ON, 13 ONH, 4 PPS, 3 peripheral sclera, and 8 choroid/RPE (Fig. 2A and Table 1 and SI Appendix, Table S1). Altogether, we recovered ~210,000 single-nucleus transcriptomes that we judged to be high quality based on the numbers of transcripts recovered and genes detected (SI Appendix, Fig. S1). Initial analysis revealed that ~60,000 of these transcriptomes were derived from the neural retina present within all samples except those from the ON; this was expected, given their proximity to other structures and limitations inherent to the manual dissection technique. We removed RPE (~10,000) and retinal cells from the dataset. Neural retinal cells were not analyzed further, and RPE was analyzed separately. We reclustered the remaining ~140,000 cells from all five tissue sources together (Fig. 2B).
Fig. 2.
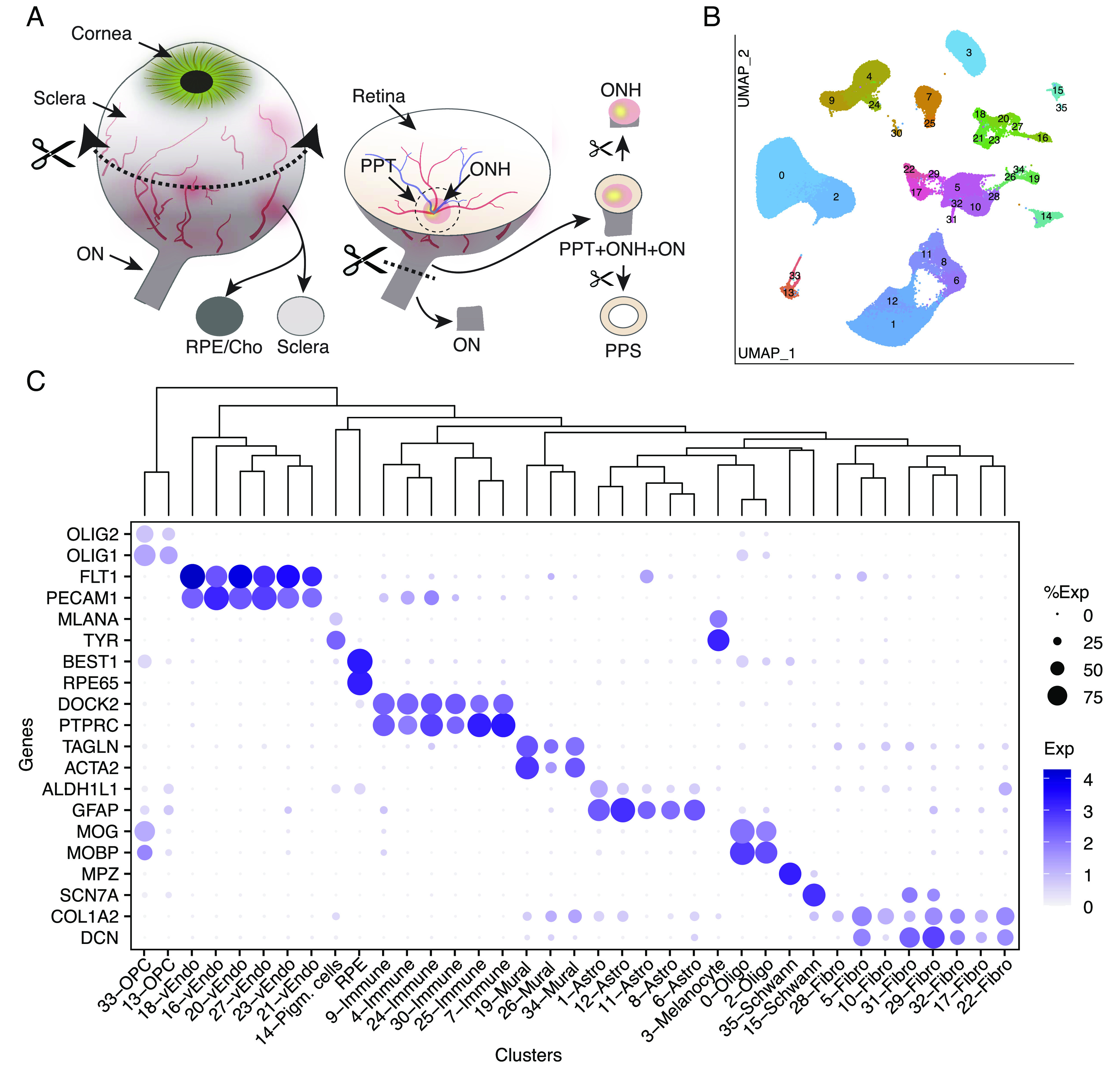
Collection of tissues from the posterior segment and initial transcriptomic analysis. (A) Schematic showing how tissues were dissected. Circumferential incision at the pars plana separated anterior and posterior segments, and tissues were dissected from the posterior segment using fine scissors and trephine tissue punches. (B) Clustering of single-nucleus expression profiles from all tissues visualized by UMAP. Each tissue was processed separately. RPE and cells from neural retina were removed before the remaining ~140 k nuclei were pooled together to generate the UMAP. (C) Dot plot showing genes selectively expressed by each cell type. In this and subsequent figures, the size of each circle is proportional to the percentage of nuclei within a cluster expressing the gene, and the color intensity depicts the average normalized transcript count in expressing cells. The dendrogram above the graph shows transcriptional relationships among cell types. ON, optic nerve; ONH, optic nerve head; PPT, peripapillary tissues; PPS, peripapillary sclera; RPE, retinal pigment epithelium; Cho, choroid; Oligo, oligodendrocytes; Endo, vascular endothelium; OPC, oligodendrocyte progenitor cell.
Table 1.
Summary of the human eye atlas “v1”
| Tissue | No. Donors | No. Cells/nuclei | No.Types* |
|---|---|---|---|
| Cornea | 5 | 37,485 | 7 |
| Lens | 5 | 13,900 | 5 |
| Iris | 4 | 57,422 | 9 |
| Ciliary body | 4 | 34,132 | 10 |
| Trabecular meshwork | 4 | 39,736 | 13 |
| Corneoscleral wedge | 4 | 36,573 | 27 |
| Foveal retina | 5 | 56,931 | 62 |
| Macular retina | 17 | 129,775 | 98 |
| Peripheral retina | 10 | 55,004 | 98 |
| Peripapillary retina | 17 | 59,001 | 98 |
| ONH | 13 | 41,178 | 32 |
| Peripapillary tissue | 4 | 4,812 | 31 |
| Sclera | 3 | 4,263 | 18 |
| Choroid | 8 | 36,122 | 22 |
| RPE | 8 | 11,422 | 1 |
| Optic nerve | 7 | 53,603 | 16 |
| Total | 29 | 671,359 | ~157 |
*Types that composite >=0.1% of total cells from the tissue.
