Abstract
Adult twin neuroimaging studies have revealed that cortical thickness (CT) and surface area (SA) are differentially influenced by genetic information, leading to their spatially distinct genetic patterning and topography. However, the postnatal origins of the genetic topography of CT and SA remain unclear, given the dramatic cortical development from neonates to adults. To fill this critical gap, this study unprecedentedly explored how genetic information differentially regulates the spatial topography of CT and SA in the neonatal brain by leveraging brain magnetic resonance (MR) images from 202 twin neonates with minimal influence by the complicated postnatal environmental factors. We capitalized on infant-dedicated computational tools and a data-driven spectral clustering method to parcellate the cerebral cortex into a set of distinct regions purely according to the genetic correlation of cortical vertices in terms of CT and SA, respectively, and accordingly created the first genetically informed cortical parcellation maps of neonatal brains. Both genetic parcellation maps exhibit bilaterally symmetric and hierarchical patterns, but distinct spatial layouts. For CT, regions with closer genetic relationships demonstrate an anterior-posterior (A-P) division, while for SA, regions with greater genetic proximity are typically within the same lobe. Certain genetically informed regions exhibit strong similarities between neonates and adults, with the most striking similarities in the medial surface in terms of SA, despite their overall substantial differences in genetic parcellation maps. These results greatly advance our understanding of the development of genetic influences on the spatial patterning of cortical morphology.
SIGNIFICANCE STATEMENT Genetic influences on cortical thickness (CT) and surface area (SA) are complex and could evolve throughout the lifespan. However, studies revealing distinct genetic topography of CT and SA have been limited to adults. Using brain structural magnetic resonance (MR) images of twins, we unprecedentedly discovered the distinct genetically-informed parcellation maps of CT and SA in neonatal brains, respectively. Each genetic parcellation map comprises a distinct spatial layout of cortical regions, where vertices within the same region share high genetic correlation. These genetic parcellation maps of CT and SA of neonates largely differ from those of adults, despite their highly remarkable similarities in the medial cortex of SA. These discoveries provide important insights into the genetic organization of the early cerebral cortex development.
Keywords: cortical thickness, genetic topography, neonates, surface area
Introduction
Cortical thickness (CT) and surface area (SA) are two essential morphologic features of the cerebral cortex and have been widely adopted in neuroimaging studies (Sowell et al., 2004; Hazlett et al., 2011; Wierenga et al., 2014; Dubois and Dehaene-Lambertz, 2015; Lyall et al., 2015; F. Wang et al., 2019b). Although both CT and SA have complex nonlinear and dynamic development, especially during early postnatal stages (Knickmeyer et al., 2008; Li et al., 2013, 2014a), each has its own distinct spatiotemporal patterns (Raznahan et al., 2011; Storsve et al., 2014; Wierenga et al., 2014; Bethlehem et al., 2022). Specifically, CT increases rapidly after birth, reaches the peak at around 14 months of age (F. Wang et al., 2019b), and then gradually decreases thereafter (Bethlehem et al., 2022). By contrast, SA expands to 69% of adult size by two years of age (Lyall et al., 2015), peaks at ∼11 years of age (Bethlehem et al., 2022), and turns out to be the principal factor that drives the growth of the cerebral cortex after CT reaching its peak (Wierenga et al., 2014). The early development of CT and SA has critical influences on behavioral and cognitive abilities as well as neurodevelopmental and neuropsychiatric disorders (Stiles and Jernigan, 2010; Brown and Jernigan, 2012; Ecker et al., 2014; Gerrits et al., 2016; Haring et al., 2016; Li et al., 2016; Vijayakumar et al., 2016). Discovering the underlying mechanisms that drive the distinct early development of CT and SA is critical for better understanding of healthy and disordered brains.
Intrinsic genetic mechanisms are widely perceived as one of the major determinants of the patterns of CT and SA (Panizzon et al., 2009; Winkler et al., 2010; Eyler et al., 2012; Jha et al., 2018; Strike et al., 2019). Previous studies have reported that the distinct genetic influences on CT and SA change from neonates to adults and exhibit regionally heterogeneous patterns (Panizzon et al., 2009; Winkler et al., 2010; Eyler et al., 2012; Schmitt et al., 2014; Jha et al., 2018; Strike et al., 2019; Teeuw et al., 2019; K. Xia et al., 2022). For example, the genetic influence on the total SA, whether in neonatal stage or in adulthood, is strong and significant. In contrast, the genetic influence on the average CT is weak and insignificant during the neonatal stage, although it increases during adulthood (Panizzon et al., 2009; Jha et al., 2018). Previous studies also revealed that the heritability estimates of CT (neonates: 0.01–0.52, adults: 0.06–0.73) and SA (neonates: 0.01–0.76, adults: 0.17–0.76) vary across cortical regions and age groups (Winkler et al., 2010; Jha et al., 2018). With reference to the above, genetic-based region-specific patterns of CT and SA may provide important information to many brain disorders. Nowadays, a series of studies on how genetic factors influence the formation of region-specific patterns of CT and SA have been performed using structural magnetic resonance imaging (MRI) of adult twin brains (Rubenstein et al., 1999; Panizzon et al., 2009; Chen et al., 2011, 2013; Schlaggar, 2011). The distinct genetic-based cortical parcellation maps of CT and SA in adults and their different organizing principles thus have been unveiled (Chen et al., 2011, 2013), providing us valuable references for understanding cortical regional specialization and potential genetic vulnerabilities associated with brain disorders. However, it remains unknown how genetic factors regulate the initial spatial patterns of CT and SA after birth, and whether they are preserved through postnatal development or evolve with age.
To fill this knowledge gap, we aimed to discover the initial genetically-regulated spatial patterns of CT and SA by taking advantage of neonatal twins. The motivation is that CT and SA in the neonatal brains are mainly genetically determined and minimally affected by the complicated postnatal environmental factors and thus are ideal candidates for discovering the postnatal origins of genetically influenced patterns. Furthermore, existing studies have demonstrated the genetic hierarchy of cortical arealization in animals (O'Leary et al., 2007) and the hierarchical transcriptional architecture of the human brain regions (Kang et al., 2011). Consistent with these findings, researchers also unveiled the hierarchical cortical genetic parcellation maps of adult CT and SA (Chen et al., 2013). We thus reasonably speculate that the cortical genetic parcellation maps of neonatal CT and SA should also be hierarchical.
To this end, brain structural MRI scans from 202 same-sex neonatal twins from the University of North Carolina (UNC) Early Brain Development Study, including monozygotic (MZ) twins and dizygotic (DZ) twins, were used in this study. The classical twin analysis model (Maes, 2005; Neale and Cardon, 2013), also termed as ACE model, was adopted to estimate the genetic and environmental influences on the variances of CT and SA, and to compute the genetic correlations between different vertices on the cortical surface. Accordingly, we leveraged the data-driven spectral clustering (Ng et al., 2001; Von Luxburg, 2007) method to generate genetic parcellation maps of CT and SA in neonatal brains. Since vertices within the same region are maximally genetically correlated, the resulting parcellation maps well represent the genetically regulated topography of CT and SA in neonates. Then, we calculated the pair-wise genetic similarity matrices among regions of discovered genetic parcellations of CT and SA, respectively. Based on these two genetic similarity matrices, we adopted the dendrogram to further explore the hierarchical genetic organization among regions of genetic parcellations of CT and SA, respectively.
Materials and Methods
Participants
The Institutional Review Boards of the University of North Carolina at Chapel Hill and Duke University Medical Center (DUMC) approved this study. Pregnant mothers, who have provided written informed consent for this research, were recruited during their second trimesters of pregnancy from prenatal diagnostic clinics at UNC Hospitals and DUMC. Subjects with abnormal fetal ultrasounds or mothers with major medical diseases or psychiatric illnesses were excluded from this study. Subjects included in this study had no congenital anomaly, significant medical illness, or MRI abnormality (Gilmore et al., 2012). Structural MR images from 202 neonates, including 112 DZ twins and 90 MZ twins with same sex, were adopted in this study. The detailed demographic information is shown in Table 1.
Table 1.
Demographic information of used neonatal twins
| All | Male | Female | |
|---|---|---|---|
| Subject numbers | 202 | 110 | 92 |
| PMW at birth | 35.48 ± 2.27 | 35.53 ± 2.26 | 35.41 ± 2.31 |
| PMW at scan | 40.80 ± 1.77 | 40.73 ± 1.64 | 40.89 ± 1.92 |
PMW: postmenstrual weeks
MRI acquisition
All T1-weighted and T2-weighted images were collected on a Siemens head-only 3T scanner. Acquired with a 3D magnetization-prepared rapid gradient echo (MPRAGE) sequence, the acquisition parameters of T1-weighted images (160 sagittal slices) are: TR/TE = 1,820/4.38 ms, flip angle = 7°, inversion time = 1,100 ms, and resolution = 1 × 1 × 1 mm3. T2-weighted images (70 transverse slices) were acquired with the turbo spin-echo (TSE) sequences and acquisition parameters are: TR/TE = 7,380/119 ms, flip angle = 150° and resolution =1.25 × 1.25 × 1.95 mm3.
