Abstract
Nrf2 is a transcription factor facilitating cells’ resilience against redox and various other forms of stress. In the absence of stressors, KEAP1 and/or βTrCP mediate the ubiquitination of Nrf2 and prevent Nrf2-dependent gene expression and detoxification. AMPK regulates cellular energy homeostasis and redox balance. Previous studies indicated a potential Nrf2-AMPK cooperativity. In line with this, our lab had previously identified three AMPK-dependent phosphorylation sites (S374/408/433) in Nrf2. Given their localization in or near the Neh6 domain, known to regulate βTrCP-mediated degradation, we examined whether they may influence the βTrCP-driven degradation of Nrf2. By employing expression plasmids for WT and triple mutant (TM)-Nrf2 (Nrf2S374/408/433→A), (co)immunoprecipitation, proximity ligation, protein half-life, knockdown, ubiquitination experiments, and qPCR in Keap1-null mouse embryonic fibroblasts, we show that TM-Nrf2S→A374/408/433 had enhanced stability due to impeded interaction with βTrCP2 and reduced ubiquitination in comparison to WT-Nrf2. In addition, TM-Nrf2 elicited higher expression of the Nrf2 target gene Gclc, potentiated in the presence of a pharmacological AMPK activator. Overall, we propose that AMPK-dependent phospho-sites of Nrf2 can favor its βTrCP2-mediated degradation and dampen the extent of Nrf2 target gene expression. Therefore, targeting AMPK might be able to diminish Nrf2-mediated responses in cells with overactive Nrf2 due to KEAP1 deficiency.
Keywords: Nrf2, AMPK, βTrCP, E3 ubiquitin ligase, transcription factor
1. Introduction
Nrf2 (Nuclear factor E2 p45-related factor 2) is a ubiquitously expressed leucine zipper transcription factor [1,2,3] that plays a critical role in defending the cell against oxidative stress, promoting detoxification, regulating inflammation, and supporting cellular resilience [4,5,6]. Nrf2 consists of different domains, including a KEAP1-binding domain (Neh2), transactivation domains (Neh3–5), a RXRα binding domain (Neh7), a βTrCP binding domain (Neh6), and the DNA binding domain (Neh1) [2]. Nrf2 activity is predominantly regulated via its abundance on a (a) transcriptional, (b) posttranscriptional, (c) epigenetic level, or (d) via ubiquitination and proteasomal degradation [7].
The major Nrf2 ubiquitination pathways include the canonical KEAP1 (Kelch-like-ECH-associated protein)-dependent and the non-canonical βTrCP (β-transducin-repeat containing protein)-mediated pathway, which usually can be pictured as a limiter and regulator valve for Nrf2 signaling, respectively [8,9,10,11]. In the absence of stressors, the redox-regulated KEAP1, an adaptor for Cul3-dependent ubiquitin ligases, interacts with the Neh2 domain of Nrf2 in a 2:1 ratio and mediates the predominant share of proteasomal degradation of Nrf2. Alternatively, βTrCP, as part of SCF ubiquitin–protein ligase complexes, can interact with the Neh6 domain of Nrf2 and also lead to the ubiquitination and degradation of the transcription factor [11,12,13,14,15,16,17,18]. In the presence of stress signals, ubiquitination of Nrf2 ceases, allowing Nrf2 to migrate into the nucleus and activate over 250 Nrf2 target genes with an ARE (antioxidant response element) sequence in their regulatory regions, thus influencing redox homeostasis, detoxification, inflammation, and metabolism [19,20,21]. Given the fundamental role of Nrf2 in cellular homeostasis, its dysregulation in the form of either too much or too little activity can lead to or aggravate diseases (inflammatory and metabolic-related diseases, aging, and cancer) [6,10,21,22,23,24,25,26,27,28].
For instance, mutations in KEAP1 can cause this dysregulation, leading to the constitutive activation of NRF2 signaling [29]. KEAP1 mutations are present in various pathological conditions, including neurological and metabolic disorders, cardiovascular diseases, and cancer [8,28,30,31,32,33], with a high prevalence in lung and pancreatic cancer [34,35]. These cancers then show hijacked NRF2 hyperactivation, which facilitates high tumor growth, therapy resistance, and metastasis [33,36,37,38] due to persistent antioxidant [39] and detoxification pathways [40], allowing cancer cells to fight stress induced by aberrant proliferation and therapy [38,41].
AMP-activated protein kinase (AMPK) is a serine/threonine kinase that is formed as a heterotrimeric complex by a catalytic α subunit and regulatory β and γ subunits [42]. AMPK activity is susceptible to regulation by different cues, including phosphorylation, inhibition of dephosphorylation at Thr172, and by allosteric modulation [43]. Its structural organization renders AMPK a sensor to changes in AMP/ATP and ADT/ATP ratios, and it activates signaling pathways resulting in cellular energy homeostasis [44]. Taking into consideration that energy metabolism affects cellular behavior, AMPK was also reported to influence proliferation, cell growth, autophagy, inflammation, and redox balance [45,46,47,48,49,50,51]. Therefore, its dysregulation can be observed in many diseases and pathological conditions such as diabetes, obesity, inflammation, cancer, and redox stress [52,53,54,55,56].
Concomitant activation by a plethora of exogenous insults and initiation of overlapping stress-relieving responses [57,58] quite early indicated potential cooperativity between Nrf2 and AMPK signaling. Meanwhile, their mutual multimodal crosstalk could be shown to occur among others via direct phosphorylation of Nrf2 at different serine residues or indirectly via the AMPK/GSK-beta/TrCP axis, the p62/KEAP1/autophagy axis, or epigenetic mechanisms [22]. In most cases, AMPK activation turned out to regulate the Nrf2-dependent response in a positive manner [11,59,60,61,62,63,64].
In one of our previous studies, we could identify AMPK-dependent phosphorylation sites at S374/408/433 in Nrf2. Their mutation to alanine reduced the expression of selected Nrf2 target genes in an AMPK-dependent manner [65]. These residues are located in and near the Neh6 domain of Nrf2, hosting the two β-TrCP binding sites at 343-DSGIS-347 and 382-DSAPGS-387 [25], which are implicated in Nrf2 degradation via the proteasome [11]. Given the existence of (a) AMPK-dependent phospho-sites in Nrf2, (b) their localization near the Neh6 domain, and (c) somatic mutations in the KEAP1 leading to an anomalous activation of Nrf2, we set out to investigate the effect of the three AMPK-dependent phosphorylation sites of Nrf2 for their role in its βTrCP-driven degradation in a KEAP1 depleted background. Delving into the interplay between AMPK and Nrf2 would provide valuable and novel insights into the network of cellular stress resilience mechanisms. It might also pave the way for the rational use of pharmacological AMPK activators in order to modulate Nrf2-mediated responses, especially in cases of aberrant Nrf2 signaling [66] pertinent to certain disease contexts caused by KEAP1 deficiency.
2. Materials and Methods
2.1. Cell Culture
KEAP1-depleted mouse embryonic fibroblasts (MEFsKeap1−/−) were kind gifts from Thomas Kensler (University of Pittsburgh, Pittsburgh, PA, USA) [67]. Media, serum, and supplements for cell culture were purchased from Sigma-Aldrich, Biowest, and Corning, respectively. MEFS were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) plus 10% heat-inactivated fetal bovine serum and 1% Penicillin/Streptomycin/L-Glutamine (P/S/G, Corning Product, Corning, NY, USA). OPTI-MEM I (1×) (Gibco) was used for transient transfections.
