Abstract
Sequential gene expression of two type 1 cytokines (interleukin 2 [IL-2] and gamma interferon), one type 2 cytokine (IL-10), two monokines (IL-6 and tumor necrosis factor alpha), and one cytokine receptor (IL-2 receptor [IL-2R]) in normal human peripheral blood mononuclear cells (PBMC) following in vitro stimulation was investigated by reverse transcription-PCR methods. Two stimuli were utilized: phytohemagglutinin (PHA), which acts on the CD2 molecule and T-cell receptors, and anti-CD3 monoclonal antibody, which acts on the CD3 molecule and on T-cell receptors. Increased expression of all studied genes occurred between 1 and 4 hours after stimulation, except for that of the gene encoding IL-10, which was delayed. Expression of all but one of the genes was transient, with a maximal mRNA accumulation at about 8 h on average. IL-2R mRNA expression was an exception, showing a prolonged increase (72 h). The general profiles of expression of the five cytokine genes were similar but not identical, suggesting some shared regulatory mechanisms. When responses to four additional stimuli (pokeweed mitogen, Candida albicans, and IL-2 at high and low doses) were compared, similar profiles of cytokine gene expression were found. Thus, the various stimuli caused induction of all cytokines with quantitative, not qualitative, differences. Altogether, the present data are useful for defining the kinetics of gene expression for key cytokines in response to standard immune-cell stimuli.
T-cell activation can be initiated by diverse agents such as antigens, plant mitogens, cytokines, and monoclonal antibodies (29, 30, 42). These stimuli cause complex series of ordered interactions and events, including activation of transmembrane signaling pathways, cytokine gene expression, transcription, and translation. Upon stimulation, properties like stability and rate of synthesis of existing RNA and proteins are altered, and synthesis of new RNA and proteins is initiated. The outcomes of the activation process are T-cell proliferation and differentiation and cytokine production.
Cytokines are important mediators in the regulation of the immune response. The kinetics of gene expression and the production of several cytokines following stimulation have been studied (3, 11, 14, 16, 26, 41, 42). Most data available concern the expression of and the relationship between interleukin 2 (IL-2), IL-2 receptor (IL-2R), and gamma interferon (IFN-γ) (13, 14, 18–20, 26). The production of cytokines and/or cytokine receptors in stimulated T cells might be regulated either by common mechanisms or by independent pathways. Conflicting data exist in the literature in support of both options. Such disparate data might be due to the fact that the different stimuli used resulted in different patterns of cytokine and/or cytokine receptor mRNA production. Furthermore, the temporal relationship of the expression of different cytokine genes after stimulation with a variety of stimuli has not been well established.
In previous reports, cytokine mRNA expression in stimulated peripheral blood mononuclear cells (PBMC) has been studied by using Northern blots or nuclear transcription assays that are now recognized as moderately sensitive. Since most cytokine genes are expressed transiently and at low levels, in our present study we used reverse transcription (RT)-PCR, which provides the most sensitive method for quantitation of mRNA. In our study, cytokine gene expression for two Th1 cytokines (IL-2 and IFN-γ), one Th2 cytokine (IL-10), two monokines (IL-6 and tumor necrosis factor alpha [TNF-α]), and one cytokine receptor (IL-2R) were compared. Since different stimuli act on different cell types and via different receptors, two stimuli were studied in detail—phytohemagglutinin (PHA), which acts on the CD2 molecule and T-cell receptors, and anti-CD3 monoclonal antibody, which acts on the CD3 molecule and on T-cell receptors. Additional stimuli, including another mitogen, a microbial antigen, and the cytokine IL-2, were also evaluated. The kinetics of response were determined by sequential testing of cytokine gene expression.
MATERIALS AND METHODS
Preparation of PBMC.
Fresh blood was obtained from healthy adult volunteers under a protocol approved by the Human Subject Protection Committee of UCLA School of Medicine. After Lymphoprep (Nyegaard and Co., Oslo, Norway) density gradient centrifugation, interface mononuclear cells were collected and washed three times with Dulbecco’s phosphate-buffered saline (Gibco, Grand Island, N.Y.). The PBMC were suspended in RPMI 1640 (Gibco) with 10% human AB serum at 2 × 106 cells/ml in endotoxin-free tubes.
Cell cultures and proliferation assays.
Proliferation assays were performed in 12 by 75-mm sterile tubes (Fisher) in triplicate. In general, PBMC at 106 cells/tube in a total volume of 500 μl of RPMI 1640 supplemented with 10% human AB serum, 100 U of penicillin per ml, 100 μg of streptomycin per ml, and 0.3 mg of glutamine per ml were incubated with stimuli in 5% CO2 at 37°C. Stimuli were added to cultures after 1 h of cell resting. Cultures with 5 μg of PHA (Sigma) per ml, 200 ng of anti-CD3 antibody (NEN Research Products, Boston, Mass.) per ml, or pokeweed mitogen (PWM; Gibco) at a 1:500 dilution were incubated for 3 days. Cultures with Candida albicans (Greer Laboratories, Lenoir, N.C.) at 8 μg/ml or recombinant IL-2 (rIL-2; DuPont) at 10 or 1,000 U/ml were incubated for 6 days. During the last 6 h of the appropriate incubation time, cultures were pulsed with 1 μCi of [3H]thymidine (ICN, Irvine, Calif.) per tube. Cells were harvested on glass fiber filters with an automatic harvester (Cambridge Technology, Watertown, Mass.), and the incorporated radioactivity was measured in a liquid scintillation counter (Beckman) after the addition of 3 ml of scintillation fluid. Proliferation data were expressed as a stimulation index: (counts per minute of stimulated cells)/(counts per minute of nonstimulated cells).
