Abstract
Breast cancer, as one of the most malignant tumors, poses a serious threat to the lives of females. Nucleotide exchange factor SIL1 is an important regulator of endoplasmic reticulum function that might have a specific role in tumor progression. In this study, we aimed to investigate the effect of SIL1 on the proliferation, apoptosis, and metastasis of human breast cancer. SIL1-specific small interfering RNA was transfected into two breast cancer cell lines, MCF7 and MDA-MB-231, to generate SIL1 knockdown cells. Clone formation and Cell Counting Kit-8 assays were performed to determine cell proliferation. Wound healing and transwell assays were used to detect the cell migration and invasion, respectively. Cell cycle and apoptosis were determined by flow cytometry. The messenger RNA and protein levels of target genes were analyzed using quantitative real-time PCR and western blot. According to the results of TCGA and GTEx database analysis, we determined that SIL1 was overexpressed in 1085 breast cancer samples compared with 291 normal samples. Knockdown of SIL1 inhibited the proliferation, migration, and invasion of MCF7 and MDA-MB-231 cells, accordingly. The cell cycle was blocked at the G1 phase following transfection of SIL1-specific small interfering RNA through the inhibition of Cyclin D1, CDK4, and CDK6. SIL1 knockdown induced apoptosis and also promoted the activity of Caspase9 and Bax. Furthermore, SIL1 was able to promote phosphorylation of ERK1/2. Based on these results, SIL1 might act as an oncogene and accelerate the progression of human breast cancer.
Keywords: Nucleotide exchange factor SIL1, breast cancer, apoptosis, migration, ERK1/2 phosphorylation
Introduction
Breast cancer is one of the most malignant tumors that commonly occurs in women and poses a threat to their lives. 1 Current treatment for breast cancer includes surgery, radiotherapy, chemotherapy, and endocrine regulation. Surgery is the most effective treatment therapy for early stage in situ breast carcinoma at present. However, a considerable number of patients are diagnosed in the advanced stage of breast cancer, making the treatment difficult. 2 Therefore, in recent years, targeted therapy has become a hot topic of research. 3 Elucidating the pathogenesis of breast cancer and finding new therapeutic targets are extremely important for improving the prognosis of patients with breast cancer.4,5
Nucleotide exchange factor SIL1 (SIL1) encodes an endoplasmic reticulum (ER) resident protein with a molecular weight of 54 kDa. 6 It is a key regulator for ER function and serves as a nucleotide exchange factor for several key proteins, including glucose regulated proteins 78 (GRP78) and 70 (GRP170).7,8 When normal ER function is disturbed, misfolded, or unfolded, proteins are deposited in the ER, increasing stress. Excessive ER stress can lead to disturbances in normal cell function, leading to apoptosis and damage to tissues or organs.9,10 Recent reports have shown that ER stress is related to apoptosis and the occurrence and progression of tumors.11,12 Therefore, we speculate that SIL1, as a key regulator of ER function, might have a specific role in tumor development.
In this study, we aimed to investigate the specific role of SIL1 in the proliferation, apoptosis, and metastasis of human breast cancer. SIL1 was selected for further study via genomic analysis of 1085 breast cancer tissues and 291 normal breast tissues in an online dataset. The results showed that downregulation of SIL1 could inhibit proliferation, migration, and invasion, but induce cell cycle block and apoptosis. Moreover, SIL1 was found to promote the phosphorylation of ERK1/2. Based on this information, we determined that SIL1 acts as an oncogene in regulating tumor progression, but the mechanism and clinical value of SIL1 in breast cancer requires further study.
Materials and methods
Cell lines
Breast cancer cell lines (MCF7, SK-BR-3, MDA-MB-468, MDA-MB-231 and MDA-MB-157) and control human breast epithelial cell line (MCF10A) were purchased from the American Type Culture Collection (ATCC, USA). Dulbecco’s Modified Eagle’s medium (DMEM) replenished with 0.1 mg/mL streptomycin, 100 U/mL penicillin, and 10% fetal bovine serum was used for cell culture. Once cell density reached 60%, SIL1-specific small interfering RNA (siRNA) was transfected into MCF7 and MDA-MB-231 cells (si-SIL1), respectively, with a nonsense siRNA as negative control (NC) using Lipofectamine2000 (Invitrogen, USA). SiRNAs were synthesized by Ruibo (Guangzhou, China). The sequences of siRNAs were as follows: SIL1-siRNA1, GGCUGGAUAUCAACACCAATTUUGGUGUUGAUAUCCAGCCTT; SIL1-siRNA2, CGGAGAAGAUGUUCGCCGATTUCGGCGAACAUCUUCUCCGTT; SIL1-siRNA3, UGGUACGGCUGAUCAACAATT UUGUUGAUCAGCCGUACCATT.
Quantitative real-time PCR (qPCR)
Trizol reagent was used for total RNA extraction. After transfection for 24 h, RNA was extracted from the cell lines, then reverse transcribed to complementary DNA (cDNA). The qPCR kit UltraSYBR Mixture (CwBio, China) was used to detect the level of SIL1 messenger RNA (mRNA). Primers were synthesized as follows (Genewiz, China): SIL1 sense, 5′ TGCTTCACCTTCTGCCTCAG 3′, and antisense, 5′ GAACACCTCCAGGACTTCGG 3′; GAPDH sense, 5′ TCAATGTCGGCGCCTATTTC 3′, and antisense, 5′ CACCCTGTTGCTGTAGCCAAA 3′. The relative quantification was identified by the method of 2-ΔΔCt after standardization to the GAPDH level. qPCR assay was performed in triplicate.
