Abstract
Presently, excessive fat deposition is the main reason to limit the development of duck industry. In the production, the methods of restricted feeding (RF) were widely used to reduce the lipid deposition of ducks. The liver (L), abdominal adipose (AA), and subcutaneous adipose (SA) were the main tissues of lipid metabolism and deposition of ducks. However, the mechanisms of lipid metabolism and deposition of ducks under RF have not been fully clarified. In this study, in order to better understand the mechanisms of lipid metabolism and deposition in ducks under RF, a total of 120 male Nonghua ducks were randomly divided into a free feeding group (FF, n = 60) and RF group (RF, n = 60), then comparative transcriptomic analysis of L, AA, and SA between FF (n = 3) and RF (n = 3) ducks was performed at 56 d of age. Phenotypically, L, AA, and SA index of FF group was higher than that in RF group. There were 279, 390, and 557 differentially expressed genes (DEGs) in L, AA, and SA. Functional enrichment analysis revealed that ECM-receptor interaction and metabolic pathways were significantly enriched in L, AA, and SA. Lipid metabolism-related pathways including fatty acid metabolism, unsaturated fatty acid synthesis, and steroidogenesis were significantly enriched in AA and SA. Moreover, through integrated analysis weighted gene coexpression network (WGCNA) and protein-protein interaction network, 10 potential candidate genes involved in the ECM-receptor interaction and lipid metabolism pathways were identified, including 3-hydroxy-3-methylglutaryl-CoA synthase 2 (HMGCS2), aldolase B (ALDOB), formimidoyltransferase cyclodeaminase(FTCD), phosphoenolpyruvate carboxykinase 1 (PCK1), tyrosine aminotransferase (TAT), stearoyl-CoA desaturase (SCD), squalene epoxidase (SQLE), phosphodiesterase 4B (PDE4B), choline kinase A (CHKA), and elongation of very-long-chain fatty acids-like 2 (ELOVL2), which could play a key role in lipid metabolism and deposition of ducks under RF. Our study reveals that the liver might regulate the lipid metabolism of abdominal adipose and subcutaneous adipose through ECM-receptor interaction and metabolic pathways (fatty acid metabolism, unsaturated fatty acid synthesis, and steroid synthesis), thus to reduce the lipid deposition of ducks under RF. These results provide novel insights into the avian lipid metabolism and will help better understand the underlying molecular mechanisms.
Key words: duck, restricted feeding, lipid metabolism and deposition, transcriptome sequencing
INTRODUCTION
At present, the meat duck industry usually adopts a high-density and intensive rearing model, and excessive fat deposition in meat ducks, which affects the further development of the meat duck industry (Na et al., 2019). In meat poultry production, restricted feeding (RF) was widely used to reduce lipid deposition in meat ducks (Saibaba et al., 2021). Previous studies have shown that RF could affect growth performance, reproduction, and metabolic function of meat poultry, especially on lipid metabolism and deposition (Pan et al., 2014; Mehus et al., 2021; Shusha et al., 2021).
In poultry, more than 90% of lipid was synthesized in liver and deposited in adipose tissues after liver metabolism (Nie et al., 2009). It has been observed that liver and adipose tissue were easily adapted to feed restriction, but their hormone levels and metabolites have changed, thus significantly affecting their lipid metabolism and deposition (Hillgartner et al., 1995; Kersten, 2001; Zaefarian et al., 2019). Compared to free feeding (FF), the expression levels of SREBP-1, ME, ACL, ACC, FAS, and SCD1 in liver were decreased under RF, and the plasma hormone levels that could induce many genes encoding lipogenic enzymes also decreased (Richards et al., 2003). Additionally, one research showed that the liver could reduce the lipid synthesis and TG secretion through transcriptional regulation to suit the energy restricted diet (Desert et al., 2018). Meanwhile, RF could increase the expression of CPT1-A in the liver of broilers, thereby reducing the content of adipose tissue (Lunedo et al., 2019). Wei et al. (2019) also found that the genes related to lipid metabolism significant changes in the liver and abdominal adipose of broilers under RF. Furthermore, Lindholm et al. (2022) further analyzed the transcriptome profiles of chicken liver between FF and RF and found that most of DEGs were related to energy metabolism. These results indicated that RF might have significant effects on lipid metabolism and deposition in liver and adipose tissue. However, until now little is known about the mechanism of lipid metabolism and deposition in duck liver and adipose tissue under RF.
With the rapid development of high-throughput sequencing technologies, RNA-seq technology has been favored by researchers for its high efficiency and speed. Therefore, this study aimed to compare and analyze the mRNA profiles of liver, abdominal adipose, and subcutaneous adipose between FF and RF ducks using RNA-seq. These results were expected to help elucidate the molecular mechanisms regulating lipid metabolism and deposition in ducks under RF.
MATERIALS AND METHODS
Ethics Statement
All animal handling procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Sichuan Agricultural University (Chengdu campus, Sichuan, China, Permit No. DKY20170913).
Animals and Sample Collection
In this study, a total of 120 one-day-old healthy Nonghua ducks hatched from the same batch and having similar body weights ranging from 52.0 g to 58.0 g were used as our experimental animals. All ducks were hatched and reared in the Waterfowl Breeding Experimental Farm of Sichuan Agricultural University (Ya'an campus, Sichuan, China). After incubation, these 1-day-old all ducks were first raised net-rearing until 14 d of age, and they were then raised floor-rearing. At 35 d of age, these ducks were randomly equally assigned to FF and RF groups. As shown in Table 1, one group was fed freely (FF), and the other group was fed 70% of the FF intake (RF). During this period, the ducks were raised under natural light and temperature and had free access to water. At 56 d of age, 18 healthy ducks with similar body weights were randomly selected from FF and RF, respectively, for weighing and collecting tissue samples. The selected ducks were slaughtered (euthanized by carbon dioxide anesthesia and exsanguination by severing the carotid artery), and the weights and organ indexes of liver, abdominal adipose, and subcutaneous adipose were measured (organ index = organ weight (g)/body weight (g) × 100%). All tissues were washed with PBS and frozen in liquid nitrogen, and then stored at −80°C until RNA extraction.
