Abstract
We established a rapid method for the identification of influenza A and B virus strains: the peroxidase-antiperoxidase (PAP) staining method with two subtype-specific murine monoclonal antibodies, C179 (H1 and H2 specific) and F49 (H3 specific), and an anti-influenza B virus rabbit polyclonal serum. The types and subtypes of 160 strains were examined, and 158 strains were identified to be the same by the hemagglutination-inhibition (HI) test and the PAP method. In contrast to the results by the HI test, two strains were revealed to be a mixture of two subtypes (H1 and H3) by the PAP method, which was confirmed by plaque cloning. We further analyzed clinical specimens by the PAP method by directly inoculating specimens into Madin-Darby canine kidney cells in microplates. After 40 h of incubation, the types and subtypes of viruses in 52 of 152 specimens were clearly identified. Since the reactivities of the two monoclonal antibodies are not influenced by the antigenic drift of influenza virus, the newly developed method should be applicable not only for rapid diagnosis but also for the epidemiological study of influenza.
In Japan, as in other industrialized countries, influenza is an important infectious disease, every year afflicting large numbers of people, sometimes fatally. Therefore, isolation of the virus from patients with influenza-like illness has been carried out extensively at public health institutes throughout the country. The data thus obtained are useful not only for epidemiological studies but also for developing suitable countermeasures against influenza.
Presently, confirmatory diagnosis of influenza involves isolation of the virus mainly from Madin-Darby canine kidney (MDCK) cells and subsequent identification by the hemagglutination-inhibition (HI) test with standard ferret sera which reacts to influenza A and B viruses in a subtype-specific manner. For virus isolation, inoculated cultures are observed daily until the cytopathic effect appears, which is usually after 1 week. For some specimens which do not show a clear cytopathic effect, a blind passage in the cells is performed. Moreover, if the infected culture fluids do not have enough hemagglutinating activity for the HI test, the viruses must be propagated until they show relatively high hemagglutinin (HA) titers.
Antisera to the HA must be prepared continuously, since antigenic drift of the HA hampers the identification of isolated viruses. To avoid such problems, several investigators have applied monoclonal antibodies which react broadly with a specific type or subtype (1, 3, 4, 9–17). However, no one has ever obtained consistent reactivity with subtype-specific monoclonal antibodies. Recently, we produced a monoclonal antibody, C179, that reacts with all H1 and H2 strains to almost the same degree (7). Moreover, in preliminary experiments, a newly produced monoclonal antibody, F49, was shown to react with all H3 strains studied. Since epidemics of influenza are usually caused by the H1 and H3 subtypes of influenza A and B viruses, rapid detection and identification are expected by the application of the two monoclonal antibodies and anti-type B-specific serum.
Here, we apply the antibodies described above to peroxidase-antiperoxidase (PAP) staining (6) and discuss the usefulness of monoclonal antibodies for the rapid diagnosis of influenza.
MATERIALS AND METHODS
Cells.
MDCK cells were used in all experiments. They were grown in Eagle’s minimal essential medium supplemented with 10% fetal bovine serum.
Viruses.
The influenza viruses used for the characterization of monoclonal antibodies have been described previously (7). A total of 160 strains of influenza virus that had been subtyped by the HI test were used. These consisted of 149 strains isolated in our laboratory (Division of Virology, Osaka Prefectural Institute of Public Health) between 1970 and 1995 and 11 strains isolated in other countries after 1968 (23 H1N1 strains, 114 H3N2 strains, and 23 B strains). Propagation was carried out in MDCK cells or embryonated hen eggs.
Antibodies.
Two monoclonal antibodies, C179 and F49, were obtained by immunizing mice with A/Okuda/57 (H2N2) and A/Aichi/2/68 (H3N2), respectively. A polyclonal antibody against influenza B virus was obtained by immunizing rabbits with B/Nagasaki/1/76.
Typing and subtyping of influenza strains by PAP staining.