Western blot analysis
After transfection for 48 h, radioimmunoprecipitation assay (RIPA) buffer containing phosphatase and protease inhibitors was used for total protein extraction. Proteins from each group were separated and transferred to a polyvinylidene fluoride membrane. The membrane was then blocked, followed by incubation with primary antibodies (1:1000) overnight at 4 °C. Next, the secondary antibodies (1:5000) were given for 1 h at room temperature. After incubation with the enhanced chemiluminescence (ECL) reagent, the expression values of proteins were normalized against GAPDH and quantified with QUANTITY ONE software. Anti-human SIL1 (1: 500, ab5639), Cyclin D1 (1: 2000, ab16663), CDK4 (1: 2000, ab108357), CDK6 (1: 1000, ab124821), ERK1/2 (1: 1000, ab17942), p-ERK1/2 (1: 500, ab223500), Acitve-Caspase9 (1: 1000, ab32539), Bcl-2 (1: 1000, ab182858), Bax (1: 1000, ab32503), and GAPDH (1: 5000, ab181602) were all purchased from Abcam (USA). Each experiment included triplicate measurements. Secondary antibodies, Goat Anti-Mouse IgG H&L (HRP) (ab205719), and Goat Anti-Rabbit IgG H&L (HRP) (ab6721) were also purchased from Abcam (USA).
Cell Counting Kit-8 (CCK8)
CCK8 assay was used to detect the proliferation of breast cancer cells. After transfection with siRNA, 2 × 103 cells/well were transferred into a 96-well plate and cultured in a 5% CO2 incubator at 37 °C. Cell activity was detected every 24 h. CCK8 reagent (Solarbio Science & Technology, China) was added to the cells and incubated at 37 °C for 2h. The optical density (OD) value was analyzed at a wavelength of 450 nm. Each experiment was performed in triplicate.
Clone formation assay
Approximately 200 cells transfected with siRNA were seeded into a 35-mm cell culture dish, then incubated for 14 to 21 days at 37 °C until visible colonies had formed. Incubation was performed for 20 min with 4% paraformaldehyde, then cells were stained with crystal violet. After 30 min, the number of clones consisting of 10 cells or more were counted under the microscope. The number of colonies = number of clones / number of inoculated cells. Clone formation assay was performed in triplicate.
Wound healing assay
Cell migration capability was evaluated via wound healing assays. After transfection, cells were transferred to a 6-well plate. When cells were cultured to 90% confluence, a straight line on the cell monolayer was scratched by a 200-µL pipette tip. The width of the wound was measured at 0 h and 24 h time points, respectively. Cell migration velocity was represented by wound closure normalized to that of the NC group. Wound healing assay was performed three times.
Transwell assay
The invasion of cancer cells was determined with a Matrigel-coated (BD Biosciences) transwell assay. The upper chamber was coated with 100 μL diluted Matrigel. After 24 h of transfection, 2 × 104 cells with fetal bovine serum (FBS)-free DMEM were transferred to the upper chamber, and the complete medium was added to the lower chamber. After 24 h, the invasive cells were fixed with 4% paraformaldehyde, then stained with 0.1% crystal violet. The number of invasive cells were counted under a microscope. Each experiment was performed in triplicate.
Flow cytometry for cell cycle detection
Cell cycle was determined by flow cytometry. Cells (3 × 106/mL) were starved for 24 h after transfection, then fixed overnight with 70% ethanol. Next, cells were stained with 400 μL propidium iodide (PI; Roche, Switzerland) in a dark environment. The results were analyzed with Flowjo software. This assay was performed three times.
Flow cytometry for apoptosis detection
Cell apoptosis was determined by flow cytometry using the Annexin V-FITC Apoptosis Detection kit. About 2 × 106 cells were suspended with 1 mL Annexin V binding buffer. Then, 100 μL cell suspension was added to 5 μL Annexin V / FITC mix for 5 min, followed by the addition of 10 μL PI dye incubation for another 5 min. The results were analyzed using Flowjo software. This assay was performed three times.
Gelatin zymography
After transfection for 24 h, cells were incubated with FBS-free DMEM for an additional 24 h. The supernatants were collected and the total protein was quantified. About 20 µg of proteins with 1 mg/mL gelatin A (Sigma Chemical Co., USA) were loaded into 10% SDS/PAGE. After electrophoresis for 1.5 h, the gel was stained with 0.25% Coomassie Blue R-250 for 4 h. Then, Image Quant TL V2003 software was utilized to measure the intensities. This assay was performed three times.
Statistical analyses
The data in this study were analyzed with SPSS 20.0 software. The comparison between two groups was performed using Student’s t test. All data were presented as mean ± standard deviation, and P < 0.05 was considered statistically significant.
