Abstract
This review explores the emerging role of hydrogen sulfide (H2S) in modulating epigenetic mechanisms involved in neurodegenerative diseases. Accumulating evidence has begun to elucidate the multifaceted ways in which H2S influences the epigenetic landscape and, subsequently, the progression of various neurodegenerative disorders, including Alzheimer’s, Parkinson’s, and Huntington’s disease. H2S can modulate key components of the epigenetic machinery, such as DNA methylation, histone modifications, and non-coding RNAs, impacting gene expression and cellular functions relevant to neuronal survival, inflammation, and synaptic plasticity. We synthesize recent research that positions H2S as an essential player within this intricate network, with the potential to open new therapeutic avenues for these currently incurable conditions. Despite significant progress, there remains a considerable gap in our understanding of the precise molecular mechanisms and the potential therapeutic implications of modulating H2S levels or its downstream targets. We conclude by identifying future directions for research aimed at exploiting the therapeutic potential of H2S in neurodegenerative diseases.
Keywords: hydrogen sulfide, epigenetic regulation, neurodegenerative diseases, DNA methylation, histone modifications, non-coding RNAs, Alzheimer’s disease, Parkinson’s disease, Huntington’s disease
1. Introduction
Neurodegenerative diseases, including Alzheimer’s, Parkinson’s, and Huntington’s, pose a formidable challenge to the modern medicine [1,2]. These conditions are characterized by the relentless and irreversible loss of neurons, leading to a gradual decline in cognitive and motor functions [3]. They have become a significant subset of non-communicable diseases, exacerbated by our longer human lifespan [4], impacting the lives of millions of individuals worldwide. Not only do these diseases cause emotional distress, but they also impose substantial economic burdens on society [5].
Delving into the biology of these complex conditions reveals an intricate web of causative factors involving a complex interplay of genetic, epigenetic, and environmental influences that collectively drive disease onset and progression [6,7,8,9]. Among these factors, epigenetic changes have emerged as critical determinants in the development and course of neurodegenerative diseases [10,11,12,13,14].
Epigenetic mechanisms play a fundamental role in gene regulation by facilitating dynamic changes in gene activity without altering the underlying DNA sequence [15,16,17,18,19,20,21,22,23,24]. Epigenetics includes DNA methylation, histone modification, and the non-coding RNAs [25,26,27,28,29,30].
Beyond their biological significance, epigenetic changes also offer an intriguing evolutionary perspective [31,32,33]. Modifying gene activity in response to environmental cues without altering the DNA sequence provides organisms with a remarkable evolutionary advantage [34,35,36,37]. It has likely allowed living organisms to adapt and thrive in diverse environments [38,39,40,41,42].
Despite its notorious association with the smell of rotten eggs and its potential for toxicity in high concentrations, H2S has emerged as a molecule of interest in studying many physiological and pathological processes [43,44,45,46,47,48,49,50,51]. H2S is endogenously produced in the brain through the enzymatic breakdown of cysteine by cystathionine β-synthase (CBS), cystathionine γ-lyase (CSE), and 3-mercaptopyruvate sulfurtransferase (3-MST) [52]. It acts as a neuromodulator and has been shown to play a pivotal role in regulating synaptic transmission, neuronal survival, and neuroinflammation [53,54].
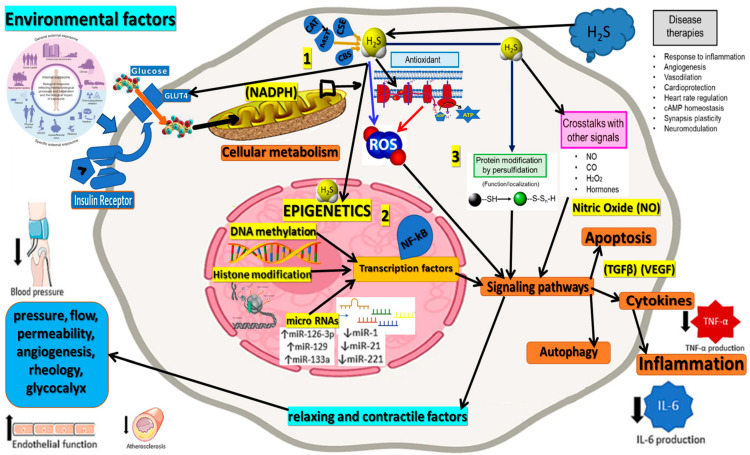
Recent evidence suggests that H2S could interact with the various epigenetic mechanisms involved in neurodegenerative diseases [55,56,57,58,59,60]. By influencing epigenetic changes, H2S could impact gene expression patterns relevant to these diseases. This potential interaction between H2S and the epigenetic landscape provides a fresh perspective into our understanding of these complex conditions and highlights the need for further research into the role and mechanisms of H2S in these diseases [61,62,63,64,65]. Therefore, this review aims to provide a comprehensive overview of the current research exploring the role of H2S in regulating epigenetic processes associated with neurodegenerative diseases (Figure 1).
