Candida dubliniensis is a recently described species of chlamydospore- and germ tube-positive yeast which has been recovered primarily from the oral cavities of human immunodeficiency virus (HIV)-infected individuals and AIDS patients. The organism has been recovered from patients in widespread geographic locations, and its phenotypic and genotypic characteristics have been described in depth in a number of recent reports (6, 9, 30, 31, 33–35). Currently, the best available data on the incidence of C. dubliniensis are from a study of Irish subjects (9). In that study, C. dubliniensis was recovered from the oral cavities of 27% of HIV-infected individuals and 32% of AIDS patients with clinical symptoms of oral candidiasis and from 19% of HIV-infected individuals and 25% of AIDS patients without clinical symptoms of oral candidiasis (9). Oral C. dubliniensis isolates have also been recovered from 5 of 150 (3%) Irish HIV-negative healthy individuals without clinical signs of oral candidiasis and from 7 of 48 (14.6%) Irish HIV-negative patients with denture-associated oral candidiasis (9). For the majority of patients, C. dubliniensis was coisolated with other Candida species, the most common of which was C. albicans (9, 31). Among the Irish HIV-infected patients, mentioned above, from whom C. dubliniensis was recovered, C. dubliniensis was the only species recovered from 23 of 94 (24.5%) of the individuals included in the study (9). Furthermore, in at least one other study, oral isolates now known to be C. dubliniensis were the only species recovered on successive occasions from two separate AIDS patients (29, 30). These findings suggest that as well as being a minor constituent of the normal oral microbial flora of humans, C. dubliniensis can cause disease independently of other Candida species, at least in HIV-infected individuals and AIDS patients. The most common clinical manifestation of candidiasis apparently caused by C. dubliniensis in the Irish HIV-infected population is erythematous candidiasis (31), which is perhaps not surprising because this is the most common manifestation of candidiasis encountered in HIV-infected individuals and AIDS patients (28).
C. albicans is generally acknowledged to be the most pathogenic Candida species and is the species most frequently implicated in oral disease in immunocompromised and immunocompetent individuals (23). However, over the past decade the incidence of infections caused by C. albicans has decreased relative to the incidence of infections caused by other species of Candida, including C. tropicalis, C. glabrata, and C. krusei (10–12, 26, 34, 37). The recent emergence of C. dubliniensis as an opportunistic pathogen appears to coincide with this apparent epidemiological shift.
The earliest Candida isolate now known to be C. dubliniensis was recovered from a postmortem lung specimen recovered from a patient who died in the United Kingdom in 1957 (31, 35). However, due to the lack of available clinical data we have no evidence to suggest that this isolate was implicated in disease. This strain (strain NCPF 3108) was originally misidentified as C. stellatoidea and was included as a reference strain for this species in the British National Collection of Pathogenic Fungi (NCPF 3108 is not ex-type, i.e., not derived from the type specimen, as originally reported by us [31]). This anomaly led us to suggest originally that isolates now known to be C. dubliniensis may have been an unusual variant of C. stellatoidea (31). Despite the fact that NCPF 3108, now known to be C. dubliniensis, had been recovered from at least one clinical sample as far back as the 1950s, it was not until the late 1980s or early 1990s that the next isolates of C. dubliniensis were identified. These isolates were recovered from HIV-infected individuals and AIDS patients in Australia, Ireland, and the United Kingdom (23, 29–31, 35); however, it was not until 1995 that it was suggested that these isolates comprised a new species (35). The name proposed for this new species was C. dubliniensis, after Dublin, the capital city of the Republic of Ireland, where the new species was first identified as such (35). Other C. dubliniensis isolates have since been identified from patients in Argentina, Australia, Belgium, Canada, France, Finland, Germany, Greece, Ireland, Spain, Switzerland, the United Kingdom, and the United States (3, 4, 14, 16, 17, 23, 27, 29, 30, 32, 33, 35).
This widespread distribution of C. dubliniensis throughout the world and its apparent involvement in oral disease warrants in-depth investigation, particularly because resistance to the commonly used antifungal drug fluconazole has been found in clinical isolates and because fluconazole-susceptible isolates have been shown to be able to rapidly develop resistance to the drug in vitro (21). Of particular interest is the determination of accurate measurements of the incidence of the organism, the occurrence of resistance to antifungal agents, and its precise role in disease. However, in order to achieve this, it is essential to be able to discriminate accurately and rapidly between C. dubliniensis and other Candida species, particularly C. albicans. The purpose of this minireview is to describe briefly the phenotypic and genotypic properties of C. dubliniensis, with particular emphasis on those traits which may be of use in the identification of the species in clinical specimens.
CHARACTERISTICS OF C. DUBLINIENSIS
Phenotypic characteristics.