The procedures used for virus inoculation and visualization of infected cells by PAP staining were those described previously (6). Briefly, MDCK cells in 96-well flat-bottom plates were inoculated with virus solution in triplet and the plates were incubated for about 16 h at 35°C. The cells were treated with two subtype-specific monoclonal antibodies (C179 and F49) and rabbit anti-mouse immunoglobulin (1:1,000; Organon Teknika, Malvern, Pa.) or B type-specific rabbit serum. The cells were treated successively with goat anti-rabbit immunoglobulin G antibody (1:500; Organon Teknika) and PAP (rabbit antiperoxidase) complex (1:5,000; Organon Teknika). Finally, a peroxidase reaction was conducted by the method described by Graham and Karnovsky (2). The stained cells were observed under an ordinary light microscope.
Rapid detection and identification of influenza viruses in clinical specimens.
Throat washings from patients with influenza-like symptoms were examined by the method described above, with slight modifications. Monolayers of MDCK cells in 24-well microplates were inoculated with the specimens for 45 min at 35°C. Three wells were used for each specimen. After removal of the specimens and washing with phosphate-buffered saline, the cells were covered with Eagle’s minimal essential medium containing 5 μg of trypsin per ml. After incubation for 40 h, the cells were fixed with absolute ethanol and were stained by using each of the three antibodies described above.
HI test.
The isolated influenza viruses were identified by the HI test in which standard ferret sera immunized against each of the influenza A and B viruses were used.
Immunoprecipitation assays.
For the preparation of labeled virus antigens, MDCK cells were infected with A/PR/8/24 (H1N1) or A/Aichi/2/68 (H3N2) and were then incubated for about 20 h in the presence of 35S-methionine (20 μCi/ml). The cells were collected by centrifugation, and the pelleted cells were solubilized with RIPA buffer (0.01 M Tris-HCl [pH 7.4], 0.15 M NaCl, 1% sodium dodecyl sulfate [SDS], 1 mM EDTA). The radiolabeled antigens were mixed with the monoclonal antibodies, and immune complexes were precipitated by using protein G-protein A agarose (Oncogene Science, Uniondale, N.Y.). The precipitated antigens were eluted from the Sepharose with sample buffer containing 2-mercaptoethanol and were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) as described previously (5).
Reverse transcription-PCR.
Viral RNAs were prepared from medium containing cultured infected cells by boiling the medium for 10 min and removing the cell debris by low-speed centrifugation. cDNA synthesis was performed with random primers and reverse transcriptase (Takara) at 42°C for 1 h. The primers used for PCR were designed according to the nucleotide sequences of the A and B regions in the HA gene conserved in a serotype-specific manner (7). For the first and second PCRs of the H1N1 subtype, two primer sets (5′-GGATGGTTACAGGACTAAGGAAC-3′ [sense] and 5′-TTTCTCGATAACAGAATT-3′ [antisense] and 5′-CCATCCATTCAATCCAGAGGT-3′ [sense] and 5′-CACCTTGTTTGTAATCCC-3′ [antisense]) were used, while for the H3N2 subtype two other primer sets (5′-TTGGCAACAGGGATGCGGAAT-3′ [sense] and 5′-CTTCTCGATTAACCAATT-3′ [antisense] and 5′-GTACCAGAAAAACAAACTAG-3′ [sense] and 5′-CAATTTCCCATTGATTTG-3′ [antisense]) were used.
RESULTS
Serological characterization of monoclonal antibodies C179 and F49.
The abilities of monoclonal antibodies C179 and F49 to detect influenza A and B viruses were examined by PAP staining (Table 1). C179, previously reported to recognize the common neutralizing epitope of influenza A virus H1 and H2 strains (7), stained the cells infected with all strains of the H1 and H2 subtypes but not the cells infected with any strain of the H3 subtype or the B type. In contrast, F49 reacted with all strains of the H3 subtype but with none of the strains of the A or B type.
TABLE 1.