For time course studies of cytokine and/or cytokine receptor gene expression, aliquots of PBMC were cultured in the same way as for the proliferation assay but without pulsing with tritiated thymidine. Cultures were centrifuged after various stimulation times, and cells were collected. At the end of the culturing period, no significant changes in cell viability were observed when the cultures were tested by the trypan blue exclusion method.
RNA isolation and cytokine and/or cytokine receptor mRNA quantitation in cultured cells.
The procedures for RNA isolation and RT-PCR quantitation, including data on linearity, reproducibility, sensitivity, etc., and an optimal assay performance were described in detail previously (15). Briefly, for RNA isolation and reverse transcription, cells were lysed by guanidinium isothiocyanate (4 M) in sodium citrate (25 mM) buffer, pH 7.0, with 0.5% sarcosyl and 0.1 M β-2-mercaptoethanol. For RNA isolation, 0.1 volume of 2 M sodium acetate was added together with 1 volume of water-saturated phenol and 0.2 volume of 49:1 chloroform-isoamyl alcohol. After centrifugation, RNA was extracted in the aqueous phase and the phenol-chloroform extraction was repeated once more. The RNA was then precipitated with isopropanol at −20°C for 1 h. After centrifugation, the pellet was washed with 70% ethanol twice and dissolved in diethyl pyrocarbonate-treated water containing 20 μmol of RNase inhibitor (9). Ten nanograms of total RNA was used for each RT-PCR.
cDNA was synthesized from oligo(dT)-primed RNA by incubation at 42°C for 15 min, and then at 99°C for 5 min and a soak at 5°C for 5 min with Moloney murine leukemia virus reverse transcriptase (GIBCO, Bethesda Research Laboratories) and 1 mM deoxynucleoside triphosphate. For the semiquantitative PCR, the reaction mixture contained 10 mM Tris-HCl, 2 mM MgCl2, 0.2 mM deoxynucleoside triphosphate, 0.2 μM 5′ and 3′ oligonucleotide primers, and 2.5 μmol of AmpliTaq DNA polymerase (Perkin-Elmer Cetus). Trace amounts (0.01 μM) of [α-32P]dATP were added. Aliquots were then amplified by 35 cycles (cytokines and cytokine receptor) or 25 cycles (β-actin) of denaturation at 95°C for 1 min and annealing and extension at 60°C for 1 min. The sequences of the primers used with cytokine-encoding genes are shown in Table 1.
TABLE 1.
Cytokine primers used for PCR
| Gene product (direction of strand) | Fragment size (bases) | Primer sequence |
|---|---|---|
| IL-2 (5′) | 222 | 5′-GAATGGAATTAATAATTACAAGAATCCC-3′ |
| IL-2 (3′) | 222 | 5′-TGTTTCAGACCCTTTAGTTCAG-3′ |
| TNF-α (5′) | 325 | 5′-CAGAGGGAAGAGTTLCCCAG-3′ |
| TNF-α (3′) | 325 | 5′-CCTTGGTCTGGATAGGAGACG-3′ |
| IL-10 (5′) | 352 | 5′-ATGCCCCAAGCTGAGAACCAAGACCCA-3′ |
| IL-10 (3′) | 352 | 5′-TCTCAAGGGGCTGGGTCAGCTATCCCA-3 |
| IFN-γ (5′) | 520 | 5′-ATGAAATATACAAGTTATAGC-3′ |
| IFN-γ (3′) | 520 | 5′-TTACTGGGATGCTCTTCGACCTCGAAACAGCAT-3′ |
| IL-6 (3′) | 190 | 5′-ATGTAGCCGCCCCACACAGA-3′ |
| IL-6 (3′) | 190 | 5′-CATCCATCTTTTTCAGCCAT-3′ |
| IL-2R (5′) | 398 | 5′-GAATTTATCATTTCGTGGTGGGGCA-3′ |
| IL-2R (3′) | 398 | 5′-TCTTCTACTCTTCCTCTGTCTCCG-3′ |
| β-Actin (5′) | 1,126 | 5′-ATGGATGATGATATCGCCGCC-3′ |
| β-Actin (3′) | 1,126 | 5′-CTAGAAGCATTTGCGGTGGACGATGGAGGGGCC-3′ |
PCR products were analyzed by electrophoresis on a Tris-borate-EDTA acrylamide gel and autoradiographed with a Hyperfilm-HP (Amersham). The radioactive product bands were cut from the dried gel and quantified by beta scintillation counting, and results were recorded as counts per minute. All of the data regarding cytokine and cytokine receptor PCR products in a sample were normalized according to the amount of β-actin detected in the same sample. The size (measured as adenosine content) of the amplicons and the levels of maximum mRNA production for different cytokines were not correlated. Thus, the measured radioactivity levels (in counts per minute) generally represented the cytokine mRNA concentrations without further corrections. For more reliable comparison, in the time course experiments the quantitation of cytokine and cytokine receptor mRNA induced by each of the two stimuli used was performed in one set of experiments.
Statistical analysis.