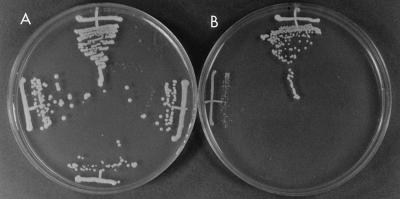
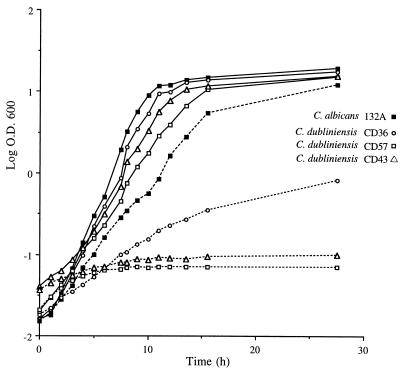
Isolates of C. dubliniensis grow well at 30 and 37°C on culture media routinely used to grow Candida species (35). Colonies formed on solid media, such as Sabouraud agar or potato dextrose agar (PDA), are a creamy white color, similar to those formed by C. albicans. Isolates of C. dubliniensis frequently appear to undergo phenotypic switching, and small petite colonies can often be observed, particularly after prolonged storage (32). However, unlike C. albicans, isolates of C. dubliniensis grow poorly or not at all at 42°C (6, 9, 33, 35). Figure 1 shows the comparative growth of three selected oral isolates of C. dubliniensis and a reference oral strain of C. albicans on PDA following growth at 37 and 42°C for a 48-h period. All three of the C. dubliniensis isolates, isolates CD57, CD43, and CD36 (type strain), and the reference strain C. albicans 132A grew well and apparently to a similar extent at 37°C (Fig. 1A). However, only C. albicans 132A grew efficiently on this medium at 42°C (Fig. 1B). C. dubliniensis CD57 and CD43 showed hardly any growth at all, while the type strain, C. dubliniensis CD36, grew but to a much lesser extent relative to the growth of C. albicans 132A. In addition to the examination of plates following incubation for 48 h, discrimination of C. dubliniensis isolates from C. albicans isolates is readily facilitated by examination of the plates following 24 h of growth. Similar results were obtained with these isolates following growth in yeast extract-peptone-dextrose (YEPD) broth medium. In these experiments, which were repeated on several separate occasions, equal inocula (2 × 105 CFU) of each isolate were added separately to YEPD medium, the cultures were incubated in an orbital incubator set at 150 rpm at either 37 or 42°C, and the optical density at 600 nm was measured at hourly intervals. The results of a typical experiment are presented in Fig. 2. All four cultures grew at similar rates at 37°C. However, at 42°C, only C. albicans 132A grew efficiently (although at a slightly reduced rate compared to growth at 37°C). C. dubliniensis CD57 and CD43 showed very little growth at 42°C over the course of 24 h, while C. dubliniensis CD36 showed moderate growth (with a mean doubling time of approximately 3.5 h, compared to a doubling time of approximately 1.5 h for C. albicans 132A).
FIG. 1.
Comparative growth of oral C. dubliniensis and C. albicans on PDA following 48 h of incubation at 37°C (A) and 42°C (B). The growth of the following isolates is shown: C. albicans 132A, C. dubliniensis CD57, C. dubliniensis CD43, and C. dubliniensis CD36 (clockwise from the top in each panel).
FIG. 2.
Growth curves of the oral isolates C. dubliniensis CD36, CD57, and CD43 and the oral reference strain C. albicans 132A in YEPD broth medium at 37°C (solid lines) and 42°C (dashed lines). O.D., optical density.
Although it is impossible to distinguish between C. albicans and C. dubliniensis colonies on conventional solid media, it is possible to distinguish between colonies of the two species following primary isolation from clinical specimens (6, 9) by using the recently developed commercially available chromogenic agar medium CHROMagar Candida (22). Following 48 h of growth at 37°C on this medium, C. albicans colonies appear light blue-green. In contrast, under the same conditions, C. dubliniensis colonies appear dark green and are easily distinguishable from other Candida species (6, 9). We stress that CHROMagar Candida agar is useful only for identifying colonies of C. dubliniensis following primary plating from clinical specimens after 48 h of incubation, because one recent study reported that C. dubliniensis isolates can lose the ability to yield dark green colonies on this medium following subculture or storage (30), a phenomenon also observed in our laboratory. The reasons for this apparent instability are unclear but may be related to the ability of C. dubliniensis isolates to exhibit the phenomenon of phenotypic switching.
It has also been recently described that C. dubliniensis isolates can be distinguished from isolates of C. albicans following growth on methyl blue-Sabouraud agar (30). On this medium C. albicans isolates fluoresce with a yellow color on exposure to long-wave UV light, while C. dubliniensis isolates fail to fluoresce under these conditions. However, that study also reported that this lack of fluorescence under these conditions may not be reproducible in isolates subjected to storage and repeated subculture. Interestingly, all C. dubliniensis isolates examined thus far belong solely to C. albicans serotype A (19, 30, 31, 33, 35), while C. albicans isolates can belong to either C. albicans serotype A or serotype B (8).
