Abstract
Salmonella Isangi is an infrequent serovar that has recently been reported in several countries due to nosocomial infections. A considerable number of reports indicate Salmonella Isangi multidrug resistance, especially to cephalosporins, which could potentially pose a risk to public health worldwide. Genomic analysis is an excellent tool for monitoring the emergence of microorganisms and related factors. In this context, the aim of this study was to carry out a genomic analysis of Salmonella Isangi isolated from poultry in Brazil, and to compare it with the available genomes from the Pathogen Detection database and Sequence Read Archive. A total of 142 genomes isolated from 11 different countries were investigated. A broad distribution of extended-spectrum beta-lactamase (ESBL) genes was identified in the Salmonella Isangi genomes examined (blaCTX-M-15, blaCTX-M-2, blaDHA-1, blaNDM-1, blaOXA-10, blaOXA-1, blaOXA-48, blaSCO-1, blaSHV-5, blaTEM-131, blaTEM-1B), primarily in South Africa. Resistome analysis revealed predicted resistance to aminoglycoside, sulfonamide, macrolide, tetracycline, trimethoprim, phenicol, chloramphenicol, and quaternary ammonium. Additionally, PMQR (plasmid-mediated quinolone resistance) genes qnr19, qnrB1, and qnrS1 were identified, along with point mutations in the genes gyrAD87N, gyrAS83F, and gyrBS464F, which confer resistance to ciprofloxacin and nalidixic acid. With regard to plasmids, we identified 17 different incompatibility groups, including IncC, Col(pHAD28), IncHI2, IncHI2A, IncM2, ColpVC, Col(Ye4449), Col156, IncR, IncI1(Alpha), IncFIB (pTU3), Col(B5512), IncQ1, IncL, IncN, IncFIB(pHCM2), and IncFIB (pN55391). Phylogenetic analysis revealed five clusters grouped by sequence type and antimicrobial gene distribution. The study highlights the need for monitoring rare serovars that may become emergent due to multidrug resistance.
Keywords: whole-genome sequencing, nontyphoidal Salmonella, multidrug resistance, ESBL, PMQR, quinolone, beta-lactamase
1. Introduction
Nontyphoidal Salmonella (NTS) remains one of the most critical enterobacteria, causing foodborne gastroenteritis worldwide (WHO, 2022). Salmonella Typhimurium and Salmonella Enteritidis continue to be the most reported NTS serovars due to their high endemicity in numerous countries [1,2]. Salmonella enterica encompasses approximately 2610 different serovars, many of which are rare and neglected [2]. However, the emergence of rare serovars in human and animal populations should not be ignored. Recently, rare serovars have emerged in African [3], American [4], and Asian [5] countries, with many exhibiting multidrug resistance to antimicrobials. Horizontal gene transfer (HGT) plays a crucial role in these emergences, primarily with plasmids, which can be easily transmitted among serovars [1]. Therefore, monitoring and surveillance of all Salmonella serovars become essential to predict and understand the dissemination trends of rare serovars and to develop prevention measures against emerging pathogens.
Salmonella Isangi is a less-known serovar with few attributed studies. Extended-spectrum beta-lactamase (ESBL)-producing Salmonella Isangi has become common in South Africa and has also been reported in several other countries, mainly in association with nosocomial infections [3,4,6,7,8]. Notably, Brazil appears to be the only country reporting Salmonella Isangi in animal production, particularly, poultry [9], which may represent a new trend in this serovar adaptation in the last few years. The emergence of this serovar in animal production represents a risk for the One Health approach.
Large-scale genomic analysis has become a fundamental epidemiological tool in collecting and exploring free available complete genomes in several public databases [10,11,12]. Critical information may go unnoticed when many samples are sequenced for surveillance purposes, and many genomes remain unexplored. Thus, large-scale genomic analysis can be a solution to better understand the distribution of genes and topographical characteristics such as location, species, and primary sources of isolation of pathogens not usually studied [12,13,14,15].
As Salmonella Isangi seems to be emerging and adapting within Brazilian poultry production, we sought to perform a genomic characterization of a recently sequenced isolate from poultry production in Brazil. In parallel, we genetically compared it with other already available Salmonella Isangi genomes in the NCBI database worldwide to bring new understanding about the genetic epidemiology of this rare serovar.
2. Material and Methods
2.1. Bacterial Isolate and Whole-Genome Sequencing
The Salmonella Isangi isolate was obtained from the reference laboratory of enterobacteria of the Oswaldo Cruz Institute (LABENT-IOC) Salmonella collection. DNA extraction was performed using a commercial kit (Dneasy Blood & Tissue, QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. The genomic DNA of the isolate was sequenced using the Illumina DNA prep library preparation kit at the Miseq platform (Illumina, San Diego, CA, USA) with 300 bp paired-end sequencing.
2.2. Acquisition of Complementary Genomic Data
The filtering tools on the Sequence Read Archive (SRA) web browser of the National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov/pathogens/ (accessed on 2 August 2022)) were used to select and download raw read sequences of Salmonella Isangi available as of 2 August 2022. The genomes were selected according to their quality (i.e., N50 and contig number) and metadata availability (i.e., isolation source, collection date, location).
2.3. Genome Assembly and Quality Filtering
Trimmomatic 0.39 [16] was used to trim raw sequence reads and remove poor-quality bases. FastQC v0.11.9 was employed to assess the quality of trimmed reads and SPAdes 3.15.4 [17] for de novo assembly. The quality of draft genomes was evaluated using QUAST v5.2.0.0 [18] and the assembled contigs were ordered using Mauve 2.4.0 [19]. Samtools 1.15.1 was used to estimate the assembled average coverage. All sequences were annotated using Prokka version 1.14.6 [20].
2.4. In Silico Genome Characterization
SISTR (Salmonella in silico Typing Resource) 1.1.1 [21] software was used to perform in silico serotyping on all the Salmonella Isangi genomes. Multi-locus sequence typing (MLST) software 2.19.0 [22] was employed for in silico genotyping. To identify antimicrobial resistance genes (AMR), plasmid replicons, and virulence factors, ABRicate software 1.0.1 (https://github.com/tseemann/abricate (accessed on 24 January 2023)) was used alongside the Resfinder [23], Plasmidfinder [24], and Virulence Factor (VFDB) (http://www.mgc.ac.cn/VFs/ (accessed on 24 January 2023)) databases, respectively. Pointfinder software 0.8.0 [23] was used to identify AMR point mutations conferring potential antimicrobial resistance within the genomes. A customized pESI database [13] was used to identify plasmids for emergent Salmonella Infantis on the Salmonella Isangi genomes. Minimum nucleotide identity and coverage thresholds of 90% and 60%, respectively, were employed for all analyses.