Linear regression analysis was used to evaluate the correlation between the proliferative stimulation index and (i) the time when the first measurable change in the amount of cytokine or cytokine receptor mRNA was found, (ii) the time when the maximum amount of mRNA was reached and (iii) the maximum amount of induced mRNA. The Kendall rank correlation method was used to assess the correlation between the proliferation stimulation index and the cytokine or cytokine receptor level.
RESULTS
Cytokine and cytokine receptor mRNA production in unstimulated PBMC.
Low levels of all cytokine genes were demonstrable by the RT-PCR method in PBMC before culturing (Fig. 1). This production of mRNA could reflect in vivo gene production or some activation during cell separation. To examine these possibilities, PBMC from two subjects were cultured for 4, 8, 24, and 72 h without any stimuli added, and cytokine mRNA production was measured. As shown in Table 2, the level of mRNA encoding all cytokines in PBMC after resting (i.e., in a culture without added stimuli) was significantly lower than the mRNA level immediately after cell separation (with the exception of IL-2R mRNA from one subject). Variations between amounts of different cytokine mRNA are evident, but none of the mRNA amounts declined to zero (Table 2). Furthermore, within 72 h, none of the cultures of unstimulated PBMC demonstrated the maximum of cytokine gene expression at 8 h that we consistently observed for PBMC stimulated with five different stimuli, including IL-2 at two doses (one low and one high). Thus, low but detectable mRNA levels, different for each cytokine and each individual, were consistently found for all six cytokine mRNAs examined.
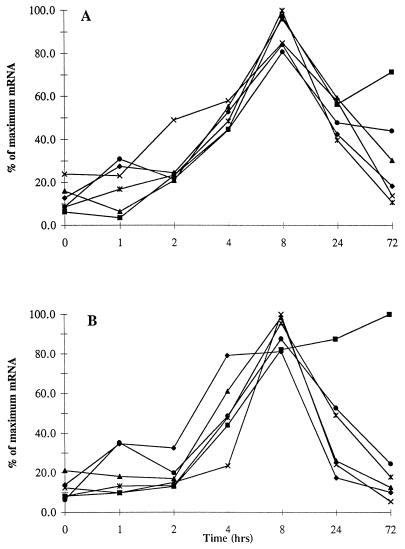
FIG. 1.
Time course of cytokine mRNA production and accumulation after stimulation. PBMC were stimulated with PHA (A) and with anti-CD3 monoclonal antibody (B). Total cellular RNA was isolated at the indicated times after activation. mRNA encoding IL-2 (⧫), IL-2R (▪), IL-6 (▴), IL-10 (×), TNF-α (*), and IFN-γ (•) was measured by the semiquantitative RT-PCR method. Data for samples from four healthy donors are presented as mean proportions of the maximal amounts of each specific mRNA.
TABLE 2.
Cytokine mRNA levels in unstimulated PBMC
| mRNA | mRNA level in PBMC from subjecta
|
|||
|---|---|---|---|---|
| DC-A
|
DC-B
|
|||
| Immediately after cell separation | After restingb | Immediately after cell separation | After restingb | |
| IL-2 | 14.6 | 5.7 | 0.4 | 0.1 |
| IL-2R | 13.5 | 18.9 | 3.3 | 2.7 |
| IL-6 | 26.0 | 21.8 | 30.0 | 13.2 |
| IL-10 | 4.9 | 3.0 | 2.0 | 0.8 |
| TNF-α | 25.7 | 13.2 | 3.2 | 2.9 |
| IFN-γ | 23.3 | 7.7 | 5.0 | 0.3 |
Percentage of the maximum mRNA expression upon stimulation with PHA shown in Fig. 1.
Minimum mRNA levels in unstimulated PBMC found within 4 to 72 h of culture.
Time course of cytokine and cytokine receptor mRNA expression following stimulation.
In order to define and compare the kinetics of IL-2, IL-2R, IL-6, IL-10, TNF-α, and IFN-γ gene expression following in vitro stimulation, time course experiments were carried out with PBMC from four healthy donors. Semiquantitative RT-PCR analyses were used to determine the level of each cytokine mRNA at each time point for each stimulus. The experimental results (in counts per minute) from each sample were adjusted for radioisotope decay and by normalizing them to the sample’s β-actin amount. The time course of changes in mRNA levels for four healthy donors is presented as a mean percent of the maximal mRNA level for each cytokine after stimulation with PHA (Fig. 1A). Stimulation with anti-CD3 monoclonal antibody is shown separately in Fig. 1B. Four features of the response were evaluated: the time of induction, the time and the level of maximal mRNA accumulation, and the rate of subsidence. The patterns of mRNA production induced by PHA and by anti-CD3 were compared as well.
The initial increase is the first measurable response to stimulation and is defined here by the earliest point in time, within the time frame utilized in this study, that a 25% or greater increase in levels of specific gene production compared to levels at time zero was found. Genes encoding IFN-γ, TNF-α, and IL-2 were the earliest induced (within 1 h of incubation), regardless of the stimuli. The time of induction of IL-6, IL-2R, and IL-10 mRNA ranged from 1 to 4 h.
Maximum expression was observed for all five cytokine genes at 8 h (Fig. 1). However, the IL-2R gene was maximally expressed at 72 h upon stimulation with anti-CD3.