Results
Human SIL1 was upregulated in breast cancer
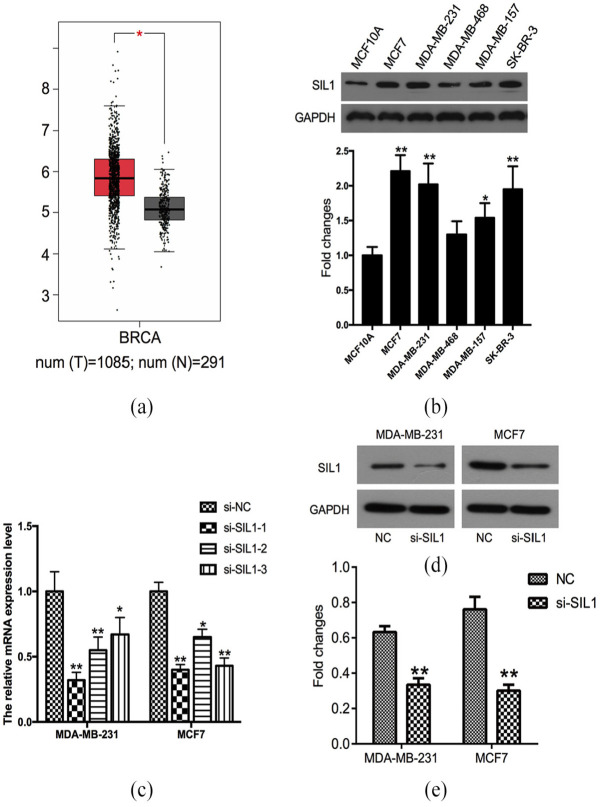
With the development of sequencing technology, cancer genomics projects have generated an overwhelming amount of cancer genomics data. This has made it possible to screen various biological indicators without collecting a large number of clinical samples. Gene Expression Profiling Interactive Analysis (GEPIA) is a website for analyzing the genomic data in The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) databases. 13 Figure 1(a) shows the boxplot generated from the GEPIA; the red and gray boxes represent breast invasive carcinoma (BRCA) and normal breast samples, respectively. According to the results of differential genes analysis between BRCA and normal breast samples, there was enhanced expression of SIL1 mRNA in BRCA (n = 1085) compared with normal samples (n = 291) (P < 0.05). In addition, SIL1 mRNA expression in some breast cancer cell lines was also significantly increased when compared with the control breast epithelial cell, MCF10A (P < 0.01; Figure 1(b)). These observations suggest that SIL1 was upregulated and might be involved in the progression of human breast cancer.
Figure 1.
Human SIL1 was upregulated in breast cancers. (a) SIL1 expression in tumor tissues (n = 1085) was significantly higher than that in normal tissues (n = 291) according to GEPIA analysis result of breast invasive carcinoma (BRCA). (b) The mRNA expression level of SIL1 in human breast epithelial cell MCF10A, and breast cancer cell lines (MCF7, MDA-MB-468, MDA-MB-231, SK-BR-3, and MDA-MB-157). (c) The mRNA expression level of SIL1 detected by qPCR in cells transfected with three SIL1-specific siRNAs (si-SIL1-1, si-SIL1-2, and si-SIL1-3) and negative control siRNA (NC). (d) The protein level of SIL1 was determined using western blot analysis after transfected with si-SIL1-1 or negative control siRNA (NC). (e) Statistical analysis of western blot result showed that the protein level of SIL1 was knocked down by si-SIL1-1.
*P < 0.05; **P < 0.01.
Knockdown of SIL1 impeded the proliferation of human breast cancer cells
MDA-MB-231 and MCF7, the two most SIL1 upregulated cell lines in Figure 1(b), were used to investigate the effect of SIL1 on tumor proliferation by RNA knockdown. Three SIL1-specific siRNAs were transfected into breast cancer cells to generate SIL1 knockdown cells (si-SIL1) with the cells transfected with a control siRNA serving as the negative control cells (NC). The qPCR results showed that the inhibited effects of si-SIL1-1 on SIL1 mRNA expression were the greatest both in MDA-MB-231 and MCF7 (P < 0.01; Figure 1(c)). Therefore, si-SIL1-1 was used in the following experiments. The western blot assay was further used to detect the effect of si-SIL1-1 on SIL1 protein expression. As shown in Figure 1(d) and (e), the protein levels of SIL1 were significantly downregulated following si-SIL1 treatment (P < 0.05).
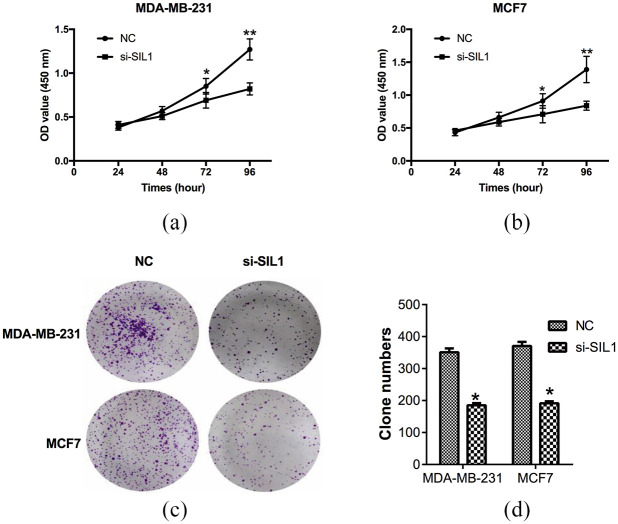
Then, the proliferation of si-SIL1 and NC cells was determined by CCK8 assay. After the transfection of si-SIL1 for 72 h, the OD values of MDA-MB-231 decreased to 0.69 ± 0.09, which was markedly lower than the NC cells at 0.85 ± 0.09 (P < 0.05, Figure 2(a)). The OD values of MDA-MB-231 after being transfected with si-SIL1 for 96 h (0.82 ± 0.07) also decreased significantly in comparison with the NC group (1.27 ± 0.12) (P < 0.01, Figure 2(a)). The OD values of MCF7 after being transfected with si-SIL1 for 72 h (0.71 ± 0.13) and 96 h (0.84 ± 0.07) declined compared with NC cells (72 h, 0.91 ± 0.11; 96 h, 1.39 ± 0.20) (P < 0.05, P < 0.01, Figure 2(b)). Clone formation assay was also used to confirm the effect of SIL1 on cell proliferation. After incubation for 14 days, clone numbers of cells transfected with si-SIL1 (MDA-MB-231, 185 ± 7; MCF7, 191 ± 6) markedly declined compared with the control cells (MDA-MB-231, 351 ± 12; MCF7, 370 ± 13), indicating that si-SIL1 inhibited cell growth in breast cancer (P < 0.05, Figure 2(c) and (d)).