2.5. Phylogenetic Analysis
The detection and prediction of the effects of whole-genome variants were carried out using the Rapid haploid variant calling and core genome alignment—Snippy v4.6.0 (https://github.com/tseemann/snippy (accessed on 24 January 2023)). As there is no complete sequence of Salmonella Isangi available as a reference in the public genome repertoires, we used a complete genome from Salmonella Infantis (Accession number GCA_029592185.1), which was also our outgroup. Gubbins v2.4.1 was used to identify and exclude recombinant regions within the core genome alignment to enhance the accuracy of phylogenetic reconstructions. Phastaf v0.1.0 (https://github.com/tseemann/phastaf (accessed on 24 January 2023)) and Barrnap v0.9 (https://github.com/tseemann/barrnap (accessed on 24 January 2023)) were used to identify phage regions and ribosomal RNA for masking purposes in the reference genome, respectively. The final high-quality core-genome SNPs were extracted from the alignment file using the software SNP-sites v2.5.1 [25]. Iqtree v2.0.3 was employed to generate a maximum likelihood phylogeny from this core genome alignment, using a GTR + F + I + G4 substitution model, with 1000 bootstraps replication to support the tree nodes.
3. Results
3.1. Data Collection and MLST Distribution
A total of 141 raw sequences of Salmonella Isangi were downloaded from the SRA web browser. By utilizing the SISTR software, we confirmed that all genomes were Isangi serovar. The genomes were obtained from strains isolated from 11 different countries: South Africa (n = 65); Uganda (n = 1); United Kingdom (n = 35); United States of America (n = 16); Brazil (n = 15); Germany (n = 1), India (n = 2), Netherlands (n = 1), Ireland (n = 2), Indonesia (n = 1), and Taiwan (n = 1) (Supplementary Table S1). Our analysis comprised 142 genomes, including the newly sequenced Salmonella Isangi genome (Sal08932019 Accession: JARETA000000000) from Brazil. Regarding the sources, 109 genomes came from human isolates, three were from animals, three from food, seven from feed, six from the environment, and 14 were not identified. We identified that genomes belonged to nine different sequence types (ST): ST216 (n = 62); ST335 (n = 59); ST1994 (n = 8); ST2261 (n = 1); ST369 (n = 1); ST3101 (n = 1); ST3390 (n = 1); ST5028 (n = 1); and ST7036 (n = 2) (Supplementary Table S1).
3.2. Resistome
The 142 genomes presented a resistome that included the following repertoire of antimicrobial genes: aminoglycoside [aac(3)-Iia_1, aac(3)-Iid, aac(3)-Via, aac(6’)-Iaa, aac(6’)-Ib-cr, aadA1, aadA2,aadA5, ant(3’’)-Ia, aph(3’’)-Ib, aph(3’’)-Ib, aph(3’)-Ia, aph(6)-Id], sulfonamide [sul1, sul2, sul3], macrolide [mph(A), mph(E)], tetracycline [tet(A)], trimethoprim [dfrA12, dfrA14, dfrA15, dfrA17, dfrA1, dfrA23], phenicol [floR], beta-lactams [blaCTX-M-15, blaCTX-M-2, blaDHA-1, blaNDM-1, blaOXA-10, blaOXA-1, blaOXA-48, blaSCO-1, blaSHV-5, blaTEM-131, blaTEM-1B], chloramphenicol [catA1, catB3], and quaternary ammonium [qacE] (Supplementary Table S1). We identified a wide distribution of extended-spectrum beta-lactamase (ESBL) genes among the genomes from South Africa and the United Kingdom (Figure 1). ESBL genes were also identified in Salmonella Isangi isolated from Ireland (blaTEM-1B) and the United States (blaCTX-M-2). Furthermore, we detected 83 genomes exhibiting plasmid-mediated quinolone resistance (PMQR) genes (Supplementary Table S1). The PMQR qnrB1 (n = 47) was the most common quinolone resistance gene observed, followed by qnrB19 (n = 36) and qnrS1 (n = 2). All 47 genomes carrying the qnrB1 gene co-produced at least one ESBL gene (blaCTX-M-15, blaDHA-1, blaNDM-1, blaOXA-10, blaOXA-1, blaOXA-48, blaSCO-1, blaSHV-5, blaTEM-131, blaTEM-1B), with many of them co-producing more than one. Ten of the genomes carrying the qnrB19 gene co-produced ESBL gene (blaCTX-M-2) and were isolated from the United States, whereas the others were positive only for qnrB19. All genomes carrying qnrS1 were positive for qnrB19 and co-produced ESBL genes (blaLAP-2, blaTEM-1B). Our sequenced genome (Sal08932019 Accession: JARETA000000000) presented only the plasmid-mediated quinolone resistance gene qnrB19, a characteristic shared with the other Salmonella Isangi isolated from Brazil [9]. The PMQR qnrB19 was identified in genomes isolated from Brazil (n = 14), the United Kingdom (n = 11), the United States (n = 10), and Ireland (n = 1). The qnrB1 was primarily identified in South Africa (n = 46) and the United Kingdom (n = 1). The qnrS1 was identified in genomes isolated from Ireland (n = 1) and the United Kingdom (n = 1). Regarding the determination of quinolone resistance, 10 isolates presented a single mutation in the genes gyrAD87N, gyrAS83F, and gyrBS464F, which conferred resistance to ciprofloxacin and nalidixic acid. These genomes were all isolated from South Africa (Table 1). Seven genomes isolated from the United Kingdom (n = 6) and India (n = 1) presented a single mutation in the gene parCT57S; however, there was no predicted phenotype.
Figure 1.
Predicted genetic traits of the Salmonella Isangi (n = 142). AMR genes presence is represented by the black spots.
Table 1.