C. dubliniensis isolates produce germ tubes and chlamydospores, features usually diagnostic for C. albicans (30, 31, 33, 35). A number of reports have described that the production of chlamydospores by isolates now known to be C. dubliniensis and by other unusual isolates which may belong to C. dubliniensis is unusual, in that the chlamydospores are produced abundantly and often in clusters or contiguous pairs (6, 9, 27, 33, 35). However, this phenomenon was not observed in one recent study of C. dubliniensis isolates recovered in Belgium (30). The ability of individual Candida isolates to assimilate a range of carbohydrate compounds as the sole source of carbon or nitrogen has been used extensively for species identification. Commercially available yeast identification kits based on these tests, such as the API ID 32C and 20C AUX systems (bioMérieux, Marcy l’Étoile, France), have been used extensively and successfully throughout the world for many years for identifying yeast isolates from clinical specimens (8). The profile of assimilation reactions generated by these kits yields a numerical code which suggests a list of species in order of probability by comparison of the profile with the profiles in a database. However, isolates of C. dubliniensis are not readily identifiable by either system because the assimilation profiles for C. dubliniensis are not currently included in the respective databases (31, 33, 35). In addition, the results of these tests are sometimes difficult to assess, with some of the reactions leading to ambiguous results which can be difficult to reproduce (30, 33).
One significant difference between isolates of C. albicans and C. dubliniensis is the inability of the latter to express β-glucosidase activity. This trait, originally identified in multilocus enzyme electrophoresis (MEE) experiments, forms the basis of a reliable assay for the discrimination of the two species. This assay, developed originally by Boerlin et al. (3), has subsequently been used successfully to discriminate between C. dubliniensis and C. albicans by two other groups (30, 33).
Other phenotypic traits have been investigated by other laboratories. Of particular note is the finding that, in comparison with isolates of C. albicans, isolates of C. dubliniensis have been demonstrated to produce increased levels of extracellular proteinase and to have an increased ability to adhere to buccal epithelial cells (17).
Genotypic characteristics.
One of the earliest clues which suggested that C. dubliniensis isolates represented a group of organisms distinct from typical C. albicans isolates arose from DNA fingerprinting studies. A group of isolates recovered from the oral cavities of Irish AIDS patients were originally identified as C. albicans on the basis of their ability to produce germ tubes and chlamydospores (31, 35). However, when genomic DNA from these isolates was digested with EcoRI and probed with the repetitive C. albicans-specific DNA fingerprinting probe 27A, the fingerprinting patterns obtained comprised fewer and fainter bands than normally expected for C. albicans (31, 35). Despite the presence of fewer hybridization bands in the C. dubliniensis patterns, the 27A fingerprinting probe is very effective at discriminating between individual isolates of C. dubliniensis (33, 35). The unusual fingerprinting patterns of C. dubliniensis isolates coupled with their unusual carbohydrate assimilation profiles led us to suggest that these organisms may have been an atypical form of C. albicans or C. stellatoidea (31). Isolates with similar unusual fingerprinting profiles (many of which have since been confirmed to be C. dubliniensis) have been described by investigators at several laboratories around the world (1, 3, 4, 17, 18, 23, 29, 30, 33). C. dubliniensis isolates also yield distinct restriction fragment length polymorphism patterns when genomic DNA is digested with the restriction enzyme HinfI (35). In addition, when subjected to analysis with other fingerprinting probes, such as oligonucleotides homologous to repetitive microsatellite sequences [i.e., (GATA)4, (GTG)5, and (GT)8], these isolates yield fingerprinting patterns easily discernible from those for conventional C. albicans and C. stellatoidea isolates (31, 35).
C. dubliniensis isolates can also be differentiated from other Candida species by a variety of other molecular biology-based techniques. Randomly amplified polymorphic DNA (RAPΔ) analysis using a wide range of oligonucleotides as primers in amplification reactions results in clearly distinct profiles for isolates of C. dubliniensis (35). In addition, primers homologous to C. albicans-specific sequences, such as OREN and CARE-2, also yield distinctive products following PCR amplification with template DNA from C. dubliniensis (17). The apparent gross genetic differences between C. dubliniensis and C. albicans are also evident when karyotypes of the two species are compared. Following pulsed-field gel electrophoresis of C. dubliniensis DNA, 10 or more chromosome-sized DNA bands can be resolved, usually with one or more bands less than 1 Mb in size (1, 35). In C. albicans seven or eight bands are usually found, although small supernumerary chromosomes or chromosomal fragments have been observed in individual isolates (36).
MEE is an excellent technique for the detection of genetic differences in populations of organisms. This technique detects at a variety of distinct loci pangenomic variation resulting from evolutionary divergence at the sequence level. In two separate studies, in which different loci were examined, one group of isolates which has since been identified definitively as C. dubliniensis (3, 33) and another group which has tentatively been identified as such (16, 27) were demonstrated to be genetically very divergent from the other Candida species examined, including C. albicans and C. stellatoidea. These data provide additional evidence for the distinct species designation of C. dubliniensis. Interestingly, despite the extensive genetic variability within C. dubliniensis as detected with the DNA fingerprinting probe 27A, MEE detects very little intraspecies divergence (3, 16, 27).