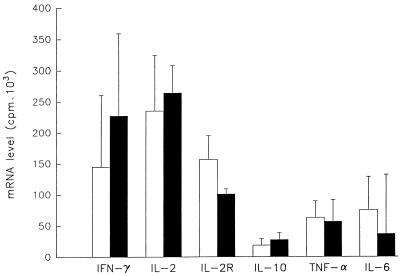
The mean maximal level of mRNA production observed upon PHA and anti-CD3 stimulation of the six genes studies is presented in Fig. 2. IL-2 and IFN-γ mRNAs show much higher absolute maximum levels than IL-10, TNF-α, and IL-6 mRNAs after PHA or anti-CD3 stimulation. When the four individuals are compared, there are some differences in the times of maximum production of specific genes (Table 3), but the general pattern is similar in most subjects.
FIG. 2.
Patterns of maximal cytokine mRNA production after stimulation. The mean (standard error) values of the maximum levels of each category of mRNA are presented for samples from four donors after stimulation with PHA (open bars) and anti-CD3 monoclonal antibody (filled bars).
TABLE 3.
Maximum level of induced cytokine mRNA and time of maximal response
| Stimulus and subject | mRNA ratioa
|
|||||
|---|---|---|---|---|---|---|
| IL-2 | IL-2R | IL-6 | IL-10 | TNF-α | IFN-γ | |
| PHA | ||||||
| DC-1 | 29 (8) | 51 (72) | 15 (24) | 2 (8) | 34 (8) | 78 (8) |
| DC-2 | 3 (8) | 18 (8) | 115 (8) | 7 (8) | 52 (8) | 22 (24) |
| DC-A | 7 (4) | 7 (4) | 4 (4) | 21 (4) | 4 (8) | 4 (4) |
| DC-B | 265 (8) | 30 (8) | 3 (8) | 49 (8) | 31 (8) | 20 (8) |
| Anti-CD3 | ||||||
| DC-1 | 12 (8) | 54 (72) | 5 (8) | 3 (8) | 19 (8) | 91 (8) |
| DC-2 | 3 (4) | 12 (72) | 69 (8) | 15 (8) | 50 (8) | 20 (24) |
| DC-A | 10 (4) | 6 (72) | 3 (8) | 43 (8) | 6 (4) | 6 (4) |
| DC-B | 277 (4) | 13 (72) | 3 (4) | 58 (8) | 11 (8) | 41 (8) |
In samples from four healthy donors, the ratio of the maximum amount of mRNA (in counts per minute) upon stimulation with PHA or anti-CD3 to the amount of mRNA (in counts per minute) in unstimulated cells was determined. The time (in hours) after stimulation when the maximum level of mRNA was reached is indicated in parentheses.
The profile of the accumulation of mRNA encoding IL-2R is different from that of the other genes studied. Within the time period (72 h) and time intervals examined, IL-2R mRNA, regardless of the stimulus, demonstrated continued expression rather than transient expression (Fig. 1). After anti-CD3 stimulation, three of four subjects showed continuous increases in IL-2R mRNA levels and maximum IL-2R mRNA levels at 72 h. Cultures from all four subjects after PHA stimulation and one of four subjects after anti-CD3 clearly demonstrated a decrease in IL-2R mRNA levels at 24 h, followed by an increase to the maximum level at 72 h (data not shown). As it has been reported that IL-2 can regulate the expression of its own receptor, such prolonged and “double peak” production of IL-2R mRNA could be a result of induction by IL-2 during the preceding 24-h period of PHA or anti-CD3 stimulation.
PBMC from two donors were used for pilot characterization of cytokine genes’ responses to additional stimuli. A microbial antigen (C. albicans), a T-cell and B-cell stimulus (PWM) and IL-2 at high (1,000-U/ml) and low (10-U/ml) doses were used in addition to PHA and anti-CD3. A measurable increase in the expression of all six genes examined was induced by each of the six stimuli except C. albicans which was not able to induce detectable IL-10 mRNA production (data not shown). The peak responses were observed at 8 h, and the kinetic patterns for the four additional stimuli were generally parallel to the patterns of responses to PHA and anti-CD3.
Relationship between cytokine and/or cytokine receptor mRNA expression and cell proliferation.
Lymphocyte proliferation is an important aspect of the cellular immune response. In parallel experiments, cell proliferation and induction of maximal levels of mRNA encoding IL-2, IL-2R, IFN-γ, IL-10, TNF-α and IL-6 with each of the six stimuli were measured. Proliferative stimulation indices were 139.1 (PHA), 13.3 (anti-CD3), 33.4 (PWM), 1.3 (C. albicans), 23.0 (rIL-2, 1,000 U/ml), and 3.3 (rIL-2, 10 U/ml). The maximum levels of IL-2 and TNF-α mRNA correlated significantly with levels of cell proliferation (Table 4). However, by linear regression analysis, no significant correlation was found between the proliferative stimulation index and the time of cytokine induction. PHA had a 10-fold-higher stimulation index than anti-CD3. This effect of PHA was associated with higher expression of IL-10 mRNA (Table 3) in all samples, which may have contributed to the reduced proliferative response to anti-CD3.
TABLE 4.
Correlation between lymphocyte proliferation and cytokine gene expression
| mRNA | Proliferative stimulation index | P valuea |
|---|---|---|
| IL-2 | 0.73 | 0.028 |
| IL-2R | 0.26 | 0.298 |
| IL-6 | 0.60 | 0.068 |
| IL-10 | −0.07 | 0.500 |
| TNF-α | 0.73 | 0.028 |
| IFN-γ | −0.07 | 0.500 |
Data represent Kendall’s coefficient of rank correlation between proliferative stimulation index and maximal value of cytokine gene expression upon stimulation.