Table 1.
The feeding methods of ducks in each group.
| Items | FF (n = 60) | RF (n = 60) |
|---|---|---|
| 0–14 d | Free feeding | Free feeding |
| 15–35 d | Free feeding | Free feeding |
| 36–42 d | 210 g | 147 g |
| 43–49 d | 220 g | 154 g |
| 50–56 d | 230 g | 161 g |
Abbreviations: d, day; FF, free feeding; g, gram; RF, restricted feeding.
RNA Isolation and Sequencing
In this study, 3 ducks with similar body weight were selected in the FF and RF groups for transcriptome studies, respectively. The RNeasy Mini Kit (QIAGEN, Beijing, China) was used to extract the total RNA of liver, abdominal adipose, and subcutaneous adipose according to the manufacturer's instructions. RNA concentration was detected by Nanodrop, and RNA integrity was checked by Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). All RNA samples were qualified and sent to Glibizzia (Beijing, China) for library construction (Additional file 1: Table S1). The mRNA libraries were sequenced by DNBSEQ-T7-PE150 (HuaDa, Shenzhen, China). The clean reads were obtained after the filtration of low-quality reads using standard quality control by FastaQC software.
Transcriptome Bioinformatics Analysis
Clean reads were mapped to the Anas platyrhynchos (assembly ZJU1.0) domestication reference genome using the HISAT2 (version 2.2.1) software (Kim et al., 2015). The output SAM (sequencing alignment/mapping) file was converted to a BAM (binary alignment/mapping) file and sorted using SAMtools (version 1.10) (Li et al., 2009). The expression values (fragments per kilobase of transcript per million fragments mapped) of each gene were calculated based on the length of the gene and the read count mapped to this gene by featureCounts (version 2.02) (Liao et al., 2014). Differentially expressed genes (DEGs) in different feeding groups were analyzed using the DEseq2 package in R and then filtered with P values <0.05 and |log2FC| > 1 (Love et al., 2014). The screened DEGs were subjected to the Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis in KOBAS 3.0 (Ai and Kong, 2018). Subsequently, the DEGs of different feeding groups were constructed the weighted gene coexpression network using the WGCNA package in R (Langfelder and Horvath, 2008). Finally, the network visualization analysis was completed using Cytoscape (version 3.2.1) (Smoot et al., 2011).
Real-Time PCR Verification
Five significantly DEGs were selected for quantitative real-time PCR (qRT-PCR) to validate the RNA-Seq results. Previously, total RNA extracted from the liver and subcutaneous adipose were reverse transcribed into cDNA using a RevertAid First Strand cDNA Synthesis Kit (Thermo, MA). Primer 5.0 was used to design the primers (Table 2). A BLAST search against the reference genome was then carried out to confirm primers were specific for the intended target genes. SYBR Green PCR Super Mix (Bio-Rad, Hercules, CA) and a Bio-Rad CFX96 real-time PCR detection system (Bio-Rad, Hercules, CA) were used for qRT-PCR, and each sample was assayed 3 times. The reaction was performed at 95°C for 10 s, 60°C for 60 s, and 95°C for 15 s, after which it was slowly heated from 60°C to 99°C. The β-ACTIN and GAPDH was used as a housekeeping gene. The 2−ΔΔCt method was used for normalization of the qRT-PCR results, after which the normalized data were used for statistical analysis, and P < 0.05 was considered significantly different (Livak and Schmittgen, 2001).
Table 2.
PCR primers used in this study.
| Genes | Forward (5′–3′) | Reverse (5′–3′) | TM (°C) |
|---|---|---|---|
| SCD | ATCTTTGATGAGACCTACCGTG | GTGGCTTTGTAGGACCGATG | 58.0 |
| CHKA | CCTTCGGCTCTCCCTCAC | GCAACTGTCTCAATGGTATCGG | 56.0 |
| ELOVL2 | ACCTTGGGATTACACTGCTCTC | GTTCAGGACACACCACCAGATA | 56.0 |
| SQLE | CAAATACAGCCTTATCACCGCA | AAGACCTTCCACTGACAACACG | 56.0 |
| PDE4B | AGAACGAGAAAGAGGAATGGAA | TGTATTGCTGAAGTAGCCGATG | 56.0 |
| GAPDH | AAGGCTGAGAATGGGAAAC | TTCAGGGACTTGTCATACTTC | 60.0 |
| β-ACTIN | GCTATGTCGCCCTGGATTTC | CACAGGACTCCATACCCAAGAA | 60.0 |
Statistical Analysis
Statistical analysis was performed using the SPSS 24.0 software (IBM, Newyork). The body weight, liver index, abdominal adipose index as well as subcutaneous adipose index were presented as mean ± SD. Data for both different groups were tested for normal distribution with SPSS 24.0, followed by ANOVA testing and Duncan's test. Differences were considered statistically significant at P < 0.05.