All of the phenotypic and genotypic data described above strongly suggested that C. dubliniensis is a species distinct from C. albicans. However, these studies did not allow a quantitative estimation of the genetic relatedness of these organisms. In order to achieve this, sequences of the rRNA genes of representative isolates were analyzed (32, 33, 35). The sequences of these genes have provided the basis for a great number of phylogenetic studies in a wide variety of organisms and were originally chosen for study because they are highly conserved in evolutionary terms and are present at high copy numbers in most eukaryotic genomes (24). In an initial study in our laboratory, we determined the sequence of a 500-bp region of the V3 variable region of the large rRNA gene from eight epidemiologically unrelated isolates of C. dubliniensis recovered in Ireland, Australia, and the United Kingdom (the latter consisting of the NCPF 3108 strain described above) and from a variety of other Candida species, including C. albicans, C. glabrata, C. kefyr, C. krusei, C. parapsilosis, C. stellatoidea, and C. tropicalis (35). When these sequences were compared, the sequences from the eight C. dubliniensis isolates were found to be identical and to be clearly distinct from those obtained from the other species studied. The C. dubliniensis sequence was approximately 2.5% divergent from that obtained for its closest relative, C. albicans. More recently the sequence of the corresponding region was obtained for other isolates of C. dubliniensis recovered from patients in Argentina, Switzerland, and the United Kingdom (33), and for each isolate the sequence was found to be identical to those for the Irish and Australian C. dubliniensis isolates described above (35). The phylogenetic relatedness of C. albicans and C. dubliniensis no doubt reflects the phenotypic and genotypic similarities between the two species. More recently we have obtained the sequence for the entire 1.8-kb small rRNA gene of C. dubliniensis (EMBL accession no. X99399). By using this sequence, comparative phylogenetic studies similar to those described above again confirmed that C. dubliniensis was clearly distinct from the other species examined, including C. albicans (32). Further analysis of the sequence of the large rRNA gene of C. dubliniensis isolates indicated the presence of a self-splicing group I intron. However, this region displays two sites with major sequence divergence from the sequences of the homologous loci in C. albicans and has been suggested to form a complex cloverleaf structure unique to C. dubliniensis (4). Another recent study, involving sequence analysis of the 5′ end of the large rRNA subunit genes from a wide variety of clinically important ascomycetous yeasts, also demonstrated the distinct phylogenetic position of C. dubliniensis and its close relationship with C. albicans (14). In this study, the C. albicans and C. dubliniensis sequences were found to differ by approximately 2.2%; this difference is similar to the sequence differences found between C. tropicalis and C. maltosa (i.e., 2.8%) and between C. parapsilosis and the ascosporic species Lodderomyces elongisporus (i.e., 3.2%). Peterson and Kurtzman (25) have previously suggested that strong evidence for a separate species exists when this region contains greater than 1% nucleotide substitution between two organisms. All of these phylogenetic studies serve to confirm the taxonomic status of C. dubliniensis as a distinct species.
THE EMERGENCE OF C. DUBLINIENSIS
Since the original report describing the identification of C. dubliniensis in 1995 (35), increasing numbers of studies have described the recovery of these and similar organisms from around the world (1, 3, 4, 14, 16–18, 27, 30, 33). Although isolates of the species have been recovered from healthy subjects, for the most part, isolates of C. dubliniensis have been recovered from the oral cavities of individuals infected with HIV or, more frequently, from patients with AIDS. A small number of isolates of C. dubliniensis (i.e., three isolates recovered from 110 individuals) have also been recovered from Irish HIV-infected and uninfected women with vaginitis (21, 32). To date there have been no reports, of which we are aware, that this species is implicated in systemic disease in either immunocompetent or immunocompromised individuals. These findings suggest that the oral cavity may be a natural ecological niche for C. dubliniensis, although it is possible that this organism occupies other anatomical sites and may also be present in other animal species. Although infections caused by C. dubliniensis in healthy immunocompetent individuals have been recorded, they are relatively rare (9). This suggests that under most circumstances the immune system, possibly in concert with the normal microbial flora, prevents overgrowth of C. dubliniensis. However, when T-cell immunity is impaired (as in individuals with HIV infection and AIDS), this ability to maintain C. dubliniensis at low levels appears to be diminished.