2.2. Reagents and Compound Treatment
Cycloheximide (CHX) was purchased from Sigma-Aldrich (North Brunswick, NJ, USA), A769662 came from ApexBio (Houston, TX, USA), and MG132 was purchased from ChemCruz (Huissen, The Netherlands). The working concentration of (a) CHX is 100 μM, (b) A769662 is 50 μM, and (c) MG132 is 10 μΜ.
2.3. Plasmids
The mammalian expression vectors pcDNA3-EGFP-C4-Nrf2 (EGFP-Myc-WT-Nrf2) #21549, pGLUE-HA-FBXW11#36969, and His-Ubiquitin (#31815) were obtained from Addgene and pcDNA3.1 vector from Invitrogen. The triple serine–alanine mutant form of Nrf2 (EGFP-MycTM-Nrf2) was designed as previously described [65].
2.4. siRNA Sequences
Mouse Fbxw11-specific smart pool siRNA was obtained from Dharmacon (#L-058886-00-0005, Lafayette, CO, USA). Non-targeting siRNA pool was obtained from Merck (#SIC007, Rahway, NJ, USA). The working concentration was 100 nM.
2.5. Transient Transfections
Transfections with plasmids in MEFsKeap1−/− cells were performed by using Lipofectamine LTX with PLUSTM Reagent (Invitrogen, Bridgewater, NJ, USA) according to the manufacturer’s instructions. Transfection with siRNA or co-transfections with siRNA and plasmids in MEFsKeap1−/− cells were performed by using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions.
2.6. Immunoblotting
MEFsKeap1−/− cells were lysed as previously described [68]. Cell lysates were separated by SDS-PAGE, and proteins were blotted to PVDF membranes. For detection, we used antibodies against the tags of the transfected proteins of interest in order to avoid the interference of endogenous proteins and the poor performance of commercially available antibodies for our purpose. Antibodies were used against Myc-Tag (#2276S, 1:1000 dilution) from Cell Signaling (Danvers, MA, USA), HA-Tag (#3724S, 1:1000 dilution) from Cell Signaling, beta-actin (#8457S, 1:1000 dilution) from Cell Signaling and FBXW11 (#13149-1-AP, 1:000 dilution) from Proteintech (Rosemont, IL, USA). Densitometric quantification was performed with ImageQuant™ TL, Cytiva (Tokyo, Japan)
2.7. Immunoprecipitation
MEFsKeap1−/− cells were transfected with the indicated constructs for 48 h, and then treated with MG132 proteasomal inhibitor 4 h before their lysis. Cells were lysed and further processed as previously described [65].
2.8. Proximity Ligation Assay (PLA) and Indirect Immunofluorescence
MEFsKeap1−/− cells were grown on 0.1% gelatin-coated glass coverslips, transfected with the indicated constructs for 48 h, and then treated with MG132 proteasomal inhibitor 4 h before their fixation with 4% Paraformaldehyde (#j19943-k2, Thermo Fischer Scientific, Vienna, Austria). After the fixation, they were permeabilized in 0.1% Triton X and blocked in 1× PLA blocking solution for 1 h at 37 °C in a humidity chamber. Then, they were subjected to PLA (#DUO92004, DUO92002, Merck). Fixed samples were incubated with HA and MYC-Tag primary antibodies (1:1000 dilution) raised in separate species (mouse and rabbit) that bind to HA-βTrCP2 and Myc-Nrf2 forms, respectively in a humidity chamber at 4 °C overnight. Then, the samples were washed twice with Duolink® Wash Buffer A for 5 min. Secondary antibodies conjugated with oligonucleotides (PLA probe MINUS and PLA probe PLUS) were diluted with the Duolink® Antibody Diluent at 1:5 PLA probe dilution and incubated in a humidity chamber at 37 °C for 1 h. The next step included the ligation and amplification process according to the manufacturer’s instructions: The DNA oligonucleotides hybridized with the two PLA probes, and the enzyme formed a ligated (closed) circle when the probes were in close proximity. Finally, one of the PLA probes acted as a primer for the polymerase, generating a repeating, concatemeric product that was still tethered to the secondary antibody. Labeled oligonucleotides were then hybridized with the amplified DNA to yield a signal that was easily detectable by fluorescence microscopy. Samples were mounted with Diamond DAPI (#P36966, Invitrogen) to counterstain the nuclei. Z-stacks were obtained with a Leica DMi8 inverted confocal microscope, and images were analyzed by LAS X (3.1.2) software.
2.9. Quantitative Chain Polymerase Reaction (qPCR)
RNA isolation, reverse transcription, and qPCR were performed as previously described [69]. mRNA levels of the genes of interest were normalized to the indicated control via the ΔΔCt method. Primers used for qPCR were Hmox1 Fw:5′-AAGCCGAGAATGCTGAGTTCA-3′, Hmox1 Rv:5′-GCCGTGTAGATATGGTACAAGGA, Gclc Fw:5′-TGGCCACTATCTGCCCAATT-3′, Gclc Rv:5′-GTCTGACACGTAGCCTCGGTAA-3′, TBP Fw:5′-TCTACCGTGAATCTTGGCTGT-3′, and TBP Rv:5′-GTCCGTGGCTCTCTTATTCTCA-3′.
2.10. In Vitro Ubiquitinylation Assay
MEFsKeap1−/− cells were transiently co-transfected with HA-βTrCP2, His ubiquitin, and EGFP-Myc-WT-Nrf2 or EGFP-Myc-TM-Nrf2 for 48 h and further processed to the in vitro ubiquitination assay as previously described [70].
2.11. Statistical Analysis
The statistical analyses were performed by Software GraphPad Prism 9. Student’s t-test or one-way ANOVA was performed for the quantification analysis of the results based on the number of comparison groups, followed by the proper ad hoc test. Data are presented as means ± SEM from at least three independent biological replicates. Regarding the statistical analysis performed using multiple comparison one-way ANOVA, it was followed with Sidak’s correction, * p < 0.05 and ** p < 0.01.
3. Results
3.1. Mutation of the AMPK-Dependent Phospho-Sites S374, S408, and S433 to Alanine Stabilizes Nrf2 in Keap1−/− Cells in a βTrCP2-Dependent Manner
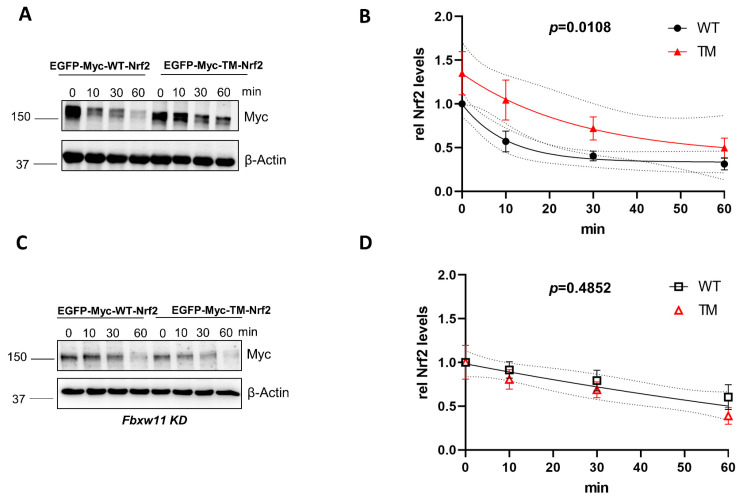
The main degradation pathways of Nrf2 are (a) the KEAP1-dependent pathway and (b) the KEAP1-independent pathways, involving the βTrCP E3 ubiquitin ligase subunit of the SCFβΤrCP complex. To address whether the previously identified phospho-sites of Nrf2 have a role in the stability of the protein in the context of βTrCP-mediated degradation, we used KEAP1-depleted mouse embryonic fibroblasts (MEFs) to overcome KEAP1 interference. Cells were transfected with a wild form of Nrf2 (GFP-MYC-WT-Nrf2) or a triple mutated version of Nrf2 (GFP-MYC-TM-NRF2) in which S374, S408, and S433 were mutated to alanine. To monitor the time-dependent decay of Nrf2 protein, de novo protein synthesis was blocked by treatment with cycloheximide. The triple mutated form of Nrf2 had a higher half-life in comparison to the wild-type form (~18 min vs. 7 min) (Figure 1A,B). This finding showed that mutation of the identified phospho-sites renders the protein more stable and less susceptible to degradation. To further investigate whether the increased stability of TM-Nrf2 was indeed related to βTrCP-mediated degradation, we knocked down βTrCP2 by siRNA and repeated the experimental setup from above. Downregulation of βTrCP2 resulted in similar kinetics of degradation between the WT-Nrf2 and the TM-Nrf2 (Figure 1C,D). These data suggest that the identified residues control the abundance of Nrf2 in a βTrCP2-dependent manner. Successful depletion of endogenous mouse βTrCP2 by Fbxw11 siRNA was confirmed by immunoblot in transfected MEFsKeap1−/− (Figure S1).