RESULTS
Phenotypic Differences of Different Tissues Between FF and RF
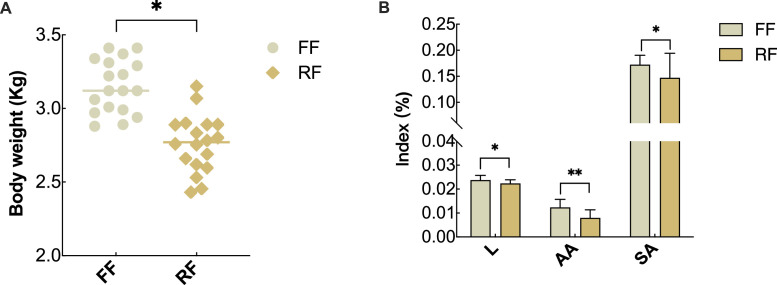
As shown in Figure 1A, ducks in the FF group had higher body weight (P < 0.05). In addition, compared with the FF group, the liver, abdominal adipose, and subcutaneous adipose index of ducks in RF group were significantly lower than that in FF group (P < 0.05, Figure 1B).
Figure 1.
Phenotypic differences of ducks between FF and RF. (A) Body weight analysis of ducks between FF and RF; (B) Organ index analysis of liver, abdominal adipose, and subcutaneous adipose of ducks between FF and RF. * Indicates P < 0.05, ⁎⁎ indicates P < 0.01. FF: free feeding; RF: restricted feeding; L: liver; AA: abdominal adipose; SA: subcutaneous adipose.
Overview of the mRNA Transcriptome Sequencing Between FF and RF
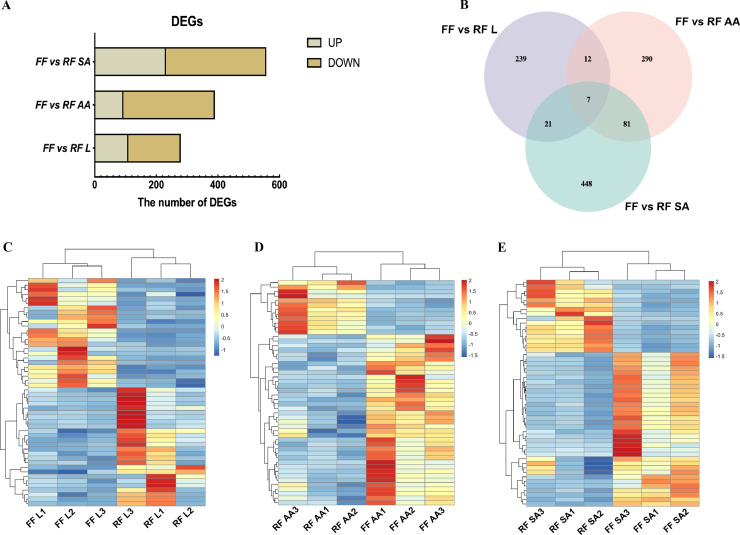
A total of 146.30 Gb raw data were obtained from 18 samples through RNA-seq, and the mapping rate of the clean reads ranged from 88.74 to 94.29% (Additional file 2: Table S2). In this study, there were 279, 390, and 557 DEGs identified in liver, abdominal adipose, and subcutaneous adipose between FF and RF, respectively (Figure 2A; Additional file 3: Table S3). Based on the identified DEGs in the liver, abdominal adipose, and subcutaneous adipose, 7 genes were common in both tissues as shown in the Venn diagram (Figure 2B). The hierarchical clustering map also recapitulated the distinct gene expression patterns in the liver, abdominal adipose, and subcutaneous adipose between FF and RF (Figure 2C–E).
Figure 2.
Identification of DEGs in the liver, abdominal adipose, and subcutaneous adipose of ducks between FF and RF. (A) The number of DEGs in different tissues; (B) Venn diagram of DEGs; (C–E) DEGs clustering heat map of each tissue.
Functional Enrichment Analysis of the DEGs in the Liver, Abdominal Adipose, and Subcutaneous Adipose Between FF and RF
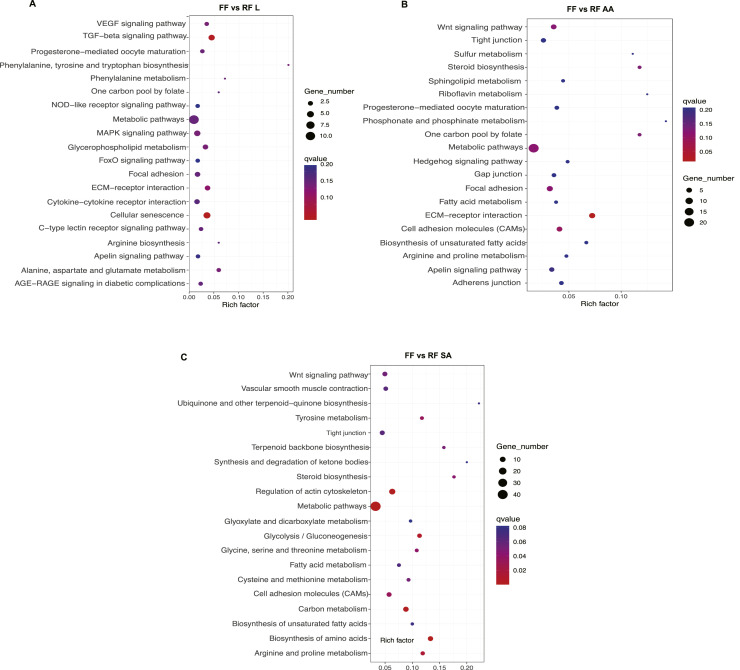
In this study, KEGG enrichment analysis showed that a total of 54, 71, and 89 KEGG pathways were enriched in liver, abdominal adipose, and subcutaneous adipose, respectively (Additional file 4: Table S4). The top 20 significantly enriched KEGG pathways were listed in Figure 3. In the liver, the 5 significantly enriched pathways were cell senescence, TGF-β signal pathway, ECM-receptor interaction, glycerol phospholipid metabolism, and alanine, aspartic acid, and glutamate metabolism pathway (Figure 3A). In the abdominal adipose, the most enriched KEGG pathways were ECM-receptor interaction, cell adhesion molecules, aggregation adhesion, metabolic pathway, and Wnt-signal pathway (Figure 3B). Furthermore, the top 5 significantly enriched KEGG pathways were metabolic pathway, amino acid biosynthesis, actin cytoskeleton regulation, and carbon metabolism, glycolysis/gluconeogenesis pathway in subcutaneous adipose (Figure 3C). Noticeably, the ECM-receptor interaction and metabolic pathways were commonly enriched by the DEGs in liver, abdominal adipose, and subcutaneous adipose between FF and RF. In addition, lipid metabolism-related pathways (fatty acid metabolism, unsaturated fatty acid synthesis, and steroid synthesis) were significantly enriched in abdominal adipose and subcutaneous adipose between FF and RF.