The oral cavity is a very complex environment and has a rich normal microbial flora, including a variety of species of Candida, which can include C. dubliniensis. The population densities of individual Candida species are likely to be dynamic and to change frequently, depending on environmental conditions, including diet, drug treatment, and state of immunocompetence (7). More than one species and/or strain of Candida is often present in patients with oral candidiasis, thus making it difficult to determine which strains are actively contributing to the symptoms of disease. As stated previously, C. albicans is the species most frequently implicated in oral candidiasis. However, in patients with recurrent disease C. albicans can be replaced as the majority species by other, less pathogenic species, such as C. dubliniensis, C. glabrata, and C. krusei (7, 31, 34). One hypothesis that may account for this epidemiological shift is that these latter species, unlike C. albicans, can persist in the oral cavity following antifungal treatment. This is certainly likely to be the case for species such as C. glabrata and C. krusei, which, when compared with C. albicans, are generally less susceptible to commonly used antifungal drugs such as fluconazole (7, 26, 34). The majority of C. dubliniensis clinical isolates examined so far are susceptible to commonly used antifungal drugs, including fluconazole (MIC range, 0.125 to 1.0 μg ml−1) (21). However, oral C. dubliniensis isolates with significantly reduced susceptibility to fluconazole (MIC range, 8 to 32 μg ml−1) have been recovered from AIDS patients who had previously been treated with fluconazole (21). Furthermore, we have been able to readily “train” fluconazole-susceptible isolates of C. dubliniensis to become resistant (MIC range, 16 to 64 μg ml−1) by subculturing the organisms on medium supplemented with progressively increasing concentrations of drug (21). The resistance of these derivatives was found to be stable following in vitro subculture in the absence of drug. Concurrent control studies with a reference oral isolate of C. albicans failed to yield fluconazole-resistant derivatives (21). In a more recent study, fluconazole-resistant derivatives of a C. albicans isolate were obtained following prolonged exposure to the drug; however, this resistance phenotype was not stable and was readily lost following in vitro subculture (5). If this ability of C. dubliniensis to develop stable fluconazole resistance rapidly does occur in vivo, this may explain, at least in part, its recent emergence as an opportunistic pathogen in the oral cavities of HIV-infected individuals and AIDS patients, who are often treated with this drug.
IDENTIFICATION OF C. DUBLINIENSIS FROM CLINICAL SPECIMENS
In order to assess fully the clinical importance of C. dubliniensis, a concerted and in-depth epidemiological analysis must be conducted with a wide range of patient populations. In order to achieve this, it is essential that we be able to identify this species in clinical samples as accurately and as rapidly as possible. To be of use in extensive studies, identification techniques must also be amenable to a large sample volume throughput, inexpensive, easy to apply, and reproducible. Of particular importance in the case of C. dubliniensis is the ability to differentiate with confidence between this species and C. albicans and C. stellatoidea. In the case of C. albicans and C. stellatoidea, these species are now considered synonymous (15, 20). However, from the data described above it is clear that C. dubliniensis shares many characteristics in common with C. albicans, particularly phenotypic properties. Our original suggestion that isolates now known to be C. dubliniensis were either an unusual form of C. albicans (on the basis of positive germ tube and chlamydospore tests) or an unusual form of C. stellatoidea (on the basis of similarity to NCPF 3108) highlights the problems in discriminating between these species. In addition, in a retrospective study of our oral C. albicans isolate collection (originally identified on the basis of germ tube and chlamydospore production), we found that approximately 2% of isolates from asymptomatic healthy individuals and approximately 17% of isolates from HIV-infected individuals which had originally been identified as C. albicans were in fact C. dubliniensis (9). Moreover, these cases of misidentification suggest that the estimated incidence of C. dubliniensis in other centers around the world may be greater than is currently understood.
Despite the many similarities between C. dubliniensis and C. albicans, there are significant differences between the two species. The most pronounced differences are genetic, as determined by DNA fingerprinting, karyotype analysis, and DNA sequence analysis of rRNA genes. However, the techniques used to detect these genetic differences are time-consuming and expensive and are not readily applicable to large numbers of isolates. Fortunately, several phenotypic tests can be used. C. albicans and C. dubliniensis can be differentiated from all other Candida species by the fact that they alone can produce germ tubes and chlamydospores. It has been reported that C. dubliniensis produces chlamydospores more abundantly than C. albicans (27, 33, 35); however, comparisons of relative abundance can be quite subjective, and this finding was not reproduced in one recent study (30). Therefore, we suggest that hyperproduction of chlamydospores should not be used for the initial identification of C. dubliniensis. Although C. dubliniensis isolates grow well at 37°C, the majority do not grow at 42°C, and those that do grow do so poorly. In tests with 112 epidemiologically unrelated isolates of C. dubliniensis, 94 showed no growth at 42°C, while 18 showed limited growth (32). In contrast, isolates of C. albicans grow well at this temperature. Even with C. dubliniensis isolates that grow at 42°C, after 24 to 48 h growth on PDA medium the growth is so poor as to allow these isolates to be readily distinguished from C. albicans isolates (Fig. 1). This characteristic can easily be assayed by comparing the growth of test and control isolates on PDA plates at 37 and 42°C.
The recent introduction of CHROMagar Candida medium has been an important additional aid in the analysis of Candida populations from clinical specimens. This is a solid medium containing chromogenic substrates, which allows the differentiation of several clinically important Candida species on the basis of colony color (22). When plated on this medium colonies of C. dubliniensis appear dark green, while those of C. albicans are a much lighter blue-green (6, 30, 33), often with one side of the colony showing little color at all. These color differences are easily distinguishable (6, 30). However, since the ability of C. dubliniensis to yield dark green colonies on CHROMagar Candida medium can be unstable following subculture or storage (30), we recommend that CHROMagar Candida be used only for the presumptive identification of C. dubliniensis colonies following primary isolation from clinical specimens. We and others have found it to be particularly useful for this purpose (6, 9, 30, 33).