Figure 1.
Mutation of the AMPK-dependent phospho-sites to alanine stabilizes Nrf2 in Keap1−/− cells in a βTrCP2-dependent manner. (A,B). Keap1−/− MEFs were transfected with EGFP-MYC tagged WT- or TM-Nrf2 expression plasmids for 48 h and then exposed to cycloheximide (50 μM) for different periods of time. Cell lysates were subjected to immunoblot analysis for MYC or β-Actin. A representative blot (A) and quantification analysis (B) of n = 9 independent experiments (relative to signal at t = 0) are depicted. One-phase exponential decay curves were fitted to the data by finding the least sum of squares using GraphPad Prism. The curves were significantly different, with a p value = 0.0108. (C,D) Keap1−/− MEFs were transfected with Fbxw11-specific siRNA and with EGFP-MYC tagged WT- or TM-Nrf2 expression plasmids for 48 h, and then exposed to cycloheximide (50 μM) for different periods of time. Cell lysates were subjected to immunoblot analysis for MYC or β-Actin. A representative blot (C) and quantification analysis (D) of n = 5 independent experiments (relative to signal at t = 0) are depicted. One-phase exponential decay curves were fitted to the data by finding the least sum of squares using GraphPad Prism. There was one curve for all data sets with a p value = 0.4852. The dotted lines in (B,D) represent the confidence bands with a 95% confidence level. Data are presented as means ± SEM.
3.2. Mutation of the AMPK-Dependent Phospho-Sites to Alanine Impedes the Interaction of Nrf2 with β-TrCP2 in Keap1−/− Cells
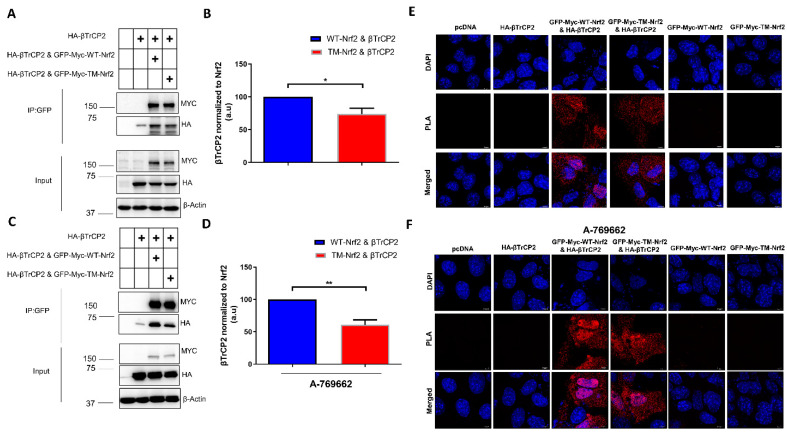
To further corroborate the role of phospho-S374, -S408, and -S433 in βTrCP2-mediated degradation of Nrf2, we tested the binding of βTrCP2 to Nrf2. We addressed this question by employing a co-immunoprecipitation and proximity ligation assay (PLA) in Keap1−/− cells transfected with GFP-Myc tagged WT or TM-Nrf2 and HA-tagged βTrCP2. Notably, in co-immunoprecipitation experiments, βTrCP2 interacted to a lower extent with TM-Nrf2 than with WT-Nrf2 (Figure 2A,B), and this impaired interaction became even more apparent in the presence of A-769662, an allosteric AMPK activator (Figure 2C,D). Accordingly, PLA experiments confirmed that Nrf2 interaction with βTrCP2 is impeded upon substitution of the identified serine residues to alanine (Figure 2E). This effect was again pronounced upon AMPK activation by A-769662: whereas WT-Nrf2-transfected cells gave a bright signal, notably predominantly in the nucleus, TM-Nrf2 transfected cells displayed rather low fluorescence (Figure 2F). To exclude experimental artifacts, cells solely expressing either WT-Nrf2, TM-Nrf2, βTrCP2, or pcDNA served as controls. Also, an interaction was exclusively detected between transfected Nrf2 and transfected β TrCP2 since the MYC-Tag of Nrf2 and the HA-Tag of β TrCP2 served as the targets of the PLA primary antibodies. These results so far supported the notion that phosphorylation of S374, S408, and S433 supports Nrf2 degradation by facilitating its interaction with βTrCP2.
Figure 2.
Mutation of the AMPK-dependent phospho-sites to alanine impedes the interaction of Nrf2 with β-TrCP2 in Keap1−/− cells. (A–D) Keap1-null MEFs were transfected with an expression plasmid for HA-tagged β-TrCP2 alone or together with constructs encoding EGFP-MYC-WT-Nrf2, the triple mutated version EGFP-MYC-TM-Nrf2, or pcDNA as indicated for 48 h followed by the absence (A,B) or presence (C,D) of the AMPK activator A769662 (50 μM, 4 h). Cells were also treated at the same time with MG132 (10 μM, 4 h) to stabilize Nrf2, which was pulled down using GFP-Trap. Precipitated proteins, as well as input controls, were immunoblotted for MYC, HA, and β-Actin. Representative blots (A,C) and quantification analysis (B,D) of 3 independent experiments are depicted. Plus symbols in the tables indicate the different constructs/chemicals used in the various experimental conditions. (E,F) Keap1-null MEFs were transfected with (i) pcDNA, (ii) HA-tagged β-TrCP2, or co-transfected with (iii) HA-tagged β-TrCP2 and EGFP-MYC-tagged WT-Nrf2, (iv) HA-tagged β-TrCP2 and EGFP-MYC-tagged TM-Nrf2 expression plasmids, or (v) EGFP-MYC-tagged WT-Nrf2 and (vi) and EGFP-MYC-tagged TM-Nrf2 alone, and then treated with MG132 (10 μM) for 4 h in the absence (E) or presence (F) of the AMPK activator A769662 (50 μM, 4 h). Cells were fixed with 4% Paraformaldehyde and subjected to PLA. Data are presented as means ± SEM. Statistical analysis was performed using Student’s t-test, * p < 0.05 and ** p < 0.01.