Figure 3.
Top 20 significantly enriched KEGG pathways. (A) FF vs. RF liver; (B) FF vs. RF abdominal adipose; (C) FF vs. RF subcutaneous adipose. The Rich factor is the ratio of the number of DEGs in the pathway and the total number of genes in the pathway. The higher the Rich factor, the higher is the degree of enrichment. The q value is the P value after multiple hypothesis test correction, in the range from 0 to 1; the closer the q value is to 0, and the more significant is the enrichment.
Construction of Coexpression Networks of the DEGs Identified Between FF and RF
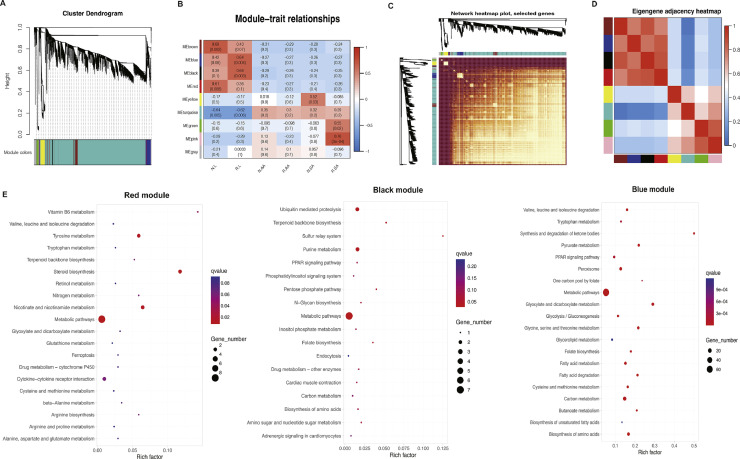
The weighted gene coexpression network (WGCNA) was performed to construct the coexpression network of DEGs identified in liver, abdominal adipose, and subcutaneous adipose between FF and RF. The results showed that genes with similar expression patterns in liver, abdominal adipose, and subcutaneous adipose were classified into 9 modules (Figure 4A, Additional file 5: Table S5). Then, correlation analysis between the phenotypes and modules indicated that genes clustered in the blue modules were strongly correlated with liver, abdominal adipose, and subcutaneous adipose phenotypes (Figure 4B). Moreover, the correlation analysis between these modules showed that the gene expression profiles in blue module was similar to that in black and red modules (Figure 4C and D).
Figure 4.
The results of weighted gene coexpression network analysis. (A) Hierarchical clustering diagram. Different colors on the abscissa represent different clustering modules; (B) Correlation between modules and traits. The abscissa represents tissues with different phenotypes, and the ordinate represents different module; (C) Visualized network heat map; (D) Correlation diagrams between modules; The redder the color of the area of different modules, the stronger the correlation; (E) KEGG analysis for red, black, and blue module DEGs.
Subsequently, KEGG enrichment analysis indicated that 35, 18, and 82 KEGG pathways were enriched in red, black, and blue modules, respectively. The significantly enriched KEGG pathways were listed in Figure 4E. Metabolic, steroid biosynthesis, nicotinate and nicotinamide metabolism, tyrosine metabolism, and vitamin B6 metabolism pathways significantly enriched by DEGs in red module. Some metabolic-related KEGG pathways were enriched only by DEGs in the black module, including metabolic, purine metabolism, pentose phosphate pathway, biosynthesis of amino acids, and PPAR signaling pathways. Moreover, DEGs in the blue module were mainly enriched in metabolic pathway, carbon metabolism, glyoxylate and dicarboxylate metabolism, fatty acid metabolism, fatty acid degradation, glycolysis/gluconeogenesis, and PPAR signaling pathways. Of note, lipid metabolism-related pathways were overlapped in the 3 modules. Therefore, we chose the DEGs in the blue, black, and red modules for subsequent analysis.