DISCUSSION
The coordinated production of cytokines following lymphocyte activation controls proliferation, differentiation, and function of cells and is crucial for regulation of the immune response (16). Detailed knowledge of these processes in normal lymphocytes provides a basis for discerning abnormalities in T-cell activation and functions of lymphocytes in disease. Indeed, the normal baseline resting levels of cytokine gene production are only now being defined. Several investigators have reported that unstimulated cells do not express cytokine mRNA (8, 23, 26). In those studies, Northern blot analyses were used. In contrast, in our present study with the highly sensitive RT-PCR method, we observed that unstimulated and resting PBMC from normal individuals express low levels of all six cytokine mRNAs. Our findings are consistent with observations for IFN-γ mRNA production in unstimulated spleen cells (24) and IL-2 and IFN-γ mRNA production in rat T-cell clones that had rested (45). In humans, spontaneous production of IL-6 and IL-2R mRNA in T cells (19, 22) and of IL-2R mRNA in monocytes (36) has been reported as well. Our results (Table 2) are compatible with the suggestion (19, 22, 24, 36, 45) that low mRNA production is caused by low constitutive expression of these genes in unstimulated PBMC that have rested. However, there may be some activation during cell separation.
We have examined the kinetics and sequence of the production of five cytokine genes upon PBMC stimulation in vitro. Our data are generally compatible with results of previous reports regarding IL-2, IL-2R, and IFN-γ (13, 14, 42). We expanded on prior studies by including TNF-α, IL-6, and IL-10 genes. By using the highly sensitive RT-PCR method, we found that gene induction occurs earlier than in previous studies. All genes studied showed increases within 1 to 4 h after stimulation. Thus, they all belong to the early gene group (41), although IFN-γ, IL-2, and TNF-α genes were the earliest to be expressed (within 1 h), and IL-2R, IL-6, and IL-10 genes were expressed later.
Maximal production of mRNA encoding IL-2, IL-6, IL-10, TNF-α, and IFN-γ occurred at nearly the same time (an average of 8 h) after PHA or anti-CD3 stimulation for all cytokine groups. Maximal PHA-stimulated IL-2R mRNA production occurred at 8 h, but production continued at an elevated level for 72 h. Stimulation with anti-CD3 induced a continuous increase in mRNA levels during the 72 h period examined. These findings differ from the reported early IL-2R induction that preceded IFN-γ and IL-2 mRNA production (26). This is most likely due to differences in stimuli used—PHA plus phorbol myristate acetate (26) versus PHA and anti-CD3 used alone here. Several reports have implicated IL-2 in expression of IL-2R and IFN-γ genes (2, 12, 25, 38, 43), as well as in the induction of IFN-γ production (7, 21, 35, 40). Our results for the time of induction and the time and level of maximal accumulation of mRNA encoding IL-2, IL-2R, and IFN-γ (Table 3) do not indicate these relationships. Our data are instead compatible with the data of Krönke et al. (26), who have demonstrated that the presence of IL-2 is not required for transcriptional activation of IL-2R and IFN-γ genes in fully stimulated cells. However, our data do not exclude an IL-2 mediated augmentation of IL-2R and IFN-γ gene expression in suboptimally stimulated cells (26) or a synergy between mitogen and low doses of IL-2 affecting IFN-γ gene expression during the early phases of a cellular immune response (2).
Our data indicate that the maximum level of production of different genes may vary when different stimuli are used (Fig. 2). Differences in these parameters were also noted when PWM was used as the stimulus (39). With PHA or anti-CD3 stimulation, amounts of mRNA encoding IL-2 and IFN-γ were much higher than those of mRNA encoding IL-6, IL-10, or TNF-α. This indicates that these two stimuli act primarily on T cells, which make up the bulk of normal PBMC, and have much less of an effect on monocytes, the major producers of IL-6, TNF-α, and IL-10.
Generally, the profile of cytokine gene expression depends at least partly on cell surface receptor binding of a ligand and activation of intracellular signaling pathways (16). Different stimuli can activate T cells in several ways: by antigen binding to the T-cell receptor–CD3 complex in association with major histocompatibility complexes (42), by modulation of other surface molecules such as CD2 and CD4 (4), and by bypassing the surface receptor signaling and directly activating protein kinase C and intracellular pathways (33). Signals from separate cell surface receptors are integrated at the level of the responsive gene (11). Previous data for T-cell lines, T-cell clones, and PBMC indicate that cells have the capacity to produce many cytokines, and our data supports the idea that the way cells are activated can alter the quantitative aspects of cytokine gene expression and protein secretion (18, 19, 45).
In our present study, some variations between individuals in the time course of cytokine mRNA production were observed. Diversity between individuals in immune-cell functional properties like proliferative, cytotoxic, and cytokine responses is well documented. Usually, such diversity within populations is presented as a reference range and is often quite broad. It should be taken into consideration when results are analyzed and interpreted. In this regard, individual variations observed in our study group do not justify the suggested classification of patterns of cytokine gene expression into type 1 (IL-2), which demonstrates rapid appearance and early maximal accumulation of mRNA, and type 2 (IFN-γ and TNF-α), which demonstrates prolonged gene production and a late peak time, a generalization based on data from a single donor (23).