3.3. Mutation of the AMPK-Dependent Phospho-Sites to Alanine on Nrf2 Impedes Its βTrcP2-Mediated Ubiquitination and Degradation in Keap1−/− Cells
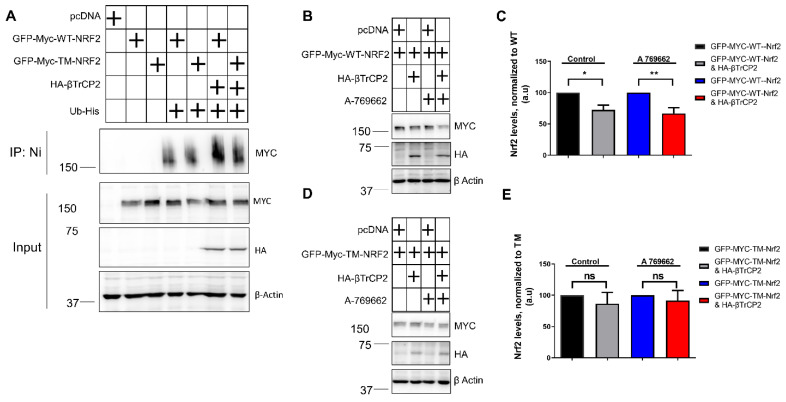
Considering that βTrCP2 is an E3 ubiquitin ligase, we further tested whether its reduced binding to TM-Nrf2 is also reflected by reduced TM-Nrf2 ubiquitination compared to WT-Nrf2. Ubiquitination experiments in KEAP1-depleted MEFs suggested that co-expression of βTrCP2 with wild-type Nrf2, but not with the triple mutated form, resulted in increased ubiquitination of Nrf2 (Figure 3A). Regarding βTrCP2-mediated regulation of Nrf2 protein levels, transient co-transfection of GFP-MYC-WT-Nrf2 with βTrCP2 in MEFsKeap1−/− led to a reduction in the protein levels of WT-Nrf2, and this downregulation was additionally enhanced in the presence of A-769662 (Figure 3B,C). However, mutation of the identified AMPK-dependent phospho-sites in TM-Nrf2 rendered the protein resistant to degradation by co-transfected βTrCP2 and insensitive to AMPK activation (Figure 3D,E). These data corroborate that AMPK-dependent S374, S408, and S433 residues favor the degradation of Nrf2 by facilitating its ubiquitination by βTrCP2.
Figure 3.
Mutation of the AMPK-dependent phospho-sites to alanine on Nrf2 impedes its βTrcP2-mediated ubiquitination and degradation in Keap1−/− cells. (A–E) Keap1-null MEFs were transfected with the indicated expression plasmids for 48 h. (A) Ubiquitination assay of EGFP-Myc tagged Nrf2 expression plasmids in Keap1-null MEFs, after co-transfection with His-tagged Ubiquitin and the indicated HA-βTrCP2 construct. (B–E) Expression levels of EGFP-Myc tagged WT-Nrf2 (B,C) or EGFP-Myc tagged TM-Nrf2 (D–E) with or without co-transfection of HA-tagged βΤrCP2 in the presence or absence of the AMPK activator A 769662 (50 μM, 4 h). Cell lysates were subjected to immunoblot analysis for MYC, HA, or actin. Representative blots (B,D) and quantification analysis (C,E) of 3 independent experiments are depicted. Plus symbols in the tables indicate the different constructs/chemicals used in the various experimental conditions. Data are presented as means ± SEM. Statistical analysis was performed using multiple comparison one-way ANOVA, with Sidak’s correction, * p < 0.05 and ** p < 0.01. ns: not significant.
3.4. The Identified AMPK-Dependent Phospho-Sites in Nrf2 Affect the Expression of Endogenous Nrf2 Target Genes
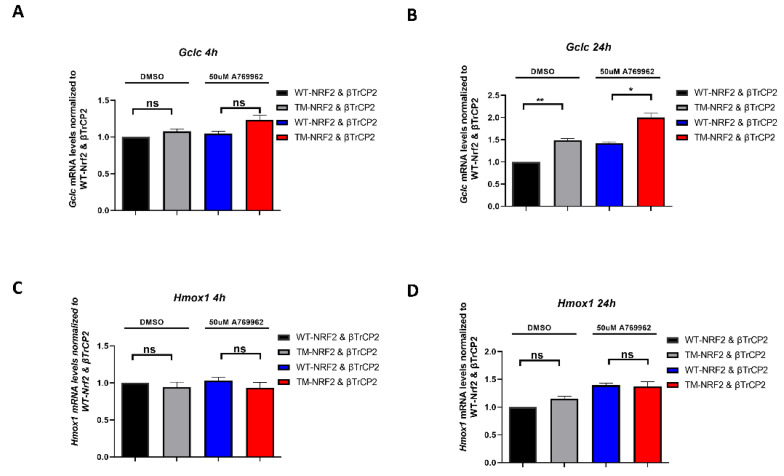
To see whether the aforementioned observations are also of relevance for Nrf2 target gene expression, we examined WT-Nrf2- and TM-Nrf2-mediated transactivation in our model system of MEFsKeap1−/− co-transfected with a βTrCP2 expression plasmid. Specifically, we investigated the transactivation of endogenous Hmox1 (heme oxygenase 1) and Gclc (Glutamate-cysteine ligase catalytic subunit) [71,72] in the presence or absence of A-769662 at two time points. Hmox1 was expressed to a similar extent by both Nrf2 forms with or without AMPK activation at 4 h (Figure 4C) and 24 h (Figure 4D). Of note, whereas mRNA levels of Gclc exhibit similar expression in the presence of either Nrf2 form irrespective of AMPK activation at 4 h (Figure 4A), Gclc mRNA was significantly elevated in the case of ΤΜ-Nrf2 in comparison to WT-Nrf2 at 24 h (Figure 4B), suggesting that the more stable TM-Nrf2 also leads to a more sustained Gclc expression. Of note, A-769662 elicited elevated mRNA levels of Gclc both in the WT- and the TM-Nrf2 setting. This could likely be attributed to other AMPK-dependent signals besides S374, S408, and S433 phosphorylation that can still positively affect the (endogenous) Nrf2-dependent gene transcription in our model, such as phosphorylation of Nrf2 on Ser558, which has been shown to induce nuclear accumulation of Nrf2 [61]. Overall, our findings suggest that AMPK-dependent phosphorylation of S374, S408, and S433 is involved in βTrCP2-mediated control of Nrf2 protein levels and activity, which may gain increased importance in a KEAP1-deficient background.
Figure 4.
The identified AMPK-dependent phospho-sites in Nrf2 affect the expression of endogenous Nrf2 target genes. (A–D) Keap1-null MEFs were transfected for 48 h with an expression plasmid for HA-tagged β-TrCP2 together with constructs encoding EGFP-MYC-WT-Nrf2 or the triple mutated version EGFP-MYC-TM-Nrf2, as indicated in the absence or presence of the AMPK activator A769662 (50 μM, 4 h and 24 h). RNA was extracted and analyzed for the abundance of Gclc (A,B) and Hmox1 (C,D) by qPCR (TBP as reference gene). Data are presented as means ± SEM. Statistical analysis was performed using multiple comparison one-way ANOVA, with Sidak’s correction, * p < 0.05 and ** p < 0.01. ns: not significant.