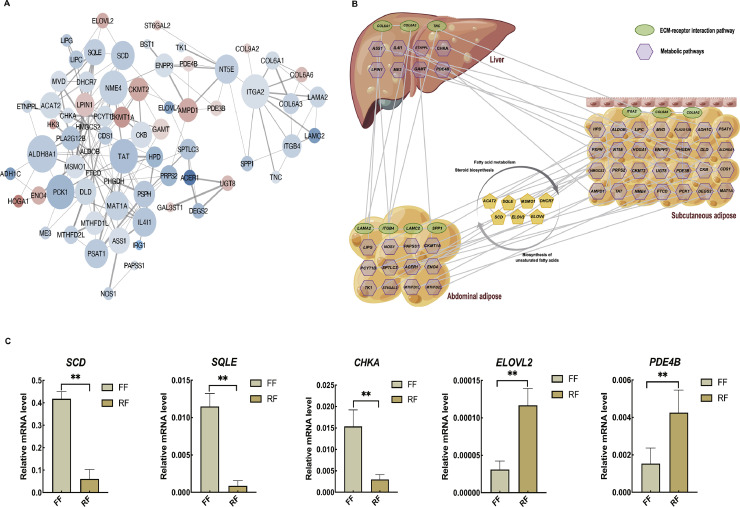
Network Analysis and qRT-PCR Validation of the DEGs Involved in Regulating Lipid Metabolism and Deposition Between FF and RF
To further identify the mechanisms regulating lipid metabolism of ducks under RF, the DEGs from the liver, abdominal adipose, and subcutaneous adipose between FF and RF were merged to construct the protein-protein interaction (PPI) network. The PPI network consisted of 69 nodes and 180 edges (Figure 5A). Network analysis found that the genes with the highest degrees included HMGCS2, ALDOB, FTCD, PCK1, TAT, SCD, SQLE, PDE4B, CHKA, and ELOVL2. Notably, our results showed that ducks under RF could reduce the lipid deposition through ECM-receptor interaction and metabolic pathways (fatty acid metabolism, unsaturated fatty acid synthesis, and steroid synthesis) (Figure 5B). Five DEGs involved in the lipid metabolism were selected for qRT-PCR validation of our RNA-seq results. These included 3 genes that were upregulated (PDE4B, CHKA, and ELOVL2), 2 genes that were upregulated (SCD and SQLE). The expression profiles of the 5 genes generated from qRT-PCR corresponded to the RNA-Seq results (Figure 5C), indicating that the RNA-seq results are reliable.
Figure 5.
Network analysis and qRT-PCR validation of DEGs in liver, abdominal adipose, and subcutaneous adipose of ducks between FF and RF. (A) PPI network of DEGs; (B) Regulation network construction involved in lipid deposition and metabolism of ducks under RF; (C) qRT-PCR validation of DEGs. “*” and “**” represent a significant difference at P < 0.05 or P < 0.01, respectively.
DICUSSION
In poultry, including ducks, the liver, abdominal adipose, and subcutaneous adipose were the main organs for lipid anabolism and deposition (He et al., 2018; Wang et al., 2019). A previous study showed that RF resulted in a lower liver and abdominal adipose index of broilers (Van der klein et al., 2017). This is similar to the results of our study which showed that in RF ducks, the liver, abdominal adipose, and subcutaneous adipose index were lower than FF ducks, indicating that RF could affect the development of liver and adipose tissue. To further understand the molecular mechanism how the FF and RF differentially affected duck lipid metabolism and deposition, we used RNA-seq to compare the transcriptomic mRNA profiles of liver, abdominal adipose, and subcutaneous adipose. As a result, a total of 279, 390, and 557 DEGs were identified in liver, abdominal adipose, and subcutaneous adipose, respectively. Subsequent analysis suggested that 7 DEGs were common in liver, abdominal adipose, and subcutaneous adipose between FF and RF. Among these genes, studies have shown that the YRDC gene plays an important role in lipid storage and metabolism processes (Liu et al., 2023). These results indicated that liver, abdominal adipose, and subcutaneous adipose might play an important role in regulating lipid metabolism and deposition of ducks under RF.
To further explain the biological roles of the DEGs, KEGG functional analysis was performed. In the liver, most DEGs were significantly enriched in TGF-β signaling, ECM-receptor interaction, glycerophospholipid metabolism, and metabolic pathways. Several studies have shown that glycerophospholipid metabolism played a crucial role in liver fatty acid metabolism (Maldonado et al., 2014), and changes of fatty acid metabolism usually leaded to the accumulation of triglycerides in liver (Alves-Bezerra and Cohen, 2017). Meanwhile, the triglycerides in liver were transported to adipose tissue as very-low-density lipoprotein (Frayn et al., 2006). Thus, these results showed that glycerophospholipid metabolism might be a potential reason for affecting lipid deposition in duck liver under RF. Furthermore, in abdominal adipose and subcutaneous adipose, our results showed that fatty acid metabolism, unsaturated fatty acid synthesis, steroid synthesis, ECM-receptor interaction, and metabolic pathways were significantly enriched. Studies have demonstrated that fatty acid metabolism, unsaturated fatty acid synthesis, and steroid synthesis could affect the lipid deposition in adipose tissue (Li et al., 2015; Torchon et al., 2017; Grabner et al., 2021). Considering the importance of these pathways on lipid deposition processes, it can be affirmed that the decrease of lipid deposition in abdominal and subcutaneous adipose in duck under RF might be related to changes in fatty acid metabolism and steroid synthesis.