Figure 2.
Knockdown of SIL1 inhibited the proliferation of human breast cancer cells. The proliferation of breast cancer cells was determined by CCK8 assay. The OD value of MDA-MB-231 (a) and MCF7 (b) cells transfected with si-SIL1 was significantly decreased compared with the NC groups. (c) Clone formation assay was performed to further detect the effect of SIL1 on cell proliferation. (d) Statistical analysis of clone numbers showed that si-SIL1 markedly inhibited cell growth.
*P < 0.05.
Knockdown of SIL1 inhibited the migration and invasion of human breast cancer cells
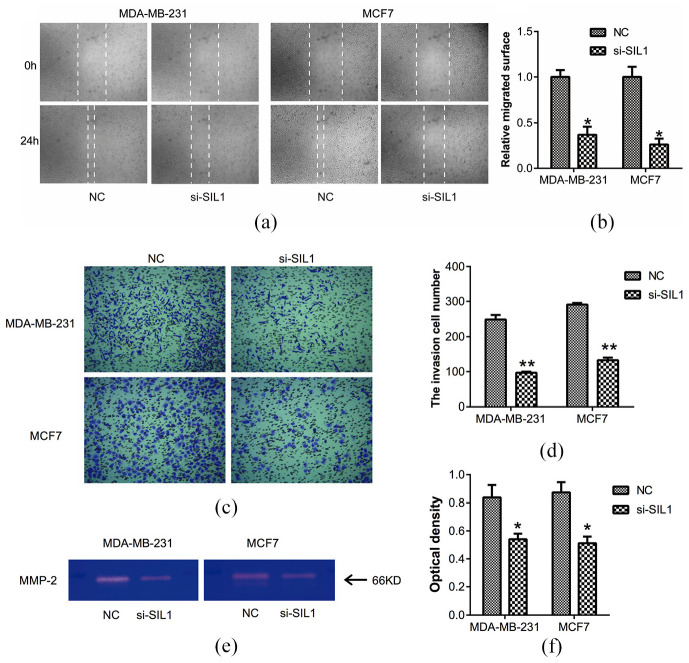
Next, we further investigated the specific role of SIL1 in the migration and invasion of breast cancer cells. As shown in Figure 3(a) and (b), the migrated distance in SIL1 downregulated MDA-MB-231 or MCF7 cells significantly declined compared with control cells in the wound healing assay (both P < 0.05). This indicates that SIL1 was involved in the migration of breast cancer cells. Similarly, using transwell assay, we also found that SIL1 knockdown could decrease the numbers of invasive cells effectively both in MDA-MB-231 and MCF7 cells (both P < 0.01, Figure 3(c) and (d)).
Figure 3.
Knockdown of SIL1 inhibited the migration and invasion of human breast cancer cells. (a) Wound healing assay was used to detect the effect of SIL1 on cell migration (magnification, ×40). (b) The relative migrated surface was showed in a column diagram, which showed a significant decline in SIL1 knockdown cells in comparison with control cells. (c) The invasion capability of cells was determined using transwell assay (magnification, ×200). (d) SIL1 knockdown inhibited the invasion of MDA-MB-231 and MCF7 cells. (e) The expression level of MMP-2 was determined using gelatin zymography. (f) SIL1 knockdown inhibited MMP-2 expression according to the result of gelatin zymography.
*P < 0.05; **P < 0.01.
To explain the reason for SIL1 involvement in the invasion of MDA-MB-231 and MCF7 cells, the expression of MMP-2 was determined using gelatin zymography (Figure 3(e)). According to the results, SIL1 knockdown could inhibit MMP-2 expression significantly when compared with control cells (both P < 0.05, Figure 3(f)). These observations indicated that suppression of SIL1 inhibited the migration and invasion through downregulation of MMP-2 expression.