4. Discussion
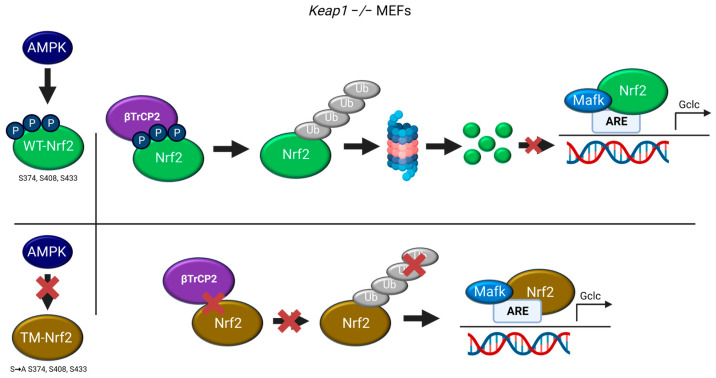
This study shows that previously identified AMPK-dependent phospho-sites in Nrf2 (S374/408/433) may participate in the regulation of Nrf2 stability by βTrCP2. Mutation of these serine residues to alanine (TM-Nrf2) reduced (i) interaction of Nrf2 with βTrCP2 and (ii) subsequent ubiquitination. Resulting sustained levels of mutated Nrf2 led to (iii) higher expression of the endogenous Nrf2 target gene Gclc in Keap1−/− cells (see Figure 5).
Figure 5.
Working model depicting the role of the AMPK-dependent S374, S408, and S433 of Nrf2 in its βTrCP2-mediated ubiquitination and degradation, regulating the expression of Nrf2 target genes (©BioRender).
In the presence of the A 769662 AMPK activator, our PLA data suggested an enhanced nuclear interaction of both wild-type and mutant Nrf2 forms with β-TrCP2, which could be explained by AMPK directly regulating other phospho-sites influencing the nuclear transport of Nrf2 [61]. One could assume that since the mutant version of Nrf2 cannot undergo β-TrCP2-mediated degradation, the only effect of AMPK on the Nrf2 mutant would be to promote its translocation into the nucleus. However, the net effect of the positive (nucleus translocation) and negative (β-TrCP2-mediated ubiquitination and degradation) influence of AMPK is observed in the case of wild-type Nrf2. These findings could indicate a possible dual role of AMPK in the regulation of Nrf2 in the absence of KEAP1.
With regard to the biological relevance of the aforementioned phospho-residues of Nrf2, we could observe that they affect the transactivating capacity in KEAP1-depleted cells. The expression of Gclc was higher in the case of the triple mutated form than in the case of wild-type Nrf2, and the presence of A 769662 enhanced the Gclc expression by both forms (compared to the DMSO conditions). These findings indicate once more the dual effect of AMPK activation in Nrf2 regulation and its target genes: higher stability of Nrf2, obtained by the conversion of the identified serine residues to alanine and the inhibition of its degradation, can result in higher transactivation of Gclc, while AMPK promotes the nuclear translocation [61] regardless of the AMPK-dependent S374/408/433 residues. Therefore, the final outcome in the expression of Gclc [72] is a net effect of the dual regulatory role of AMPK on Nrf2. However, this difference in Gclc gene expression was only observed at a 24 h time point. Thus, it is even tempting to speculate that the sites play a major role in signal termination or duration (via removal of nuclear Nrf2 after target gene expression has started) rather than initiation (nuclear translocation and start of transcription).
Contrary to the Gclc results, the analysis of Hmox1 [71] mRNA expression levels revealed no significant alterations between the Nrf2 forms. A possible explanation of this outcome may lie in the suppressive role of Bach1 in Hmox1 expression [73]. Downregulation of Bach1 might unmask any possible differences in Hmox1 gene expression between the Nrf2 forms.
Notably, the same phosphorylation sites that elicited an apparently negative impact on Nrf2 target gene expression in Keap1−/− cells did the opposite in a previous study using cells with a wild-type background, at least for a subset of target genes [65]. This suggests that the sites might act in a dual fashion, namely (a) boosting gene-selective transactivation (by a so far unknown mechanism), but also (b) boosting decay of Nrf2 by β-TrCP2. The latter effect becomes only obvious when Nrf2 is unleashed from the control by its dominant inhibitor, KEAP1.
Degradation of Nrf2 by β-TrCP is known to require preceding priming before phosphorylation by GSK3 in Neh6 to create the necessary phospho-degron, and AMPK activity is known to inhibit GSK3 signaling. Thus, one would assume that activation of AMPK would impede the β-TrCP-mediated destabilization of Nrf2. We, however, propose the acceleration of β-TrCP-conferred degradation via the three AMPK-dependent phospho-sites S374, 408, and 433. A possible resolution of this conflict could lie in the fact that our experimental approaches were conducted in a serum-supplemented medium (10% serum). Under these conditions, GSK3 was already inhibited, and thus GSK3-dependent Neh6 phospho-degron was silenced [74]. It is very likely that we tackled an Nrf2 decay pathway that functions in cells that are depleted from KEAP1 and deprived of GSK3beta activity (as, e.g., possibly found in cancers with KEAP1 mutation and overactive PI3K signaling). One may speculate that the second βTrCP-binding degron in Neh6 (382-DSAPGS-387), which is not phosphorylated by GSK3 [12], could be the one under the influence of the AMPK-dependent phospho-sites S374, 408, and 433 [11]. This hypothesis needs further investigation, e.g., regarding the role of the individual phospho-sites as potential priming or docking sites for βTrCP2. Moreover, it remains to be deciphered why AMPK activation negatively affects the GSK3-dependent 343-DSGIS-347 degron but positively affects the 382-DSAPGS-387 degron, and whether AMPK is truly the one and only kinase directly adding phosphates to serine 374, 408, and 433.
Our study is based on in vitro experiments that use genetically modified mouse embryonic fibroblasts, transiently transfected with fusion proteins. Although this model system allows straightforward and comparably fast investigation of the (biochemical) question of interest, it bears some risks. These include that (i) overexpression of transfected proteins may disrupt endogenous protein stoichiometry, (ii) fusion proteins may behave somewhat differently compared to native proteins, (iii) detection of transfected fusion proteins by antibodies directed against the tag only misses the interference of endogenous counterparts, and (iv) cells in culture cannot recapitulate the situation in vivo (e.g., cell–cell competition for fuel, oxygen tension, and organ–organ interaction). With this in mind, we still put forward a novel addition on how AMPK signaling could intersect with the Nrf2-mediated stress response, namely by triggering faster Nrf2 destabilization via phosphorylation, facilitated βTrCP2 binding, and ubiquitination, for further investigation in advanced test models. Our data further support the notion that AMPK activation with its pleiotropic effects can have a positive or negative impact on NRF2 signaling: on the one hand, it can boost Nrf2 signaling (e.g., by increasing nuclear accumulation or autophagic degradation of KEAP1) and on the other hand, it can also dampen the Nrf2 signal (e.g., by accelerated degradation). To which side the balance tips may highly depend on cellular environment, genetic background, Nrf2 target gene, signal hierarchy, and integration. Our findings shed light on the regulatory mechanisms governing Nrf2 and provide new insights into the multifaceted roles of AMPK in cellular signaling. A better understanding of the intricate interplay between AMPK and NRF2 signaling with all possible parameters by future studies, including in vivo and bioinformatics approaches, will hopefully allow safe and reliable exploitation of pharmacological AMPK activation in conditions of aberrant Nrf2 signaling.