Furthermore, when we tried to explore the molecular mechanisms in regulating lipid metabolisms and deposition, results of network analysis suggested that ECM-receptor interaction and metabolic pathways could play a critical role in affecting lipid metabolisms and deposition of ducks under RF. In this study, ECM-receptor interaction and metabolic pathways were significantly enriched in liver, abdominal adipose, and subcutaneous adipose. It has been observed that ECM-receptor interaction pathway was an important part of cell microenvironment (Rowe and Weiss, 2008), which provided physical support for adipocyte proliferation, differentiation, and migration (Schaefer and Schaefer, 2010; Jiang et al., 2013, 2014). Meanwhile, another study showed that ECM-receptor interaction pathway affected fatty acid metabolism by regulating key transcription factors (Schaffer and Lodish, 1994). Moreover, studies on chickens with different dietary quantities reported that ECM-receptor interaction pathway played an important role in lipid metabolism in liver and abdominal adipose (Chen et al., 2019; Ma et al., 2020). A previous study has also demonstrated that the metabolic pathway significantly changed under RF, so as to adapt to the decline of nutrient supply and coordinate a series of physiological processes (Chaix et al., 2019). Furthermore, PPI network analysis showed that almost all DEGs enriched in ECM-receptor interaction and metabolic pathways, including COL6A1, COL6A2, TNC, ASSI1, IL4I1, ETNPL, CHKA, LPIN1, ME3, GAMT, PDE4B, ACAT2, SQLE, MSMO1, DHCR7, SCD, ELOVL2, and ELOVL4, were significantly up- or downregulated in the Liver, abdominal adipose, and subcutaneous adipose between FF and RF groups. Previous studies have shown that LPIN1 could regulate fatty acid utilization in the triglyceride biosynthetic pathway (Kajimoto et al., 2016), while CHKA, ME3, GAMT, and PDE4B play an important role in regulating liver lipid metabolism and energy homeostasis (Barcelos et al., 2016; Liang et al., 2018; Xu et al., 2021, 2022). Meanwhile, SQLE and DHCR7 were involved in steroid synthesis (Jiang et al., 2020; Zhang et al., 2020), while ACAT2, MSMO1, SCD, ELOVL2, and ELOVL4 were key genes for fatty acid anabolism (Wang et al., 2017; Mohammadi et al., 2020; Guo et al., 2021). Moreover, Wei et al. (2019) found that the lipid metabolism of broilers liver changed significantly under RF, thus regulating the lipid deposition of abdominal adipose. Taken together, our results indicated that liver, abdominal adipose, and subcutaneous adipose might regulated lipid anabolism through ECM-receptor interaction and metabolic-related pathways (fatty acid metabolism, unsaturated fatty acid synthesis, and steroid synthesis), thus to reduce adipose tissue lipid deposition of ducks under RF.
In conclusion, we constructed the first expression profiles of liver, abdominal adipose, and subcutaneous adipose in ducks between FF and RF groups. The ECM-receptor interaction and metabolic-related pathways were all activated in liver, abdominal adipose, and subcutaneous adipose. Notably, the liver might regulate the lipid metabolism of abdominal adipose and subcutaneous adipose through ECM-receptor interaction and metabolic pathways (fatty acid metabolism, unsaturated fatty acid synthesis, and steroid synthesis), thus to reduce the lipid deposition of ducks under RF. These results provide novel insights into the avian lipid metabolism and will help better understand the underlying molecular mechanisms.
ACKNOWLEDGMENTS
This research was supported by China Agriculture Research System of MOF and MARA (CARS-42-4), School Cooperation Project of Ya’ an (21SXHZ0028), and Key Technology Support Program of Sichuan Province (2021YFYZ0014).
DISCLOSURES
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2023.102963.
Appendix. Supplementary materials
REFERENCES
- Ai C., Kong L. CGPS: a machine learning-based approach integrating multiple gene set analysis tools for better prioritization of biologically relevant pathways. J. Genet. Genom. 2018;45:489–504. doi: 10.1016/j.jgg.2018.08.002. [DOI] [PubMed] [Google Scholar]
- Alves-Bezerra M., Cohen D.E. Triglyceride metabolism in the liver. Compr. Physiol. 2017;8:1–8. doi: 10.1002/cphy.c170012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcelos R.P., Stefanello S.T., Mauriz J.L., Gonzalez-Gallego J., Soares F.A. Creatine and the liver: metabolism and possible interactions. Mini Rev. Med. Chem. 2016;16:12–18. doi: 10.2174/1389557515666150722102613. [DOI] [PubMed] [Google Scholar]
- Chaix A., Manoogian E.N.C., Melkani G.C., Panda S. Time-restricted eating to prevent and manage chronic metabolic diseases. Annu. Rev. Nutr. 2019;39:291–315. doi: 10.1146/annurev-nutr-082018-124320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Ren X., Li L., Lu S., Chen T., Tan L., Liu M., Luo Q., Liang S., Nie Q., Zhang X., Luo W. Integrative analyses of mRNA expression profile reveal the involvement of IGF2BP1 in chicken adipogenesis. Int. J. Mol. Sci. 2019;20:2923. doi: 10.3390/ijms20122923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desert C., Baéza E., Aite M., Boutin M., Cam A.L., Montfort J., Houee-Bigot M., Blum Y., Roux P.F., Hennequet-Antier C., Berri C., Metayer-Coustard S., Collin A., Allais S., Bihan E.L., Causeur D., Gondret F., Duclos M.J., Lagarrigue S. Multi-tissue transcriptomic study reveals the main role of liver in the chicken adaptive response to a switch in dietary energy source through the transcriptional regulation of lipogenesis. BMC Genom. 2018;19:187. doi: 10.1186/s12864-018-4520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frayn K.N., Arner P., Yki-Järvinen H. Fatty acid metabolism in adipose tissue, muscle and liver in health and disease. Essays Biochem. 2006;42:89–103. doi: 10.1042/bse0420089. [DOI] [PubMed] [Google Scholar]