Figure 1.
This figure illustrates the central role of Hydrogen Sulfide (H2S) in epigenetic regulation and its interactions with various processes associated with neurodegenerative diseases. H2S affects epigenetic mechanisms, including histone modification, DNA methylation, and non-coding RNAs, which modulate gene expression and cellular functions relevant to neurodegeneration. H2S influences reactive oxygen species (ROS) production and oxidative stress levels, which play a critical role in the pathogenesis of neurodegenerative disorders. By reducing ROS production (blue arrow) and inhibiting ROS-generating processes at the mitochondrial level (red arrows), H2S impacts a broad spectrum of biological functions, as depicted in this figure for illustrative purposes. The interplay between H2S, epigenetic processes, and oxidative stress offers valuable insights into the molecular mechanisms underlying neurodegeneration and highlights the potential for therapeutic interventions to restore epigenetic balance and mitigate oxidative stress to combat neurodegenerative diseases effectively. The figure corroborates 3 major parts: 1. external sources and internal production of H2S; 2. the epigenetic role of H2S; and 3. the physiological connections presented in a simplified way.
One of the key epigenetic mechanisms regulated by H2S is histone modification. H2S has been shown to modify histone proteins through sulfhydration, a process by which a sulfur atom is added to specific cysteine residues of histones [45]. Sulfhydration of histones can modulate chromatin structure and gene expression, ultimately influencing various cellular processes in neurons [66]. For instance, H2S-mediated sulfhydration of histones has been reported to affect the expression of genes involved in synaptic plasticity, memory formation, and neuronal survival. Dysregulation of this process has been implicated in the pathogenesis of neurodegenerative diseases (NDs) [67], including Alzheimer’s disease (AD) [68], Parkinson’s disease (PD), and Huntington’s disease (HD).
DNA methylation is another critical epigenetic mechanism influenced by H2S [69]. DNA methylation involves the addition of a methyl group to cytosine residues in CpG dinucleotides, leading to transcriptional repression of target genes. H2S has been shown to regulate the activity of DNA methyltransferases (DNMTs), the enzymes responsible for DNA methylation. Changes in DNMT activity due to H2S dysregulation have been associated with altered DNA methylation patterns in neurons, contributing to the aberrant gene expression observed in NDs. For instance, H2S-mediated changes in DNA methylation have been linked to the dysregulation of genes involved in neuroinflammation, oxidative stress, and neuronal survival [70,71,72,73].
In addition to histone modification and DNA methylation, H2S interacts with non-coding RNAs, including microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) [74]. miRNAs are small non-coding RNAs that post-transcriptionally regulate gene expression by targeting mRNAs for degradation or translational repression. The dysregulation of miRNAs has been implicated in various aspects of neurodegeneration, including protein aggregation, neuroinflammation, and synaptic dysfunction. H2S has been shown to modulate the expression and activity of specific miRNAs, leading to altered gene expression profiles in neurons [75,76].
Furthermore, lncRNAs, a class of non-coding RNAs longer than 200 nucleotides, have also been found to play crucial roles in neurodegenerative processes. H2S can influence the expression and function of lncRNAs, thereby affecting gene expression and cellular processes in neurons. Dysregulation of specific lncRNAs has been associated with NDs, and their interaction with H2S further highlights the significance of epigenetic regulation in the neurodegeneration [77,78].
2. Methods
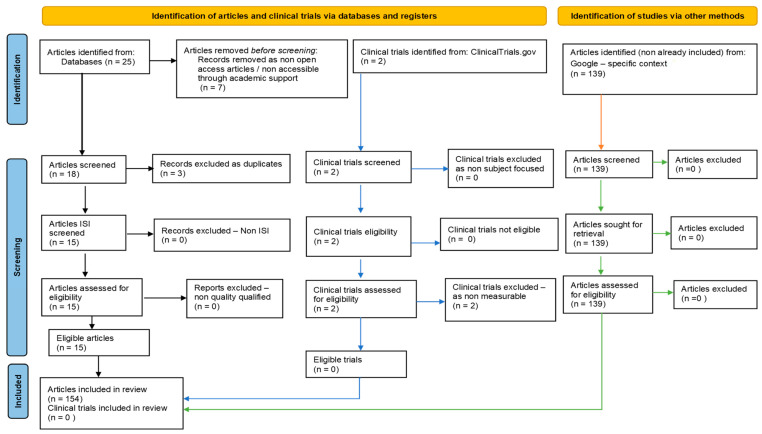
Our systematic review followed PRISMA guidelines and registered on PROSPERO ID: 449843. To ensure comprehensive coverage of the relevant literature, we searched multiple databases, including PubMed, Scopus/Elsevier, Web of Science, and Google Search. The search strategy involved the use of specific keywords and medical subject headings (MeSH) related to hydrogen sulfide, epigenetic regulation (DNA methylation, histone modifications, non-coding RNAs), and neurodegenerative diseases (such as Alzheimer’s, Parkinson’s, and Huntington’s diseases). After removing non-eligible and duplicate references, our review included 115 relevant studies. In addition to the databases mentioned above, we also searched https://clinicaltrials.gov/ (accessed on 1 June 2023) to identify potential clinical trials related to our topic (Figure 2).