Image processing and cortical surface reconstruction
All T1-weighted and T2-weighted MR images were processed using an infant-dedicated computational pipeline (http://www.ibeat.cloud/) described previously (Li et al., 2014a, 2015, 2019; L. Wang et al., 2018, 2023). The processing procedure contains the following main steps: (1) correction of intensity inhomogeneity of both T1-weighted and T2-weighted MR images using the N3 method (Sled et al., 1998); (2) linear alignment of each T2-weighted image onto its corresponding T1-weighted image using FLIRT (Smith et al., 2004); (3) removal of brain skull, brainstem, and cerebellum using a deep learning-based method (Zhang et al., 2019); (4) segmentation of brain tissues into white matter, gray matter and cerebrospinal fluid through a deep learning-based segmentation method (L. Wang et al., 2018); (5) separation of each brain into left and right hemispheres with noncortical regions filled with white matter.
After that, cortical surfaces of each MRI scan were reconstructed by the following steps: (1) correction of topological defects of white matter using a learning-based topological correction method (Sun et al., 2019); (2) reconstruction of the inner cortical surface of each hemisphere by tessellating the corrected white matter as a triangular mesh; (3) reconstruction of the outer cortical surface of each hemisphere by deforming the inner cortical surface outwards while avoiding mesh self-intersection (Li et al., 2012, 2014a); and generation of the middle cortical surface as the geometric average of the inner and outer cortical surfaces; (4) mapping the inner cortical surface onto a standard sphere while minimizing the geometric distortion (Fischl et al., 1999). To enable the vertex-wise intersubject comparison and analysis of CT and SA, we further registered each mapped spherical surface onto the age-matched template in the UNC 4D Neonatal and Infant Cortical Surface Atlases (Wu et al., 2019) using Spherical Demons (Yeo et al., 2009) and accordingly resampled each surface to a 163,842 standard mesh tessellation to build the vertex-to-vertex cortical correspondences across subjects. For each vertex, CT was computed as the minimum distance between the inner and outer surfaces, while SA was computed as one-third the sum of areas of all triangles associated with that vertex on the resampled middle cortical surface. The 163,842 vertex-wise CT and SA maps were then smoothed using iterative nearest neighbor averaging with 2560 iterations as previously described (Chen et al., 2013; F. Wang et al., 2019b). To reduce the computational cost of discovering the genetic regions, we further resampled CT and SA maps to have 2562 vertices on each hemisphere.
Twin analysis
The common model in twin analysis is the standard ACE model, which assumes that the variation in a phenotype is caused by additive genetic effects (A), common environmental effects (C), and individual-specific environmental effects (E; Maes, 2005; Neale and Cardon, 2013). In the univariate ACE model, the genotypes of MZ twins are identical and thus share 100% of their genes, while DZ twins share 50% of their genes. The common environmental effects among members of a twin pair are equal, despite their zygosity. As for the individual-specific environmental effects, they are independent across members of any twin pairs.
The bivariate ACE model is an extension of the univariate ACE model (Maes, 2005; Neale and Cardon, 2013). It can not only decompose the phenotypic variance into genetic and environmental variances, but also decompose the covariance between phenotypes into genetic and environmental covariances. Therefore, the genetic and environmental correlations between phenotypes can be estimated according to the genetic and environmental covariances. Of note, genetic correlations reflect the degree to which two phenotypes share genetic variance based on twin data. Mathematically, the genetic correlation between two phenotypes is calculated as the genetic covariance of two phenotypes divided by the square root of the product of their separate genetic variances (Neale and Cardon, 2013).
Since we aimed to, respectively, obtain the genetic correlations of CT and SA between any two vertices on the cortical surface, we adopted the standard bivariate ACE model, as in previous studies of genetic influence on CT (Panizzon et al., 2009; Rimol et al., 2010; Chen et al., 2013).
To avoid additional confounding sex effects within twins, we focused on twin pairs with the same sex. Thus, the adopted 202 subjects in this study are all same-sex twins. Before model fitting, the mean CT and total SA of the whole cerebral cortex were, respectively, regressed out from the CT and SA at each vertex through the linear regression as in (Chen et al., 2012, 2013; Jha et al., 2018).
Discovering genetic parcellations with spectral clustering
To discover the genetic parcellations of CT and SA in neonates, after obtaining their genetic correlation matrices, we leveraged the spectral clustering method (Ng et al., 2002; Von Luxburg, 2007) to group cortical vertices into a set of distinct regions. The main motivation of using spectral clustering is that it is flexible to discover clusters with arbitrary shapes according to the realistic distribution of data similarity (Meila, 2016). Specifically, spectral clustering transforms the data similarity matrix into eigenspace and represents the similarity matrix with top decomposed eigenvectors, thus can better capture the distributions of the data similarity for identifying clusters. Given a similarity matrix , where denotes the total vertex numbers of both left and right hemispheres, and the predefined region number , the spectral clustering method will: (1) calculate a diagonal degree matrix with ; (2) compute the normalized Laplacian , where is the identity matrix; (3) compute the top eigenvectors of , and arrange them in columns to form a matrix ; (4) normalize each row of to norm 1 to form the matrix with ; (5) treat each row of as a point in and perform K-means method to cluster the points into clusters.
In this study, corresponds to , which represents the genetic correlation between vertices and of CT or SA. To take advantage of strong positive correlation between vertices, before feeding the genetic correlation matrix into spectral clustering, we empirically processed the matrix by keeping the top 20% of the correlations and setting others to 0, similar to previously published work (Yeo et al., 2011). After performing the spectral clustering, the vertices on the cortical surface were grouped into regions, each with distinct genetic influence, leading to a genetic parcellation map.
To determine an appropriate region number of genetic parcellation, we applied the widely used silhouette coefficients (Rousseeuw, 1987) adopted in many previous studies (Chen et al., 2012, 2013; F. Wang et al., 2019a,b; J. Xia et al., 2019). The silhouette coefficients of the vertex denoted as based on both intraregion and inter-region dissimilarity is calculated as: . Herein, denotes the average intra-region dissimilarity between vertex and all other vertices in the same region, while denotes the minimum average inter-region dissimilarity of vertex with vertices in other regions. The dissimilarity between any two vertices was computed as where is genetic similarity between vertices and . The average silhouette coefficient from all vertices is used. High average silhouette coefficients indicate better clustering results with large inter-region dissimilarity and small intraregion dissimilarity.
Since the previous adult twin studies (Chen et al., 2013) determined 12 as the appropriate region number under genetic influence, we set the region number ranging from 2 to 15 in this study for comparison. Then the average silhouette coefficient was used to determine the most appropriate region number to further investigate the genetic parcellation maps in neonates. Finally, we explored the genetic relatedness between discovered distinct regions and constructed a dendrogram accordingly.