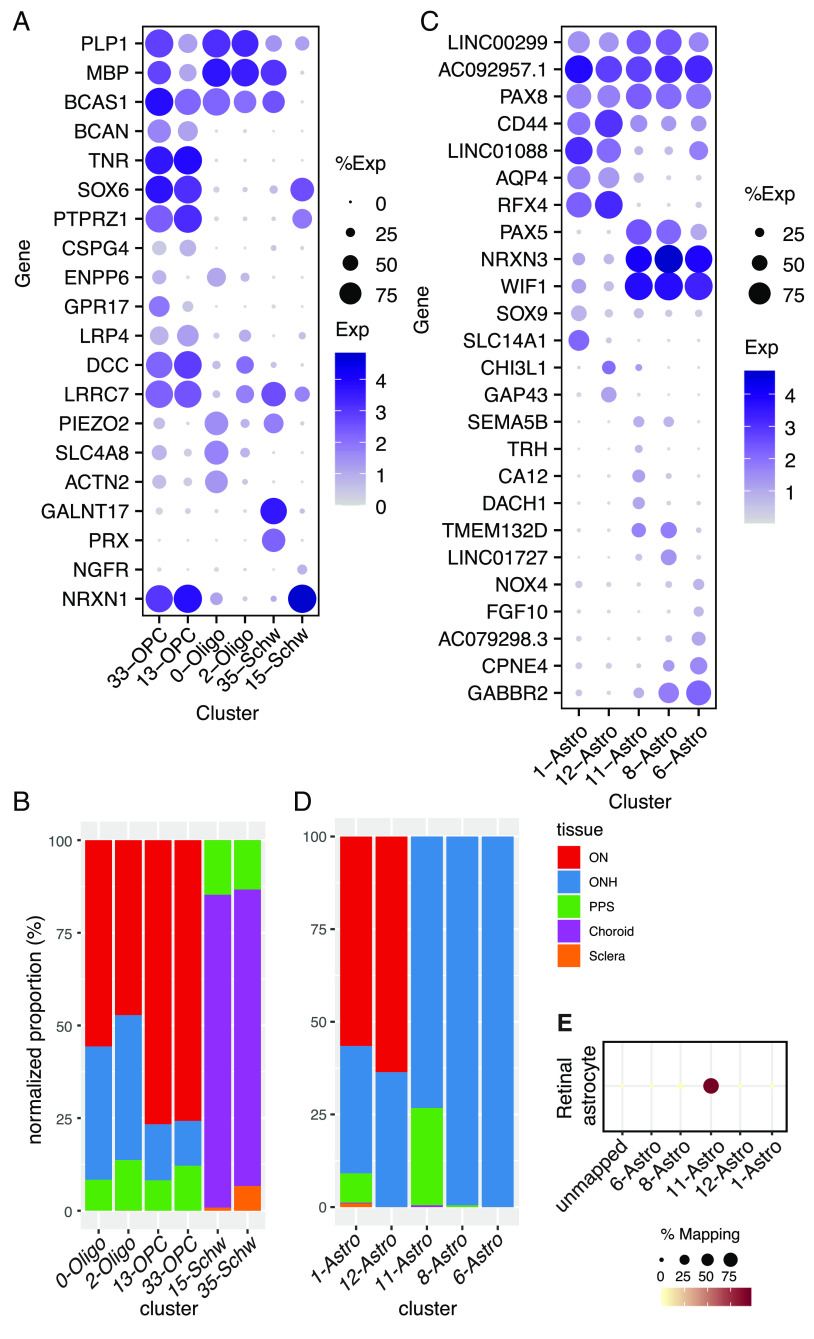
Altogether, we identified 37 clusters, all derived from multiple samples (SI Appendix, Table S2). All but RPE clusters are shown in a Uniform Manifold Approximation and Projection (UMAP) representation in Fig. 2B and numbered in the order of size (C0 has the most cells, C35 the fewest). We assigned them cell class labels based on selective expression of known markers (Fig. 2C) identifying five clusters as astrocytes, two each as oligodendrocytes, oligodendrocyte precursors, and Schwann cells, eight as fibroblasts, six as vascular endothelium, three as mural (blood vessel–associated) cells, six as immune-related cells (microglia, macrophages, and lymphocytes), and three as pigmented cells. As shown by a dendrogram representing transcriptomic similarity, types within a class were close transcriptomic relatives (Fig. 2 C, Top).
Oligodendrocytes and Their Precursors.
Two closely related clusters (C0 and C2) corresponded to mature oligodendrocytes. Both expressed markers of myelinating oligodendrocytes such as MOBP and MOG (Fig. 2C) (32, 33). They were, however, distinguishable by selective expression of multiple genes, including ACTN2 and SLC4A8, enriched in C0, and LRRC7 and DCC, enriched in C2 (Fig. 3A). Note, however, that for these cell types and others described below, genes that reliably distinguish types within a class are often expressed at similar or higher levels in other classes or structures (SI Appendix, Fig. S2). A comparison of the two oligodendrocyte populations to those described in human brain white matter by Jakël et al. (34) showed that C0 and C2 resembled their mature oligodendrocyte subtypes 1 and 5, respectively.
Fig. 3.
Glial cells. (A) Dot plot showing the expression of selected genes in oligodendrocyte, oligodendrocyte precursor cell (OPC), and Schwann cell clusters based on canonical markers (Fig. 2C). (B) Histogram showing the relative abundance of oligodendrocytes (Oligo), OPC, and Schwann clusters in five tissues. (C) Dot plot showing genes selectively expressed by all or some astrocyte types. (D) Histogram showing the relative abundance of each type of astrocyte in each tissue. (E) Transcriptional relationship of the astrocyte types identified in this study to retinal astrocytes profiled in van Zyl et al. (9). The color and size of each dot reflect the percentage of cells in retinal astrocytes (column) mapped to a corresponding type of astrocyte in the ON/ONH (rows).
As expected, both types were most abundant in optic nerve samples and nearly absent from the sclera and choroid (Fig. 3B). However, 33% of C0 oligodendrocytes and 39% of C2 oligodendrocytes were derived from ONH samples, likely because those samples extended into the intraorbital portion of the ON (Fig. 2A). To test this idea, we stained sections of the optic nerve for myelin basic protein (MBP) which is an oligodendrocyte marker. Antibodies against this protein exclusively stained the myelinated portion of the ON (Fig. 1B), supporting the idea that oligodendrocytes recovered from ONH samples were derived from the ON. A small proportion of the C0 and C2 oligodendrocytes also appeared to be derived from the PPS samples, likely a result from the difficulty of completely separating the tightly apposed ONH and PPS during dissection.
Cells in the other two clusters (C13 and C33) expressed known markers of oligodendrocyte precursor cells (OPCs) such as OLIG1 and OLIG2 (33) (Fig. 2C). Although they were each other’s closest relatives, C33 was distinguishable from C13 by higher expression of several genes, including GPR17, ENPP6, and PLP1, which have been shown to be expressed by premyelinating oligodendrocytes (32) (Fig. 3A). Thus, C13 cells may be conventional OPCs, while C33 may correspond to cells beginning to differentiate from OPCs to oligodendrocytes. Like oligodendrocytes, OPCs were most abundant in the ON. OPCs were less abundant than mature oligodendrocytes in the ONH (Fig. 3B), suggesting that they may be concentrated more distally along the ON.
Schwann Cells.
Clusters C15 and C35 corresponded to Schwann cells, the glia of the peripheral nervous system. Both were predominantly derived from the choroid, with minor contributions from the PPS and sclera (Fig. 3B). This distribution reflects the innervation of choroidal vessels by autonomic neurons (2). We identify C15 and C35 as comprising nonmyelinating and myelinating Schwann cells, respectively. C35 but not C15 expressed high levels of PRX, MBP, and MPZ which encode myelin proteins expressed by Schwann cells but not oligodendrocytes (Figs. 2C and 3A). In contrast, C15 is rich in SCN7A, NCAM1, L1CAM, and NGFR (Figs. 2C and 3A and Dataset S1), a profile known to represent nonmyelinating Schwann cells (35, 36).
Astrocytes.
Five clusters (C1, 6, 8, 11, and 12) were close transcriptomic relatives and expressed known astrocyte markers including GFAP and ALDH1L1 (Fig. 2C). Additional markers that distinguished astrocytes from other cell classes included PAX8 and the noncoding RNAs AC092957.1, LINC00299, and JAKMIP2-AS1 (Fig. 3C and SI Appendix, Fig. S3A and Dataset S1).
Comparisons among astrocyte clusters revealed distinct expression patterns and locations. The largest, C1, comprised ~64% of all astrocytes. C1 and C12 were both most abundant in the optic nerve and expressed AQP4 at high levels (Fig. 3 C and D). They were present at far higher levels in the myelinated parts of the ON than ONH (SI Appendix, Fig. S3B), consistent with a previous report on AQP4 expression (37). Despite similarities in their transcriptomic profile and tissue distribution, they were molecularly distinct; C1 selectively expressed SLC14A1 and SOX9, whereas C12 expressed higher levels of GAP43 and CHI3L1 (Fig. 3C).