5. Conclusions
In this study, we elucidated the role of the AMPK-dependent phosphorylation sites (S374, S408, and S433) of Nrf2 in its βTrCP2-driven degradation in a KEAP1-depleted background. Mutation of the three serines to alanine impeded the interaction of Nrf2 with βTrCP2 and its βTrCP2-mediated ubiquitination and degradation. Consequently, higher stability of Nrf2 positively affected gene expression of Nrf2 targets. Thus, modulating the AMPK-Nrf2 crosstalk could decrease cellular stress resilience via diminished Nrf2 stability, which would be of use in the context of diseases with overactive Nrf2 signaling. This hypothesis warrants further investigation in appropriate model systems.
Acknowledgments
This work was supported by the Austrian Science Fund (FWF P33778). The authors would like to thank the University of Vienna for its support, access to infrastructure, and general funding. In addition, we thank Verena Dirsch and the entire Molecular Target Group for fruitful discussions and input, and especially the technicians for continuous laboratory support. Furthermore, we acknowledge T. Kensler for kindly providing us with Keap1−/− MEFs and N. Wakabayashi for generating this KEAP1-depleted cell line in T.Kensler’s laboratoty. Open Access Funding by the Austrian Science Fund (FWF).
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antiox12081586/s1, Figure S1: Validation of the Fbxw11 knock down in Keap1−/− cells.
Author Contributions
E.P. designed and performed the experiments, acquired and analyzed the data, prepared all figures, and drafted the manuscript. S.E. and S.N.S.C. helped with sample preparation, data acquisition, or illustration of the working model. E.H.H. conceived the study, provided funding, supervised the experimental design, work, and data interpretation, and revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available upon request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by the Austrian Science Fund (FWF P33778 to EHH).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Moi P., Chan K., Asunis I., Cao A., Kan Y.W. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc. Natl. Acad. Sci. USA. 1994;91:9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tonelli C., Chio I.I.C., Tuveson D.A. Transcriptional Regulation by Nrf2. Antioxid. Redox Signal. 2018;29:1727–1745. doi: 10.1089/ars.2017.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matzinger M., Fischhuber K., Heiss E.H. Activation of Nrf2 signaling by natural products-can it alleviate diabetes? Biotechnol. Adv. 2018;36:1738–1767. doi: 10.1016/j.biotechadv.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Itoh K., Chiba T., Takahashi S., Ishii T., Igarashi K., Katoh Y., Oyake T., Hayashi N., Satoh K., Hatayama I., et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 5.Chorley B.N., Campbell M.R., Wang X., Karaca M., Sambandan D., Bangura F., Xue P., Pi J., Kleeberger S.R., Bell D.A. Identification of novel NRF2-regulated genes by ChIP-Seq: Influence on retinoid X receptor alpha. Nucleic Acids Res. 2012;40:7416–7429. doi: 10.1093/nar/gks409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobayashi E.H., Suzuki T., Funayama R., Nagashima T., Hayashi M., Sekine H., Tanaka N., Moriguchi T., Motohashi H., Nakayama K., et al. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat. Commun. 2016;7:11624. doi: 10.1038/ncomms11624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayes J.D., Dinkova-Kostova A.T. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014;39:199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 8.Cuadrado A., Rojo A.I., Wells G., Hayes J.D., Cousin S.P., Rumsey W.L., Attucks O.C., Franklin S., Levonen A.L., Kensler T.W., et al. Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat. Rev. Drug Discov. 2019;18:295–317. doi: 10.1038/s41573-018-0008-x. [DOI] [PubMed] [Google Scholar]
- 9.Hayes J.D., Chowdhry S., Dinkova-Kostova A.T., Sutherland C. Dual regulation of transcription factor Nrf2 by Keap1 and by the combined actions of beta-TrCP and GSK-3. Biochem. Soc. Trans. 2015;43:611–620. doi: 10.1042/BST20150011. [DOI] [PubMed] [Google Scholar]
- 10.Cuadrado A., Manda G., Hassan A., Alcaraz M.J., Barbas C., Daiber A., Ghezzi P., Leon R., Lopez M.G., Oliva B., et al. Transcription Factor NRF2 as a Therapeutic Target for Chronic Diseases: A Systems Medicine Approach. Pharmacol. Rev. 2018;70:348–383. doi: 10.1124/pr.117.014753. [DOI] [PubMed] [Google Scholar]
- 11.Chowdhry S., Zhang Y., McMahon M., Sutherland C., Cuadrado A., Hayes J.D. Nrf2 is controlled by two distinct beta-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene. 2013;32:3765–3781. doi: 10.1038/onc.2012.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rada P., Rojo A.I., Chowdhry S., McMahon M., Hayes J.D., Cuadrado A. SCF/beta-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol. Cell. Biol. 2011;31:1121–1133. doi: 10.1128/MCB.01204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Itoh K., Wakabayashi N., Katoh Y., Ishii T., Igarashi K., Engel J.D., Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kensler T.W., Wakabayashi N., Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen T., Sherratt P.J., Huang H.C., Yang C.S., Pickett C.B. Increased protein stability as a mechanism that enhances Nrf2-mediated transcriptional activation of the antioxidant response element. Degradation of Nrf2 by the 26 S proteasome. J. Biol. Chem. 2003;278:4536–4541. doi: 10.1074/jbc.M207293200. [DOI] [PubMed] [Google Scholar]
- 16.Zhang D.D., Lo S.C., Cross J.V., Templeton D.J., Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell. Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi A., Kang M.I., Okawa H., Ohtsuji M., Zenke Y., Chiba T., Igarashi K., Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baird L., Yamamoto M. The Molecular Mechanisms Regulating the KEAP1-NRF2 Pathway. Mol. Cell. Biol. 2020;40:e00099-20. doi: 10.1128/MCB.00099-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi M., Li L., Iwamoto N., Nakajima-Takagi Y., Kaneko H., Nakayama Y., Eguchi M., Wada Y., Kumagai Y., Yamamoto M. The antioxidant defense system Keap1-Nrf2 comprises a multiple sensing mechanism for responding to a wide range of chemical compounds. Mol. Cell. Biol. 2009;29:493–502. doi: 10.1128/MCB.01080-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McMahon M., Lamont D.J., Beattie K.A., Hayes J.D. Keap1 perceives stress via three sensors for the endogenous signaling molecules nitric oxide, zinc, and alkenals. Proc. Natl. Acad. Sci. USA. 2010;107:18838–18843. doi: 10.1073/pnas.1007387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He F., Antonucci L., Karin M. NRF2 as a regulator of cell metabolism and inflammation in cancer. Carcinogenesis. 2020;41:405–416. doi: 10.1093/carcin/bgaa039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petsouki E., Cabrera S.N.S., Heiss E.H. AMPK and NRF2: Interactive players in the same team for cellular homeostasis? Free Radic. Biol. Med. 2022;190:75–93. doi: 10.1016/j.freeradbiomed.2022.07.014. [DOI] [PubMed] [Google Scholar]