Features of Salmonella Isangi (n = 15) displaying point mutation in gyrA and parC genes, conferring resistance to ciprofloxacin and nalidixic acid.
| Isolate ID | Location | Year of Isolation | Resistome | Gene | Mutation | Predicted Phenotype |
|---|---|---|---|---|---|---|
| ERR4399363 | South Africa | 2001 | aac(3)-IIa, ant(3″)-Ia, aph(3″)-Ib, aph(6)-Id, ARR-3, blaOXA-10, blaSCO-1, blaTEM-63, cmlA1, dfrA23, floR, gyrA (D87N), qacE, sul1, sul2, tet(A) | gyrA (D87N) | GAC→AAC (D→N) | ciprofloxacin I/R, nalidixic acid |
| ERR4400265 | South Africa | 2001 | aac(3)-IIa, ant(3″)-Ia, aph(3″)-Ib, aph(6)-Id, ARR-3, blaOXA-10, blaSCO-1, blaTEM-63, cmlA1, dfrA23, floR, gyrA (D87N), qacE, sul1, sul2, tet(A) | gyrA (D87N) | GAC→AAC (D→N) | ciprofloxacin I/R, nalidixic acid |
| ERR4400834 | Tawain | 2007 | gyrA (S83F) | TCC→TTC (S→F) | ciprofloxacin I/R, nalidixic acid | |
| ERR4400884 | South Africa | 2001 | aac(3)-IIa, ant(3″)-Ia, aph(3″)-Ib, aph(6)-Id, ARR-3, blaOXA-10, blaSCO-1, blaTEM-63, cmlA1, dfrA23, floR, gyrA (D87N), qacE, sul1, sul2, tet(A) | gyrA (D87N) | GAC→AAC (D→N) | ciprofloxacin I/R, nalidixic acid |
| ERR4401297 | South Africa | 2001 | aac(3)-IIa, ant(3″)-Ia, aph(3″)-Ib, aph(6)-Id, ARR-3, blaOXA-10, blaSCO-1, blaTEM-63, cmlA1, dfrA23, floR, gyrA (D87N), qacE, sul1, sul2, tet(A) | gyrA (D87N) | GAC→AAC (D→N) | ciprofloxacin I/R, nalidixic acid |
| ERR4401298 | South Africa | 2001 | aac(3)-IIa, ant(3″)-Ia, aph(3″)-Ib, aph(6)-Id, ARR-3, blaOXA-10, blaSCO-1, blaTEM-63, cmlA1, dfrA23, floR, gyrA (D87N), qacE, sul1, sul2, tet(A) | gyrA (D87N) | GAC→AAC (D→N) | ciprofloxacin I/R, nalidixic acid |
| ERR4401299 | South Africa | 2001 | aac(3)-IIa, ant(3″)-Ia, aph(3″)-Ib, aph(6)-Id, ARR-3, blaOXA-10, blaSCO-1, blaTEM-63, cmlA1, dfrA23, floR, gyrA (D87N), qacE, sul1, sul2, tet(A) | gyrA (D87N) | GAC→AAC (D→N) | ciprofloxacin I/R, nalidixic acid |
| ERR4401598 | South Africa | 2001 | aac(3)-IIa, ant(3″)-Ia, aph(3″)-Ib, aph(6)-Id, ARR-3, blaOXA-10, blaSCO-1, blaTEM-63, cmlA1, dfrA23, floR, gyrA (D87N), qacE, sul1, sul2, tet(A) | gyrA (D87N) | GAC→AAC (D→N) | ciprofloxacin I/R, nalidixic acid |
| ERR6806075 | South Africa | 2020 | aac(3)-IIa, aph(3″)-Ib, aph(6)-Id, blaCTX-M-15, blaTEM-1B, catB3, gyrB (S464F), sul2 | gyrB (S464F) | TCT→TTC (S→F) | ciprofloxacin I/R |
| SRR1106479 | Unknown | gyrA (S83F) | TCC→TTC (S→F) | ciprofloxacin I/R, nalidixic acid | ||
| SRR1264936 | India | 2011 | parC (T57S) | parC (T57S) | ACC→AGC (T→S) | None |
| SRR13633613 | United Kingdom | 2021 | parC (T57S) | parC (T57S) | ACC→AGC (T→S) | None |
| SRR13633625 | United Kingdom | 2021 | parC (T57S) | parC (T57S) | ACC→AGC (T→S) | None |
| SRR13633627 | United Kingdom | 2021 | parC (T57S) | parC (T57S) | ACC→AGC (T→S) | None |
| SRR13633629 | United Kingdom | 2021 | parC (T57S) | parC (T57S) | ACC→AGC (T→S) | None |
| SRR13645903 | United Kingdom | 2021 | parC (T57S) | parC (T57S) | ACC→AGC (T→S) | None |
| SRR3002515 | United States | 2009 | parC (T57S) | parC (T57S) | ACC→AGC (T→S) | None |
3.3. Virulome
The analyzed genomes presented the core virulence genes responsible for Salmonella’s pathogenicity (Supplementary Table S1). Moreover, three genomes from South Africa (n = 2) and the United States (n = 1) presented the yersiniabactin siderophore (ybtAPQSTUX, fyuA, irp1, and irp2), which increases iron uptake. We also evaluated the presence of pESI using a customized database with the pESI backbone genes. A single genome from India presented 99 out of 113 genes from the pESI core genome. This genome did not display any antimicrobial resistance genes or other essential genes that make up the pESI megaplasmid. Beforehand, we ruled out the hypothesis that this genome had a specific pESI structure.
The most frequent plasmid incompatibility group identified among the analyzed Salmonella Isangi was IncC [n = 66], followed by Col(pHAD28) [n = 39], IncHI2 [n = 37], IncHI2A [n = 28], IncM2 [n = 12], ColpVC [n = 6], Col(Ye4449) [n = 5], Col156 [n = 5], IncR [n = 5], IncI1(Alpha) [n = 4], IncFIB (pTU3) [n = 3], Col(B5512) [n = 2], IncQ1 [n = 2], IncL [n = 2], IncN [n = 2], IncFIB(pHCM2) [n = 1], and IncFIB (pN55391) [n = 1].
All genomes carrying the PMQR qnrB19 mapped to the Col(pHAD28) incompatibility group, with different distributions of other incompatibility groups [Col(Ye4449), ColpVC, and IncL] (Supplementary Table S2). Among the genomes that co-produced the ESBL gene blaCTX-M-2, we identified the incompatibility groups IncHI2, IncHI2A, and IncQ1. All genomes carrying the PMQR qnrB1 displayed the IncC incompatibility group, with different distributions of other incompatibility groups [IncCM2, IncHI2, IncHI2A, IncI1(Alpha), IncL, and IncFIB (pHCM2)]. The genomes carrying the qnrS1 displayed the same distribution of incompatibility groups as the genomes carrying the qnrB19 gene (Supplementary Table S2).