Three clusters, C6, 8, and 11, were derived largely from the ONH. All expressed WIF1 and PAX5 (Fig. 3C and SI Appendix, Fig. S3D) but differentially expressed other genes that marked them as distinct types or states. For example, DACH1 was expressed at the highest levels in C11, LINC01727 in C8, and FGF10 in C6 (Fig. 3C).
Whereas ~99% of cells in C6 and C8 were derived from the ONH, 26% of C11 cells were derived from the PPS after sample-size normalization (Fig. 3D). Based on transcriptomic similarity, we identified C11 as most similar to retinal astrocytes (12) (Fig. 3E), indicating that this cluster might be derived from retinal contamination, which was prominent in both ONH and PPS samples.
Fibroblasts.
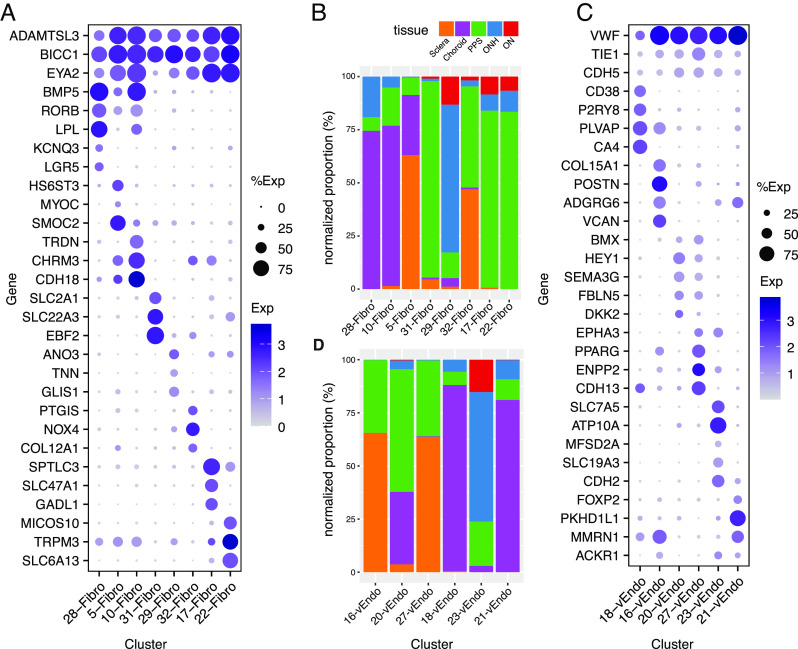
Eight transcriptomically related clusters (C5, 10, 17, 22, 28, 29, 31, and 32) were classified as fibroblasts, based on their expression of known markers such as COL1A1 and DCN (Fig. 2C) (38). This heterogeneity is consistent with numerous reports that have characterized multiple fibroblast types in single tissues (9, 39). The majority of cells in seven of the clusters were derived from a single tissue: C10 and 28 from the choroid, C17, 22, and 31 from the PPS, C5 from the sclera, and C29 from the ONH; the eighth, C32, was derived to a similar extent from the peripheral and PPS (Fig. 4B and SI Appendix, Table S3).
Fig. 4.
Fibroblasts and vascular endothelial cells. (A) Dot plot showing genes selectively expressed by all or each fibroblast cluster. (B) Histogram showing the relative abundance of each type of fibroblasts in each tissue. (C) Dot plot showing genes selectively expressed by all or each vascular endothelial cell type. (D) Histogram showing the abundance of each endothelial cell type in each tissue.
Differentially expressed genes included HS6ST3 and SMOC2 by C5; TRDN by C10; SLC47A1 and GADL1 by C17; TRPM3 and SLC6A13 by C22; KCNQ3 and LGR5 by C28; ANO3 and TNN by C29; SLC22A3 and EBF2 by C31; and PTGIS and COL12A1 by C32 (Fig. 4A). Because fibroblasts are major sources of extracellular matrix (ECM), we mapped expression of ECM genes in these clusters, with emphasis on those reported to be present in the LC (SI Appendix, Fig. S5B; Discussion). Comparison of posterior segment fibroblasts to those of the anterior segment (9) suggested that all except C31 were most similar to the anterior segment’s ciliary and/or scleral fibroblasts (SI Appendix, Fig. S4A). This similarity may reflect the fact that the ciliary body and choroid are contiguous subdivisions of the uveal layer, and the scleral tissue encases the globe from its anterior-most aspect at the limbus to its posterior-most aspect encircling the optic nerve. C31 mapped to a type derived from the corneoscleral wedge, which we had termed “Fibro x” (9).
Vascular Endothelium.
Six clusters (C16, 18, 20, 21, 23, and 27) were identified as vascular endothelial cells based on the expression of markers such as PECAM1 and FLT1 (Fig. 2C). Using immunohistochemistry, we localized Von Willebrand factor (VWF)+ endothelial cells in all tissues (SI Appendix, Fig. S4 B and C). C16 and C27 were derived predominantly from the sclera (including the PPS); C18 and C21 were derived primarily from choroid; and C20 and C23 were derived from multiple tissues. C23 was the only cluster with a major contribution from the ONH (>60%) and ON (>15%).
Based on expressed markers, we tentatively assign these cells to different vessel types. C16 cells expressed markers of venules including ADGRG6, MMRN1, and POSTN; C20 and C27 cells were enriched in the expression of artery/arteriole markers such as SEMA3G, HEY1, and BMX; and C18 and C21 were enriched in the expression of genes previously shown to be associated with fenestrated capillaries, including PLVAP and CA4 in C18, and postcapillary venules including PKHD1L1 and MMRN1 in C21 (38, 40–42). Mixed expression patterns in C21 and C23 (for example, expression of the venule marker, ACKR1) suggested that these cells might be derived from both venules and capillaries (42) (Fig. 4C and SI Appendix, Fig. S4C).
Mural Cells.
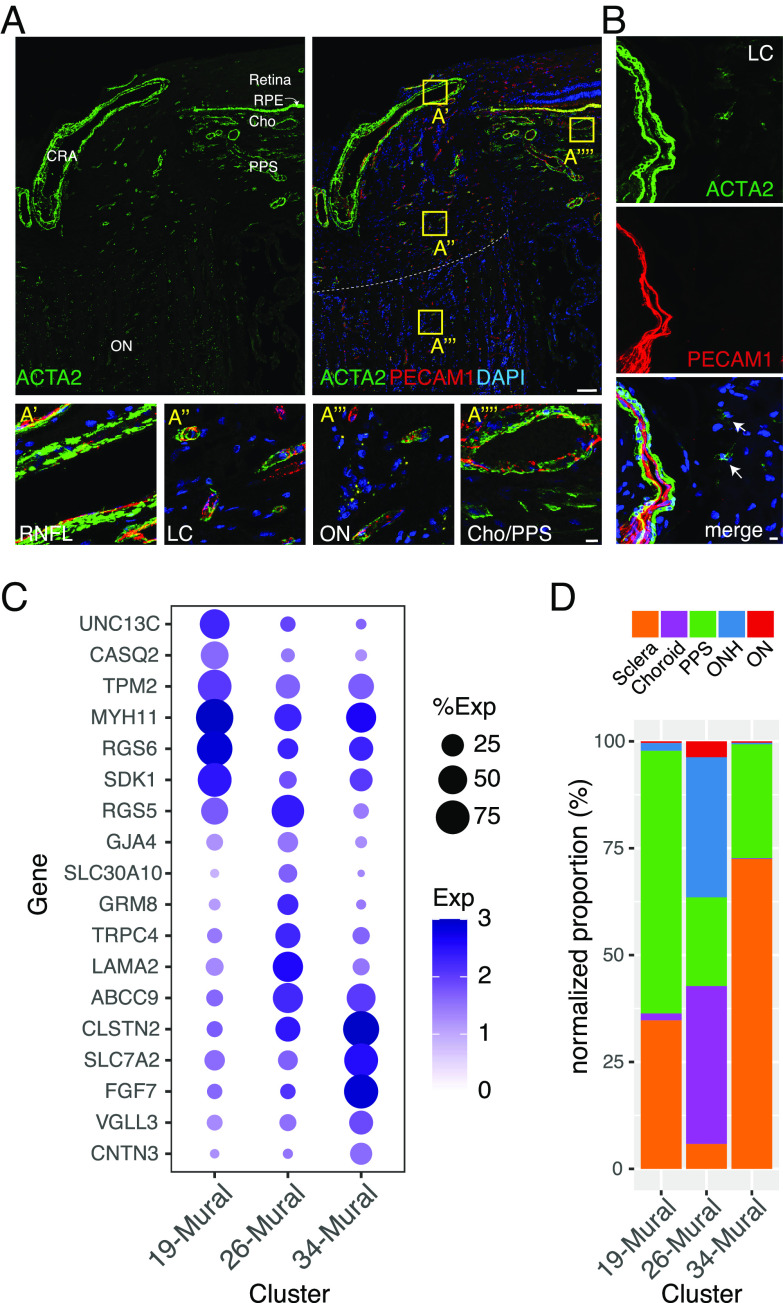
Smooth muscle cells of the microvasculature, which primarily ensheath arterioles, and pericytes, which ensheath capillaries and venules, are collectively called mural cells (43). C19, 26, and 34 were identified as mural cells based on their expression of known markers such as TAGLN and ACTA2 (40, 44–46) (Fig. 2C). Consistent with this assignment, ACTA2+ cells were closely associated with PECAM+ vascular endothelial cells (Fig. 5 A and B).