Knockdown of SIL1 induced cell cycle block in human breast cancer cells
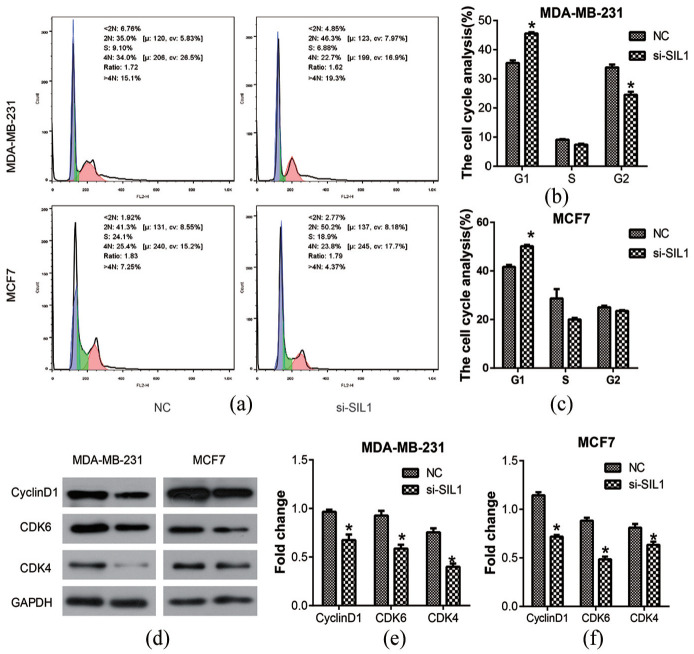
Since knocking down SIL1 could inhibit cell proliferation, we further studied whether it affected the cell cycle using flow cytometry analysis. The results showed that the rate of cells at the G1 phase was enhanced after transfection of si-SIL1 in both breast cancer cell lines (P < 0.05, Figure 4(a)–(c)). Moreover, the rate of cells at the G2 phase also increased in MDA-MB-231 cells that were transfected with si-SIL1 (P < 0.05). Then, western blot assay was used to investigate the expression of some cell cycle-related proteins, Cyclin D1, CDK4, and CDK6 (Figure 4(d)). The results indicated that all the expressions of Cyclin D1, CDK4, and CDK6 in both MDA-MB-231 and MCF7 cells decreased significantly after si-SIL1 transfection (P < 0.05, Figure 4(e) and (f)). These results suggest that knockdown of SIL1 might inhibit the proliferation of breast cancer cells by stopping the cell cycle at the G1 phase.
Figure 4.
Knockdown of SIL1 induced cell cycle block in human breast cancer cells. (a) Flow cytometry analysis was used to detect the role of SIL1 on cell cycle. Statistical analysis of flow cytometry result showed that the rate of cells on G1 phase was enhanced after the transfection of si-SIL1 both in MDA-MB-231 (b) and MCF7 cell lines (c). (d) Western blot analysis was performed to investigate the expression of cell cycle related protein, Cyclin D1, CDK4, and CDK6. Statistical analysis of western blot showed that the expressions of Cyclin D1, CDK4, and CDK6 were decreased after the transfection of si-SIL1 both in MDA-MB-231 (e) and MCF7 cell lines (f).
*P < 0.05.
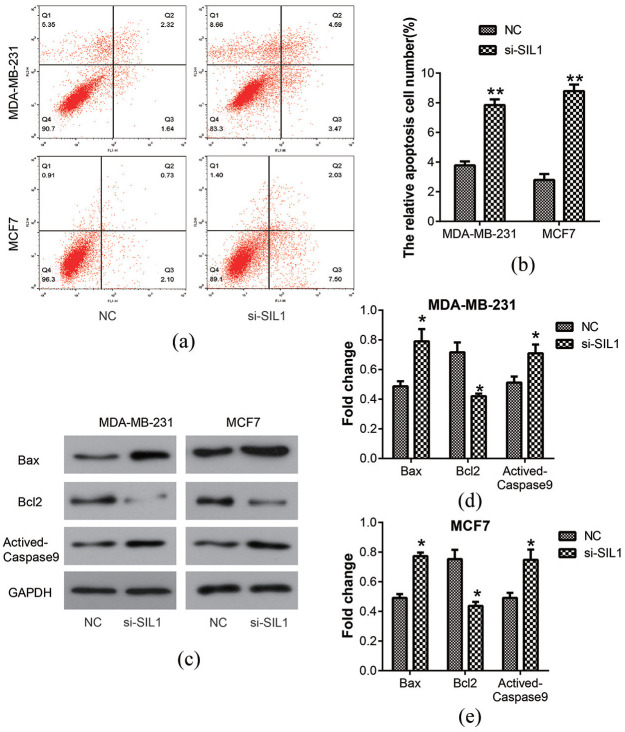
Knockdown of SIL1 promoted the apoptosis of breast cancer cells
The apoptosis of MDA-MB-231 and MCF7 cells was detected using flow cytometry analysis (Figure 5(a)). The percentage of apoptotic cells was shown in a column diagram, which showed that it was enhanced in the si-SIL1 cells (MDA-MB-231, 7.85% ± 0.38%; MCF7, 8.78% ± 0.45%) compared with the NC cells (MDA-MB-231, 3.78% ± 0.27%; MCF7, 2.80% ± 0.40%) (P < 0.01; Figure 5(b)). The expression of apoptosis-related proteins, such as Bax, Bcl2, and Actived-Caspase9, were analyzed by western blot assay (Figure 5(c)). The results showed an increase in the expression of Bax and Actived-Caspase9, but a decrease in the expression of Bcl2 after transfection of si-SIL1 in MDA-MB-231 and MCF7 cells (all P < 0.05, Figure 5(d) and (e)). These observations suggested that knockdown of SIL1 could promote cell apoptosis.