3.4. Phylogenetics and Characterization of Salmonella Isangi Lineages
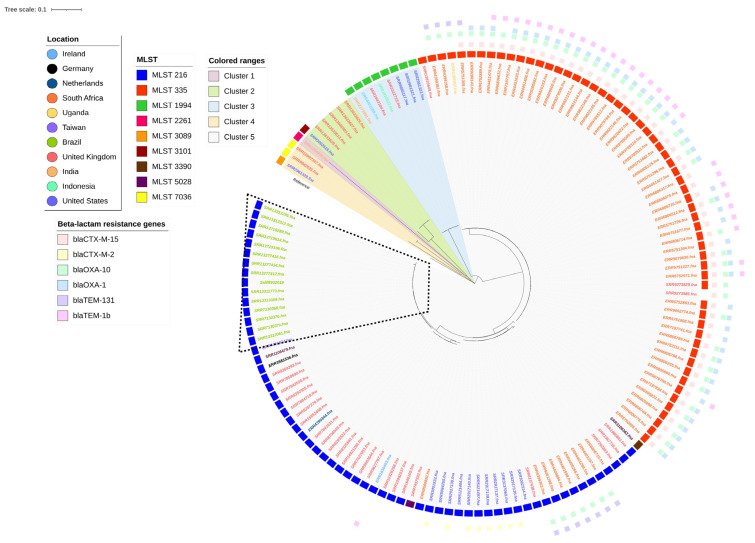
We conducted a whole-genome variant detection on the set of 142 Salmonella Isangi genomes. The phylogenetic tree that was generated showed five well-defined clusters, mostly grouped according to country, ESBL gene distribution, and MLST (Figure 2). Cluster 1 included a single genome isolated from India (ST2261) with no antimicrobial resistance gene. Cluster 2 differed from the other clusters by 13,601 SNPs and consisted of six genomes: one from the United States and five from the United Kingdom. Cluster 3 differed from the other clusters by 22,877 SNPs and was composed of three genomes: one from the United States (ST3059) and two from the United Kingdom (ST7036). Cluster 4 comprised eight genomes within the ST1994 and differed from the other clusters by 28,422 SNPs. Three came from the United States, two from the United Kingdom, and one from each of India, Ireland, and Indonesia. Cluster 5 was defined on each from ST216 and ST335, the most frequent MLST among the studied genomes, and differed from the other clusters by 22,877 SNPs. They were grouped by country, and the ESBL distribution also contributed to the genome clustering. All the genomes from South Africa presented at least one ESBL gene, with blaCTX-M-15 being the most frequent. A single ST335 genome from the United Kingdom was grouped with South African genomes and did not present the ESBL gene. The ST216 genomes from the United States were grouped beside and presented blaCTX-M-2. The ST216 genomes from South Africa showed the ESBL genes blaOXA-10 and blaTEM-1B. All the genomes from Brazil belonged to the ST216 and possessed no ESBL genes. However, Brazilian genomes, including ours, presented the qnrB19 quinolone resistance gene.
Figure 2.
Maximum-likelihood SNP-based phylogeny of 142 Salmonella Isangi genomes using, as reference, Salmonella Infantis (accession number GCA_029592185.1). The circular representation of the phylogeny was obtained using iTOL (http://itol.embl.de/ (accessed on 8 February 2023)), ignoring branch lengths. Colors of the isolates’ ID indicate different countries of origin. Colored squares indicate the genome sequence type (MLST) and the beta-lactam resistance gene presence. Isolates from Brazil are detached.
4. Discussion
Our literature review identified very few studies on Salmonella Isangi in Brazil and worldwide [3,4,9,26,27]. Recently, this serovar has been associated with foodborne disease outbreaks in China and nosocomial infections in the United States and South Africa [3,4,5,9,26,27], particularly those caused by Salmonella Isangi with ESBL genes. The emergence of rare serovars with antimicrobial resistance or multidrug resistance poses a risk to public health, as they tend to be neglected due to inadequate monitoring and prevention efforts. In this study, we performed genomic characterization of a Salmonella Isangi strain isolated from poultry in Brazil in association with other Salmonella Isangi genomes available in the NCBI database, to provide new insights from a global perspective.
The majority of the genomes analyzed in this study were isolated from South Africa (Figure 2). According to the Enteric Diseases Reference Unit of the National Institute for Communicable Diseases in South Africa, there has been an increase in Salmonella Isangi infections in the country since 2000 [6], which explains this high prevalence. We identified genomes isolated from 2001 to 2020, proving that the widespread dissemination of Salmonella Isangi is still occurring in the country. It is worth noting, however, that our results are limited to genomes that have been sequenced and deposited in the NCBI database. Nevertheless, these results may reflect a broader reality of emergence of Salmonella Isangi in South Africa. South Africa has the most significant number of HIV-positive patients worldwide [28], who tend to develop severe non-typhoidal Salmonella infection due to their immunodeficient status [29]. The high demand for last-choice antimicrobials, including extended-spectrum cephalosporins [6] to treat these severe infections, may contribute to the emergence of ESBL-positive Salmonella Isangi in the South African population.
Of the South African genomes analyzed, 54 (83.1%) displayed the blaCTX-M-15 gene. Salmonella Isangi containing this resistance gene [4] has been previously reported in a hospital outbreak in the United States. Initially, blaCTX-M-15 was associated exclusively with E. coli and Klebsiella spp. in the human population. However, recent epidemiological studies have reported the CTX-M-15 variant as one of the most widely dominant CTX-M β-lactamases [30,31]. The pandemic dissemination of CTX-M-15 among Enterobacteriaceae originated from a highly virulent E. coli O25:H4-ST131 [32,33], and their widespread occurrence is linked to the insertion element ISEcpI responsible for the mobilization of bla genes [34]. We evaluated the presence of ISEcpI in the South African genomes with blaCTX-M-15. All presented this mobile element, confirming that the horizontal gene transfer ability among the Salmonella Isangi is already established in the South African population. The blaCTX-M-15 gene is currently associated with the blaTEM-1 and blaOXA-1 resistance genes [32], and was also detected in most South African genomes (Figure 1). Although this association is often related to plasmids belonging to the IncF group [32], we observed a strong association of these genes with the IncC group in our study. Previous research conducted on E. coli demonstrated that the IncF CTX-M-15 plasmid is better adapted to this species due to its lower fitness cost compared to the IncC CTX-M-15 plasmid [31]. However, selective pressure may have been the crucial factor for the rapid adaptation of IncC plasmid among bacteria [31], and may have occurred in Salmonella Isangi genomes from South Africa.
Genomes belonging to ST335 presented qnrB1 that confers low-level quinolone resistance [35]. The Qnr family of proteins is known to protect DNA gyrase and topoisomerase from quinolone antimicrobial activity [35] and to facilitate the selection of high-level quinolone-resistant mutants [35]. The presence of PMQR genes such as qnrB1 has raised concerns among health authorities in recent years precisely because of their potential to increase resistance among bacteria. Because they are plasmodial genes, their spread is facilitated by horizontal gene transfer [36]. Therefore, active surveillance of these genes is essential to prevent the spread and emergence of serovars, such as Salmonella Isangi, which are not commonly associated with severe infection episodes. The high incidence of ST335 in South Africa, with multiple ESBL genes alongside the PMQR, may justify its endemic status across Africa. We also identified Salmonella Isangi ST335 in Uganda and the United Kingdom, which were positive for ESBL genes and more closely related to the South African genomes (Figure 1). These results reiterate the possibility of rare serovars becoming epidemic and that, therefore, they should not be neglected.