- Grabner G.F., Xie H., Schweiger M., Zechner R. Lipolysis: cellular mechanisms for lipid mobilization from fat stores. Nat. Metab. 2021;3:1445–1465. doi: 10.1038/s42255-021-00493-6. [DOI] [PubMed] [Google Scholar]
- Guo L., Wei C., Yi L., Yang W., Geng Z., Chen X. Transcriptional insights into key genes and pathways underlying Muscovy duck subcutaneous fat deposition at different developmental stages. Animals (Basel) 2021;11:2099. doi: 10.3390/ani11072099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Zheng H., Pan D., Liu T., Sun Y., Cao J., Wu Z., Zeng X. Effects of aging on fat deposition and meat quality in Sheldrake duck. Poult. Sci. 2018;97:2005–2010. doi: 10.3382/ps/pey077. [DOI] [PubMed] [Google Scholar]
- Hillgartner F.B., Salati L.M., Goodridge A.G. Physiological and molecular mechanisms involved in nutritional regulation of fatty acid synthesis. Physiol. Rev. 1995;75:47–76. doi: 10.1152/physrev.1995.75.1.47. [DOI] [PubMed] [Google Scholar]
- Jiang K., Ma Z., Wang Z., Li H., Wang Y., Tian Y., Li D., Liu X. Evolution, expression profile, regulatory mechanism, and functional verification of EBP-like gene in cholesterol biosynthetic process in chickens (Gallus Gallus) Front. Genet. 2020;11 doi: 10.3389/fgene.2020.587546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z., Sun J., Dong H., Luo O., Zheng X., Obergfell C., Tang Y., Bi J., O'Neill R., Ruan Y., Chen J., Tian X.C. Transcriptional profiles of bovine in vivo pre-implantation development. BMC Genom. 2014;15:756. doi: 10.1186/1471-2164-15-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Wei H., Song T., Yang Y., Peng J., Jiang S. Transcriptome comparison between porcine subcutaneous and intramuscular stromal vascular cells during adipogenic differentiation. PLoS One. 2013;8:e77094. doi: 10.1371/journal.pone.0077094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimoto K., Suemitsu E., Sato Y., Sakurai Y., Harashima H. Liver-specific silencing of lipin1 reduces fat mass as well as hepatic triglyceride biosynthesis in mice. Biol. Pharm. Bull. 2016;39:1653–1661. doi: 10.1248/bpb.b16-00353. [DOI] [PubMed] [Google Scholar]
- Kersten S. Mechanisms of nutritional and hormonal regulation of lipogenesis. EMBO Rep. 2001;2:282–286. doi: 10.1093/embo-reports/kve071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Papadopoulos V., Vihma V. Steroid biosynthesis in adipose tissue. Steroids. 2015;103:89–104. doi: 10.1016/j.steroids.2015.03.016. [DOI] [PubMed] [Google Scholar]
- Liang J.Q., Teoh N., Xu L., Pok S., Li X., Chu E.S.H., Chiu J., Dong L., Arfianti E., Haigh W.G., Yeh M.M., Ioannou G.N., Sung J.J.Y., Farrell G., Yu J. Dietary cholesterol promotes steatohepatitis related hepatocellular carcinoma through dysregulated metabolism and calcium signaling. Nat. Commun. 2018;9:4490. doi: 10.1038/s41467-018-06931-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Lindholm C., Batakis P., Altimiras J., Lees J. Intermittent fasting induces chronic changes in the hepatic gene expression of Red Jungle Fowl (Gallus gallus) BMC Genom. 2022;23:304. doi: 10.1186/s12864-022-08533-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Chen X., Ran J., Yin J., Zhang L., Yang Y., Cen J., Dai H., Zhou J., Gao K., Zhang J., Liu L., Chen Z., Wang H. Investigating the change in gene expression profile of blood mononuclear cells post-laparoscopic sleeve gastrectomy in Chinese obese patients. Front. Endocrinol. (Lausanne) 2023;14 doi: 10.3389/fendo.2023.1049484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunedo R., Furlan L.R., Fernandez-Alarcon M.F., Squassoni G.H., Campos D.M.B., Perondi D., Macari M. Intestinal microbiota of broilers submitted to feeding restriction and its relationship to hepatic metabolism and fat mass: fast-growing strain. J. Anim. Physiol. Anim. Nutr. (Berl.) 2019;103:1070–1080. doi: 10.1111/jpn.13093. [DOI] [PubMed] [Google Scholar]
- Ma X., Sun J., Zhu S., Du Z., Li D., Li W., Li Z., Tian Y., Kang X., Sun G. MiRNAs and mRNAs analysis during abdominal preadipocyte differentiation in chickens. Animals (Basel) 2020;10:468. doi: 10.3390/ani10030468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado E.N., Delgado I., Furland N.E., Buqué X., Iglesias A., Aveldaño M.I., Zubiaga A., Fresnedo O., Ochoa B. The E2F2 transcription factor sustains hepatic glycerophospholipid homeostasis in mice. PLoS One. 2014;9 doi: 10.1371/journal.pone.0112620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehus A.A., Rust B., Idso J.P., Hanson B., Zeng H., Yan L., Bukowski M.R., Picklo M.J. Time-restricted feeding mice a high-fat diet induces a unique lipidomic profile. J. Nutr. Biochem. 2021;88 doi: 10.1016/j.jnutbio.2020.108531. [DOI] [PubMed] [Google Scholar]