Figure 5.
Knockdown of SIL1 promoted the apoptosis in human breast cancer cells. (a) The apoptosis of MDA-MB-231 and MCF7 cells was determined using flow cytometry analysis. (b) The percentage of apoptosis cell numbers was shown in a column diagram, which showed that it was enhanced by the knockdown of SIL1 both in MDA-MB-231 and MCF7 cells. (c) The expressions of apoptosis-related proteins, Bax, Bcl2, and Actived-Caspase9 were determined by western blot analysis. The expressions of Bax and Actived-Caspase9 were increased, but Bcl2 level was decreased after the transfection of si-SIL1 in MDA-MB- 231 (d) and MCF7 (e) cells.
*P < 0.05; **P < 0.01.
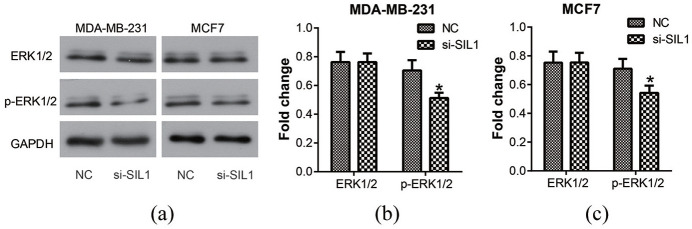
Knockdown of SIL1 inhibited the activity of ERK1/2 in breast cancer cells
Previous studies have shown significant activation of the ERK1/2 signaling pathway in breast cancer, which is involved in regulating cell proliferation, migration, and apoptosis.14,15 Therefore, we performed western blot assay to detect the activity of ERK1/2 (Figure 6(a)). According to the results, the phosphorylation of ERK1/2 decreased significantly after transfection of si-SIL1 both in MDA-MB-231 and MCF7 cells (P < 0.05, Figure 6(b) and (c)). These results suggested that the role of SIL1 in breast cancer progression might be produced via regulating the activity of the ERK1/2 signaling pathway.
Figure 6.
Knockdown of SIL1 inhibited the activity of ERK1/2 signaling pathway in human breast cancer cells. (a) Western blot analysis was used to determine the activity of ERK1/2 signaling pathway. According to the result, the phosphorylation level of ERK1/2 decreased significantly after the transfection of si-SIL1 both in MDA-MB-231 (b) and MCF7 (c) cells.
*P < 0.05.
Discussion
SIL1, also known as BAP, is one of the nucleotide exchange factors for the mammalian HSP70 chaperone protein, GRP78 (also known as BIP). SIL1 regulates the activity of the GRP78 ATPase. SIL1 mutations can cause a rare neurodegenerative disease, Marinesco-Sjögren syndrome, which is also considered to be an ER dysfunction disease.16–18 At the same time, the loss of SIL1 function causes abnormalities in the adenosine triphosphate (ATP) cycle of GRP78 and aggregation of proteins.19,20
In this study, we found that SIL1 was upregulated in human breast cancer according to the result of online analysis. As a regulatory protein of the ER, SIL1 overexpression may result in disrupted cellular functions. SIL1 is primarily responsible for GRP78 activity regulation in the ER. Glucose-regulated proteins (GRP) are a class of stress proteins produced by cells in order to adapt to UPR, with the function of assisting the correct folding and assembly of proteins.21,22 The binding and release of GRP78 to the substrate protein is accomplished by its ATP cycle, and the ATP bounded to GRP78 is hydrolyzed to adenosine diphosphate (ADP), which promotes the stable binding of GRP78 to the unfolded protein. SIL1 can directly bind to the adenosine triphosphate (ATPase) domain on GRP78, which changes the spatial conformation of GRP78, opens its ATPase domain and destroys its binding to ADP, thereby promoting release of ADP and recombination with ATP. 19 Therefore, overexpression of SIL1 may cause cellular dysfunction by altering GRP78 activity and may be related to abnormal biological behavior of tumor cells.
Our results demonstrate that SIL1 promoted the progression of breast cancer by increasing proliferation and decreasing apoptosis of tumor cells. From the CCK8 and clone formation assays, SIL1 knockdown impeded the proliferation of MDA-MB-231 and MCF7 cells. Migration and invasion capability were also reduced by the transfection of SIL1-specific siRNA. Furthermore, suppression of SIL1 stopped the cell cycle in the G1 phase by regulating the expression of Cyclin D1, CDK4, and CDK6. According to flow cytometry analysis, SIL1 knockdown increased the activity of Caspase9 and induced cell apoptosis. In recent years, the study of the relationship between SIL1 and pathological processes has been limited to the nervous system. In 2018, the role of SIL1 in cancer was reported for the first time. Xu et al. 23 proved that si-SIL1 inhibited the progression of human glioma through the AKT/mTOR signaling pathway. Our study indicated that SIL1 could alter the phosphorylation of ERK1/2. ERK1/2 is the key protein of the mitogen-activated protein kinase (MAPK) signaling pathway, which is an important regulatory pathway for cell proliferation or differentiation and is involved in the development of a variety of tumors.24,25 Persistent activation of ERK can promote the degradation of STAT1, a signal transduction and transcription activating factor, thus promoting cell proliferation and inhibiting apoptosis.26,27 Our results are consistent with the above results, suggesting that SIL1 promotes the growth of breast cancer cells by activating ERK1/2 signaling pathway.