Results
Genetic parcellation maps of neonatal CT and SA
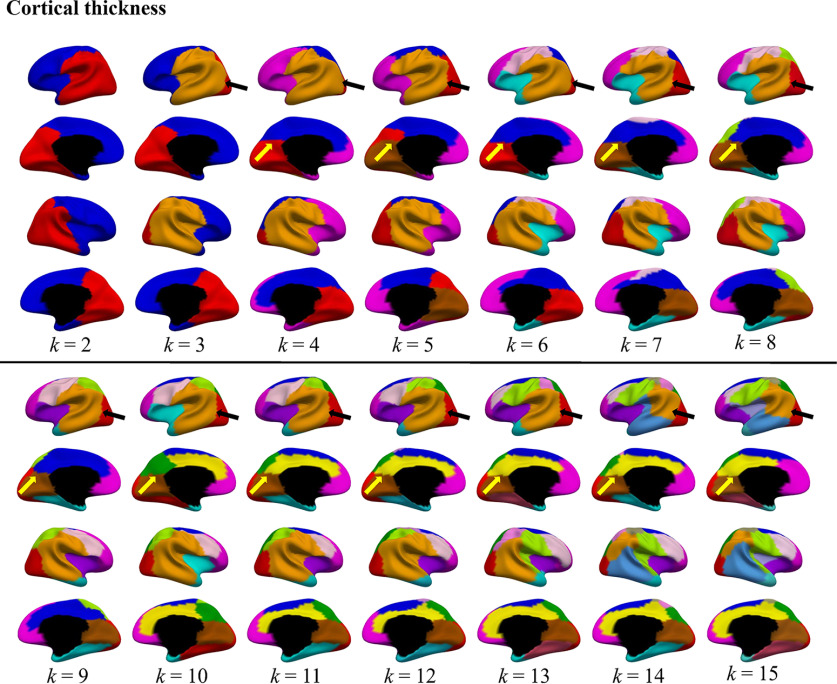
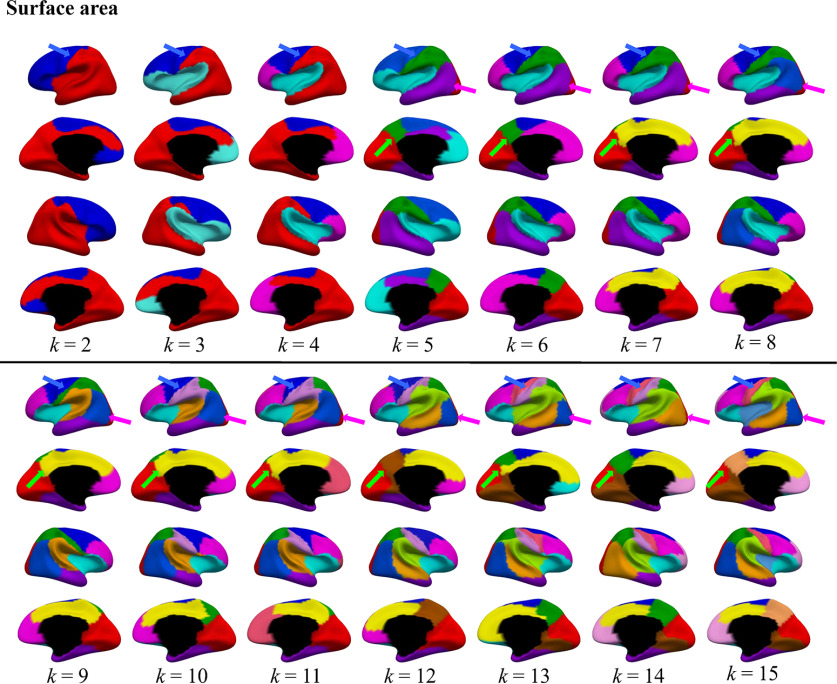
We first identified the genetic parcellation maps of CT and SA on both left and right hemispheres (Figs. 1, 2), with region numbers k increasing from 2 to 15, respectively. As can be seen, the discovered genetic parcellation maps of both CT and SA appear to be bilaterally symmetric across most regions, although we did not impose any constraint for hemispheric symmetry. Moreover, with the region number increasing, newly emerged regions largely follow the previously formed region boundaries and yield the meaningful hierarchical genetic parcellations of CT and SA. Meanwhile, the discovered genetic parcellation maps of CT differ remarkably from SA.
Figure 1.
Genetic parcellation maps of CT on both left and right hemispheres of neonates, with region numbers increasing from 2 to 15. The black and yellow arrows indicate some region boundaries consistently preserved across different region numbers.
Figure 2.
Genetic parcellation maps of SA on both left and right hemispheres of neonates, with region numbers increasing from 2 to 15. The arrows indicate some region boundaries consistently preserved across different region numbers.
As for CT (Fig. 1), when the region number equals to 2, the unveiled two-region basic genetic parcellation exhibits an anterior-posterior (A-P) division, which separates the frontal and insular cortices from other cortices. When increasing the region number to 4, the medial parietal-occipital boundary, indicated by yellow arrows, emerges and is well preserved till the 15-region partition. Besides, the boundary of the lateral occipital cortex, indicated by black arrows, appears when region number is three and exists until the region number reaches 15.
Regarding SA (Fig. 2), the identified two-region basic genetic parcellation roughly demonstrates an A-P division on the lateral surface, but a lateral-medial division on the medial surface. The lateral occipital boundary indicated by magenta arrows consistently exists with region numbers from 5 to 15. Moreover, the medial occipital boundary indicated by green arrows also appears when the region number is 5 and is well preserved until the region reaches 15. Meanwhile, the boundary, which separates the frontal and nonfrontal parts and is indicated by blue arrows, is consistently well-preserved from 2 to 15 regions.
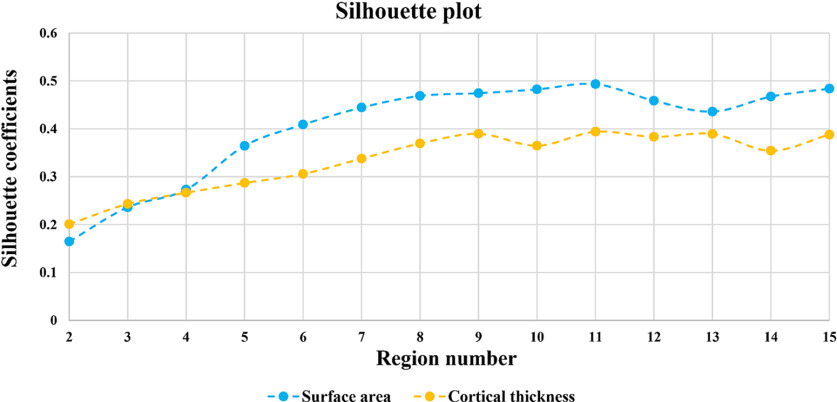
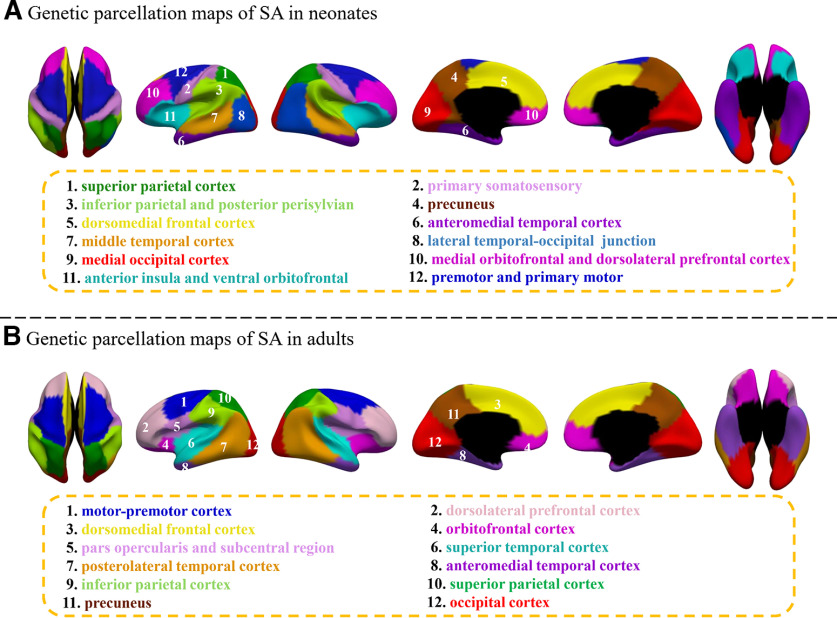
Then we estimated the average silhouette coefficients of genetic parcellation maps with different region numbers to help discover the appropriate region number. Accordingly, the appropriate region number was determined based on two criteria: (1) having a relatively high value of silhouette coefficient, which indicates better quality of a parcellation; (2) having relatively stable silhouette coefficients across the neighboring region numbers. The silhouette coefficient of CT (Fig. 3) increases from to and then becomes relatively stable from to . While the silhouette coefficient of SA keeps increasing from to and then gradually decreases to reach a local minimum at . Although the local maximal values were reached when =11 for both CT and SA, to make a direct comparison between our neonatal parcellation results and the published 12-region parcellation results of adults (Chen et al., 2013) and meanwhile consider the above criteria, we took 12-region as the final parcellation results of CT and SA in neonates (Figs. 4A, 5A). Since the region order has no influence on our results, to make the order number of each column of the pair-wise genetic similarity matrix to be continuous, we re-ordered the original region order directly derived from the spectral clustering. Therefore, regions in Figures 4A and 5A were labeled according to the order numbers of columns in the pair-wise genetic similarity matrices of CT and SA, respectively. In addition, to make a better visual comparison between neonates and adults, we labeled the relatively similar regions in their corresponding parcellation maps with the same color.
Figure 3.
Average silhouette coefficients of genetic parcellations with different numbers of regions. The yellow and blue colors represent silhouette coefficients of CT and SA, respectively.
Figure 4.