ST216 was the most widespread sequence type in the United States, the United Kingdom, South Africa, and Brazil. This sequence type has recently been found in Africa and Asia [5,37]. Among the 62 ST216 genomes, 56.5% (35) presented the PMQR gene qnrB19. The presence of qnrB19 dated from 2012 is relatively recent compared to genomes isolated before (2001–2008) (Supplementary Table S1). These results are compatible with the first description of qnrB19 in E. coli isolated from pigs in China in 2008 [38]. Quinolone is a widespread group of antimicrobials used to treat human and livestock animal infection [39]. Therefore, the cross-transfer of resistance genes from animals to humans could be facilitated. Several studies have linked the presence of qnrB19 to the increase of quinolone resistance in some South American countries, the United States, and Europe [40,41,42]. Salmonella Isangi presenting this PMQR gene in Brazil has been already studied [9], although the presence of qnrB19 in other Salmonella serovars has also been reported [43,44,45]. Among the ST216, only the Brazilian genomes were isolated from poultry or poultry environment. Our sequenced genome (Sal08932019 Accession: JARETA000000000) displayed the same genotype as the other genomes previously sequenced from Brazil, indicating that Salmonella Isangi in Brazil is likely emerging recently from abroad. The presence of qnrB19 in Salmonella Isangi is a concern for health authorities and threatens One Health maintenance. The gradual reduction of endemic Salmonella, such as S. Typhimurium and S. Enteritidis, in animal production as a result of vaccination may have created opportunities for the emergence of rare Salmonella serovars, such as Salmonella Isangi, in the food production chain, as observed in Brazil. Hence, monitoring resistance genes and plasmids is strategic to prevent and control their emergence in rare serovars.
Previous studies have identified an increased phenotypic and genotypic resistance to quinolones in Salmonella Isangi isolated from poultry [9,46]. Most of the analysis relates the quinolone resistance to two determinants: multiples point mutations on the quinolone resistance-determining region (QRDR) of DNA gyrase (gyrA) and the topoisomerase C (parC) (predicted to provide ciprofloxacin and nalidixic acid resistance) [47], and PMQR. In our study, only 10 of the analyzed genomes presented QNDR point mutations (gyrAD87N, gyrAS83F, and gyrBS464F), all of which were isolated from South Africa in 2001. In contrast, seven genomes from the United Kingdom and India presented point mutation (parCT57S) and were isolated in recent years, indicating that this specific point mutation is more widespread within recent Salmonella Isangi. No relation between QRDR and PMQR was observed.
Eight Salmonella Isangi genomes from the United States belonging to ST216 displayed blaCTX-M-2. In the United States, blaCTX-M-producing Salmonella Isangi were associated with hospital outbreaks [4]. However, to our knowledge, this is the first report of blaCTX-M-2 in Salmonella Isangi isolated from the United States. The genomes described in this study were available in 2015, but, so far, we have not identified studies concerning the CTX-M-2 in Salmonella spp. in the United States. However, several recent studies in Brazil, Colombia, and South Africa have identified CTX-M-2 in various Salmonella serovars associated with the poultry industry [48,49,50,51]. As reviewed by Bevan et al. (2017) [30], CTX-M-2 enzyme was the primary genotype in South America and was the only one identified before the 1990s [52]. This incidence remains high in the Americas, as evidenced by studies conducted mainly in E. coli [53,54,55]. CTX-M-2 seems to be highly associated with the poultry industry, which may pose a field condition for their rapid spread among other countries through horizontal gene transfer. Our results demonstrate the importance of constantly monitoring genomes deposited in databases such as the NCBI.
Salmonella Isangi genomes were grouped based on the country and ST profile (Figure 2). The phylogenetic analysis revealed five different clades. The main difference among these clades is the distribution of beta-lactam resistance genes, which we observed to be more widespread in the South African population. The most phylogenetically distant genome was from India and was clustered alone in Cluster 1. It displayed a differentiated distribution of virulence genes, with 99 out of 113 genes from the pESI megaplasmid core genome. According to Hall et al. (2022) [56] forming megaplasmids such as pESI requires the imposition of selective pressure from different niches until the megaplasmid is developed, allowing the bacteria to survive in this challenging environment. Although our analysis demonstrated no pESI in the Indian genome, these results suggest only the beginning of the acquisition of genes, due to the different selective pressures imposed in the Indian isolate. Further genomic studies are required to corroborate this hypothesis.
Regarding virulence genes, three genomes from South Africa (2020) and the United States (2010) presented the complete yersiniabactin siderophore (ybtAPQSTUX, fyuA, irp1, and irp2) that increases iron uptake. These genes are part of the Yersinia High Pathogenicity Island (HPI) [57], which is commonly found in Yersinia, Salmonella spp., and E. coli [57,58]. The presence of HPI in enterobacteria has been shown to increase environmental persistence and fitness [59]. This study suggests that the acquisition of HPI may be a determinant for Salmonella Isangi dissemination and persistence across these countries and the established population.
5. Conclusions
In summary, our study highlights the endemic success of Salmonella Isangi in South Africa and how it could be related to the acquisition of multiple ESBL genes associated with PMQR genes, mainly qnrB1. These results reaffirm the need to constantly monitor Salmonella Isangi and other rare serovar dissemination across continents, as MDR Salmonella spp. pose a significant risk to public health. Moreover, the presence of antimicrobial-resistant Salmonella Isangi in the Brazilian poultry industry could significantly impact One Health, given the international importance of Brazil in the production and export of poultry products and their potential risk of transmission to humans. Lastly, our results highlight the importance of large-scale genomic analysis as an epidemiological and monitoring tool for the surveillance of Salmonella and other microorganisms.
Acknowledgments
The authors acknowledge all individuals and institutions that contributed to this study.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics12081309/s1, Supplementary Table S1: Features of all analyzed Salmonella Isangi (n = 142), comprising information about location, isolation source, isolation type, year of isolation, SNPcluster, AMR genotype and predicted phenotype, plasmids, cgMLST, and virulence genes’ presence and absence (number 1 represents presence and 0 represents absence); Supplementary Table S2: Features of Salmonella Isangi, displaying PMQR genes and distribution of their incompatibility groups.