Production of multiple cytokines in stimulated T-cell populations might be regulated either by a common mechanism or by independent pathways. Conflicting data in the literature support both options. Data indicating that various stimulations lead to the synthesis of both IL-2 and IFN-γ mRNA (13, 14, 20) support the option of common mechanisms. This option is additionally supported by sequence data which show that IL-2 and IFN-γ genes have sequence homology at their 5′ end (17, 31). Our present data are compatible with this interpretation and extend them by including TNF-α, IL-6, and IL-10 mRNA, as our time course curves for the five cytokines are essentially parallel (Fig. 1). Data in support of diverse modes of regulation by independent pathways have included quantitative differences among IFN-γ, IL-2, and IL-2R produced by PBMC stimulated by anti-CD3 or by PHA (18, 19). We also observed quantitative differences in expression of genes for these cytokines and those for IL-6, IL-10, and TNF-α using the RT-PCR method. Each of the five stimuli were able to induce expression of the six genes studied (with one exception—C. albicans did not induce a detectable IL-10 response). Our observations therefore suggest mechanisms whereby cytokines are regulated independently but their transcriptions are induced simultaneously.
Several specifics should be considered when data from different studies are compared. PBMC populations are mixed populations of cells. Various stimuli may cause preferential stimulation of T cells or monocytes in PBMC. Quantitative differences in gene expression in CD4+ and CD8+ T-cell subsets have also been observed (6, 27), as has the interaction between T-cell subsets which influence IL-2 and IFN-γ gene expression (1). Furthermore, three types of helper T-cell clones are recognized, according to the pattern of lymphokine production (3, 11, 32). Th1 cells but not Th2 cells produce IL-2 and IFN-γ. In contrast, Th2 cells but not Th1 cells produce IL-4. In addition, a Th0 cell subset that produces almost all cytokines has been described (32). In humans, the majority of CD4+ T-cell clones obtained from healthy donors produce IFN-γ, IL-2, and IL-4 upon stimulation with various mitogens (34). In addition, besides regulation at the transcriptional level, a posttranscriptional regulation that controls the stability of mRNA also exists and is considered important (28, 44). This may depend on the mode of stimulation, and the IL-2 mRNA degradation rate can vary, in contrast to IL-2R mRNA which remains stable (5). IL-2R mRNA stability may contribute to the prolonged presence of IL-2R mRNA seen in our studies. In spite of this complexity, our data should be useful as a basis for defining the change in cytokine gene expression in PBMC responding to commonly used stimuli in normal and disease states.
ACKNOWLEDGMENTS
We thank T. Nguyen for laboratory support, S. Stehn and J. Chung for assistance with data management and statistical help, and J. Moore and D. Mathieson for help with manuscript preparation.
This work was supported by NIH grants AI36086, AI35040, and TW00003.
REFERENCES
- 1.Arad G, Ketzinel M, Tal C, Nussinovich R, Deutsch E, Schlesinger M, Gerez L, Kaempfer R. Transient expression of human interleukin-2 and interferon-γ genes is regulated by interaction between distinct cell subsets. Cell Immunol. 1995;160:240–247. doi: 10.1016/0008-8749(95)80034-g. [DOI] [PubMed] [Google Scholar]
- 2.Arad G, Nussinovich R, Kaempfer R. Interleukin-2 induces an early step in the activation of interferon-γ gene expression. Immunol Lett. 1995;44:213–216. doi: 10.1016/0165-2478(94)00217-f. [DOI] [PubMed] [Google Scholar]
- 3.Arai K-I, Lee F, Miyajima A, Miyatake S, Arai N, Yokota T. Cytokines: coordinators of immune and inflammatory responses. Annu Rev Biochem. 1990;59:783–836. doi: 10.1146/annurev.bi.59.070190.004031. [DOI] [PubMed] [Google Scholar]
- 4.Bierer B E, Burakoff S J. T-lymphocyte activation: the biology and function of CD2 and CD4. Immunol Rev. 1989;111:267–294. doi: 10.1111/j.1600-065x.1989.tb00549.x. [DOI] [PubMed] [Google Scholar]
- 5.Bill O, Garlisi C G, Grove D S, Holt G E, Mastro A M. IL-2 mRNA levels and degradation rates change with mode of stimulation and phorbol ester treatment of lymphocytes. Cytokine. 1994;6:102–110. doi: 10.1016/1043-4666(94)90015-9. [DOI] [PubMed] [Google Scholar]
- 6.Breen E C, Salazar-Gonzalez J F, Shen L P, Kolberg J A, Urdea M, Martinez-Maza O, Fahey J L. Circulating CD8 T cells show increased interferon-gamma (IFN-γ) mRNA expression in HIV infection. Cell Immunol. 1997;178:91–98. doi: 10.1006/cimm.1997.1115. [DOI] [PubMed] [Google Scholar]