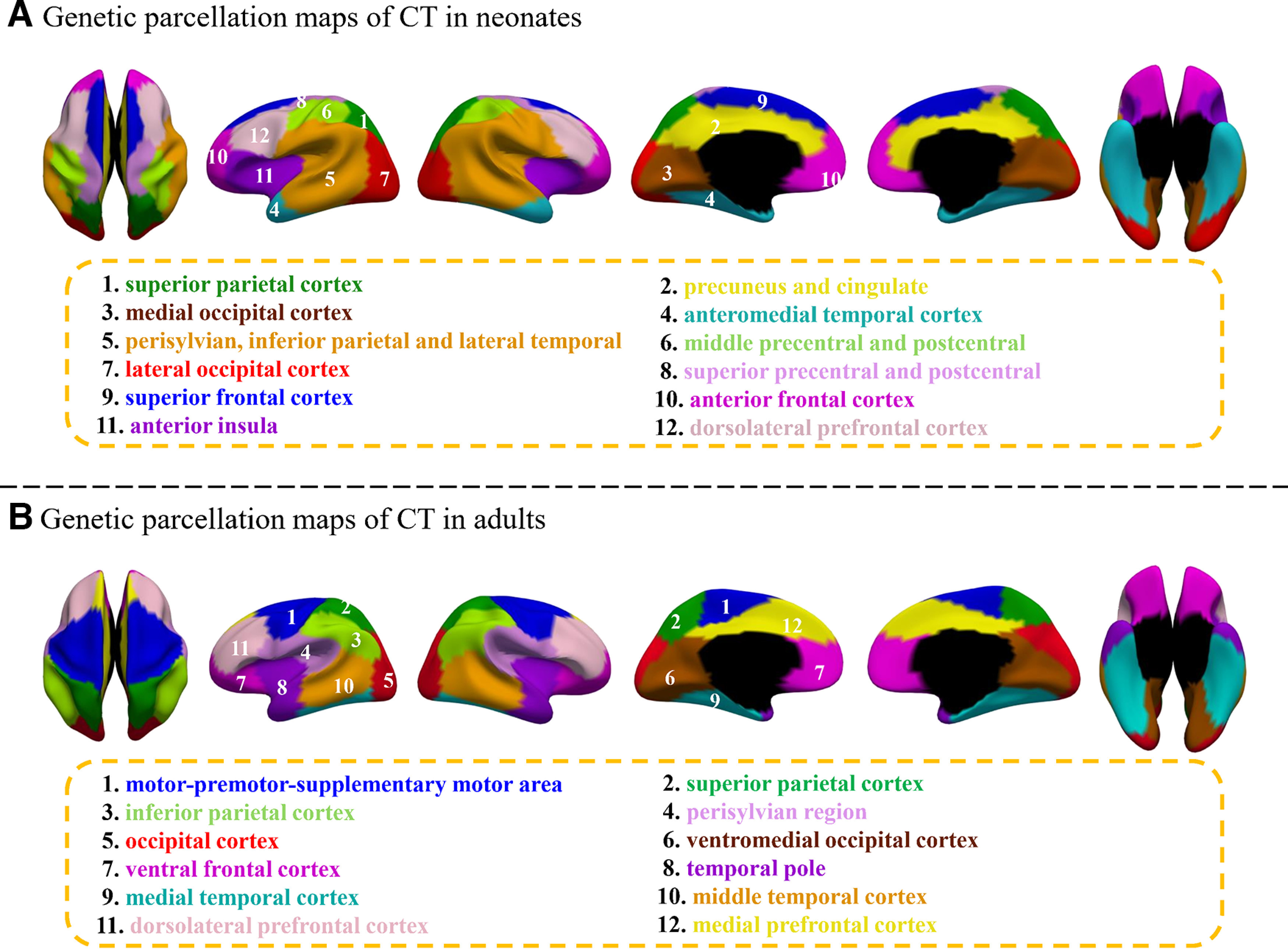
Comparison of genetic parcellation maps of CT in neonates and adults. A, Our discovered genetic parcellation map with 12 regions in neonates. B, The published genetic parcellation map with 12 regions in adults (Chen et al., 2013).
Figure 5.
Comparison of genetic parcellation maps of SA in neonates and adults. A, Our discovered genetic parcellation map with 12 regions in neonates. B, The published genetic parcellation map with 12 regions in adults (Chen et al., 2013).
The parcellation maps with 12 regions of CT and SA in neonates (Figs. 4A, 5A) approximately correspond to the structurally or functionally meaningful regions. With approximate names shown below the parcellation map, discovered genetic regions of CT approximately correspond to: (1) superior parietal cortex, (2) precuneus and cingulate, (3) medial occipital cortex, (4) anteromedial temporal cortex, (5) perisylvian, inferior parietal and lateral temporal cortex, (6) middle precentral and postcentral, (7) lateral occipital cortex, (8) superior precentral and postcentral, (9) superior frontal cortex, (10) anterior frontal cortex, (11) anterior insula, and (12) dorsolateral prefrontal cortex. The genetic parcellation map of CT in neonates (Fig. 4A) is largely different from that in adults (Fig. 4B), except for a few regions in the posterior and medial surfaces. For example, region 1 (superior parietal cortex), region 3 (medial occipital cortex), region 4 (anteromedial temporal cortex), region 7 (lateral occipital cortex), and region 10 (anterior frontal cortex) in neonates are roughly consistent with region 2, region 6, region 9, region 5, and region 7 of adults, respectively.
The discovered genetic regions of SA in neonates (Fig. 5A) approximately correspond to: (1) superior parietal cortex, (2) primary somatosensory, (3) inferior parietal and posterior perisylvian, (4) precuneus, (5) dorsomedial frontal cortex, (6) anteromedial temporal cortex, (7) middle temporal cortex, (8) lateral temporal-occipital junction, (9) medial occipital cortex, (10) medial orbitofrontal and dorsolateral prefrontal cortex, (11) anterior insula and ventral orbitofrontal, and (12) premotor and primary motor. Strikingly, genetic parcellation maps of SA in neonates (Fig. 5A) and adults (Fig. 5B) seem to resemble each other in many regions, especially for regions in the medial surface. For example, region 1 (superior parietal cortex), region 12 (premotor and primary motor), region 4 (precuneus), region 5 (dorsomedial frontal cortex), region 6 (anteromedial temporal cortex), and region 9 (medial occipital cortex) in neonates, respectively, correspond well to region 10, region 1, region 11, region 3, region 8, and region 12 in adults (Fig. 5B).
Hierarchical organization of genetic regions
After discovering the above genetically distinct regions, we tried to reveal the hierarchical organization between these regions. First, to visualize the genetic proximity between these regions, we built their pair-wise genetic similarity matrix, with each row and column corresponding to the sorted region number. Specifically, based on the genetic correlation matrix obtained from twin analysis, each cell in the pair-wise genetic similarity matrix is the average of genetic correlations within and between regions as described previously (Chen et al., 2011, 2013). Then, based on the pair-wise genetic similarity matrix of CT or SA, we performed the hierarchical clustering method to group the genetic regions into a multilevel cluster tree or dendrogram to represent the genetic organization among regions. Since the dendrogram shows the hierarchical relationship between regions, it can help us to make a direct comparison of the hierarchical organizations of CT and SA. The color bars of heatmaps represent the mean genetic correlations within and between regions of CT (SA), where higher values indicate closer genetic relatedness.
As can be observed from the dendrograms (Fig. 6A,B), which are, respectively, derived from the pair-wise genetic similarity matrix of CT and SA, the hierarchical relationship between genetic regions of CT differs remarkably from that of SA. Specifically, genetic correlations of SA (Fig. 6B) between regions within the same lobe are higher than those from different lobes, except for the precuneus cortex (region 4), dorsolateral frontal cortex (region 5), and medial occipital cortex (region 9). Regions that are genetically more similar in SA can be roughly grouped as: (1) parietal lobe, including the superior parietal cortex, primary somatosensory cortex, inferior parietal and posterior perisylvian, as well as precuneus cortex (regions 1–4); (2) temporal lobe, including the anteromedial temporal cortex, middle temporal cortex and the lateral temporal-occipital junction (regions 6–8); (3) frontal lobe, including the medial orbitofrontal and dorsolateral prefrontal, anterior insula and ventral orbitofrontal, and premotor and primary motor cortex (regions 10–12).
Figure 6.