Author Contributions
A.M.P.d.S. and P.P. contributed to the conception and design of the study. A.M.P.d.S. wrote the paper. A.M.P.d.S., A.C.S.d.J., A.B.P. and A.C.O. carried out the experiments and sample preparation. R.G.F. and P.P. contributed to the interpretation of results. D.d.P.R. provided samples and analytical tools. C.A.C.-J. supervised the project. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on the institutional websites cited in the Section 2 and supplementary material.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ) Brazil—grant number [E-26/200.891/2021], the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq)—grant number [313119/2020-1], and the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) Brazil—FinanceCode001.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Tankson J.D., Fedorka-Cray P.J., Jackson C.R., Headrick M. Genetic relatedness of a rarely isolated Salmonella: Salmonella enterica serotype Niakhar from NARMS animal isolates. J. Antimicrob. Chemother. 2006;57:190–198. doi: 10.1093/jac/dki439. [DOI] [PubMed] [Google Scholar]
- 2.Hendriksen R.S., Vieira A.R., Karlsmose S., Lo Fo Wong D.M.A., Jensen A.B., Wegener H.C., Aarestrup F.M. Global Monitoring of Salmonella Serovar Distribution from the World Health Organization Global Foodborne Infections Network Country Data Bank: Results of Quality Assured Laboratories from 2001 to 2007. Foodborne Pathog. Dis. 2011;8:887–900. doi: 10.1089/fpd.2010.0787. [DOI] [PubMed] [Google Scholar]
- 3.Govinden U., Mocktar C., Moodley P., Sturm A., Essack S. CTX-M-37 in Salmonella enterica serotype Isangi from Durban, South Africa. Int. J. Antimicrob. Agents. 2006;28:288–291. doi: 10.1016/j.ijantimicag.2006.05.028. [DOI] [PubMed] [Google Scholar]
- 4.Suleyman G., Perri M., Vager D., Samuel L., Zervos M.J., Alangaden G., Tibbetts R.J. Characterization of Salmonella Isangi possessing a CTX-M15 ESBL associated with an outbreak in a US Hospital. Diagn. Microbiol. Infect. Dis. 2016;85:386–390. doi: 10.1016/j.diagmicrobio.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 5.Li X.-P., Gao R.-H., Hou P.-B., Ren Y.-Y., Zhang H.-N., Jiang K.-Y., Chen Y.-Z., Qi Z.-G., Xu M., Bi Z.-W. Characterization of the Salmonella enterica Serotype Isangi Isolated from Patients for the First Time in China. Foodborne Pathog. Dis. 2017;14:427–431. doi: 10.1089/fpd.2016.2269. [DOI] [PubMed] [Google Scholar]
- 6.Kruger T., Szabo D., Keddy K.H., Deeley K., Marsh J.W., Hujer A.M., Bonomo R.A., Paterson D.L. Infections with Nontyphoidal Salmonella Species Producing TEM-63 or a Novel TEM Enzyme, TEM-131, in South Africa. Antimicrob. Agents Chemother. 2004;48:4263–4270. doi: 10.1128/AAC.48.11.4263-4270.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hasman H., Mevius D., Veldman K., Olesen I., Aarestrup F.M. β-Lactamases among extended-spectrum β-lactamase (ESBL)-resistant Salmonella from poultry, poultry products and human patients in The Netherlands. J. Antimicrob. Chemother. 2005;56:115–121. doi: 10.1093/jac/dki190. [DOI] [PubMed] [Google Scholar]
- 8.Kulkarni R., Ajantha G., Shubhada C., Jain P. Isolation of salmonella enterica serotype isangi from a suspected case of enteric encephalopathy. Indian J. Med. Microbiol. 2009;27:65–66. doi: 10.1016/S0255-0857(21)01759-X. [DOI] [PubMed] [Google Scholar]
- 9.Monte D.F., Nethery M.A., Barrangou R., Landgraf M., Fedorka-Cray P.J. Whole-genome sequencing analysis and CRISPR genotyping of rare antibiotic-resistant Salmonella enterica serovars isolated from food and related sources. Food Microbiol. 2021;93:103601. doi: 10.1016/j.fm.2020.103601. [DOI] [PubMed] [Google Scholar]
- 10.Giardine B., Riemer C., Hardison R.C., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., et al. Galaxy: A platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang B., Ramazzotti D., De Sano L., Zhu J., Pierson E., Batzoglou S. SIMLR: A Tool for Large-Scale Genomic Analyses by Multi-Kernel Learning. Proteomics. 2018;18:1700232. doi: 10.1002/pmic.201700232. [DOI] [PubMed] [Google Scholar]
- 12.dos Santos A.M., Panzenhagen P., Ferrari R.G., Conte-Junior C.A. Large-scale genomic analysis reveals the pESI-like megaplasmid presence in Salmonella Agona, Muenchen, Schwarzengrund, and Senftenberg. Food Microbiol. 2022;108:104112. doi: 10.1016/j.fm.2022.104112. [DOI] [PubMed] [Google Scholar]
- 13.dos Santos A.M., Panzenhagen P., Ferrari R.G., Rodrigues G.L., Conte-Junior C.A. The pESI megaplasmid conferring virulence and multiple-drug resistance is detected in a Salmonella Infantis genome from Brazil. Infect. Genet. Evol. 2021;95:104934. doi: 10.1016/j.meegid.2021.104934. [DOI] [PubMed] [Google Scholar]
- 14.Rodrigues G.L., Panzenhagen P., Ferrari R.G., dos Santos A., Paschoalin V.M.F., Conte-Junior C.A. Frequency of Antimi-crobial Resistance Genes in Salmonella From Brazil by in silico Whole-Genome Sequencing Analysis: An Overview of the Last Four Decades. Front. Microbiol. 2020;11:1864. doi: 10.3389/fmicb.2020.01864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Panzenhagen P., Portes A.B., dos Santos A.M.P., Duque S.d.S., Junior C.A.C. The Distribution of Campylobacter jejuni Virulence Genes in Genomes Worldwide Derived from the NCBI Pathogen Detection Database. Genes. 2021;12:1538. doi: 10.3390/genes12101538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gurevich A., Saveliev V., Vyahhi N., Tesler G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rissman A.I., Mau B., Biehl B.S., Darling A.E., Glasner J.D., Perna N.T. Reordering contigs of draft genomes using the Mauve Aligner. Bioinformatics. 2009;25:2071–2073. doi: 10.1093/bioinformatics/btp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seemann T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 21.Yoshida C.E., Kruczkiewicz P., Laing C.R., Lingohr E.J., Gannon V.P.J., Nash J.H.E., Taboada E.N. The Salmonella In Silico Typing Resource (SISTR): An Open Web-Accessible Tool for Rapidly Typing and Subtyping Draft Salmonella Genome Assemblies. PLoS ONE. 2016;11:e0147101. doi: 10.1371/journal.pone.0147101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pearce M.E., Alikhan N.