- Mohammadi V., Sharifi S.D., Sharafi M., Mohammadi-Sangcheshmeh A., Abedheydari E., Alizadeh A. Dietary L-carnitine affects the expression of genes involved in apoptosis and fatty acid metabolism in rooster testes. Andrologia. 2020;52:e13876. doi: 10.1111/and.13876. [DOI] [PubMed] [Google Scholar]
- Na W., Yu J.Q., Xu Z.C., Zhang X.Y., Yang L.L., Cao Z.P., Li H., Zhang H. Important candidate genes for abdominal fat content identified by linkage disequilibrium and fixation index information. Poult. Sci. 2019;98:581–589. doi: 10.3382/ps/pey426. [DOI] [PubMed] [Google Scholar]
- Nie Q., Fang M., Xie L., Peng X., Xu H., Luo C., Zhang D., Zhang X. Molecular characterization of the ghrelin and ghrelin receptor genes and effects on fat deposition in chicken and duck. J. Biomed. Biotechnol. 2009;2009 doi: 10.1155/2009/567120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y.E., Liu Z.C., Chang C.J., Huang Y.F., Lai C.Y., Walzem R.L., Chen S.E. Feed restriction ameliorates metabolic dysregulation and improves reproductive performance of meat-type country chickens. Anim. Reprod. Sci. 2014;151:229–236. doi: 10.1016/j.anireprosci.2014.10.003. [DOI] [PubMed] [Google Scholar]
- Richards M.P., Poch S.M., Coon C.N., Rosebrough R.W., Ashwell C.M., McMurtry J.P. Feed restriction significantly alters lipogenic gene expression in broiler breeder chickens. J. Nutr. 2003;133:707–715. doi: 10.1093/jn/133.3.707. [DOI] [PubMed] [Google Scholar]
- Rowe R.G., Weiss S.J. Breaching the basement membrane: who, when and how? Trends Cell Biol. 2008;18:560–574. doi: 10.1016/j.tcb.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Saibaba G., Ruzal M., Shinder D., Yosefi S., Druyan S., Arazi H., Froy O., Sagi D., Friedman-Einat M. Time-restricted feeding in commercial layer chickens improves egg quality in old age and points to lack of adipostat activity in chickens. Front. Physiol. 2021;12 doi: 10.3389/fphys.2021.651738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer J.E., Lodish H.F. Expression cloning and characterization of a novel adipocyte long chain fatty acid transport protein. Cell. 1994;79:427–436. doi: 10.1016/0092-8674(94)90252-6. [DOI] [PubMed] [Google Scholar]
- Schaefer L., Schaefer R.M. Proteoglycans: from structural compounds to signaling molecules. Cell Tissue Res. 2010;339:237–246. doi: 10.1007/s00441-009-0821-y. [DOI] [PubMed] [Google Scholar]
- Shusha E., Ahmed S., Ali E., Sabek A. Effect of different feed restriction regimens on performance, behaviors, blood cortisol, and carcass parameters of growing Sasso broilers. Trop. Anim. Health Prod. 2021;53:461. doi: 10.1007/s11250-021-02890-1. [DOI] [PubMed] [Google Scholar]
- Smoot M.E., Ono K., Ruscheinski J., Wang P.L., Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchon E., Ray R., Hulver M.W., McMillan R.P., Voy B.H. Fasting rapidly increases fatty acid oxidation in white adipose tissue of young broiler chickens. Adipocyte. 2017;6:33–39. doi: 10.1080/21623945.2016.1263777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Klein S.A., Silva F.A., Kwakkel R.P., Zuidhof M.J. The effect of quantitative feed restriction on allometric growth in broilers. Poult. Sci. 2017;96:118–126. doi: 10.3382/ps/pew187. [DOI] [PubMed] [Google Scholar]
- Wang Y.J., Bian Y., Luo J., Lu M., Xiong Y., Guo S.Y., Yin H.Y., Lin X., Li Q., Chang C.C.Y., Chang T.Y., Li B.L., Song B.L. Cholesterol and fatty acids regulate cysteine ubiquitylation of ACAT2 through competitive oxidation. Nat. Cell Biol. 2017;19:808–819. doi: 10.1038/ncb3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Jin L., Li Y., Tang Q., Hu S., Xu H., Gill C.A., Li M., Wang J. Transcriptomic analysis between normal and high-intake feeding geese provides insight into adipose deposition and susceptibility to fatty liver in migratory birds. BMC Genom. 2019;20:372. doi: 10.1186/s12864-019-5765-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z., Li P., Huang S., Lkhagvagarav P., Zhu M., Liang C., Jia C. Identification of key genes and molecular mechanisms associated with low egg production of broiler breeder hens in ad libitum. BMC Genom. 2019;20:408. doi: 10.1186/s12864-019-5801-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N.Y., Si W., Li M., Gong M., Larivière J.M., Nanaei H.A., Bian P.P., Jiang Y., Zhao X. Genome-wide scan for selective footprints and genes related to cold tolerance in Chantecler chickens. Zool. Res. 2021;42:710–720. doi: 10.24272/j.issn.2095-8137.2021.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Wang Y., Li Z., Hua Q., Jiang M., Fan X. LncRNA GAS5 knockdown mitigates hepatic lipid accumulation via regulating MiR-26a-5p/PDE4B to activate cAMP/CREB pathway. Front. Endocrinol. (Lausanne) 2022;13 doi: 10.3389/fendo.2022.889858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaefarian F., Abdollahi M.R., Cowieson A., Ravindran V. Avian liver: the forgotten organ. Animals (Basel) 2019;9:63. doi: 10.3390/ani9020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D.D., Wang D.D., Wang Z., Wang Y.B., Li G.X., Sun G.R., Tian Y.D., Han R.L., Li Z.J., Jiang R.R., Liu X.J., Kang X.T., Li H. Estrogen abolishes the repression role of gga-miR-221-5p targeting ELOVL6 and SQLE to promote lipid synthesis in chicken liver. Int. J. Mol. Sci. 2020;21:1624. doi: 10.3390/ijms21051624. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.