Fig. 5.
Mural cells. (A and B) Immunostaining for ACTA2 (green) and/or PECAM1 (red) in the whole ONH (A) and LC region (B). Boxed areas in A are shown at higher magnification in lower panels. (C) Dot plot showing genes selectively expressed by cells in each mural cell cluster. (D) Histogram showing the abundance of the three mural types in each tissue. CRA, central retinal artery; RNFL, retinal nerve fiber layer. Bars show 100 µm in top panel A and 10 µm in B and lower panels of A.
The three mural cell types were further distinguishable by selective expression of multiple genes including UNC13C, RGS6, and CASQ2 in C19, TRPC4, RGS5, and LAMA2 in C26, and FGF7, SLC7A2, and CLSTN2 in C34 (Fig. 5C and SI Appendix, Fig. S5A). The expression profiles of the mural cells suggest that C19 and C34 cells are vascular smooth muscle cells, and C26 cells are pericytes. Consistent with these assignments, C19 and C34 were derived primarily from the PPS and sclera, similar to that of the putative arteriolar endothelial cells, and C26 was derived in large part from the choroid and ONH, similar to that of the putative capillary and venule-associated endothelial types (Figs. 4D and 5D). C26 corresponds to a LAMA2+, TRPC4+ pericyte type we recently identified in the anterior segment (9) (Fig. 5C).
Immune Cells and Microglia.
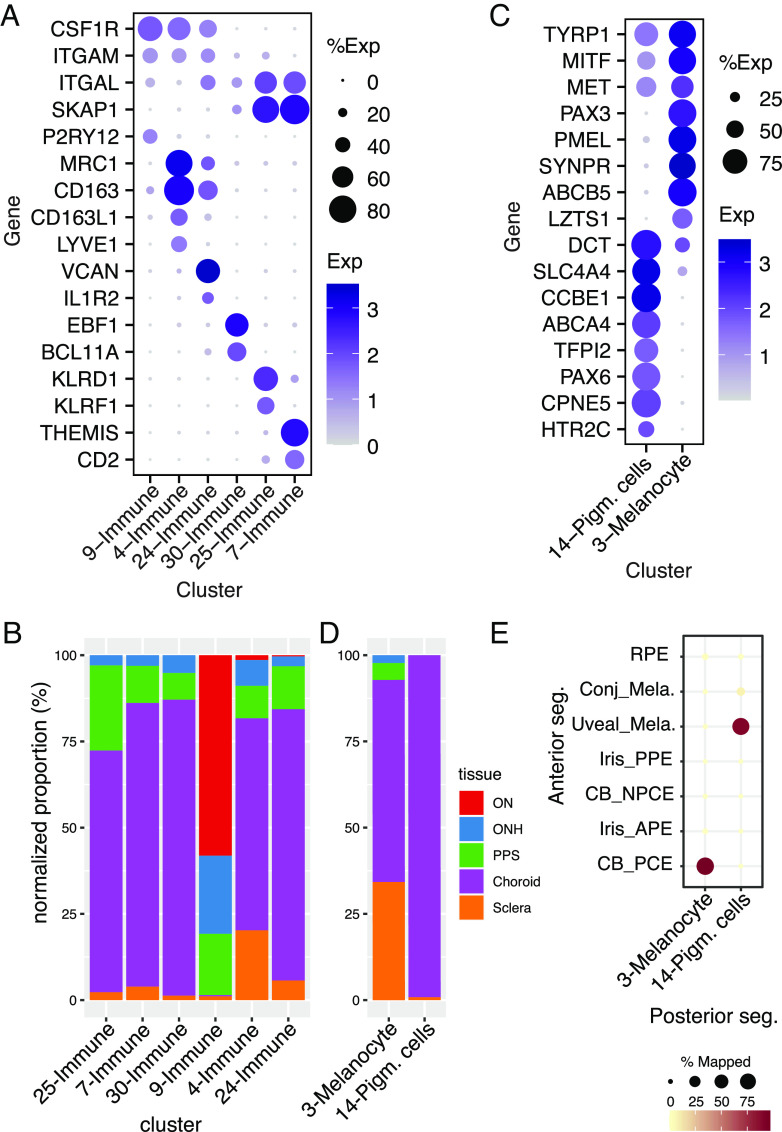
Six clusters (C4, 7, 9, 24, 25, and 30) expressed markers of immune cells including PTPRC and DOCK2 (47, 48) (Fig. 2C). Cluster 9 expressed numerous markers of microglia (e.g., P2RY12, and ITGAM; Fig. 6A). C4 and C24 expressed markers of macrophages (e.g., MRC1 and CD163) but could be differentiated from each other by selective expression of IL1R2, and VCAN in C24, and CD163L1, and LYVE1 in C4 (49, 50) (Fig. 6A). We identified C7 as T lymphocytes (e.g., CD2+ and THEMIS+), C25 as Natural Killer (NK) cells (e.g., KLRD1+ and KLRF1+), and C30 as B lymphocytes (e.g., EBF1+ and BCL11A+) (51, 52) (Fig. 6A).
Fig. 6.
Immune and pigmented cells. (A) Dot plot showing genes selectively expressed by each of the immune-related cell types. (B) Histogram showing the relative abundance of each immune-related cluster in each tissue. (C) Dot plot showing genes selectively expressed by each type of melanocytes. (D) Histogram showing the abundance of the pigmented cell clusters C3 and C14 in each tissue. (E) Confusion matrix showing transcriptional correspondence of C3 and C14 in the current dataset to those in the anterior segment described in van Zyl et al. (9) and to RPE.
Most (80%) of the microglia were derived from the ON and ONH, as expected from their known association with neural tissues. Consistent with this assignment, AIF1+ cells were abundant in the retina, ONH, and ON but rare in other tissues (SI Appendix, Fig. S5C). In contrast, >95% of the other types were derived from peripheral and PPS and choroid (Fig. 6B).
Pigmented Cells.
Two cell types, C3 and C14, expressed genes characteristic of pigmented cells such as TYR and MLANA (Fig. 2C). They could, however, be distinguished from each other by the expression of genes including PMEL and PAX3 in C3 and PAX6 and CCBE1 in C14 (53) (Figs. 2C and 6C and SI Appendix, Fig. S6). Almost all C14 cells and 71% of C3 cells were derived from choroid (Fig. 6D).