- 7.Chan S H, Perussia B, Gupta J W, Kobayashi M, Pospisil M, Young H A, Wolf S F, Young D, Clark S C, Trinchieri G. Induction of interferon γ production by natural killer cell stimulatory factor: characterization of the responder cells and synergy with other inducers. J Exp Med. 1991;173:869–879. doi: 10.1084/jem.173.4.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cherwinski H M, Schumacher J H, Brown K D, Mosmann T R. Two types of mouse helper T cell clone III. Further differences in lymphokine synthesis between Th1 and Th2 clones revealed by RNA hybridization, functionally monospecific bioassays, and monoclonal antibodies. J Exp Med. 1987;166:1229–1244. doi: 10.1084/jem.166.5.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 10.Coffman R L, Seymour B W P, Lehman D A, Hiraki D D, Christiansen J A, Shrader B, Cherwinski H M, Savelkoul H F J, Finkelman F D, Bond M W, Mosmann T R. The role of helper T cell products in mouse B cell differentiation and isotype regulation. Immunol Rev. 1988;102:5–28. doi: 10.1111/j.1600-065x.1988.tb00739.x. [DOI] [PubMed] [Google Scholar]
- 11.Crabtree G R. Contingent genetic regulatory events in T lymphocyte activation. Science. 1989;243:355–361. doi: 10.1126/science.2783497. [DOI] [PubMed] [Google Scholar]
- 12.Depper J M, Leonard W J, Drogula C, Krönke M, Waldmann T A, Greene W C. Interleukin 2 (IL-2) augments transcription of the IL-2 receptor gene. Proc Natl Acad Sci USA. 1985;82:4230–4234. doi: 10.1073/pnas.82.12.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Efrat S, Kaempfer R. Control of biologically active interleukin 2 messenger RNA formation in induced human lymphocytes. Proc Natl Acad Sci USA. 1984;81:2601–2605. doi: 10.1073/pnas.81.9.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Efrat S, Pilo S, Kaempfer R. Kinetics of induction and molecular size of mRNAs encoding human interleukin-2 and γ-interferon. Nature. 1982;297:236–239. doi: 10.1038/297236a0. [DOI] [PubMed] [Google Scholar]
- 15.Fan J, Bass H Z, Fahey J L. Elevated IFN-γ and decreased IL-2 gene expression are associated with HIV infection. J Immunol. 1993;151:5031–5040. [PubMed] [Google Scholar]
- 16.Fraser J D, Straus D, Weiss A. Signal transduction events leading to T-cell lymphokine gene expression. Immunol Today. 1993;14:357–362. doi: 10.1016/0167-5699(93)90236-E. [DOI] [PubMed] [Google Scholar]
- 17.Fujita T, Takaoka C, Matsui H, Taniguchi T. Structure of the human interleukin 2 gene. Proc Natl Acad Sci USA. 1983;80:7437–7441. doi: 10.1073/pnas.80.24.7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gauchat J-F, Walker C, De Weck A L, Stadler B M. Stimulation-dependent lymphokine mRNA levels in human mononuclear cells. Eur J Immunol. 1985;18:1441–1446. doi: 10.1002/eji.1830180921. [DOI] [PubMed] [Google Scholar]
- 19.Gauchat J-F, Gauchat D, De Weck A L, Stadler B M. Cytokine mRNA levels in antigen-stimulated peripheral blood mononuclear cells. Eur J Immunol. 1989;19:1079–1085. doi: 10.1002/eji.1830190618. [DOI] [PubMed] [Google Scholar]
- 20.Granelli-Piperno A, Andrus L, Steinman R M. Lymphokine and nonlymphokine mRNA levels in stimulated human T cells. J Exp Med. 1986;163:922–937. doi: 10.1084/jem.163.4.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Handa K, Suzuki R, Matsui H, Shimizu Y, Kumagai K. Natural killer (NK) cells as a responder to interleukin 2 (IL-2). II. IL-2-induced interferon-γ production. J Immunol. 1983;130:988–992. [PubMed] [Google Scholar]
- 22.Haskill S, Johnson C, Eierman D, Becker S, Warren K. Adherence induces selective mRNA expression of monocyte mediators and proto-oncogenes. J Immunol. 1988;140:1690–1694. [PubMed] [Google Scholar]
- 23.Hayashi S, Okamura S, Kawasaki C, Harada M, Niho Y. Sequential expression of lymphokine genes during phytohemagglutinin-stimulated mitogenesis of normal human peripheral mononuclear cells. Biomed Pharmacother. 1993;47:155–160. doi: 10.1016/0753-3322(93)90006-7. [DOI] [PubMed] [Google Scholar]
- 24.Iizawa Y, Brown J F, Czuprynski C J. Early expression of cytokine mRNA in mice infected with Listeria monocytogenes. Infect Immun. 1992;60:4068–4073. doi: 10.1128/iai.60.10.4068-4073.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kasahara T, Hooks J J, Dougherty S F, Oppenheim J J. Interleukin 2-mediated immune interferon (IFN-γ) production by human T cells and T cell subsets. J Immunol. 1983;130:1784–1789. [PubMed] [Google Scholar]
- 26.Krönke M, Leonard W J, Depper J M, Greene W C. Sequential expression of genes involved in human T lymphocyte growth and differentiation. J Exp Med. 1985;161:1593–1598. doi: 10.1084/jem.161.6.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leung J C K, Lai C K W, Chul Y L, Ho R T H, Chan C H S, Lai K N. Characterization of cytokine gene expression in CD4+ and CD8+ T cells after activation with phorbol myristate acetate and phytohaemagglutinin. Clin Exp Immunol. 1992;90:147–153. doi: 10.1111/j.1365-2249.1992.tb05847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindstein T, June C H, Ledbetter J A, Stella G, Thompson C B. Regulation of lymphokine messenger RNA stability by a surface-mediated T cell activation pathway. Science. 1989;224:339–343. doi: 10.1126/science.2540528. [DOI] [PubMed] [Google Scholar]