-F., Dallman T.J., Zhou Z., Grant K., Maiden M.C. Comparative analysis of core genome MLST and SNP typing within a European Salmonella serovar Enteritidis outbreak. Int. J. Food Microbiol. 2018;274:1–11. doi: 10.1016/j.ijfoodmicro.2018.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zankari E., Allesøe R., Joensen K.G., Cavaco L.M., Lund O., Aarestrup F.M. PointFinder: A novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 2017;72:2764–2768. doi: 10.1093/jac/dkx217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carattoli A., Zankari E., Garcìa-Fernandez A., Larsen M., Lund O., Voldby Villa L., Møller Aarestrup F., Hasman H. In Silico Detection and Typing of Plasmids. Antimicrob using PlasmidFinder and plasmid multilocus sequence typing. Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Page A.J., Taylor B., Delaney A.J., Soares J., Seemann T., Keane J.A., Harris S.R. SNP-sites: Rapid efficient extraction of SNPs from multi-FASTA alignments. Microb. Genom. 2016;2:038190. doi: 10.1099/mgen.0.000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suleyman G., Tibbetts R., Perri M.B., Vager D., Xin Y., Reyes K., Samuel L., Chami E., Starr P., Pietsch J., et al. Nosocomial Outbreak of a Novel Extended-Spectrum β-Lactamase Salmonella enterica Serotype Isangi Among Surgical Patients. Infect. Control. Hosp. Epidemiol. 2016;37:954–961. doi: 10.1017/ice.2016.85. [DOI] [PubMed] [Google Scholar]
- 27.Yadava R., Prasad M., Narayan K.G., Jayasheela M., John P.C., Mago M.L., Saxena S.N. Isolation of Salmonella isangi (6, 7, 14:d:l, 5) for the first time in India. Indian J. Med. Res. 1986;84:20–21. [PubMed] [Google Scholar]
- 28.Fassin D., Schneider H. The politics of AIDS in South Africa: Beyond the controversies. BMJ. 2003;326:495–497. doi: 10.1136/bmj.326.7387.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldberg M.B., Rubin R.H. The Spectrum of Salmonella Infection. Infect. Dis. Clin. N. Am. 1988;2:571–598. doi: 10.1016/S0891-5520(20)30212-9. [DOI] [PubMed] [Google Scholar]
- 30.Bevan E.R., Jones A.M., Hawkey P.M. Global epidemiology of CTX-M β-lactamases: Temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 2017;72:2145–2155. doi: 10.1093/jac/dkx146. [DOI] [PubMed] [Google Scholar]
- 31.Mahérault A.-C., Kemble H., Magnan M., Gachet B., Roche D., Le Nagard H., Tenaillon O., Denamur E., Branger C., Landraud L. Advantage of the F2:A1:B- IncF Pandemic Plasmid over IncC Plasmids in In Vitro Acquisition and Evolution of blaCTX-M Gene-Bearing Plasmids in Escherichia coli. Antimicrob. Agents Chemother. 2019;63:e01130-19. doi: 10.1128/AAC.01130-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carattoli A. Resistance Plasmid Families in Enterobacteriaceae. Antimicrob. Agents Chemother. 2009;53:2227–2238. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leflon-Guibout V., Blanco J., Amaqdouf K., Mora A., Guize L., Nicolas-Chanoine M.-H. Absence of CTX-M Enzymes but High Prevalence of Clones, Including Clone ST131, among Fecal Escherichia coli Isolates from Healthy Subjects Living in the Area of Paris, France. J. Clin. Microbiol. 2008;46:3900–3905. doi: 10.1128/JCM.00734-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poirel L., Bonnin R.A., Nordmann P. Analysis of the Resistome of a Multidrug-Resistant NDM-1-Producing Escherichia coli Strain by High-Throughput Genome Sequencing. Antimicrob. Agents Chemother. 2011;55:4224–4229. doi: 10.1128/AAC.00165-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Briales A., Rodríguez-Martínez J.M., Velasco C., de Alba P.D., Domínguez-Herrera J., Pachón J., Pascual A. In Vitro Effect of qnrA1, qnrB1, and qnrS1 Genes on Fluoroquinolone Activity against Isogenic Escherichia coli Isolates with Mutations in gyrA and parC. Antimicrob. Agents Chemother. 2011;55:1266–1269. doi: 10.1128/AAC.00927-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tran J.H., Jacoby G.A. Mechanism of plasmid-mediated quinolone resistance. Proc. Natl. Acad. Sci. USA. 2002;99:5638–5642. doi: 10.1073/pnas.082092899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paglietti B., Falchi G., Mason P., Chitsatso O., Nair S., Gwanzura L., Uzzau S., Cappuccinelli P., Wain J., Rubino S. Diversity among human non-typhoidal salmonellae isolates from Zimbabwe. Trans. R. Soc. Trop. Med. Hyg. 2013;107:487–492. doi: 10.1093/trstmh/trt046. [DOI] [PubMed] [Google Scholar]
- 38.Yue L., Jiang H.-X., Liao X.-P., Liu J.-H., Li S.-J., Chen X.-Y., Chen C.-X., Lü D.-H., Liu Y.-H. Prevalence of plasmid-mediated quinolone resistance qnr genes in poultry and swine clinical isolates of Escherichia coli. Veter. Microbiol. 2008;132:414–420. doi: 10.1016/j.vetmic.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 39.Singer A.C., Shaw H., Rhodes V., Hart A. Review of Antimicrobial Resistance in the Environment and Its Relevance to Environmental Regulators. Front. Microbiol. 2016;7:1728. doi: 10.3389/fmicb.2016.01728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferrari R., Galiana A., Cremades R., Rodriguez J.C., Magnani M., Tognim M.C.B., Oliveira T.C.R.M., Royo G. Plasmid-mediated quinolone resistance by genes qnrA1 and qnrB19 in Salmonella strains isolated in Brazil. J. Infect. Dev. Ctries. 2011;5:496–498. doi: 10.3855/jidc.1735. [DOI] [PubMed] [Google Scholar]
- 41.García-Fernández A., Fortini D., Veldman K., Mevius D., Carattoli A. Characterization of plasmids harbouring qnrS1, qnrB2 and qnrB19 genes in Salmonella. J. Antimicrob. Chemother. 2009;63:274–281. doi: 10.1093/jac/dkn470. [DOI] [PubMed] [Google Scholar]