One of the earliest indications that C. dubliniensis represented a distinct group of organisms came from the observation of their unusual carbohydrate assimilation profiles determined with commercially available systems such as the API ID 32C and API 20C AUX kits (31). However, although C. dubliniensis isolates generate distinct assimilation patterns with these kits, the results of these assays can be difficult to interpret and those for some substrates (particularly trehalose) are sometimes not reproducible (30, 33). Another characteristic of C. dubliniensis which could potentially be applied to its rapid identification is its lack of fluorescence under long-wave UV light following growth on methyl blue-Sabouraud agar (30). However, just as the dark green color of C. dubliniensis colonies on CHROMagar Candida appears to be unstable following subculture or storage, it has been reported that this trait of nonfluorescence can also be lost under similar conditions (30). From MEE studies, it is evident that C. dubliniensis, unlike C. albicans, is unable to express β-glucosidase activity (3). A rapid assay, based on this finding, has been described recently, and the assay can effectively differentiate between the two species with confidence (3, 30, 33). By this assay, all C. albicans isolates tested generate fluorescence when cells are mechanically disrupted and incubated with the substrate methylumbelliferyl-β-glucoside. Under the same conditions isolates of C. dubliniensis show no fluorescence (3, 30, 33). The results of this assay for discriminating C. dubliniensis from C. albicans are sufficiently definitive.
Even though molecular biology-based techniques are generally more cumbersome and difficult to apply than methods based on phenotypic properties, sequence differences between C. albicans and C. dubliniensis could potentially be exploited in the design of oligonucleotide primers for use in diagnostic PCR tests, an approach which is increasingly being used in clinical laboratories. The advantages of PCR over other molecular biology-based techniques include its speed, reproducibility, and high sample volume throughput. Already, differences between the sequences of the large and small rRNA subunits and the actin and phospholipase C genes from C. dubliniensis and other Candida species have been determined (2, 4, 14, 32, 35), and it is likely that as the sequences of other genes and intergenic regions are determined other candidate target loci for use in diagnostic analyses will be detected. DNA fingerprinting probes which are specific for C. dubliniensis (i.e., Cd1 and Cd4) have been isolated and characterized recently (13). As well as having immense potential for the epidemiological investigation of C. dubliniensis isolates, these species-specific sequences should also prove to be excellent markers for simple molecular biology-based diagnostic and identification assays.
CONCLUSIONS
C. dubliniensis is a recently identified opportunistic yeast pathogen associated with oral candidiasis, particularly in HIV-infected individuals and AIDS patients. In-depth epidemiological studies are required to determine the precise clinical importance of this organism, particularly because of its ability to rapidly develop fluconazole resistance in vitro. In order for this to occur it is essential that investigators develop techniques which allow C. dubliniensis to be easily distinguished from other Candida species, especially C. albicans. Although closely related to C. albicans, C. dubliniensis can be readily distinguished from this and other Candida species on the basis of a variety of phenotypic and genetic characteristics. In our laboratory we routinely screen clinical samples by initially plating the samples on CHROMagar Candida medium at 37°C. A selection of colonies which are dark green in color after 48 h of incubation are tested for their ability to produce germ tubes and chlamydospores and are assayed for their ability to grow at 42°C on PDA. Reference strains of C. dubliniensis and C. albicans are included in all of these tests. Chlamydospore- and germ tube-positive clinical isolates which fail to grow or which grow poorly at 42°C are tentatively identified as C. dubliniensis. Definitive identification can be obtained by testing for a lack of intracellular β-glucosidase activity, a stable phenotypic trait (3, 30, 33). This rapid assay has recently been recommended as the most reliable means of easily discriminating between C. albicans and C. dubliniensis (30). However, if required, further confirmation can be obtained by performing any of a number of DNA fingerprinting techniques. We routinely probe genomic DNA with the C. albicans-specific DNA fingerprinting probe 27A; however, restriction fragment length polymorphism analysis with HinfI digestion and RAPD analysis are also effective and are more rapid and easier to perform.
It is still only a little more than 2 years since C. dubliniensis was first identified as a distinct taxon, and much concerning its biology and epidemiology remains to be elucidated. Further detailed studies will certainly lead to a clearer picture regarding the clinical relevance of this species and to the development of improved means for its identification. In the meantime, results from our laboratory suggest that the incidence of C. dubliniensis is continuing to increase. This combined with the findings that a significant proportion of isolates have reduced susceptibility to fluconazole and that susceptible isolates have the propensity to generate resistant derivatives suggests that the emergence of these organisms may have implications for antifungal drug treatment regimens.
The C. dubliniensis type strain has been lodged with the British National Collection for Pathogenic Fungi, Mycology Reference Laboratory, Bristol Public Health Laboratory, Kingsdown, Bristol, United Kingdom (accession no. NCPF 3949), and with the Centraalbureau voor Schimmelcultures, Baarn, The Netherlands (accession no. CBS 7987).
ACKNOWLEDGMENTS
The work performed in our laboratory was supported by Irish Health Research Board grants 134/95 and 41/96 and by grant 047204 from The Wellcome Trust.