- 29.Meuer S C, Hussey R E, Cantrell D A, Hogdon J D, Schlossman S F, Smith K A, Reinherz E L. Triggering of the T3-Ti antigen receptor complex results in clonal T cell proliferation through an interleukin 2-dependent autocrine pathway. Proc Natl Acad Sci USA. 1984;81:1509–1513. doi: 10.1073/pnas.81.5.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meuer S C, Hussey R E, Fabbi M, Fox D A, Acuto O, Fitzgerald K A, Hogdon J C, Protentis J P, Schlossman S F, Reinherz E L. An alternative pathway of T cell activation: a functional role for the 50 kD T11 sheep erythrocyte receptor protein. Cell. 1984;36:897–906. doi: 10.1016/0092-8674(84)90039-4. [DOI] [PubMed] [Google Scholar]
- 31.Mita S, Maeda S, Obaru K, Nishino N, Shimada K, Hirano T, Onoue K, Ogawa T, Ogawa H. Isolation and characterization of a human interleukin 2 gene. Biochem Biophys Res Commun. 1983;117:114–121. doi: 10.1016/0006-291x(83)91548-6. [DOI] [PubMed] [Google Scholar]
- 32.Mosmann T R, Coffman R L. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol. 1989;7:145–173. doi: 10.1146/annurev.iy.07.040189.001045. [DOI] [PubMed] [Google Scholar]
- 33.Nishizuka Y. The role of protein kinase C in cell surface signal transduction and tumor promotion. Nature. 1984;308:693–698. doi: 10.1038/308693a0. [DOI] [PubMed] [Google Scholar]
- 34.Paliard X, De Waal Malefijt R, Yssel H, Blanchard D, Chretien I, Abrams J, De Vries J, Spits H. Simultaneous production of IL-2, IL-4, and IFN-γ by activated human CD4+ and CD8+ T cell clones. J Immunol. 1988;141:849–855. [PubMed] [Google Scholar]
- 35.Pestka S, Langer J A. Interferons and their actions. Annu Rev Biochem. 1987;56:727–777. doi: 10.1146/annurev.bi.56.070187.003455. [DOI] [PubMed] [Google Scholar]
- 36.Rambaldi A, Young D C, Herrmann F, Cannistra S A, Griffin J D. Interferon-γ induces expression of the interleukin 2 receptor gene in human monocytes. Eur J Immunol. 1987;17:153–156. doi: 10.1002/eji.1830170127. [DOI] [PubMed] [Google Scholar]
- 37.Reed J C, Alpers J D, Nowell P C, Hoover R G. Sequential expression of proto-oncogenes during lectin-stimulated mitogenesis of normal human lymphocytes. Proc Natl Acad Sci USA. 1986;83:3982–3986. doi: 10.1073/pnas.83.11.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reem G, Yeh N H. Interleukin 2 regulates expression of its receptor and synthesis of gamma interferon by human T lymphocytes. Science. 1984;225:429–430. doi: 10.1126/science.6429853. [DOI] [PubMed] [Google Scholar]
- 39.Toyoda M, Zhang X, Petrosian A, Galera O A, Wang S-J, Jordan S C. Modulation of immunoglobulin production and cytokine mRNA expression in peripheral blood mononuclear cells by intravenous immunoglobulin. J Clin Immunol. 1994;14:178–189. doi: 10.1007/BF01533367. [DOI] [PubMed] [Google Scholar]
- 40.Trinchieri G, Matsumoto-Kobayashi M, Clark S C, Seehra J, London L, Perussia B. Response of resting human peripheral blood natural killer cells to interleukin 2. J Exp Med. 1984;160:1147–1169. doi: 10.1084/jem.160.4.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ullman K S, Northrop J P, Verweij C L, Crabtree G R. Transmission of signals from the T lymphocyte antigen receptor to the genes responsible for cell proliferation and immune function: the missing link. Annu Rev Immunol. 1990;8:421–452. doi: 10.1146/annurev.iy.08.040190.002225. [DOI] [PubMed] [Google Scholar]
- 42.Weiss A, Imboden J, Hardy K, Manager B, Terhorst C, Stobo J. The role of the T3/antigen receptor complex in T-cell activation. Annu Rev Immunol. 1986;4:593–619. doi: 10.1146/annurev.iy.04.040186.003113. [DOI] [PubMed] [Google Scholar]
- 43.Welte K, Andreff M, Platzer E, Holloway K, Rubin B Y, Moore M A S, Mertelsmann R. Interleukin 2 regulates the expression of Tac antigen on peripheral blood T lymphocytes. J Exp Med. 1984;160:1390–1403. doi: 10.1084/jem.160.5.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wiskocil R, Weiss A, Imboden J, Kamin-Lewis R, Stobo J. Activation of a human T cell line: a two-stimulus requirement in the pretranslational events involved in the coordinate expression of interleukin 2 and γ-interferon genes. J Immunol. 1985;134:1599–1603. [PubMed] [Google Scholar]
- 45.Zhao Z, Calder V L, McLauchlan M, Lightman S L. Differential lymphokine expression by rat antigen-specific CD4+ T cell lines with antigen and mitogen. Cell Immunol. 1994;159:220–234. doi: 10.1006/cimm.1994.1309. [DOI] [PubMed] [Google Scholar]