- 23.Liu J., Xu Y., Yan M., Yu Y., Guo Y. 18beta-Glycyrrhetinic acid suppresses allergic airway inflammation through NF-kappaB and Nrf2/HO-1 signaling pathways in asthma mice. Sci. Rep. 2022;12:3121. doi: 10.1038/s41598-022-06455-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao Y.Y., Wang H.L., Cheng X.L., Wei F., Bai X., Lin R.C., Vaziri N.D. Metabolomics analysis reveals the association between lipid abnormalities and oxidative stress, inflammation, fibrosis, and Nrf2 dysfunction in aristolochic acid-induced nephropathy. Sci. Rep. 2015;5:12936. doi: 10.1038/srep12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang P., Geng J., Gao J., Zhao H., Li J., Shi Y., Yang B., Xiao C., Linghu Y., Sun X., et al. Macrophage achieves self-protection against oxidative stress-induced ageing through the Mst-Nrf2 axis. Nat. Commun. 2019;10:755. doi: 10.1038/s41467-019-08680-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H.X., Chen Y., Xu R., He Q.Y. Nrf2 mediates the resistance of human A549 and HepG2 cancer cells to boningmycin, a new antitumor antibiotic, in vitro through regulation of glutathione levels. Acta Pharmacol. Sin. 2018;39:1661–1669. doi: 10.1038/aps.2018.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adinolfi S., Patinen T., Jawahar Deen A., Pitkanen S., Harkonen J., Kansanen E., Kublbeck J., Levonen A.L. The KEAP1-NRF2 pathway: Targets for therapy and role in cancer. Redox Biol. 2023;63:102726. doi: 10.1016/j.redox.2023.102726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sayin V.I., LeBoeuf S.E., Singh S.X., Davidson S.M., Biancur D., Guzelhan B.S., Alvarez S.W., Wu W.L., Karakousi T.R., Zavitsanou A.M., et al. Activation of the NRF2 antioxidant program generates an imbalance in central carbon metabolism in cancer. Elife. 2017;6:e28083. doi: 10.7554/eLife.28083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pillai R., Hayashi M., Zavitsanou A.M., Papagiannakopoulos T. NRF2: KEAPing Tumors Protected. Cancer Discov. 2022;12:625–643. doi: 10.1158/2159-8290.CD-21-0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quinti L., Dayalan Naidu S., Trager U., Chen X., Kegel-Gleason K., Lleres D., Connolly C., Chopra V., Low C., Moniot S., et al. KEAP1-modifying small molecule reveals muted NRF2 signaling responses in neural stem cells from Huntington’s disease patients. Proc. Natl. Acad. Sci. USA. 2017;114:E4676–E4685. doi: 10.1073/pnas.1614943114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uruno A., Yamamoto M. The KEAP1-NRF2 System and Neurodegenerative Diseases. Antioxid. Redox Signal. 2023;38:974–988. doi: 10.1089/ars.2023.0234. [DOI] [PubMed] [Google Scholar]
- 32.Taguchi K., Yamamoto M. The KEAP1-NRF2 System in Cancer. Front. Oncol. 2017;7:85. doi: 10.3389/fonc.2017.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Romero R., Sayin V.I., Davidson S.M., Bauer M.R., Singh S.X., LeBoeuf S.E., Karakousi T.R., Ellis D.C., Bhutkar A., Sanchez-Rivera F.J., et al. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat. Med. 2017;23:1362–1368. doi: 10.1038/nm.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohta T., Iijima K., Miyamoto M., Nakahara I., Tanaka H., Ohtsuji M., Suzuki T., Kobayashi A., Yokota J., Sakiyama T., et al. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res. 2008;68:1303–1309. doi: 10.1158/0008-5472.CAN-07-5003. [DOI] [PubMed] [Google Scholar]
- 35.Qin J.J., Cheng X.D., Zhang J., Zhang W.D. Dual roles and therapeutic potential of Keap1-Nrf2 pathway in pancreatic cancer: A systematic review. Cell Commun. Signal. 2019;17:121. doi: 10.1186/s12964-019-0435-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Purohit V., Wang L., Yang H., Li J., Ney G.M., Gumkowski E.R., Vaidya A.J., Wang A., Bhardwaj A., Zhao E., et al. ATDC binds to KEAP1 to drive NRF2-mediated tumorigenesis and chemoresistance in pancreatic cancer. Genes Dev. 2021;35:218–233. doi: 10.1101/gad.344184.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rojo de la Vega M., Chapman E., Zhang D.D. NRF2 and the Hallmarks of Cancer. Cancer Cell. 2018;34:21–43. doi: 10.1016/j.ccell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Best S.A., De Souza D.P., Kersbergen A., Policheni A.N., Dayalan S., Tull D., Rathi V., Gray D.H., Ritchie M.E., McConville M.J., et al. Synergy between the KEAP1/NRF2 and PI3K Pathways Drives Non-Small-Cell Lung Cancer with an Altered Immune Microenvironment. Cell Metab. 2018;27:935–943.e934. doi: 10.1016/j.cmet.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 39.Scalera S., Mazzotta M., Cortile C., Krasniqi E., De Maria R., Cappuzzo F., Ciliberto G., Maugeri-Sacca M. KEAP1-Mutant NSCLC: The Catastrophic Failure of a Cell-Protecting Hub. J. Thorac. Oncol. 2022;17:751–757. doi: 10.1016/j.jtho.2022.03.011. [DOI] [PubMed] [Google Scholar]
- 40.Ghareghomi S., Moosavi-Movahedi F., Saso L., Habibi-Rezaei M., Khatibi A., Hong J., Moosavi-Movahedi A.A. Modulation of Nrf2/HO-1 by Natural Compounds in Lung Cancer. Antioxidants. 2023;12:735. doi: 10.3390/antiox12030735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lv Y., Lv X., Zhang J., Cao G., Xu C., Zhang B., Lin W. BRD4 Targets the KEAP1-Nrf2-G6PD Axis and Suppresses Redox Metabolism in Small Cell Lung Cancer. Antioxidants. 2022;11:661. doi: 10.3390/antiox11040661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Young L.H. A crystallized view of AMPK activation. Cell Metab. 2009;10:5–6. doi: 10.1016/j.cmet.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 43.Steinberg G.R., Carling D. AMP-activated protein kinase: The current landscape for drug development. Nat. Rev. Drug Discov. 2019;18:527–551. doi: 10.1038/s41573-019-0019-2. [DOI] [PubMed] [Google Scholar]
- 44.Lin S.C., Hardie D.G. AMPK: Sensing Glucose as well as Cellular Energy Status. Cell Metab. 2018;27:299–313. doi: 10.1016/j.cmet.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 45.Jansen T., Kvandova M., Daiber A., Stamm P., Frenis K., Schulz E., Munzel T., Kroller-Schon S. The AMP-Activated Protein Kinase Plays a Role in Antioxidant Defense and Regulation of Vascular Inflammation. Antioxidants. 2020;9:525. doi: 10.3390/antiox9060525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jeon S.M. Regulation and function of AMPK in physiology and diseases. Exp. Mol. Med. 2016;48:e245. doi: 10.1038/emm.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salminen A., Hyttinen J.M., Kaarniranta K. AMP-activated protein kinase inhibits NF-kappaB signaling and inflammation: Impact on healthspan and lifespan. J. Mol. Med. 2011;89:667–676. doi: 10.1007/s00109-011-0748-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin C.H., Cheng Y.C., Nicol C.J., Lin K.H., Yen C.H., Chiang M.C. Activation of AMPK is neuroprotective in the oxidative stress by advanced glycosylation end products in human neural stem cells. Exp. Cell Res. 2017;359:367–373. doi: 10.1016/j.yexcr.2017.08.019. [DOI] [PubMed] [Google Scholar]
- 49.Yamauchi T., Kamon J., Minokoshi Y., Ito Y., Waki H., Uchida S., Yamashita S., Noda M., Kita S., Ueki K., et al. Adiponectin stimulates glucose utilization and fatty-acid oxidation by activating AMP-activated protein kinase. Nat. Med. 2002;8:1288–1295. doi: 10.1038/nm788. [DOI] [PubMed] [Google Scholar]