We identify C3 cells as melanocytes, based on their tissue source and expression pattern (Fig. 6 and SI Appendix, Table S3 and Dataset S1). Two types of melanocytes have been characterized in the anterior segment—a uveal type that resides in the iris and ciliary body and a conjunctival type that populates the conjunctiva (9). The C3 melanocytes were more closely related to the uveal than to the conjunctival type (Fig. 6E). C14 cells, in contrast, were more similar to pigmented epithelial cells of the ciliary body than to melanocytes (9), both in terms of overall transcriptomic similarity and differentially expressed genes (Fig. 6 C and E). To our knowledge, cells of this type have not been described in the posterior segment. However, because they were obtained from peripheral samples, it is possible that they are derived from a transitional zone between the contiguous RPE and ciliary body pigmented epithelial layer.
A third pigmented cell type, RPE, was derived largely from choroid/RPE samples but was also present as contaminants in sclera and ONH samples. RPE pigment is generated by tyrosinase, but its levels are known to be low (54). The RPE dataset included >10,000 single nucleus transcriptomes from peripheral samples. Their transcriptomes generally resembled those reported in recent single-cell studies (15, 17–19, 55–57), although our dataset is substantially larger. Along with known markers, we found several previously unreported markers that are selectively expressed by RPE even when compared to all known ocular cell types (SI Appendix, Fig. S6 and Table S3).
We also profiled >1,200 RPE nuclei from a macular sample and mapped genes differentially expressed between the macula and periphery (SI Appendix, Table S4). They include WFDC1, CXCL14, and CALCB enriched in macular RPE and PMEL and IGFBP5 enriched in peripheral RPE. Many of these genes were also reported to be differentially expressed in previous studies (15, 17–19, 55–58).
Expression of Glaucoma-Associated Genes.
We previously generated cell atlases of the human neural retina and ocular anterior segment (9, 10, 12, 59). Together with the results presented above, they comprise a whole human eye cell atlas (Table 1).
A major motivation for generating this atlas was to map the expression of genes implicated in ocular diseases. As an illustration, we focus on glaucoma for two reasons. First, the most common form of glaucoma, POAG, is a leading cause of irreversible blindness worldwide (20). Second, as detailed in the Introduction, cells and structures throughout the eye have been implicated in its pathogenesis. Over 200 genomic loci have been associated with POAG risk and/or IOP levels by GWAS analyses, together nominating >212 genes in the pathogenesis of glaucoma, increased IOP (which is easily measured), or both (25, 30, 31, 60). Here, we mapped the expression of the nearest gene/s to the GWAS loci for POAG risk or IOP regulation, along with rare Mendelian causes of POAG, in all ocular cell types that we have identified.
As shown in SI Appendix, Fig. S7, at least some genes are expressed at significant levels in every ocular cell type, and most are expressed in multiple types. To seek expression patterns that correlate with disease features, we focused on cell types suspected to be involved in glaucoma and assessed the expression of four groups of genes: 1) genes implicated in POAG susceptibility but not IOP regulation (“POAG only”), 2) genes implicated in IOP regulation but not POAG susceptibility (“IOP only”), 3) genes implicated in normal-tension glaucoma, in which glaucoma progresses in the absence of increased IOP (normal tension glaucoma, NTG), and 4) genes associated with early onset or congenital glaucoma (“early onset”) (SI Appendix, Table S5).
We calculated two disease gene scores: one that assesses the cell type specificity of each disease gene group in each cell type (z score, Fig. 7A) and another that evaluates the enrichment of genes from each disease group in each cell type compared to all glaucoma-related genes (ratio, Fig. 7B). (Expression of the 44 genes associated with both POAG and IOP regulation is not displayed in these graphs but is provided in SI Appendix, Fig. S7B). For 14 of the 17 cell types (or small groups of related types), the two methods gave consistent results (Fig. 7). Genes in the early-onset group, many being monogenic causes of large effect, were expressed at the highest levels in the three main cell types of the outflow pathways, which we have called trabecular meshwork, iris, and ciliary fibroblasts. (The names “ciliary” and “iris” reflect the presence of these types in adjacent structures as well as the trabecular meshwork.)
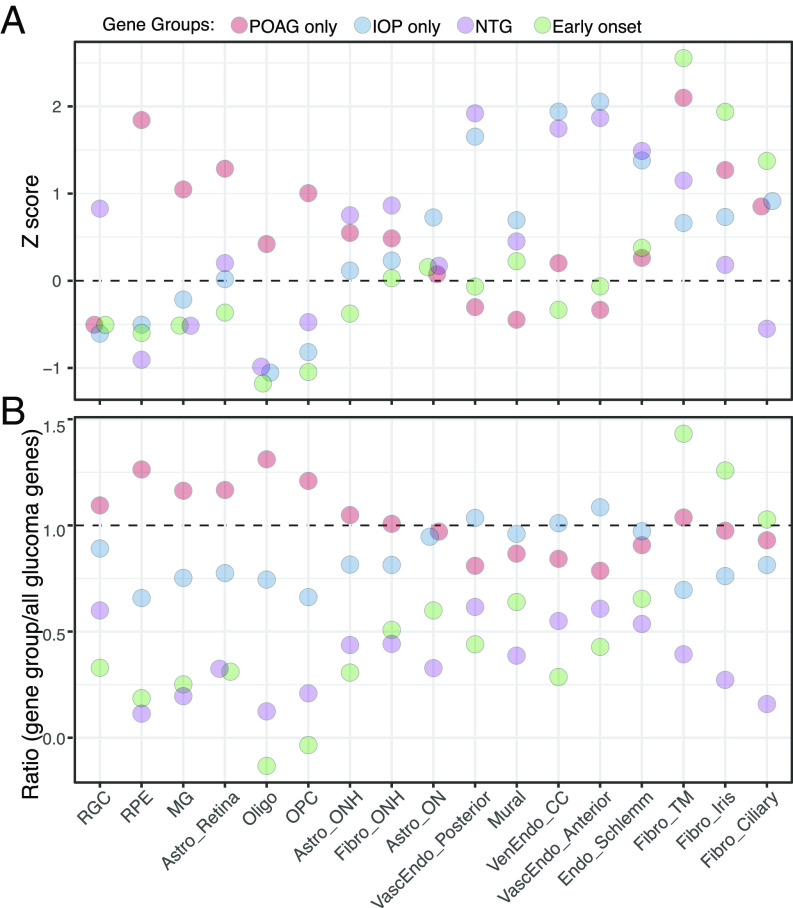
Fig. 7.
Expression of genes implicated in glaucoma. (A) Cell-type specific enrichment z scores of groups of genes that have been associated with POAG but not IOP, IOP but not POAG, normal tension glaucoma (NTG), or early-onset/congenital glaucoma. (B) The ratio of cell-type specific enrichment scores of each gene group to cell-type specific enrichment scores of all glaucoma-associated genes (shown in SI Appendix, Fig. S7B and Table S5). CC, collector channel; MG, Müller glia, Vasc, vascular; Ven, venous; Endo, endothelium.
Genes in the IOP-only group demonstrated the highest cell type specificity in vessel-associated cells (three vascular endothelial types and mural cells) as well as in the endothelium of Schlemm’s canal and in astrocytes of the optic nerve. Genes in the NTG group were also expressed at high levels in endothelial types as assessed by z score. Genes in the POAG-only group were expressed at highest levels, compared to other gene groups, in retinal cells—RPE, retinal astrocytes, and Müller glia—as well as in oligodendrocytes, oligodendrocyte precursors, and astrocytes of the optic nerve. However, POAG-only genes also demonstrated high cell type specificity in fibroblasts of the trabecular meshwork.