- 42.Tyson G.H., Li C., Hsu C.-H., Bodeis-Jones S., McDermott P.F. Diverse Fluoroquinolone Resistance Plasmids From Retail Meat E. coli in the United States. Front. Microbiol. 2019;10:2826. doi: 10.3389/fmicb.2019.02826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wasyl D., Hoszowski A., Zając M. Prevalence and characterisation of quinolone resistance mechanisms in Salmonella spp. Veter. Microbiol. 2014;171:307–314. doi: 10.1016/j.vetmic.2014.01.040. [DOI] [PubMed] [Google Scholar]
- 44.González F., Araque M. Association of Transferable Quinolone Resistance Determinant qnrB19 with Extended-Spectrumβ-Lactamases in Salmonella Give and Salmonella Heidelberg in Venezuela. Int. J. Microbiol. 2013;2013:628185. doi: 10.1155/2013/628185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karczmarczyk M., Martins M., McCusker M., Mattar S., Amaral L., Leonard N., Aarestrup F.M., Fanning S. Characterization of antimicrobial resistance in Salmonella enterica food and animal isolates from Colombia: Identification of a qnrB19-mediated quinolone resistance marker in two novel serovars. FEMS Microbiol. Lett. 2010;313:10–19. doi: 10.1111/j.1574-6968.2010.02119.x. [DOI] [PubMed] [Google Scholar]
- 46.Jibril A.H., Okeke I.N., Dalsgaard A., Menéndez V.G., Olsen J.E. Genomic Analysis of Antimicrobial Resistance and Resistance Plasmids in Salmonella Serovars from Poultry in Nigeria. Antibiotics. 2021;10:99. doi: 10.3390/antibiotics10020099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Veldman K., Cavaco L.M., Mevius D., Battisti A., Franco A., Botteldoorn N., Bruneau M., Perrin-Guyomard A., Cerny T., Escobar C.D.F., et al. International collaborative study on the occurrence of plasmid-mediated quinolone resistance in Salmonella enterica and Escherichia coli isolated from animals, humans, food and the environment in 13 European countries. J. Antimicrob. Chemother. 2011;66:1278–1286. doi: 10.1093/jac/dkr084. [DOI] [PubMed] [Google Scholar]
- 48.Fitch F.M., Carmo-Rodrigues M.S., Oliveira V.G.S., Gaspari M.V., dos Santos A., de Freitas J.B., Pignatari A.C. β-Lactam Resistance Genes: Characterization, Epidemiology, and First Detection of blaCTX-M-1 and blaCTX-M-14 in Salmonella spp. Isolated from Poultry in Brazil—Brazil Ministry of Agriculture’s Pathogen Reduction Program. Microb. Drug Resist. 2016;22:164–171. doi: 10.1089/mdr.2015.0143. [DOI] [PubMed] [Google Scholar]
- 49.Fernandes S.A., Camargo C.H., Francisco G.R., Bueno M.F.C., Garcia D.O., Doi Y., Casas M.R.T. Prevalence of Extended-Spectrum β-Lactamases CTX-M-8 and CTX-M-2-Producing Salmonella Serotypes from Clinical and Nonhuman Isolates in Brazil. Microb. Drug Resist. 2017;23:580–589. doi: 10.1089/mdr.2016.0085. [DOI] [PubMed] [Google Scholar]
- 50.Castellanos L.R., Van Der Graaf-Van Bloois L., Donado-Godoy P., León M., Clavijo V., Arévalo A., Bernal J.F., Mevius D.J., Wagenaar J.A., Zomer A., et al. Genomic Characterization of Extended-Spectrum Cephalosporin-Resistant Salmonella enterica in the Colombian Poultry Chain. Front. Microbiol. 2018;9:2431. doi: 10.3389/fmicb.2018.02431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perin A.P., Martins B.T.F., Barreiros M.A.B., Yamatogi R.S., Nero L.A., Bersot L.d.S. Occurrence, quantification, pulse types, and antimicrobial susceptibility of Salmonella sp. isolated from chicken meat in the state of Paraná, Brazil. Braz. J. Microbiol. 2019;51:335–345. doi: 10.1007/s42770-019-00188-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bartoloni A., Pallecchi L., Riccobono E., Mantella A., Magnelli D., Di Maggio T., Villagran A., Lara Y., Saavedra C., Strohmeyer M., et al. Relentless increase of resistance to fluoroquinolones and expanded-spectrum cephalosporins in Escherichia coli: 20 years of surveillance in resource-limited settings from Latin America. Clin. Microbiol. Infect. 2013;19:356–361. doi: 10.1111/j.1469-0691.2012.03807.x. [DOI] [PubMed] [Google Scholar]
- 53.Leão C., Clemente L., Moura L., Seyfarth A.M., Hansen I.M., Hendriksen R.S., Amaro A. Emergence and Clonal Spread of CTX-M-65-Producing Escherichia coli From Retail Meat in Portugal. Front. Microbiol. 2021;12:653595. doi: 10.3389/fmicb.2021.653595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Furlan J.P.R., Lopes R., Ramos M.S., dos Santos L.D.R., Rosa R.d.S., Savazzi E.A., Stehling E.G. Colistin-resistant mcr-1-positive Escherichia coli ST1775-H137 co-harboring blaCTX-M-2 and blaCMY-2 recovered from an urban stream. Infect. Genet. Evol. 2021;96:105156. doi: 10.1016/j.meegid.2021.105156. [DOI] [PubMed] [Google Scholar]
- 55.Palmeira J.D., Ferreira H., Madec J.-Y., Haenni M. Draft genome of a ST443 mcr-1—And bla CTX-M-2 -carrying Escherichia coli from cattle in Brazil. J. Glob. Antimicrob. Resist. 2018;13:269–270. doi: 10.1016/j.jgar.2018.05.010. [DOI] [PubMed] [Google Scholar]
- 56.Hall J.P.J., Botelho J., Cazares A., Baltrus D.A. What makes a megaplasmid? Philos. Trans. R. Soc. B Biol. Sci. 2021;377:20200472. doi: 10.1098/rstb.2020.0472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Petermann S.R., Sherwood J.S., Logue C.M. The Yersinia high pathogenicity island is present in Salmonella enterica Subspecies I isolated from turkeys. Microb. Pathog. 2008;45:110–114. doi: 10.1016/j.micpath.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 58.dos Santos A.M., Ferrari R.G., Panzenhagen P., Rodrigues G.L., Conte-Junior C.A. Virulence genes identification and characterization revealed the presence of the Yersinia High Pathogenicity Island (HPI) in Salmonella from Brazil. Gene. 2021;787:145646. doi: 10.1016/j.gene.2021.145646. [DOI] [PubMed] [Google Scholar]
- 59.Oelschlaeger T.A., Zhang D., Schubert S., Carniel E., Rabsch W., Karch H., Hacker J. The High-Pathogenicity Island Is Absent in Human Pathogens of Salmonella enterica Subspecies I but Present in Isolates of Subspecies III and VI. J. Bacteriol. 2003;185:1107–1111. doi: 10.1128/JB.185.3.1107-1111.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on the institutional websites cited in the Section 2 and supplementary material.