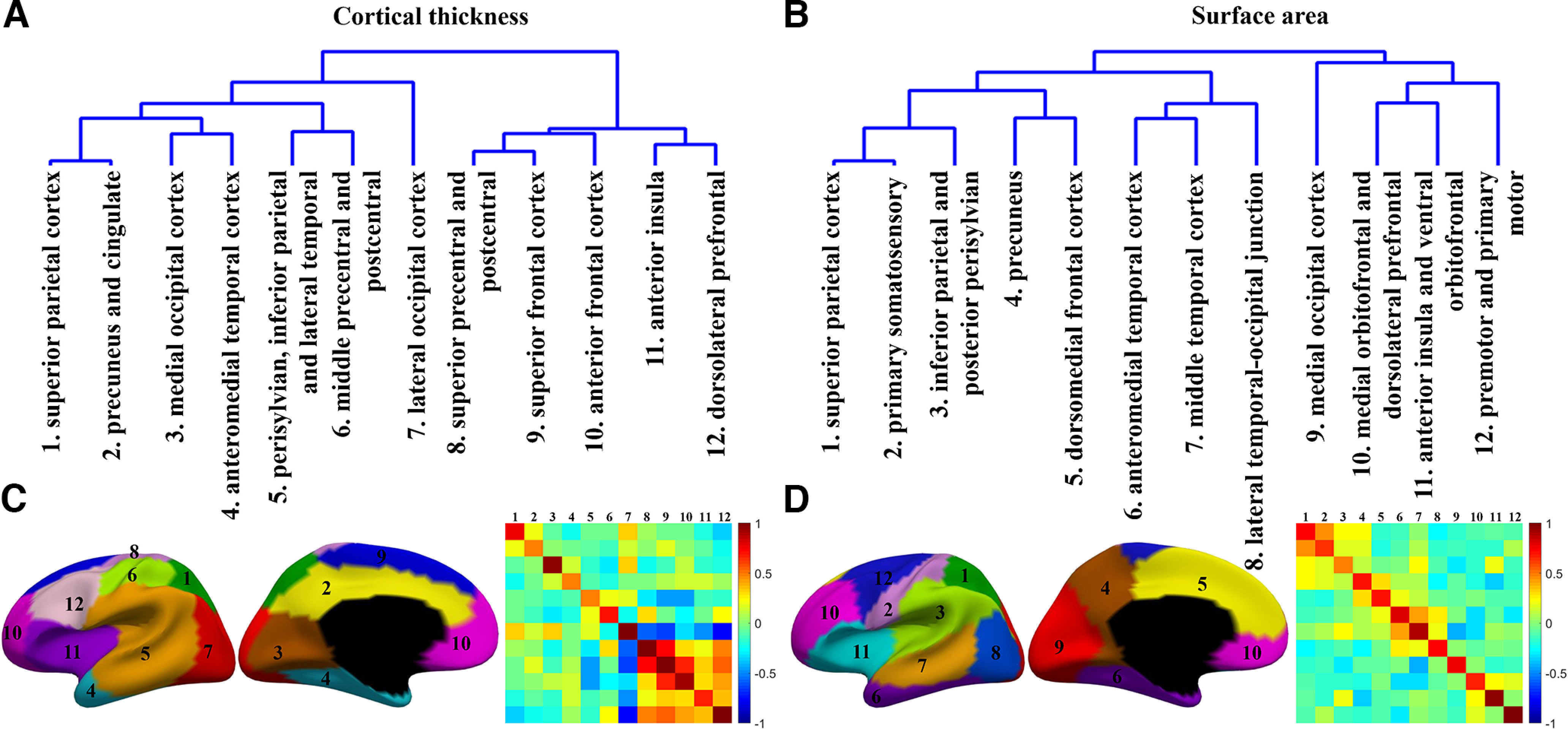
Hierarchical organization of our discovered genetic regions in neonates. A, B, Dendrograms derived from 12 regions based on the pair-wise genetic similarity matrix among clusters of CT and SA, respectively. C, D, Corresponding genetic parcellation maps with region locations labeled in number. Heatmaps represent the pair-wise genetic similarity matrix among regions of CT (C) and SA (D).
The genetic organization of CT (Fig. 6A) among regions differs remarkably from that of SA (Fig. 6B). The organization among genetic regions of CT (Fig. 6A) can be roughly divided into frontal and nonfrontal parts, which can also be considered as an A-P division. Specifically, the frontal parts approximately contain the superior precentral and postcentral (region 8), superior frontal cortex (region 9), anterior frontal cortex (region 10), anterior insula (region 11), and dorsolateral prefrontal cortex (region 12). While the nonfrontal parts cover the superior parietal cortex (region 1), precuneus and cingulate (region 2), medial occipital cortex (region 3), anteromedial temporal cortex (region 4), perisylvian, inferior parietal and lateral temporal cortex (region 5), middle precentral and postcentral (region 6), and the lateral occipital cortex (region 7). Meanwhile, in the nonfrontal parts, regions with strong genetic correlation are typically spatially adjacent, but come from different lobes, e.g., regions 1 and 2, regions 3 and 4, and regions 5 and 6.
Discussion
To our knowledge, this is the first study to comprehensively explore the early postnatal genetic topography of CT and SA using neonatal twins. We found that the genetic parcellations of both CT and SA are bilaterally symmetric, and the discovered regions largely correspond to existing structural or functional parcels (Tzourio-Mazoyer et al., 2002; Desikan et al., 2006; Gordon et al., 2016). Of note, we performed the spectral clustering purely based on the genetic correlation matrix without any restrictions for hemispheric symmetry or anatomic knowledge, thus the naturally-formed bilaterally symmetric regions may reflect the intrinsic constraint of genetic regulation on cortical regionalization (formation of structurally and functionally distinct cortical regions) of both hemispheres, consistent with the observation in previous adult research (Wilkinson et al., 1989; Fraser et al., 1990; Puelles and Rubenstein, 2003; Tomancak et al., 2007).
We found that when the region number is 2, the basic genetic parcellations of CT and SA are similar to some extent on the lateral surface, as both roughly exhibit an anterior-posterior (A-P) division. As initial patterns of CT and SA are mainly determined by genetic factors before birth, these findings suggest that genetic influence in forming the initial patterns of CT and SA may be similar on the lateral surface on a coarse scale. Such results are generally consistent with previous studies that significant genetic overlap exists between CT and SA in infants (Jha et al., 2018). Studies in rodent models show that the initiation of cortical regionalization is predominantly driven by intrinsic genetic mechanisms involving morphogen gradients along the A-P, dorsal-ventral (D-V), and medial-lateral (M-L) axes of the developing cortex (O'Leary et al., 2007; Sansom and Livesey, 2009). These morphogens, which play critical roles in neurogenesis and progenitor cell proliferation (Tiberi et al., 2012), may serve as the common factors affecting the development of both CT and SA. Unlike in neonates, very little genetic correlation is found between CT and SA in adults (Chen et al., 2013). Moreover, in adults, the basic genetic regions of SA follow an A-P division, while the basic genetic regions of CT follow a D-V division (Chen et al., 2013). A possible explanation is that CT and SA may share similar genetic influences in neonates on a coarse scale, which subsequently diverge over time, leading to differences in basic genetic patterns between neonates and adults (K. Xia et al., 2022).
When the region number is 12, the genetic parcellations of CT and SA demonstrate significantly distinct patterns. As in the radial unit hypothesis of cerebral cortex development (Rakic, 1988, 2000), SA is determined by the number of radial columns, whereas CT is under the influence of the number of cells within a column. The two different processes may be manipulated by their unique regulatory genes and thus leading to distinct genetic patterns between CT and SA. In fact, a large-scale human study reveals that CT and SA are associated with largely distinct common genetic variants (Grasby et al., 2020). For example, SA is associated with common genetic variants affecting the Wnt signaling pathway (Nusse and Varmus, 1992; Nusse, 2005), which regulates progenitor cell expansion. On the contrary, CT is associated with common genetic variants affecting genes functioning in neural differentiation, migration, and myelination.
Moreover, since the patterns of CT and SA in neonates are mainly genetically determined and inevitably suffer from complicated postnatal environmental influence during development, the similar patterns of SA and different patterns of CT between neonates and adults may suggest that SA is under strong genetic influence, while CT has weaker genetic influences and is more likely influenced by environmental factors (Jha et al., 2018, 2019). Previous studies have revealed that SA is mainly determined by the shape of cortical folding (Mota and Herculano-Houzel, 2015; Garcia et al., 2018), which features the greatest development during the third trimester of pregnancy and has been well formed at term birth, especially for the primary and secondary cortical folds (Armstrong et al., 1995; White et al., 2010; Li et al., 2014b). Although SA undergoes more growth than CT from birth to adulthood, the SA expansion of the medial surface is less than the lateral surface (Li et al., 2013). Therefore, SA may intrinsically suffer from less postnatal environmental influences, especially for the medial surface, and keep relatively stable genetic patterns across the lifespan. Furthermore, though the peaking time of the average CT is around 14 months of age (F. Wang et al., 2019b), rather earlier than the total SA peaking at around 11 years of age (Bethlehem et al., 2022), CT still shows age-related changes across the lifespan and tends to thin after reaching its peak (Wierenga et al., 2014; Bethlehem et al., 2022). This processing is driven by age-related biological procedures, e.g., synaptic pruning (Stiles and Jernigan, 2010; Lyall et al., 2015) and myelination (Natu et al., 2019), which are reported to be influenced by environmental factors (Tooley et al., 2021). Moreover, compared with SA, CT is more sensitive (Hutton et al., 2009; Winkler et al., 2012) and vulnerable to postnatal environmental influences, likely leading to different genetic topography between neonates and adults.