For the three remaining cell types—RGCs, ONH astrocytes, and ONH fibroblasts—the two methods give different results. When compared among cell types, the genes implicated in NTG exhibited cell type specificity in these three cell types. In contrast, when compared to the expression of all glaucoma-related genes, those in the POAG-only group were expressed at the highest relative levels in all three cell types. The significance of these patterns is discussed below.
Discussion
Our study was motivated in large part by the knowledge that defects in the posterior segment have been implicated in the pathogenesis of many ocular diseases (5, 7, 21, 23). However, although the posterior segment of primates, including humans, has been studied intensively using histological and histochemical methods (2, 61–66), knowledge of the cell types it contains and the genes that each type expresses remains incomplete. We therefore used a high-throughput single nucleus RNA sequencing method to generate an atlas of cell types in six tissues of the posterior segment: ON, ONH, PPS, peripheral sclera, choroid, and RPE. In conjunction with our previous single-cell analyses of the anterior segment and neural retina (9, 10, 12, 59) and those of others (11–19, 55, 67–70), data presented here result in a first-pass cell atlas of the entire human eye (Table 1).
Lamina Cribrosa.
The LC region is a specialized region of the ONH that provides channels through which RGC axons enter the ON (21). Its two main compartments are bundles of axons that are unmyelinated proximal to the ON and instead ensheathed by astrocytes, and interstitial areas rich in extracellular matrix within which fibroblast-like cells (LC cells) are embedded (21, 23, 71). As a structure contiguous with the wall of the eye, it is exposed to different pressures from inside and outside the globe. Increased IOP leads to increased strain in the LC and the PPS with which it is continuous, leading to alterations in astrocytes, remodeling of the extracellular matrix, and possible deleterious effects on the vasculature that supplies the region (21). Normal tension glaucoma, in which there is no measurable abnormality of IOP, may result from increased susceptibility to the basal level of pressure (24).
Human LC cells have been tentatively identified in cultures prepared from the ONH and studied intensively because of their importance in the pathogenesis of glaucoma (23, 64, 71–73). However, few markers have been found that label putative LC cells in situ, and we were unable to definitively annotate any cluster in our dataset as LC cells. Additionally, although previous reports proposed that ACTA2 is a marker for LC cells (71, 72), most ONH-derived ACTA2+ cells in the ONH are vessel-associated mural cells (pericytes and smooth muscle; Fig. 5A). Weakly ACTA2+ cells were also present (Fig. 5B), but they were predominantly present in the sclera (SI Appendix, Fig. S5D).
We therefore assessed the distribution of cells in our dataset, based on the presumption that if LC cells comprised a unique population, we would expect them to be largely confined to the ONH or, given the likelihood of contamination, present at low abundance in the PPS. Three cell types stood out in this respect. Two were astrocytes (C6 and 8; Fig. 3D), which are, by definition, not LC cells. The third, fibroblast type C29, was enriched in but not confined to the ONH: nearly 70% of the cells in this cluster derived from the ONH, with another ~10% from the PPS and ~20% from the ON (Fig. 4B).
Two additional results make C29 an intriguing candidate. First, following the suggestion of Paula et al. (23) that LC cells in the ONH share numerous structural and functional features with juxtacanalicular cells in the trabecular meshwork, we compared them to types characterized in our cell atlas of the anterior segment (9, 10). Indeed, C29 cells resembled ciliary fibroblasts, which are present in the trabecular meshwork as well as the ciliary body (SI Appendix, Fig. S4A). Second, we compared the expression of genes in cell types of the ONH with those documented in cultured LC cells (71, 72, 74) (SI Appendix, Fig. S5B). We relied on culture cells because of the paucity of analysis in vivo. C29 fibroblasts expressed most of the genes encoding known LC-associated molecules (SI Appendix, Fig. S5B). Notably, however, in neither respect—gene expression or relationship to anterior segment—were C29 fibroblasts distinguished from those in other clusters.
Together, these results are consistent with two hypotheses. First, C29 fibroblasts may be the elusive LC cells. Alternatively, what have been referred to as LC cells may in fact be not a single type but rather a heterogeneous group that includes fibroblasts, pericytes, astrocytes, and endothelial cells. Indeed, these cell types are likely to have been present in LC cultures, and, to our knowledge, no studies of such cultures have shown that any gene or protein marks the entire population.
Glaucoma.
Genetic studies of POAG have identified numerous loci associated with disease risk, most of which are associated with increased IOP, which is a major risk factor for glaucoma (25, 30, 31, 60). We asked whether expression patterns of genes in these loci could provide insight into subsets of glaucoma, including normal tension and early-onset (generally familial and monogenic) glaucoma. We also queried the expression of two subsets of genes identified by GWAS: those associated with POAG but not increased IOP and those associated with increased IOP but not POAG. Remarkably, genes in these four groups were expressed preferentially by different cell types.
Genes implicated in congenital glaucoma (e.g., LTBP2, CYP1B1, and PITX2) were expressed at the highest levels in three fibroblast types that are present in the iridocorneal angle and trabecular meshwork. This pattern is consistent with the current understanding of the contribution of these cells to angle and outflow architecture.
Genes implicated in IOP but not POAG susceptibility were expressed at the highest levels in vascular endothelial and mural cells as well as the endothelium of Schlemm’s canal. Together with the evidence that elevated IOP resulting from interference with outflow pathways leads to glaucoma, this pattern raises the question of whether some sources of increased IOP may be less damaging than others. Alternatively, these may be genes of small effect, so their signal in POAG GWAS fails to reach genome-wide significance.
Genes implicated in POAG susceptibility but not IOP regulation were expressed at the highest relative levels in retinal cells—RPE, retinal astrocytes, and Müller glia—as well as in oligodendrocytes, oligodendrocyte precursors, and astrocytes of the optic nerve. Although all three of the non-neural retinal cell types have been discussed as contributors to glaucoma pathogenesis (24, 75), mechanisms remain to be elucidated. Alterations in the ON in glaucoma have generally been viewed as consequences of axonal damage and loss, but our results support the hypothesis (76) that it may also play a causal role. The genes in this list provide entry points for further analysis.
Finally, genes implicated in POAG but not IOP were highly expressed in RGCs, ONH astrocytes, and ONH fibroblasts by one metric (Fig. 7B), and NTG genes showed the highest specificity in these cell types by the other metric (Fig. 7A). These results are satisfying, in that dysfunction and death of RGCs are the cause of vision loss in glaucoma. Genes implicated in normal tension glaucoma were also enriched in endothelial cells, consistent with the idea that endothelial dysfunction can contribute to the compression of RGC axons in the ONH (75).
In summary, our analysis of the cell types in which glaucoma susceptibility and causal genes are expressed supports long-held views of the importance of the trabecular meshwork and the ONH in glaucoma pathogenesis but also draws attention to other cell types that have been less studied, such as astrocytes, oligodendrocytes, and oligodendrocyte precursors in the ON. It will also be important to seek differences in gene expression between control and glaucomatous eyes. However, such changes could be either causes or results of disease processes. Therefore, knowing expression patterns in normal tissue of genes implicated in the disease is an essential complement to such studies.
Materials and Methods
Tissue Acquisition, Dissection, and Processing.
Human eyes were obtained postmortem at a median of 6 h from death either from the Massachusetts General Hospital via the Rapid Autopsy Program or the Steele Center for Translational Medicine at the John A. Moran Eye Center (SCTM), University of Utah (SI Appendix, Table S1). Acquisition and use of human tissue were approved by the Human Studies Committees at Harvard (Dana Farber/Harvard Cancer Center Protocol No. 13-416) and University of Utah (Protocol IRB_00010201). Whole globes were transported either to Harvard University or the University of Utah in a humid chamber on ice and processed within an hour of enucleation. For details of dissection, see SI Appendix, Materials and Methods. Single-nucleus libraries were generated, sequenced, and analyzed by methods described in van Zyl et al. (9), modified as detailed in SI Appendix, Materials and Methods.
Glaucoma Gene Expression Analysis.