SIL1 may also affect cell apoptosis by regulating GRP78 activity. Recent studies have shown that GRP78 is overexpressed in some tumor cells and is positively correlated with malignancy.28–30 Overexpression of GRP78 can reduce the apoptosis of tumor cells and block the apoptosis induced by chemical drugs. 31 High GRP78 expression contributes to tumor development as well as the production of drug resistance, and even induces immune tolerance of tumor cells. 32 Therefore, SIL1 may inhibit cell apoptosis by activating GRP78, but this potential mechanism requires further study.
Since we detected high expression of SIL1 in breast cancer, we only investigated the effect of knocking down SIL1 on cell function. However, it is still necessary to explore the role of SIL1 overexpression in breast cancer cells. At the same time, it is urgent to verify the carcinogenic effect of SIL1 in an in vivo environment. Therefore, exploring the role of SIL1 overexpression and the SIL1/GRP78 axis in breast cancer in vivo and in vitro is the focus for our future research.
In conclusion, we have proven that SIL1 is upregulated in breast cancer, and that knockdown of SIL1 inhibits the progression of breast cancer cells by inducing cell cycle retardation and cell apoptosis. Moreover, SIL1 is involved in the regulation of the ERK1/2 signaling pathway. Based on the results of our study, we propose that SIL1 has potential as a therapeutic target for breast cancer treatment in the future.
Author biographies
Zhi-feng Li, Associate Professor, graduated from Nantong University in 2008 with a master’s degree in medicine. After graduation, he worked in the clinical and basic research of breast cancer in Nantong Maternal and Child Health Hospital. His research interests include immunotherapy of breast cancer, nanomedicine, and the effect of puerarin on apoptosis of breast cancer cells. Li Zhifeng may be contacted at lizhifeng1203@hotmail.com
Wei-wei Xu, graduated from Nantong Medical College in 2004 with a bachelor’s degree in medicine. After graduation, she was engaged in comprehensive treatment of malignant tumors at Nantong Cancer Hospital. She specializes in the comprehensive treatment of breast cancer. Her main research direction is targeted therapy for breast cancer.
Ji-dan Li, obtained Master’s degree in general surgery from Southeast University. His research focuses on the therapy of granulomatous lobular mastitis and the immunotherapy of breast cancer. He may be contacted at Leejidan_88@hotmail.com
Feng-ling Tao, graduated from Yangzhou University Medical College in 2003 and obtained a bachelor’s degree in medicine. She is mainly engaged in the rehabilitation of cancer patients in the Nantong Maternal and Child Health Hospital.
Jian-xin Chen, Professor, graduated from Xuzhou Medical College in 1989 with a bachelor’s degree in medicine. His research interests include comprehensive treatment of breast cancer, sentinel lymph node biopsy, and postoperative reconstruction of breast cancer.
Jin-hua Xu, Professor, graduated from Yangzhou University. He is currently the chief physician and vice president of Nantong Tumour Hospital, the committee member of Jiangsu Basic Theory and Literature Research Committee. He has been engaged in acupuncture clinical work for more than 20 years. He is currently focusing on immunotherapy for breast cancer. He may be contacted at ntxujinhua@outlook.com
Footnotes
Availability of data and materials: The data and materials are available from the corresponding author on reasonable request.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by Nantong City-level Science and Technology Plan (No. JCZ18055).
ORCID iD: Jin-hua Xu  https://orcid.org/0000-0002-7181-5413
https://orcid.org/0000-0002-7181-5413
References
- 1.Jemal A.Cancer statistics, 2015. CA Cancer J Clin 2015; 60: 277–300. [DOI] [PubMed] [Google Scholar]
- 2.Veronesi U, Boyle P, Goldhirsch A, et al. Breast cancer. Lancet 2005; 365: 1727–1741. [DOI] [PubMed] [Google Scholar]
- 3.Yiannakopoulou E.Pharmacogenomics of breast cancer targeted therapy: focus on recent patents. Recent Pat DNA Gene Seq 2012; 6(1): 33–46. [DOI] [PubMed] [Google Scholar]
- 4.Yang SE, Zhao B.Research progress in the use of drugs for breast cancer targeted therapy. Cancer Biol Med 2008; 5: 320–325. [Google Scholar]
- 5.The Lancet. Breast cancer targeted therapy: successes and challenges. Lancet 2017; 389(10087): 2350. [DOI] [PubMed] [Google Scholar]
- 6.Stevens KLP, Black AL, Wells KM, et al. Diminished Ost3-dependent N-glycosylation of the BiP nucleotide exchange factor Sil1 is an adaptive response to reductive ER stress. Proc Natl Acad Sci USA 2017; 114(47): 12489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Behnke J, Feige MJ, Hendershot LM.BiP and its nucleotide exchange factors Grp170 and Sil1: mechanisms of action and biological functions. J Mol Biol 2015; 427(7): 1589–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bracher A, Verghese J.GrpE, Hsp110/Grp170, HspBP1/Sil1 and BAG domain proteins: nucleotide exchange factors for Hsp70 molecular chaperones. Subcell Biochem 2015; 78: 1–33. [DOI] [PubMed] [Google Scholar]