These findings are also consistent with previous studies of genetic influences on CT and SA at different developmental stages. For example, in a neonatal twin study, genetic influences on the total SA (0.78) are significant and stronger than that on the average CT (0.29; Jha et al., 2018). Meanwhile, with twins between zero and two years of age, researchers found relatively stronger genetic influences on SA (0.59 in neonates, 0.74 in one-year olds, and 0.73 in two-year olds) than on CT (0.48 in neonates, 0.37 in one-year olds, and 0.44 in two-year olds) across all three examined age groups (K. Xia et al., 2022). Additionally, studies with participants between three and 20 years of age indicated that shared environmental influences (socioeconomic status) are larger on changes of CT than on changes of SA (Piccolo et al., 2016).
We also found that the hierarchical organization of CT among the 12 regions is different from that of SA. Roughly organized according to four cortical lobes, the discovered genetic regions of SA within the same lobe are genetically more correlated than regions across lobes, consistent with the organization principle of SA in adults (Chen et al., 2013). This suggests relatively little postnatal environmental influences on the genetic organization principle of SA. Nevertheless, the organization of CT in neonates does not always follow this principle. Specifically, for CT in neonates, clusters with closer genetic correlation roughly correspond to an A-P division, which separates the cerebral cortex into frontal and nonfrontal parts. Moreover, in the nonfrontal parts, regions with strong genetic correlations are typically from different lobes. According to the previous study on the developmental topography of CT during infancy (F. Wang et al., 2019b), this potentially reflects the differential maturation patterns of CT. For example, most regions in nonfrontal cortices reach their peaks of CT during the second postnatal year, while many regions in the frontal cortex show a monotonical increase of CT during the first two postnatal years, e.g., regions 8, 9, and 12 in Figure 4A. Previous adult studies also suggested that regions with close genetic similarity in CT tend to have similar maturational timing (Chen et al., 2013). Taken together, these findings suggest CT and SA have distinct organizational principles for genetic similarity among discovered regions since birth, which differentially evolve from neonates to adults.
It is also worth noting that the A-P division of the genetic organization of CT in neonates in our study aligns with prior studies investigating the evolution of the cerebral cortex. For example, previous studies discovered that both genetic correlation and structural covariance of CT in adults exhibited an A-P pattern (Valk et al., 2020). A comparable organization of structural covariance of CT in macaques is also present, potentially indicating that the A-P division is phylogenetically conserved during evolution. Further studies of the genetic organization of CT in macaques are needed to confirm this.
Although the revealed genetic parcellations of CT and SA in neonates are neurobiologically meaningful, some limitations in our study should be noted. First, our study only contains neonatal twins, thus further studies with twin data from different developing age groups is needed to fill the gap of genetic parcellations of CT and SA between neonates and adults. Second, genetic factors affect human cortical structures, which can be characterized not only by CT and SA, but also by other cortical features, e.g., cortical folding, gyrification, and functional connectivity. Future studies might be performed on how genetic factors influence other cortical features to complement the research on genetically influenced cortical parcellations during early brain development. Third, since many neurodevelopmental disorders are rooted during infancy, future research should be performed to explore whether genetic parcellations can help identify early biomarkers for early interventions.
In summary, this study has three contributions. First, leveraging the neonatal twins, for the first time, we revealed the postnatal origins of the genetic topography of CT and SA. The genetically informed parcellation maps of CT and SA are distinct, but both tend to be hierarchically organized and bilaterally symmetric and correspond well to existing structurally or functionally meaningful regions. Second, by comparing our revealed genetic parcellations in neonates with those in adults, we found their larger differences in most regions, but also striking similarities in many regions in terms of surface area, especially on the medial surface. Third, we showed that CT and SA have distinct organizational principles of genetic similarity among regions in the generated parcellation maps since birth, which differentially evolve from neonates to adults. These results help us better understand how genetic factors shape the initial postnatal organizational patterns of CT and SA and provide important references for future studies of the genetic contribution to the development of CT and SA. Our generated genetically informed cortical parcellation maps of CT and SA will be publicly available on our website.
Footnotes
This work was partially supported by National Institutes of Health Grants MH116225, MH117943, MH123202, MH116527, MH070890, and HD053000.
The authors declare no competing financial interests.
References
- Armstrong E, Schleicher A, Omran H, Curtis M, Zilles K (1995) The ontogeny of human gyrification. Cereb Cortex 5:56–63. 10.1093/cercor/5.1.56 [DOI] [PubMed] [Google Scholar]
- Bethlehem RAI, et al. (2022) Brain charts for the human lifespan. Nature 604:525–533. 10.1038/s41586-022-04554-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown TT, Jernigan TL (2012) Brain development during the preschool years. Neuropsychol Rev 22:313–333. 10.1007/s11065-012-9214-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Panizzon MS, Eyler LT, Jernigan TL, Thompson W, Fennema-Notestine C, Jak AJ, Neale MC, Franz CE, Hamza S, Lyons MJ, Grant MD, Fischl B, Seidman LJ, Tsuang MT, Kremen WS, Dale AM (2011) Genetic influences on cortical regionalization in the human brain. Neuron 72:537–544. 10.1016/j.neuron.2011.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Gutierrez ED, Thompson W, Panizzon MS, Jernigan TL, Eyler LT, Fennema-Notestine C, Jak AJ, Neale MC, Franz CE, Lyons MJ, Grant MD, Fischl B, Seidman LJ, Tsuang MT, Kremen WS, Dale AM (2012) Hierarchical genetic organization of human cortical surface area. Science 335:1634–1636. 10.1126/science.1215330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Fiecas M, Gutiérrez ED, Panizzon MS, Eyler LT, Vuoksimaa E, Thompson WK, Fennema-Notestine C, Hagler DJ, Jernigan TL, Neale MC, Franz CE, Lyons MJ, Fischl B, Tsuang MT, Dale AM, Kremen WS (2013) Genetic topography of brain morphology. Proc Natl Acad Sci U S A 110:17089–17094. 10.1073/pnas.1308091110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, Albert MS, Killiany RJ (2006) An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 31:968–980. 10.1016/j.neuroimage.2006.01.021 [DOI] [PubMed] [Google Scholar]
- Dubois J, Dehaene-Lambertz G (2015) Fetal and postnatal development of the cortex: MRI and genetics. Brain Mapping: An Encyclopedic Reference 2:11–19. [Google Scholar]
- Ecker C, Shahidiani A, Feng Y, Daly E, Murphy C, D'Almeida V, Deoni S, Williams SC, Gillan N, Gudbrandsen M, Wichers R, Andrews D, Van Hemert L, Murphy DGM (2014) The effect of age, diagnosis, and their interaction on vertex-based measures of cortical thickness and surface area in autism spectrum disorder. J Neural Transm (Vienna) 121:1157–1170. 10.1007/s00702-014-1207-1 [DOI] [PubMed] [Google Scholar]
- Eyler LT, Chen C-H, Panizzon MS, Fennema-Notestine C, Neale MC, Jak A, Jernigan TL, Fischl B, Franz CE, Lyons MJ, Grant M, Prom-Wormley E, Seidman LJ, Tsuang MT, Fiecas MJA, Dale AM, Kremen WS (2012) A comparison of heritability maps of cortical surface area and thickness and the influence of adjustment for whole brain measures: a magnetic resonance imaging twin study. Twin Res Hum Genet 15:304–314. 10.1017/thg.2012.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Dale AM (1999) Cortical surface-based analysis: II: inflation, flattening, and a surface-based coordinate system. Neuroimage 9:195–207. 10.1006/nimg.1998.0396 [DOI] [PubMed] [Google Scholar]
- Fraser S, Keynes R, Lumsden A (1990) Segmentation in the chick embryo hindbrain is defined by cell lineage restrictions. Nature 344:431–435. 10.1038/344431a0 [DOI] [PubMed] [Google Scholar]