Staining titers of monoclonal antibodies (C179 and F49) and anti-B type rabbit serum against influenza A and B viruses
| Virus type and strain | Staining titer
|
||
|---|---|---|---|
| C179 | F49 | Anti-B type | |
| H1N1 | |||
| A/PR/8/34 | 1,638,400 | <400 | <400 |
| A/Bangkok/10/83 | 1,638,400 | <400 | <400 |
| A/Yamagata/120/86 | 409,600 | <400 | <400 |
| A/Osaka/930/88 | 409,600 | <400 | <400 |
| A/Suita/1/89 | 409,600 | <400 | <400 |
| H2N2 | |||
| A/Okuda/57 | 1,638,400 | <400 | <400 |
| A/Adachi/2/57 | 1,638,400 | <400 | <400 |
| A/Kumamoto/1/65 | 409,600 | <400 | <400 |
| A/Kaizuka/2/65 | 409,600 | <400 | <400 |
| A/Izumi/5/65 | 409,600 | <400 | <400 |
| H3N2 | |||
| A/Aichi/2/68 | <400 | 409,600 | <400 |
| A/Fukuoka/C29/85 | <400 | 102,400 | <400 |
| A/Sichuan/2/87 | <400 | 102,400 | <400 |
| A/Ibaraki/1/90 | <400 | 102,400 | <400 |
| A/Suita/1/90 | <400 | 102,400 | <400 |
| B, B/Nagasaki/1/87 | <400 | <400 | 25,600 |
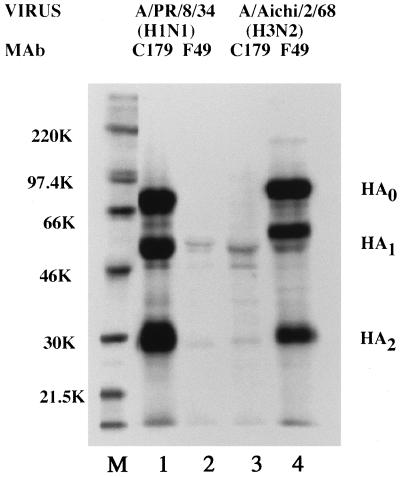
These monoclonal antibodies were further analyzed by an immunoprecipitation assay (Fig. 1). C179 immunoprecipitated the HA protein of A/PR/8/34 (H1N1) but not that of A/Aichi/2/68 (H3N2). On the other hand, F49 reacted only with A/Aichi/2/68 (H3N2). The results confirmed the staining data, that is, that both monoclonal antibodies recognize the HA protein in a subtype-specific manner.
FIG. 1.
SDS-PAGE analysis of influenza virus polypeptides immunoprecipitated by monoclonal antibodies (MAb) against influenza HA proteins. 35S-methionine-labeled influenza virus-infected MDCK cell extracts were immunoprecipitated with monoclonal antibodies C179 (lanes 1 and 3) and F49 (lanes 2 and 4). Lanes 1 and 2, A/PR/8/34-infected cells; lanes 3 and 4, A/Aichi/2/68-infected cells; M, molecular weight markers (K, thousands).
Typing and subtyping of influenza viruses by PAP staining.
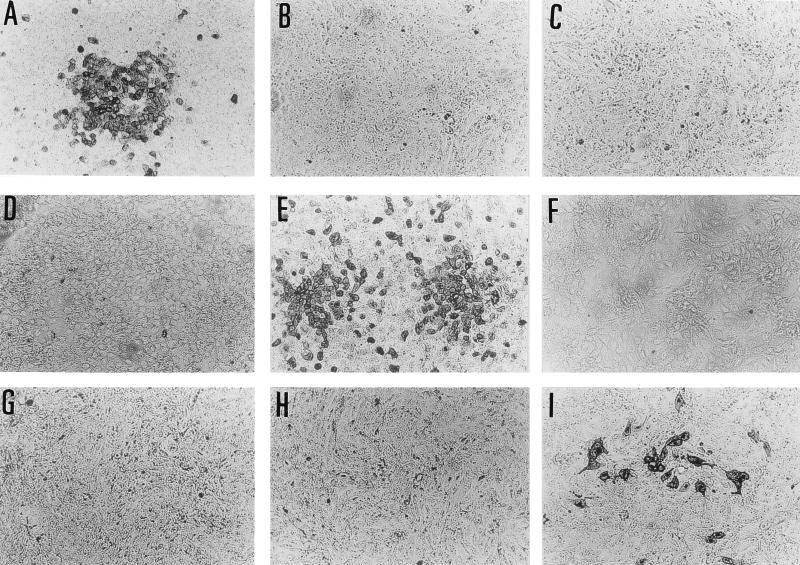
Figure 2 shows that the infected MDCK cells were stained by only one of the three antibodies. Consequently, we could determine the virus type or subtype relatively quickly.