- 9.Hetz C, Saxena S.ER stress and the unfolded protein response in neurodegeneration. Nat Rev Neurol 2017; 13(8): 477–491. [DOI] [PubMed] [Google Scholar]
- 10.Yoshida H.ER stress and diseases. FEBS J 2007; 274: 630–658. [DOI] [PubMed] [Google Scholar]
- 11.Kim B, Kim HS, Jung EJ, et al. Curcumin induces ER stress-mediated apoptosis through selective generation of reactive oxygen species in cervical cancer cells. Mol Carcinog 2016; 55(5): 918–928. [DOI] [PubMed] [Google Scholar]
- 12.Ryu S, Lim W, Bazer FW, et al. Chrysin induces death of prostate cancer cells by inducing ROS and ER stress. J Cell Physiol 2017; 232(12): 3786. [DOI] [PubMed] [Google Scholar]
- 13.Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017; 45(W1): W98–W102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomes LR.TGF-β1 modulates the homeostasis between MMPs and MMP inhibitors through p38 MAPK and ERK1/2 in highly invasive breast cancer cells. BMC Cancer 2012; 12: 26–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang M, Song S, Yi Z, et al. Human biliverdin reductase promotes EMT through the ERK1/2 signal pathway in breast cancer. Eur J Pharmacol 2016; 788: 45–53. [DOI] [PubMed] [Google Scholar]
- 16.Howes J, Shimizu Y, Feige MJ, et al. C-terminal mutations destabilize SIL1/BAP and can cause Marinesco-Sjögren syndrome. J Biol Chem 2012; 287: 8552–8560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nair P, Hamzeh AR, Mohamed M, et al. Marinesco-Sjögren syndrome in an Emirati child with a novel mutation in SIL1 affecting the 5′ untranslated region. Med Princ Pract 2016; 25: 580–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kashimada A, Hasegawa S, Isagai T, et al. Targeting the enhanced ER stress response in Marinesco-Sjögren syndrome. J Neurol Sci 2018; 385: 49–56. [DOI] [PubMed] [Google Scholar]
- 19.Rosam M, Krader D, Nickels C, et al. Bap (SIL1) regulates the molecular chaperone BiP by coupling release of nucleotide and substrate. Nat Struct Mol Biol 2018; 25(1): 90–100. [DOI] [PubMed] [Google Scholar]
- 20.Chung KT, Shen Y, Hendershot LM.BAP, a mammalian BiP-associated protein, is a nucleotide exchange factor that regulates the ATPase activity of BiP. J Biol Chem 2002; 277(49): 47557–47563. [DOI] [PubMed] [Google Scholar]
- 21.Daneshmand S, Quek ML, Lin E, et al. Glucose-regulated protein GRP78 is up-regulated in prostate cancer and correlates with recurrence and survival. Hum Pathol 2007; 38(10): 1547–1552. [DOI] [PubMed] [Google Scholar]
- 22.Sharma SH, Rajamanickam V, Nagarajan S.Antiproliferative effect of p-coumaric acid targets UPR activation by downregulating Grp78 in colon cancer. Chem Biol Interact 2018; 291: 16–28. [DOI] [PubMed] [Google Scholar]
- 23.Xu H, Xu S, Zhang R, et al. SIL1 functions as an oncogene in glioma by AKT/mTOR signaling pathway. Onco Targets Ther 2018; 11: 3775–3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang C, Li P, Xuan J, et al. Cholesterol enhances colorectal cancer progression via ROS elevation and MAPK signaling pathway activation. Cell Physiol Biochem 2017; 42(2): 729–742. [DOI] [PubMed] [Google Scholar]
- 25.Gu XD, Xu LL, Zhao H, et al. Cantharidin suppressed breast cancer MDA-MB-231 cell growth and migration by inhibiting MAPK signaling pathway. Braz J Med Biol Res 2017; 50(7): e5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spudich A, Frigg R, Kilic E, et al. Aggravation of ischemic brain injury by prion protein deficiency: role of ERK-1/-2 and STAT-1. Neurobiol Dis 2005; 20(2): 442–449. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Chen Y, Liu Z, et al. ERK is a negative feedback regulator for IFN-γ/STAT1 signaling by promoting STAT1 ubiquitination. BMC Cancer 2018; 18: 613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsing J, Mcconkey D, Logsdon C.GRP78 and HSP90 are overexpressed in pancreatic cancer. Cancer Res 2007; 67: 708. [Google Scholar]
- 29.Chiu CC, Cheng AJ.Grp78 is over-expressed in head neck cancer and is a potential molecular target for inhibition of oncogenesis. Cancer Res 2007; 67: 4282. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, Wu H, Li Z, et al. A positive feedback loop between GRP78 and VPS34 is critical for GRP78-mediated autophagy in cancer cells. Exp Cell Res 2017; 351(1): 24–35. [DOI] [PubMed] [Google Scholar]
- 31.Lee AS.GRP78 induction in cancer: therapeutic and prognostic implications. Cancer Res 2007; 67(8): 3496–3499. [DOI] [PubMed] [Google Scholar]
- 32.Abdel Malek MA, Jagannathan S, Malek E, et al. Molecular chaperone GRP78 enhances aggresome delivery to autophagosomes to promote drug resistance in multiple myeloma. Oncotarget 2015; 6(5): 3098–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]