- Garcia K, Kroenke C, Bayly P (2018) Mechanics of cortical folding: stress, growth and stability. Philos Trans R Soc Lond B Biol Sci 373:20170321. 10.1098/rstb.2017.0321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrits NJ, van Loenhoud AC, van den Berg SF, Berendse HW, Foncke EM, Klein M, Stoffers D, van der Werf YD, van den Heuvel OA (2016) Cortical thickness, surface area and subcortical volume differentially contribute to cognitive heterogeneity in Parkinson's disease. PLoS One 11:e0148852. 10.1371/journal.pone.0148852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore JH, Shi F, Woolson SL, Knickmeyer RC, Short SJ, Lin W, Zhu H, Hamer RM, Styner M, Shen D (2012) Longitudinal development of cortical and subcortical gray matter from birth to 2 years. Cereb Cortex 22:2478–2485. 10.1093/cercor/bhr327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon EM, Laumann TO, Adeyemo B, Huckins JF, Kelley WM, Petersen SE (2016) Generation and evaluation of a cortical area parcellation from resting-state correlations. Cereb Cortex 26:288–303. 10.1093/cercor/bhu239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasby KL, et al. (2020) The genetic architecture of the human cerebral cortex. Science 367:eaay6690. 10.1126/science.aay6690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring L, Müürsepp A, Mõttus R, Ilves P, Koch K, Uppin K, Tarnovskaja J, Maron E, Zharkovsky A, Vasar E, Vasar V (2016) Cortical thickness and surface area correlates with cognitive dysfunction among first-episode psychosis patients. Psychol Med 46:2145–2155. 10.1017/S0033291716000684 [DOI] [PubMed] [Google Scholar]
- Hazlett HC, Poe MD, Gerig G, Styner M, Chappell C, Smith RG, Vachet C, Piven J (2011) Early brain overgrowth in autism associated with an increase in cortical surface area before age 2 years. Arch Gen Psychiatry 68:467–476. 10.1001/archgenpsychiatry.2011.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton C, Draganski B, Ashburner J, Weiskopf N (2009) A comparison between voxel-based cortical thickness and voxel-based morphometry in normal aging. Neuroimage 48:371–380. 10.1016/j.neuroimage.2009.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha SC, Xia K, Schmitt JE, Ahn M, Girault JB, Murphy VA, Li G, Wang L, Shen D, Zou F, Zhu H, Styner M, Knickmeyer RC, Gilmore JH (2018) Genetic influences on neonatal cortical thickness and surface area. Hum Brain Mapp 39:4998–5013. 10.1002/hbm.24340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha SC, Xia K, Ahn M, Girault JB, Li G, Wang L, Shen D, Zou F, Zhu H, Styner M, Gilmore JH, Knickmeyer RC (2019) Environmental influences on infant cortical thickness and surface area. Cereb Cortex 29:1139–1149. 10.1093/cercor/bhy020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, et al. (2011) Spatio-temporal transcriptome of the human brain. Nature 478:483–489. 10.1038/nature10523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knickmeyer RC, Gouttard S, Kang C, Evans D, Wilber K, Smith JK, Hamer RM, Lin W, Gerig G, Gilmore JH (2008) A structural MRI study of human brain development from birth to 2 years. J Neurosci 28:12176–12182. 10.1523/JNEUROSCI.3479-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Nie J, Wu G, Wang Y, Shen D; Alzheimer's Disease Neuroimaging Initiative (2012) Consistent reconstruction of cortical surfaces from longitudinal brain MR images. Neuroimage 59:3805–3820. 10.1016/j.neuroimage.2011.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Nie J, Wang L, Shi F, Lin W, Gilmore JH, Shen D (2013) Mapping region-specific longitudinal cortical surface expansion from birth to 2 years of age. Cereb Cortex 23:2724–2733. 10.1093/cercor/bhs265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Nie J, Wang L, Shi F, Gilmore JH, Lin W, Shen D (2014a) Measuring the dynamic longitudinal cortex development in infants by reconstruction of temporally consistent cortical surfaces. Neuroimage 90:266–279. 10.1016/j.neuroimage.2013.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Wang L, Shi F, Lyall AE, Lin W, Gilmore JH, Shen D (2014b) Mapping longitudinal development of local cortical gyrification in infants from birth to 2 years of age. J Neurosci 34:4228–4238. 10.1523/JNEUROSCI.3976-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Wang L, Shi F, Gilmore JH, Lin W, Shen D (2015) Construction of 4D high-definition cortical surface atlases of infants: methods and applications. Med Image Anal 25:22–36. 10.1016/j.media.2015.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Wang L, Shi F, Lyall AE, Ahn M, Peng Z, Zhu H, Lin W, Gilmore JH, Shen D (2016) Cortical thickness and surface area in neonates at high risk for schizophrenia. Brain Struct Funct 221:447–461. 10.1007/s00429-014-0917-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Wang L, Yap P-T, Wang F, Wu Z, Meng Y, Dong P, Kim J, Shi F, Rekik I, Lin W, Shen D (2019) Computational neuroanatomy of baby brains: a review. Neuroimage 185:906–925. 10.1016/j.neuroimage.2018.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyall AE, Shi F, Geng X, Woolson S, Li G, Wang L, Hamer RM, Shen D, Gilmore JH (2015) Dynamic development of regional cortical thickness and surface area in early childhood. Cereb Cortex 25:2204–2212. 10.1093/cercor/bhu027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes HH (2005) ACE model. In: Encyclopedia of statistics in behavioral science. Chichester, UK: John Wiley & Sons, Ltd. [Google Scholar]
- Meila M (2016) Spectral clustering: a tutorial for the 2010's. In: Handbook of cluster analysis, pp 1–23. Boca Raton: CRC Press. [Google Scholar]
- Mota B, Herculano-Houzel S (2015) Cortical folding scales universally with surface area and thickness, not number of neurons. Science 349:74–77. 10.1126/science.aaa9101 [DOI] [PubMed] [Google Scholar]
- Natu VS, Gomez J, Barnett M, Jeska B, Kirilina E, Jaeger C, Zhen Z, Cox S, Weiner KS, Weiskopf N, Grill-Spector K (2019) Apparent thinning of human visual cortex during childhood is associated with myelination. Proc Natl Acad Sci U S A 116:20750–20759. 10.1073/pnas.1904931116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale M, Cardon LR (2013) Methodology for genetic studies of twins and families. Vol 67. Berlin: Springer Science and Business Media. [Google Scholar]
- Ng AY, Jordan MI, Weiss Y (2001) On spectral clustering: Analysis and an algorithm. Advances in neural information processing systems, Vancouver, Canada, July 03, 2001. Proceedings of the 14th International Conference on Neural Information Processing Systems: Natural and Synthetic, 2001:849–856. [Google Scholar]
- Nusse R (2005) Wnt signaling in disease and in development. Cell Res 15:28–32. 10.1038/sj.cr.7290260 [DOI] [PubMed] [Google Scholar]
- Nusse R, Varmus HE (1992) Wnt genes. Cell 69:1073–1087. 10.1016/0092-8674(92)90630-u [DOI] [PubMed] [Google Scholar]
- O'Leary DD, Chou SJ, Sahara S (2007) Area patterning of the mammalian cortex. Neuron 56:252–269. 10.1016/j.neuron.2007.10.010 [DOI] [PubMed] [Google Scholar]
- Panizzon MS, Fennema-Notestine C, Eyler LT, Jernigan TL, Prom-Wormley E, Neale M, Jacobson K, Lyons MJ, Grant MD, Franz CE, Xian H, Tsuang M, Fischl B, Seidman L, Dale A, Kremen WS (2009) Distinct genetic influences on cortical surface area and cortical thickness. Cereb Cortex 19:2728–2735. 10.1093/cercor/bhp026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo LR, Merz EC, He X, Sowell ER, Noble KG; Pediatric Imaging, Neurocognition, Genetics Study (2016) Age-related differences in cortical thickness vary by socioeconomic status. PLoS One 11:e0162511. 10.1371/journal.pone.0162511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puelles L, Rubenstein JL (2003) Forebrain gene expression domains and the evolving prosomeric model. Trends Neurosci 26:469–476. 10.1016/S0166-2236(03)00234-0 [DOI] [PubMed] [Google Scholar]
- Rakic P (1988) Specification of cerebral cortical areas. Science 241:170–176. 10.1126/science.3291116 [DOI] [PubMed] [Google Scholar]
- Rakic P (2000) Radial unit hypothesis of neocortical expansion. Evolutionary Developmental Biology of the Cerebral Cortex: Novartis Foundation Symposium, Vol 228, pp 30–45. Chichester, UK: John Wiley & Sons, Ltd. [DOI] [PubMed] [Google Scholar]
- Raznahan A, Shaw P, Lalonde F, Stockman M, Wallace GL, Greenstein D, Clasen L, Gogtay N, Giedd JN (2011) How does your cortex grow? J Neurosci 31:7174–7177. 10.1523/JNEUROSCI.0054-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimol LM, Panizzon MS, Fennema-Notestine C, Eyler LT, Fischl B, Franz CE, Hagler DJ, Lyons MJ, Neale MC, Pacheco J, Perry ME, Schmitt JE, Grant MD, Seidman LJ, Thermenos HW, Tsuang MT, Eisen SA, Kremen WS, Dale AM (2010) Cortical thickness is influenced by regionally specific genetic factors. Biol Psychiatry 67:493–499. 10.1016/j.biopsych.2009.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseeuw PJ (1987) Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math 20:53–65. 10.1016/0377-0427(87)90125-7 [DOI] [Google Scholar]