FIG. 2.
The two monoclonal antibodies (C179 and F49) and anti-influenza B virus rabbit serum stained the infected cells in a subtype- or type-specific manner. The MDCK cells in 96-well flat bottom plates were infected with A/Osaka/219/91 (A, B, and C), A/Osaka/174/85 (D, E, and F), and B/Osaka/753/93 (G, H, and I) and were stained with C179 (A, D and G), F49 (B, E and H), and anti-influenza B virus rabbit serum (C, F, and I). The cells infected with A/Osaka/219/91 were stained with C179 but not with F49 or anti-influenza B virus rabbit serum, which confirmed that the strain was H1N1. Similarly, A/Osaka/174/85 and B/Osaka/753/93 were identified to be H3N2 and the B type, respectively.
In this study, 160 strains of influenza virus which had been identified by the HI test were examined by PAP staining. The types or subtypes of all the strains were shown to be identical by both methods, with two exceptions. Two strains (A/Osaka/29/91 and A/Osaka/107/93) which reacted with both C179 and F49 in the staining test were further analyzed by PCR and plaque cloning. The results of PCR agreed with those of the HI test, in which the subtypes of A/Osaka/29/91 and A/Osaka/107/93 were H1N1 and H3N2, respectively. However, further investigation by plaque cloning revealed that both strains are mixtures of two strains, with the ratios of H1N1:H3N2 being as follows: 21:1 for A/Osaka/29/91 and 17:104 for A/Osaka/107/93. Consequently, the HI test and PCR detected only the major population in the virus specimens, while PAP staining accurately detected both major and minor populations.
Detection and typing and subtyping of influenza viruses in clinical specimens by PAP staining.
Throat washings obtained from patients with influenza-like illness were split into two, and virus isolation and PAP staining were performed. We examined 152 specimens obtained at the peaks of the influenza seasons of 1993–1994 and 1994–1995 (Table 2), since relatively high percentages of virus isolation were expected at the peak periods. All positive specimens were identified by both methods to be of the H3N2 subtype, with slightly higher positive ratios by virus detection than by PAP staining in both influenza seasons.
TABLE 2.
Comparison of virus isolation and staining test for detection of influenza virus strains in throat washings
| Epidemic season | Method for detection | No. of specimens | No. of positive specimens | Rate of positivity (%) |
|---|---|---|---|---|
| 1993–1994 | Virus isolation | 72 | 33 | 45.8 |
| Staining test | 72 | 25 | 34.7 | |
| 1994–1995 | Virus isolation | 80 | 34 | 42.5 |
| Staining test | 80 | 27 | 33.8 |
DISCUSSION
Monoclonal antibodies have been extensively applied to the rapid detection and identification of influenza viruses (1, 3, 4, 9–17). By using monoclonal antibodies against the nucleoprotein or matrix protein of influenza virus, type A viruses were clearly distinguishable from type B viruses (1, 10, 14–17). However, to discriminate between the H1 and H3 subtypes of influenza A virus, monoclonal antibodies against HA which determines subtype specificity are needed. Since the HA manifests frequent antigenic drift, HA specific monoclonal antibodies do not show constant reactivity against strains collected over a long period of time. Therefore, pools of monoclonal antibodies against the HA have been applied (13).
We formerly reported on a monoclonal antibody, C179, which reacts to all H1 and H2 subtype strains to almost the same degree (7). Here, we report on another monoclonal antibody, F49, which reacts only to H3-subtype strains. The reactogenicities of C179 and F49 in the staining test suggested their applicability for the subtyping of influenza A virus strains (Table 1). The results of the immunoprecipitation assays suggest that the two antibodies recognize the conserved epitope in the HA of each subtype (Fig. 1).