After cell type annotation, gene expression (SI Appendix, Table S5) was assessed as described in van Zyl et al. (9). For mapping the expressions of ocular disease–related genes, the POAG gene list was taken from Supplementary Data 4 in Gharahkhani et al. (30) and the IOP list from Khawaja et al. (31). Only genes that were expressed in at least 5% of cells in one or more clusters were included. Analytical methods are described in SI Appendix, Materials and Methods.
Histology.
Whole globes were fixed, immunostained, and imaged as previously described (9). Antibodies used are listed in SI Appendix, Table S6.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Acknowledgments
This work was supported by the Chan-Zuckerberg Initiative (CZF-2019-002459), NIH (5K12EY016335, EY028633, EY031424, and U01 MH105960), an unrestricted grant from Research to Prevent Blindness to the Department of Ophthalmology and Visual Sciences, University of Utah, and by charitable donations to the Sharon Eccles Steele Center for Translational Medicine. A.M. was supported by T32NS007473, F32EY031930, and K99EY033457. We thank Lisa Nichols and Jennifer Mohler for assistance in acquiring and processing ocular tissues and the tissue donors and their families for their generosity.
Author contributions
A.M., W.Y., T.v.Z., and J.R.S. designed research; A.M., W.Y., C.P., and K.A.O. performed research; C.P. and G.S.H. contributed new reagents/analytic tools; A.M., W.Y., Z.H., A.V.S., T.v.Z., G.S.H., and J.R.S. analyzed data; and J.R.S. and A.M. wrote the paper.
Competing interests
A.V.S. coauthored a paper (30) that included data provided by a consortium of which J.J.S. and D.J.Z. were members.
Footnotes
Reviewers: J.J.S., Wills Eye Hospital; and D.J.Z., Johns Hopkins University School of Medicine.
Data, Materials, and Software Availability
The accession number at Gene Expression Omnibus for the raw data reported in this paper is GSE236566. Data can be visualized at https://singlecell.broadinstitute.org/single_cell/study/SCP2298 (77).
Supporting Information
References
- 1.Goel M., Picciani R. G., Lee R. K., Bhattacharya S. K., Aqueous humor dynamics: A review. Open Ophthalmol. J. 4, 52 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nickla D. L., Wallman J., The multifunctional choroid. Prog. Retin. Eye Res. 29, 144–168 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forrester J., et al. , Anatomy of the Eye and Orbit, the Eye Basic Sciences in Practice (Elsevier, 2016). [Google Scholar]
- 4.Steinmetz J. D., et al. , Causes of blindness and vision impairment in 2020 and trends over 30 years, and prevalence of avoidable blindness in relation to VISION 2020: the Right to Sight: An analysis for the Global Burden of Disease Study. Lancet Glob Health 9, e144–e160 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freund K. B., Sarraf D., Mieler W. F., Yannuzzi L. A., The retinal Atlas E-book (Elsevier Health Sciences, 2016). [Google Scholar]
- 6.Hartong D. T., Berson E. L., Dryja T. P., Retinitis pigmentosa. Lancet 368, 1795–1809 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Quigley H., Glaucoma. Lancet 377, 1367–1377 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Mitchell P., Liew G., Gopinath B., Wong T. Y., Age-related macular degeneration. Lancet 392, 1147–1159 (2018). [DOI] [PubMed] [Google Scholar]
- 9.van Zyl T., et al. , Cell atlas of the human ocular anterior segment: Tissue-specific and shared cell types. Proc. Natl. Acad. Sci. U.S.A. 119, e2200914119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Zyl T., et al. , Cell atlas of aqueous humor outflow pathways in eyes of humans and four model species provides insight into glaucoma pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 117, 10339–10349 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng Y.-R., et al. , Molecular classification and comparative taxonomics of foveal and peripheral cells in primate retina. Cell 176, 1222–1237.e1222 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan W., et al. , Cell Atlas of The Human Fovea and Peripheral Retina. Sci. Rep. 10, 9802 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lukowski S. W., et al. , A single-cell transcriptome atlas of the adult human retina. EMBO J. 38, e100811 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang Q., et al. , Single-nuclei RNA-seq on human retinal tissue provides improved transcriptome profiling. Nat. Commun. 10, 5743 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Voigt A. P., et al. , Single-cell transcriptomics of the human retinal pigment epithelium and choroid in health and macular degeneration. Proc. Natl. Acad. Sci. U.S.A. 116, 24100–24107 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Menon M., et al. , Single-cell transcriptomic atlas of the human retina identifies cell types associated with age-related macular degeneration. Nat. Commun. 10, 1–9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orozco L. D., et al. , Integration of eQTL and a single-cell atlas in the human eye identifies causal genes for age-related macular degeneration. Cell Rep. 30, 1246–1259.e1246 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Huang L., et al. , Dynamic human retinal pigment epithelium (RPE) and choroid architecture based on single-cell transcriptomic landscape analysis. Genes Dis. (2022), in press, 10.1016/j.gendis.2022.11.007. [DOI] [PMC free article] [PubMed]
- 19.Collin J., et al. , Single cell RNA sequencing reveals transcriptional changes of human choroidal and retinal pigment epithelium cells during fetal development, in healthy adult and intermediate age-related macular degeneration. Hum. Mol. Genet. 32, 1698–1710 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allison K., Patel D., Alabi O., Epidemiology of glaucoma: The past, present, and predictions for the future. Cureus 12, e11686 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strickland R. G., Garner M. A., Gross A. K., Girkin C. A., Remodeling of the lamina cribrosa: Mechanisms and potential therapeutic approaches for glaucoma. Int. J. Mol. Sci. 23, 8068 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Safa B. N., Wong C. A., Ha J., Ethier C. R., Glaucoma and biomechanics. Curr. Opin. Ophthalmol. 33, 80–90 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paula J. S., O’Brien C., Stamer W. D., Life under pressure: The role of ocular cribriform cells in preventing glaucoma. Exp Eye Res. 151, 150–159 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calkins D. J., Critical pathogenic events underlying progression of neurodegeneration in glaucoma. Prog. Retin. Eye Res. 31, 702–719 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiggs J. L., Pasquale L. R., Genetics of glaucoma. Hum. Mol. Genet. 26, R21–R27 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sears N. C., Boese E. A., Miller M. A., Fingert J. H., Mendelian genes in primary open angle glaucoma. Exp Eye Res. 186, 107702 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collantes E. R. A., et al. , EFEMP1 rare variants cause familial juvenile-onset open-angle glaucoma. Hum. Mutat. 43, 240–252 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiggs J. L., CPAMD8, a new gene for anterior segment dysgenesis and childhood glaucoma. Ophthalmology 127, 767–768 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Fu H., et al. , Thrombospondin 1 missense alleles induce extracellular matrix protein aggregation and TM dysfunction in congenital glaucoma. J. Clin. Invest. 132, e156967 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gharahkhani P., et al. , Genome-wide meta-analysis identifies 127 open-angle glaucoma loci with consistent effect across ancestries. Nat. Commun. 12, 1258 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khawaja A. P., et al. , Genome-wide analyses identify 68 new loci associated with intraocular pressure and improve risk prediction for primary open-angle glaucoma. Nat. Genet. 50, 778–782 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hughes E. G., Stockton M. E., Premyelinating oligodendrocytes: Mechanisms underlying cell survival and integration. Front. Cell Dev. Biol. 9, 714169 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yaqubi M., et al. , Regional and age-related diversity of human mature oligodendrocytes. Glia 70, 1938–1949 (2022). [DOI] [PubMed] [Google Scholar]