REFERENCES
- 1.Anthony R M, Midgley J, Sweet S P, Howell S A. Multiple strains of Candida albicans in the oral cavity of HIV-positive and HIV-negative patients. Microb Ecol Health Dis. 1995;8:23–30. [Google Scholar]
- 2.Bennett D E, McCreary C E, Coleman D C. Genetic characterization of a phospholipase C gene from Candida albicans: presence of homologous sequences in Candida species other than C. albicans. Microbiology. 1998;144:55–72. doi: 10.1099/00221287-144-1-55. [DOI] [PubMed] [Google Scholar]
- 3.Boerlin P, Boerlin-Petzold F, Durussel C, Addo M, Pagani J-L, Chave J-P, Bille J. Cluster of atypical Candida isolates in a group of human immunodeficiency virus-positive drug users. J Clin Microbiol. 1995;33:1129–1135. doi: 10.1128/jcm.33.5.1129-1135.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boucher H, Mercure S, Montplaisir S, Lemay G. A novel group I intron in Candida dubliniensis is homologous to a Candida albicans intron. Gene. 1996;180:189–196. doi: 10.1016/s0378-1119(96)00453-2. [DOI] [PubMed] [Google Scholar]
- 5.Calvet H M, Yeaman M R, Filler S G. Reversible fluconazole resistance in Candida albicans: a potential in vitro model. Antimicrob Agents Chemother. 1997;41:535–539. doi: 10.1128/aac.41.3.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coleman, D., D. Sullivan, K. Haynes, M. Henman, D. Shanley, D. Bennett, G. Moran, C. McCreary, L. O’Neill, and B. Harrington. 1997. Molecular and phenotypic analysis of Candida dubliniensis: a recently identified species linked with oral candidosis in HIV-infected and AIDS patients. Oral Dis. 3(Suppl. 1):S96–S101. [DOI] [PubMed]
- 7.Coleman D C, Bennett D E, Gallagher P J, Flint S R, Nolan A, Mulcahy F M, Sullivan D J, Henman M C, Russell R J, Shanley D B. Oral candidiasis and HIV infection: antifungal drug resistance and changes in Candida population dynamics. In: Greenspan J S, Greenspan D, editors. Oral manifestations of HIV infection. Chicago, Ill: Quintessence Publishing Co., Ltd.; 1995. pp. 112–118. [Google Scholar]
- 8.Coleman D C, Bennett D E, Sullivan D J, Gallagher P J, Henman M C, Shanley D B, Russell R J. Oral Candida in HIV infection and AIDS: new perspectives new approaches. Crit Rev Microbiol. 1993;19:61–82. doi: 10.3109/10408419309113523. [DOI] [PubMed] [Google Scholar]
- 9.Coleman D C, Sullivan D J, Bennett D E, Moran G P, Barry H J, Shanley D B. Candidiasis: the emergence of a novel species, Candida dubliniensis. AIDS. 1997;11:557–567. doi: 10.1097/00002030-199705000-00002. [DOI] [PubMed] [Google Scholar]
- 10.Fridkin S K, Jarvis W R. Epidemiology of nosocomial fungal infections. Clin Microbiol Rev. 1996;9:499–511. doi: 10.1128/cmr.9.4.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hazen K C. New and emerging yeast pathogens. Clin Microbiol Rev. 1995;8:462–478. doi: 10.1128/cmr.8.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jarvis W R. Epidemiology of nosocomial fungal infections, with emphasis on Candida species. Clin Infect Dis. 1995;20:1526–1530. doi: 10.1093/clinids/20.6.1526. [DOI] [PubMed] [Google Scholar]
- 13.Joly, S., and D. R. Soll. 1997. Personal communication.