It has been reported that the amino acid substitutions in the HA occur mainly in the globular head region (18, 19). On the other hand, conserved amino acid sequences were observed in the HA2, which constitutes the stem region of the HA (18, 19). These observations suggest that C179 recognizes the middle of the stem region conformationally (7). Moreover, in preliminary experiments we confirmed that F49 recognizes an epitope in the stem region. In those experiments CV-1 cells transfected with a vector containing a gene encoding headless HA of the H3N2 subtype were stained by F49 (data not shown). The fact that F49 does not have neutralizing activity suggests that it recognizes a sequential epitope of the HA.
The usefulness of PAP staining for the subtyping of clinical isolates was clearly shown in the present study, which compared the PAP staining technique with the conventional HI test. With two exceptions, the types or subtypes of the strains were identical in both tests. This further supports our supposition that epitopes recognized by the two monoclonal antibodies have been conserved in nature. It is noteworthy that mixed infections with the H1N1 and H3N2 subtypes were detectable only by staining and not by the HI test or PCR. Thus, PAP staining may be more sensitive and reliable than the other methods, and the next step is to conduct detailed studies with fresh clinical specimens.
In the season when influenza is epidemic, which is usually from December to March, many specimens from the monitoring hospitals in Osaka Prefecture are sent to our institute (Osaka Prefectural Institute of Public Health). The clinical specimens described in Table 2 were obtained at the peak of the influenza season, and that explains the relatively high virus isolation rates in both seasons. The virus isolation rates throughout the year for various years are as follows: 11.0% (number of specimens positive for virus/total number of specimens = 70/636) in 1993, 36.0% (561/1,559) in 1994, and 15.8% (153/966) in 1995. The virus detection rate by the staining test was slightly lower than that by virus isolation. To evaluate the detection rates by the two tests for significance, trials with large numbers of clinical specimens are now under consideration. However, a detection time of as short as 40 h is considerable, especially at the beginning of the influenza season, when rapid detection and typing of the influenza virus are needed. Similarly, when mass outbreaks of influenza-like illness occur in schools or local communities, virological examinations are vital. A direct detection method which could identify influenza viruses in clinical specimens within 2 days would meet public health requirements.
When the influenza A virus H2 strains recur in the future, it becomes impossible to distinguish H1 strains from H2 strains with C179. However, the similarity of the DNA sequences of the genes for the H1 and H2 stem regions is so high (7) that it is hard to raise monoclonal antibodies which recognize H1 stem regions but not H2 stem regions by immunizing mice with viral proteins. Therefore, we are now planning to raise H1 strain-specific monoclonal antibodies by immunizing mice with the DNA coding for the stem region of H1 · HA. On the other hand, we have produced monoclonal antibodies against influenza type B virus which react to all type B strains. Applications for anti-B monoclonal antibodies in newly developed methods are under investigation, and it should facilitate the clearer detection and identification of viruses in clinical specimens.
ACKNOWLEDGMENTS
This work was supported by grants from the Ministry of Education, Japan, and from Daido Insurance Company, Osaka, Japan.