- Rubenstein JL, Anderson S, Shi L, Miyashita-Lin E, Bulfone A, Hevner R (1999) Genetic control of cortical regionalization and connectivity. Cereb Cortex 9:524–532. 10.1093/cercor/9.6.524 [DOI] [PubMed] [Google Scholar]
- Sansom SN, Livesey FJ (2009) Gradients in the brain: the control of the development of form and function in the cerebral cortex. Cold Spring Harb Perspect Biol 1:a002519. 10.1101/cshperspect.a002519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaggar BL (2011) Mapping genetic influences on cortical regionalization. Neuron 72:499–501. 10.1016/j.neuron.2011.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt JE, Neale MC, Fassassi B, Perez J, Lenroot RK, Wells EM, Giedd JN (2014) The dynamic role of genetics on cortical patterning during childhood and adolescence. Proc Natl Acad Sci U S A 111:6774–6779. 10.1073/pnas.1311630111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC (1998) A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging 17:87–97. 10.1109/42.668698 [DOI] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE (2004) Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage 23:S208–S219. 10.1016/j.neuroimage.2004.07.051 [DOI] [PubMed] [Google Scholar]
- Sowell ER, Thompson PM, Leonard CM, Welcome SE, Kan E, Toga AW (2004) Longitudinal mapping of cortical thickness and brain growth in normal children. J Neurosci 24:8223–8231. 10.1523/JNEUROSCI.1798-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiles J, Jernigan TL (2010) The basics of brain development. Neuropsychol Rev 20:327–348. 10.1007/s11065-010-9148-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storsve AB, Fjell AM, Tamnes CK, Westlye LT, Overbye K, Aasland HW, Walhovd KB (2014) Differential longitudinal changes in cortical thickness, surface area and volume across the adult life span: regions of accelerating and decelerating change. J Neurosci 34:8488–8498. 10.1523/JNEUROSCI.0391-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strike LT, Hansell NK, Couvy-Duchesne B, Thompson PM, de Zubicaray GI, McMahon KL, Wright MJ (2019) Genetic complexity of cortical structure: differences in genetic and environmental factors influencing cortical surface area and thickness. Cereb Cortex 29:952–962. 10.1093/cercor/bhy002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Zhang D, Lian C, Wang L, Wu Z, Shao W, Lin W, Shen D, Li G; UNC/UMN Baby Connectome Project Consortium (2019) Topological correction of infant white matter surfaces using anatomically constrained convolutional neural network. Neuroimage 198:114–124. 10.1016/j.neuroimage.2019.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeuw J, Brouwer RM, Koenis MM, Swagerman SC, Boomsma DI, Hulshoff Pol HE (2019) Genetic influences on the development of cerebral cortical thickness during childhood and adolescence in a Dutch longitudinal twin sample: the brainscale study. Cereb Cortex 29:978–993. 10.1093/cercor/bhy005 [DOI] [PubMed] [Google Scholar]
- Tiberi L, Vanderhaeghen P, Van Den Ameele J (2012) Cortical neurogenesis and morphogens: diversity of cues, sources and functions. Curr Opin Cell Biol 24:269–276. 10.1016/j.ceb.2012.01.010 [DOI] [PubMed] [Google Scholar]
- Tomancak P, Berman BP, Beaton A, Weiszmann R, Kwan E, Hartenstein V, Celniker SE, Rubin GM (2007) Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biol 8:R145. 10.1186/gb-2007-8-7-r145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooley UA, Bassett DS, Mackey AP (2021) Environmental influences on the pace of brain development. Nat Rev Neurosci 22:372–384. 10.1038/s41583-021-00457-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M (2002) Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 15:273–289. 10.1006/nimg.2001.0978 [DOI] [PubMed] [Google Scholar]
- Valk SL, Xu T, Margulies DS, Masouleh SK, Paquola C, Goulas A, Kochunov P, Smallwood J, Yeo BT, Bernhardt BC (2020) Shaping brain structure: genetic and phylogenetic axes of macroscale organization of cortical thickness. Sci Adv 6:eabb3417. 10.1126/sciadv.abb3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayakumar N, Allen NB, Youssef G, Dennison M, Yücel M, Simmons JG, Whittle S (2016) Brain development during adolescence: a mixed-longitudinal investigation of cortical thickness, surface area, and volume. Hum Brain Mapp 37:2027–2038. 10.1002/hbm.23154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Luxburg U (2007) A tutorial on spectral clustering. Stat Comput 17:395–416. 10.1007/s11222-007-9033-z [DOI] [Google Scholar]
- Wang F, Lian C, Wu Z, Wang L, Lin W, Gilmore JH, Shen D, Li G (2019a) Revealing developmental regionalization of infant cerebral cortex based on multiple cortical properties. International Conference on Medical Image Computing and Computer-Assisted Intervention, Shenzhen, China, October 13–17, 2019. Proceedings, Part II 22. Springer International Publishing, 2019: 841–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Lian C, Wu Z, Zhang H, Li T, Meng Y, Wang L, Lin W, Shen D, Li G (2019b) Developmental topography of cortical thickness during infancy. Proc Natl Acad Sci U S A 116:15855–15860. 10.1073/pnas.1821523116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Li G, Shi F, Cao X, Lian C, Nie D, Liu M, Zhang H, Li G, Wu Z (2018) Volume-based analysis of 6-month-old infant brain MRI for autism biomarker identification and early diagnosis. International Conference on Medical Image Computing and Computer-Assisted Intervention, Granada, Spain, September 16-20, 2018. Proceedings, Part III 11. Springer International Publishing, 2018: 411–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wu Z, Chen L, Sun Y, Lin W, Li G (2023) iBEAT V2. 0: a multisite-applicable, deep learning-based pipeline for infant cerebral cortical surface reconstruction. Nat Protoc 18:1488–1509. 10.1038/s41596-023-00806-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T, Su S, Schmidt M, Kao CY, Sapiro G (2010) The development of gyrification in childhood and adolescence. Brain Cogn 72:36–45. 10.1016/j.bandc.2009.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierenga LM, Langen M, Oranje B, Durston S (2014) Unique developmental trajectories of cortical thickness and surface area. Neuroimage 87:120–126. 10.1016/j.neuroimage.2013.11.010 [DOI] [PubMed] [Google Scholar]
- Wilkinson DG, Bhatt S, Cook M, Boncinelli E, Krumlauf R (1989) Segmental expression of Hox-2 homoeobox-containing genes in the developing mouse hindbrain. Nature 341:405–409. 10.1038/341405a0 [DOI] [PubMed] [Google Scholar]
- Winkler AM, Kochunov P, Blangero J, Almasy L, Zilles K, Fox PT, Duggirala R, Glahn DC (2010) Cortical thickness or grey matter volume? The importance of selecting the phenotype for imaging genetics studies. Neuroimage 53:1135–1146. 10.1016/j.neuroimage.2009.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler AM, Sabuncu MR, Yeo BT, Fischl B, Greve DN, Kochunov P, Nichols TE, Blangero J, Glahn DC (2012) Measuring and comparing brain cortical surface area and other areal quantities. Neuroimage 61:1428–1443. 10.1016/j.neuroimage.2012.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Wang L, Lin W, Gilmore JH, Li G, Shen D (2019) Construction of 4D infant cortical surface atlases with sharp folding patterns via spherical patch-based group-wise sparse representation. Hum Brain Mapp 40:3860–3880. 10.1002/hbm.24636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Wang F, Benkarim OM, Sanroma G, Piella G, González Ballester MA, Hahner N, Eixarch E, Zhang C, Shen D, Li G (2019) Fetal cortical surface atlas parcellation based on growth patterns. Hum Brain Mapp 40:3881–3899. 10.1002/hbm.24637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia K, Schmitt JE, Jha SC, Girault JB, Cornea E, Li G, Shen D, Styner M, Gilmore JH (2022) Genetic influences on longitudinal trajectories of cortical thickness and surface area during the first 2 years of life. Cereb Cortex 32:367–379. 10.1093/cercor/bhab213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo BT, Sabuncu MR, Vercauteren T, Ayache N, Fischl B, Golland P (2009) Spherical demons: fast diffeomorphic landmark-free surface registration. IEEE Transactions on Medical Imaging 29:650–668. 10.1109/TMI.2009.2030797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo BT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, Smoller JW, Zöllei L, Polimeni JR, Fischl B, Liu H, Buckner RL (2011) The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol 106:1125–1165. 10.1152/jn.00338.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Wang L, Zong X, Lin W, Li G, Shen D (2019) FRNET: flattened residual network for infant MRI skull stripping. IEEE 16th International Symposium on Biomedical Imaging, Venice, Italy, April 8–11, 2019. Proceedings IEEE International Symposium on Biomedical Imaging, 2019: 999–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]