- 14.Kurtzman C P, Robnett C J. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J Clin Microbiol. 1997;35:1216–1223. doi: 10.1128/jcm.35.5.1216-1223.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwon-Chung K J, Riggsby W S, Uphoff R A, Hicks J B, Whelan W L, Reiss E, Magee B B, Wickes B L. Genetic differences between type I and type II Candida stellatoidea. Infect Immun. 1989;57:527–532. doi: 10.1128/iai.57.2.527-532.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Guennec R, Reynes J, Mallié M, Pujol C, Janbon F, Bastide J-M. Fluconazole- and intraconazole-resistant Candida albicans strains from AIDS patients: multilocus enzyme electrophoresis analysis and antifungal susceptibilities. J Clin Microbiol. 1995;33:2732–2737. doi: 10.1128/jcm.33.10.2732-2737.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCullough M, Ross B, Reade P. Characterization of genetically distinct subgroup of Candida albicans strains isolated from oral cavities of patients infected with human immunodeficiency virus. J Clin Microbiol. 1995;33:696–700. doi: 10.1128/jcm.33.3.696-700.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCullough M J, Ross B C, Dwyer B D, Reade P C. Genotype and phenotype of oral Candida albicans from patients infected with the human immunodeficiency virus. Microbiology. 1994;140:1195–1202. doi: 10.1099/13500872-140-5-1195. [DOI] [PubMed] [Google Scholar]
- 19.Mercure S, Senechal S, Auger P, Lemay G, Montplaisir S. Candida albicans serotype analysis by flow cytometry. J Clin Microbiol. 1996;34:2106–2112. doi: 10.1128/jcm.34.9.2106-2112.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyer S A, Ahearn D G, Yarrow D. Genus 4. Candida Berkhout. In: Kreger van Rij N J W, editor. The yeasts: a taxonomic study. Amsterdam, The Netherlands: Elsevier; 1984. pp. 585–844. [Google Scholar]
- 21.Moran G P, Sullivan D J, Henman M C, McCreary C E, Harrington B J, Shanley D B, Coleman D C. Antifungal drug susceptibility of oral Candida dubliniensis isolates from HIV-infected and non-HIV-infected subjects and generation of stable fluconazole-resistant derivatives in vitro. Antimicrob Agents Chemother. 1997;41:617–623. doi: 10.1128/aac.41.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Odds F C, Bernaerts R. CHROMagar Candida, a new differential isolation medium for presumptive identification of clinically important Candida species. J Clin Microbiol. 1994;32:1923–1929. doi: 10.1128/jcm.32.8.1923-1929.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Odds F C, Schmid J, Soll D R. Epidemiology of Candida infection in AIDS. In: Vanden Bossche H, et al., editors. Mycoses in AIDS patients. New York, N.Y: Plenum Press; 1990. pp. 67–74. [Google Scholar]
- 24.Olsen G J, Woese C R. Ribosomal RNA: a key to phylogeny. FASEB J. 1993;7:113–123. doi: 10.1096/fasebj.7.1.8422957. [DOI] [PubMed] [Google Scholar]
- 25.Peterson S W, Kurtzman C P. Ribosomal RNA sequence divergence among sibling species of yeasts. Syst Appl Microbiol. 1991;14:124–129. [Google Scholar]
- 26.Pfaller, M. A. 1996. Nosocomial candidiasis: emerging species, reservoirs, and modes of transmission. Clin Infect. Dis. 22(Suppl. 2):S89–S94. [DOI] [PubMed]
- 27.Pujol C, Renaud F, Mallié M, de Meeûs T, Bastide J-M. Atypical strains of Candida albicans recovered from AIDS patients. J Med Vet Mycol. 1997;35:115–121. [PubMed] [Google Scholar]
- 28.Samaranayake L P. Oral mycoses in HIV infection. Oral Surg Oral Med Oral Pathol. 1992;73:171–180. doi: 10.1016/0030-4220(92)90191-r. [DOI] [PubMed] [Google Scholar]
- 29.Schmid J, Odds F C, Wiselka M J, Nicholson K G, Soll D R. Genetic similarity and maintenance of Candida albicans strains from a group of AIDS patients, demonstrated by DNA fingerprinting. J Clin Microbiol. 1992;30:935–941. doi: 10.1128/jcm.30.4.935-941.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schoofs A, Odds F C, Colebunders R, Ieven M, Goosens H. Use of specialised isolation media for recognition and identification of Candida dubliniensis isolates from HIV-infected patients. Eur J Clin Infect Dis. 1997;16:296–300. doi: 10.1007/BF01695634. [DOI] [PubMed] [Google Scholar]
- 31.Sullivan D, Bennett D, Henman M, Harwood P, Flint S, Mulcahy F, Shanley D, Coleman D. Oligonucleotide fingerprinting of isolates of Candida species other than C. albicans and of atypical Candida species from human immunodeficiency virus-positive and AIDS patients. J Clin Microbiol. 1993;31:2124–2133. doi: 10.1128/jcm.31.8.2124-2133.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sullivan, D., and D. Coleman. Unpublished data.
- 33.Sullivan D J, Haynes K, Bille J, Boerlin P, Rodero L, Lloyd S, Henman M, Coleman D. Widespread geographic distribution of oral Candida dubliniensis strains in human immunodeficiency virus-infected individuals. J Clin Microbiol. 1997;35:960–964. doi: 10.1128/jcm.35.4.960-964.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sullivan D J, Henman M C, Moran G P, O’Neill L C, Bennett D E, Shanley D B, Coleman D C. Molecular genetic approaches to identification, epidemiology and taxonomy of non-albicans Candida species. J Med Microbiol. 1996;44:399–408. doi: 10.1099/00222615-44-6-399. [DOI] [PubMed] [Google Scholar]
- 35.Sullivan D J, Westerneng T J, Haynes K A, Bennett D E, Coleman D C. Candida dubliniensis sp. nov.: phenotypic and molecular characterisation of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology. 1995;141:1507–1521. doi: 10.1099/13500872-141-7-1507. [DOI] [PubMed] [Google Scholar]
- 36.Wickes B L, Petter R. Genomic variation in Candida albicans. Curr Top Med Mycol. 1996;7:71–86. [PubMed] [Google Scholar]
- 37.Wingard J R. Importance of Candida species other than C. albicans as pathogens in oncology patients. Clin Infect Dis. 1995;20:115–125. doi: 10.1093/clinids/20.1.115. [DOI] [PubMed] [Google Scholar]