REFERENCES
- 1.Espy M J, Smith T F, Harmon M W, Kendal A P. Rapid detection of influenza virus by shell vial assay with monoclonal antibodies. J Clin Microbiol. 1986;24:677–679. doi: 10.1128/jcm.24.4.677-679.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graham R C, Karnovsky M J. The early stages of adsorption of injected horseradish peroxidase in the proximal tubules of monkey kidney: ultrastructural cytochemistry by a new technique. J Histochem Cytochem. 1966;14:291–302. doi: 10.1177/14.4.291. [DOI] [PubMed] [Google Scholar]
- 3.McQuillin J, Madeley C R, Kendal A P. Monoclonal antibodies for the rapid diagnosis of influenza A and B virus infections by immunofluorescence. Lancet. 1985;ii:911–914. doi: 10.1016/s0140-6736(85)90849-9. [DOI] [PubMed] [Google Scholar]
- 4.Mills R D, Cain K J, Woods F L. Detection of influenza virus by centrifugal inoculation of MDCK cells and staining with monoclonal antibodies. J Clin Microbiol. 1989;27:2505–2508. doi: 10.1128/jcm.27.11.2505-2508.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakagawa N, Mukai T, Sakamoto J, Hata A, Okuno T, Takeda K, Yamanishi K. Antigenic analysis of human herpesvirus 7 (HHV-7) and HHV-6 using immune sera and monoclonal antibodies against HHV-7. J Gen Virol. 1997;78:1131–1137. doi: 10.1099/0022-1317-78-5-1131. [DOI] [PubMed] [Google Scholar]
- 6.Okuno Y, Tanaka K, Baba K, Maeda A, Kunita N, Ueda S. Rapid focus reduction neutralization test of influenza A and B viruses in microtiter system. J Clin Microbiol. 1990;28:1308–1313. doi: 10.1128/jcm.28.6.1308-1313.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okuno Y, Isegawa Y, Sasao F, Ueda S. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J Virol. 1993;67:2552–2558. doi: 10.1128/jvi.67.5.2552-2558.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sagawa H, Ohshima A, Kato I, Okuno Y, Isegawa Y. The immunological activity of a deletion mutant of influenza virus haemagglutinin lacking the globular region. J Gen Virol. 1996;77:1483–1487. doi: 10.1099/0022-1317-77-7-1483. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt N J, Ota M, Gallo D, Fox V L. Monoclonal antibodies for rapid, strain-specific identification of influenza virus isolates. J Clin Microbiol. 1982;16:763–765. doi: 10.1128/jcm.16.4.763-765.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shalit I, Mckee P A, Beauchamp H, Waner J L. Comparison of polyclonal antiserum versus monoclonal antibodies for the rapid diagnosis of influenza A virus infection by immunofluorescence in clinical specimens. J Clin Microbiol. 1985;22:877–879. doi: 10.1128/jcm.22.5.877-879.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stokes C E, Bernstein J M, Kyger S A, Hayden F G. Rapid diagnosis of influenza A and B by 24-h fluorescent focus assays. J Clin Microbiol. 1988;26:1263–1266. doi: 10.1128/jcm.26.7.1263-1266.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swenson P D, Kaplan M H. Rapid detection of influenza virus in cell culture by indirect immunoperoxidase staining with type-specific monoclonal antibodies. Diagn Microbiol Infect Dis. 1987;7:265–268. doi: 10.1016/0732-8893(87)90142-8. [DOI] [PubMed] [Google Scholar]
- 13.Tkác̆ová M, Varec̆ková E, Baker I C, Love J M, Ziegler T. Evaluation of monoclonal antibodies for subtyping of currently circulating human type A influenza viruses. J Clin Microbiol. 1997;35:1196–1198. doi: 10.1128/jcm.35.5.1196-1198.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walls H H, Harmon M W, Slagle J J, Stocksdale C, Kendal A P. Characterization and evaluation of monoclonal antibodies developed for typing influenza A and B viruses. J Clin Microbiol. 1986;23:240–245. doi: 10.1128/jcm.23.2.240-245.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walls H H, Johansson K H, Harmon M W, Halonen P E, Kendal A P. Time-resolved fluoroimmunoassay with monoclonal antibodies for rapid diagnosis of influenza infections. J Clin Microbiol. 1986;24:907–912. doi: 10.1128/jcm.24.6.907-912.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waner J L, Todd S J, Shalaby H, Murphy P, Wall L V. Comparison of directigen FLU-A with viral isolation and direct immunofluorescence for the rapid detection and identification of influenza A virus. J Clin Microbiol. 1991;29:479–482. doi: 10.1128/jcm.29.3.479-482.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waris M, Ziegler T, Kivivirta M, Ruuskanen O. Rapid detection of respiratory syncytial virus and influenza A virus in cell culture by immunoperoxidase staining with monoclonal antibodies. J Clin Microbiol. 1990;28:1159–1162. doi: 10.1128/jcm.28.6.1159-1162.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiley D C, Skehel J J. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu Rev Biochem. 1987;56:365–394. doi: 10.1146/annurev.bi.56.070187.002053. [DOI] [PubMed] [Google Scholar]
- 19.Wiley D C, Wilson I A, Skehel J J. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature (London) 1981;289:373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]