- 34.Jäkel S., et al. , Altered human oligodendrocyte heterogeneity in multiple sclerosis. Nature 566, 543–547 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Z., et al. , Specific marker expression and cell state of Schwann cells during culture in vitro. PloS One 10, e0123278 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watanabe E., Hiyama T. Y., Kodama R., Noda M., Nax sodium channel is expressed in non-myelinating Schwann cells and alveolar type II cells in mice. Neurosci. Lett. 330, 109–113 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Kimball E., et al. , The role of aquaporin-4 in optic nerve head astrocytes in experimental glaucoma. PloS One 16, e0244123 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muhl L., et al. , Single-cell analysis uncovers fibroblast heterogeneity and criteria for fibroblast and mural cell identification and discrimination. Nat Commun 11, 1–18 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng C.-C., et al. , Single-cell RNA-seq reveals fibroblast heterogeneity and increased mesenchymal fibroblasts in human fibrotic skin diseases. Nat. Commun. 12, 1–16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garcia F. J., et al. , Single-cell dissection of the human brain vasculature. Nature 603, 893–899 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hageman G. S., Zhu X. L., Waheed A., Sly W. S., Localization of carbonic anhydrase IV in a specific capillary bed of the human eye. Proc. Natl. Acad. Sci. U.S.A. 88, 2716–2720 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Voigt A. P., et al. , Choroidal endothelial and macrophage gene expression in atrophic and neovascular macular degeneration. Hum. Mol. Genet. 31, 2406–2423 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Attwell D., et al. , What is a pericyte?. J. Cereb. Blood Flow Metab. 36, 451–455 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baek S.-H., et al. , Single cell transcriptomic analysis reveals organ specific pericyte markers and identities. Front. Cardiovasc. Med. 9, 876591 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winkler E. A., et al. , A single-cell atlas of the normal and malformed human brain vasculature. Science 375, eabi7377 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mitchell T. S., et al. , RGS5 expression is a quantitative measure of pericyte coverage of blood vessels. Angiogenesis 11, 141–151 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Ji L., Xu S., Luo H., Zeng F., Insights from DOCK2 in cell function and pathophysiology. Front. Mol. Biosci. 9, 997659 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rheinländer A., Schraven B., Bommhardt U., CD45 in human physiology and clinical medicine. Immunol. Lett. 196, 22–32 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Masuda T., Sankowski R., Staszewski O., Prinz M., Microglia heterogeneity in the single-cell era. Cell Rep 30, 1271–1281 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Amor S., et al. , White matter microglia heterogeneity in the CNS. Acta Neuropathol. 143, 125–141 (2021). [DOI] [PubMed] [Google Scholar]
- 51.Freud A. G., Mundy-Bosse B. L., Yu J., Caligiuri M. A., The broad spectrum of human natural killer cell diversity. Immunity 47, 820–833 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morgan D., Tergaonkar V., Unraveling B cell trajectories at single cell resolution. Trends Immunol. 43, 210–229 (2022). [DOI] [PubMed] [Google Scholar]
- 53.Belote R. L., et al. , Human melanocyte development and melanoma dedifferentiation at single-cell resolution. Nat. Cell Biol. 23, 1035–1047 (2021). [DOI] [PubMed] [Google Scholar]
- 54.Smith-Thomas L., et al. , Human ocular melanocytes and retinal pigment epithelial cells differ in their melanogenic properties in vivo and in vitro. Curr. Eye Res. 15, 1079–1091 (1996). [DOI] [PubMed] [Google Scholar]
- 55.Hu Y., et al. , Dissecting the transcriptome landscape of the human fetal neural retina and retinal pigment epithelium by single-cell RNA-seq analysis. PLoS Biol. 17, e3000365 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu Z., et al. , A single-cell transcriptome atlas of the human retinal pigment epithelium. Front. Cell Dev. Biol. 9, 802457 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mullin N. K., et al. , Transcriptomic and chromatin accessibility analysis of the human macular and peripheral retinal pigment epithelium at the single cell level. Am. J. Pathol. (2023), 10.1016/j.ajpath.2023.01.012. [DOI] [PMC free article] [PubMed]
- 58.Whitmore S. S., et al. , Transcriptomic analysis across nasal, temporal, and macular regions of human neural retina and RPE/choroid by RNA-Seq. Exp Eye Res. 129, 93–106 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hahn J., et al. , Evolution of neuronal cell classes and types in the vertebrate retina. bioRxiv [Preprint] (2023), 10.1101/2023.04.07.536039 (Accessed 8 April 2023). [DOI] [PMC free article] [PubMed]
- 60.Craig J. E., et al. , Multitrait analysis of glaucoma identifies new risk loci and enables polygenic prediction of disease susceptibility and progression. Nat. Genet. 52, 160–166 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Radius R. L., Gonzales M., Anatomy of the lamina cribrosa in human eyes. Arch. Ophthalmol. 99, 2159–2162 (1981). [DOI] [PubMed] [Google Scholar]
- 62.Elkington A., Inman C., Steart P., Weller R., The structure of the lamina cribrosa of the human eye: An immunocytochemical and electron microscopical study. Eye 4, 42–57 (1990). [DOI] [PubMed] [Google Scholar]
- 63.Lockwood H., et al. , Lamina cribrosa microarchitecture in normal monkey eyes part 1: Methods and initial results. Invest. Ophthalmol. Vis. Sci. 56, 1618–1637 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tovar-Vidales T., Wordinger R. J., Clark A. F., Identification and localization of lamina cribrosa cells in the human optic nerve head. Exp Eye Res. 147, 94–97 (2016). [DOI] [PubMed] [Google Scholar]
- 65.Szeto J., et al. , Regional differences and physiologic behaviors in peripapillary scleral fibroblasts. Invest. Ophthalmol. Vis. Sci. 62, 27–27 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boote C., et al. , Scleral structure and biomechanics. Prog. Retin. Eye Res. 74, 100773 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sridhar A., et al. , Single-cell transcriptomic comparison of human fetal retina, hPSC-derived retinal organoids, and long-term retinal cultures. Cell Rep. 30, 1644–1659.e1644 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Collin J., et al. , A single cell atlas of human cornea that defines its development, limbal progenitor cells and their interactions with the immune cells. Ocul. Surf. 21, 279–298 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gautam P., et al. , Multi-species single-cell transcriptomic analysis of ocular compartment regulons. Nat. Commun. 12, 5675 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Patel G., et al. , Molecular taxonomy of human ocular outflow tissues defined by single-cell transcriptomics. Proc. Natl. Acad. Sci. U.S.A. 117, 12856–12867 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hernandez M. R., Igoe F., Neufeld A. H., Cell culture of the human lamina cribrosa. Invest. Ophthalmol. Vis. Sci. 29, 78–89 (1988). [PubMed] [Google Scholar]
- 72.Lopez N. N., Clark A. F., Tovar-Vidales T., Isolation and characterization of human optic nerve head astrocytes and lamina cribrosa cells. Exp Eye Res. 197, 108103 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wallace D. M., O’Brien C. J., The role of lamina cribrosa cells in optic nerve head fibrosis in glaucoma. Exp Eye Res. 142, 102–109 (2016). [DOI] [PubMed] [Google Scholar]
- 74.Rogers R., Dharsee M., Ackloo S., Flanagan J. G., Proteomics analyses of activated human optic nerve head lamina cribrosa cells following biomechanical strain. Invest. Ophthalmol. Vis. Sci. 53, 3806–3816 (2012). [DOI] [PubMed] [Google Scholar]
- 75.Moore D., et al. , Dysfunctional regulation of ocular blood flow: A risk factor for glaucoma? Clin. Ophthalmol. 2, 849–861 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xue J., et al. , Demyelination of the optic nerve: An underlying factor in glaucoma? Front. Aging Neurosci. 13, 701322 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Monavarfeshani A., et al. , Transcriptomic analysis of the ocular posterior segment completes a cell atlas of the human eye. Broad Single Cell Portal. https://singlecell.broadinstitute.org/single_cell/study/SCP2298. Deposited 5 July 2023. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Data Availability Statement
The accession number at Gene Expression Omnibus for the raw data reported in this paper is GSE236566. Data can be visualized at https://singlecell.broadinstitute.org/single_cell/study/SCP2298 (77).