- 50.Bujak A.L., Crane J.D., Lally J.S., Ford R.J., Kang S.J., Rebalka I.A., Green A.E., Kemp B.E., Hawke T.J., Schertzer J.D., et al. AMPK activation of muscle autophagy prevents fasting-induced hypoglycemia and myopathy during aging. Cell Metab. 2015;21:883–890. doi: 10.1016/j.cmet.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao M., Klionsky D.J. AMPK-dependent phosphorylation of ULK1 induces autophagy. Cell Metab. 2011;13:119–120. doi: 10.1016/j.cmet.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Day E.A., Ford R.J., Steinberg G.R. AMPK as a Therapeutic Target for Treating Metabolic Diseases. Trends Endocrinol. Metab. 2017;28:545–560. doi: 10.1016/j.tem.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 53.Dasgupta B., Chhipa R.R. Evolving Lessons on the Complex Role of AMPK in Normal Physiology and Cancer. Trends Pharmacol. Sci. 2016;37:192–206. doi: 10.1016/j.tips.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang B.B., Zhou G., Li C. AMPK: An emerging drug target for diabetes and the metabolic syndrome. Cell Metab. 2009;9:407–416. doi: 10.1016/j.cmet.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 55.Eichner L.J., Brun S.N., Herzig S., Young N.P., Curtis S.D., Shackelford D.B., Shokhirev M.N., Leblanc M., Vera L.I., Hutchins A., et al. Genetic Analysis Reveals AMPK Is Required to Support Tumor Growth in Murine Kras-Dependent Lung Cancer Models. Cell Metab. 2019;29:285–302.e287. doi: 10.1016/j.cmet.2018.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shao D., Oka S., Liu T., Zhai P., Ago T., Sciarretta S., Li H., Sadoshima J. A redox-dependent mechanism for regulation of AMPK activation by Thioredoxin1 during energy starvation. Cell Metab. 2014;19:232–245. doi: 10.1016/j.cmet.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Habib S.L., Yadav A., Kidane D., Weiss R.H., Liang S. Novel protective mechanism of reducing renal cell damage in diabetes: Activation AMPK by AICAR increased NRF2/OGG1 proteins and reduced oxidative DNA damage. Cell Cycle. 2016;15:3048–3059. doi: 10.1080/15384101.2016.1231259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kukidome D., Nishikawa T., Sonoda K., Imoto K., Fujisawa K., Yano M., Motoshima H., Taguchi T., Matsumura T., Araki E. Activation of AMP-activated protein kinase reduces hyperglycemia-induced mitochondrial reactive oxygen species production and promotes mitochondrial biogenesis in human umbilical vein endothelial cells. Diabetes. 2006;55:120–127. doi: 10.2337/diabetes.55.01.06.db05-0943. [DOI] [PubMed] [Google Scholar]
- 59.Mo C., Wang L., Zhang J., Numazawa S., Tang H., Tang X., Han X., Li J., Yang M., Wang Z., et al. The crosstalk between Nrf2 and AMPK signal pathways is important for the anti-inflammatory effect of berberine in LPS-stimulated macrophages and endotoxin-shocked mice. Antioxid. Redox Signal. 2014;20:574–588. doi: 10.1089/ars.2012.5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zimmermann K., Baldinger J., Mayerhofer B., Atanasov A.G., Dirsch V.M., Heiss E.H. Activated AMPK boosts the Nrf2/HO-1 signaling axis—A role for the unfolded protein response. Free Radic. Biol. Med. 2015;88:417–426. doi: 10.1016/j.freeradbiomed.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Joo M.S., Kim W.D., Lee K.Y., Kim J.H., Koo J.H., Kim S.G. AMPK Facilitates Nuclear Accumulation of Nrf2 by Phosphorylating at Serine 550. Mol. Cell. Biol. 2016;36:1931–1942. doi: 10.1128/MCB.00118-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lv H., Liu Q., Wen Z., Feng H., Deng X., Ci X. Xanthohumol ameliorates lipopolysaccharide (LPS)-induced acute lung injury via induction of AMPK/GSK3beta-Nrf2 signal axis. Redox Biol. 2017;12:311–324. doi: 10.1016/j.redox.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cuadrado A. Structural and functional characterization of Nrf2 degradation by glycogen synthase kinase 3/beta-TrCP. Free Radic. Biol. Med. 2015;88:147–157. doi: 10.1016/j.freeradbiomed.2015.04.029. [DOI] [PubMed] [Google Scholar]
- 64.Rada P., Rojo A.I., Evrard-Todeschi N., Innamorato N.G., Cotte A., Jaworski T., Tobon-Velasco J.C., Devijver H., Garcia-Mayoral M.F., Van Leuven F., et al. Structural and functional characterization of Nrf2 degradation by the glycogen synthase kinase 3/beta-TrCP axis. Mol. Cell. Biol. 2012;32:3486–3499. doi: 10.1128/MCB.00180-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matzinger M., Fischhuber K., Poloske D., Mechtler K., Heiss E.H. AMPK leads to phosphorylation of the transcription factor Nrf2, tuning transactivation of selected target genes. Redox Biol. 2020;29:101393. doi: 10.1016/j.redox.2019.101393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hellyer J.A., Padda S.K., Diehn M., Wakelee H.A. Clinical Implications of KEAP1-NFE2L2 Mutations in NSCLC. J. Thorac. Oncol. 2021;16:395–403. doi: 10.1016/j.jtho.2020.11.015. [DOI] [PubMed] [Google Scholar]
- 67.Wakabayashi N., Dinkova-Kostova A.T., Holtzclaw W.D., Kang M.I., Kobayashi A., Yamamoto M., Kensler T.W., Talalay P. Protection against electrophile and oxidant stress by induction of the phase 2 response: Fate of cysteines of the Keap1 sensor modified by inducers. Proc. Natl. Acad. Sci. USA. 2004;101:2040–2045. doi: 10.1073/pnas.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heiss E.H., Schachner D., Zimmermann K., Dirsch V.M. Glucose availability is a decisive factor for Nrf2-mediated gene expression. Redox Biol. 2013;1:359–365. doi: 10.1016/j.redox.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bahiraii S., Brenner M., Yan F., Weckwerth W., Heiss E.H. Sulforaphane diminishes moonlighting of pyruvate kinase M2 and interleukin 1beta expression in M1 (LPS) macrophages. Front. Immunol. 2022;13:935692. doi: 10.3389/fimmu.2022.935692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Petsouki E., Gerakopoulos V., Szeto N., Chang W., Humphrey M.B., Tsiokas L. FBW7 couples structural integrity with functional output of primary cilia. Commun. Biol. 2021;4:1066. doi: 10.1038/s42003-021-02504-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Loboda A., Damulewicz M., Pyza E., Jozkowicz A., Dulak J. Role of Nrf2/HO-1 system in development, oxidative stress response and diseases: An evolutionarily conserved mechanism. Cell. Mol. Life Sci. 2016;73:3221–3247. doi: 10.1007/s00018-016-2223-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.He F., Ru X., Wen T. NRF2, a Transcription Factor for Stress Response and Beyond. Int. J. Mol. Sci. 2020;21:4777. doi: 10.3390/ijms21134777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kassovska-Bratinova S., Yang G., Igarashi K., Dennery P.A. Bach1 modulates heme oxygenase-1 expression in the neonatal mouse lung. Pediatr. Res. 2009;65:145–149. doi: 10.1203/PDR.0b013e318191eedc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sutherland C., Leighton I.A., Cohen P. Inactivation of glycogen synthase kinase-3 beta by phosphorylation: New kinase connections in insulin and growth-factor signalling. Pt 1Biochem. J. 1993;296:15–19. doi: 10.1042/bj2960015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available upon request from the corresponding author.