Figure 2.
Adapted PRISMA flow diagram, customized for our study.
3. Results and Discussion
3.1. Hydrogen Sulfide and Neurodegenerative Diseases: An Overview
Hydrogen sulfide (H2S) is a multifaceted gasotransmitter, a gas molecule that occurs naturally in organisms and has been recognized for its diverse roles in physiological and pathological processes [79,80,81]. Despite its association with the unpleasant smell of rotten eggs, H2S has garnered increasing attention in biology [48,82]. H2S has been found to influence neuroinflammation, oxidative stress, and mitochondrial dysfunction, all of which are implicated in the pathogenesis of neurodegenerative disorders. Its significance in neurodegeneration is an intriguing area of investigation, with the potential to reveal new therapeutic strategies [54,83,84]. By contextualizing the study of H2S within the framework of evolution and epigenetics, we obtain a deeper understanding of its complex physio-pathology [45,85,86,87].
This scientific exploration highlights the importance of interdisciplinary research, bridging evolutionary biology and molecular medicine, and may pave the way for improved treatments and better quality of life for individuals impacted by neurodegenerative disorders and potentially other health conditions.
In the central nervous system (CNS), H2S regulates vasodilation, protects against oxidative stress-induced damage, and modulates inflammatory responses, vital for maintaining neuronal health and adequate brain function [88,89]. H2S also influences proper immune system functioning, as it acts as an anti-inflammatory agent, dampening excessive inflammation and promoting immune balance [90]. However, it is essential to tightly control H2S levels, as high concentrations can become toxic, leading to cellular damage and death [54,91]. Enzymes such as CBS, CSE, and 3-MST regulate H2S levels to prevent harmful accumulation while allowing for its beneficial signaling functions [84,92]. The study of H2S in biology is placed within the context of evolution, suggesting that its role in cellular processes has likely evolved to help organisms adapt to changing environments and cope with environmental challenges, thus influencing gene expression and cellular functions in various ways [32,93,94].
In Alzheimer’s disease (AD), H2S has been shown to modulate the activity of enzymes involved in amyloid-beta (Aβ) production and tau protein phosphorylation, critical processes in AD pathogenesis [54,95]. Specifically, H2S can promote the production of Aβ through its influence on the enzyme beta-secretase (BACE1) and the gamma-secretase complex [96,97]. Additionally, H2S has been shown to induce tau phosphorylation, leading to the aggregation of hyperphosphorylated tau into neurofibrillary tangles [98]. Furthermore, H2S can contribute to neuroinflammation and oxidative stress, exacerbating neurodegeneration in AD [99].
In Parkinson’s disease (PD), the aggregation of alpha-synuclein protein into Lewy bodies is a central feature of the disease [100]. H2S has been implicated in the assembly and misfolding of alpha-synuclein, promoting its neurotoxicity and contributing to the progression of PD. Moreover, H2S can affect mitochondrial function and induce oxidative stress, both associated with dopaminergic neuronal death in PD [101,102,103]. H2S-induced inflammation and microglial activation may also affect disease pathogenesis [83,103,104].
In Huntington’s disease (HD), accumulating mutant huntingtin protein with an expanded polyglutamine repeat is critical in the disease process [105]. H2S has been shown to influence the aggregation and toxicity of mutant huntingtin, contributing to the degeneration of neurons in the striatum and other brain regions affected in HD. Additionally, H2S can exacerbate mitochondrial dysfunction and oxidative stress, further contributing to neuronal damage in HD [106,107].
Emerging research also suggests that H2S may be involved in the dysregulation of autophagy, a cellular process crucial for removing misfolded proteins and damaged organelles. Dysfunctional autophagy has been implicated in the pathogenesis of neurodegenerative diseases, and H2S may contribute to autophagic impairments in these conditions [108,109,110,111]. Although the precise mechanisms by which H2S exerts its effects in neurodegenerative diseases are still under investigation, the emerging evidence highlights its potential as a promising therapeutic target.
3.2. Epigenetic Regulation in Neurodegenerative Diseases
Epigenetic regulation plays a pivotal role in the pathogenesis of neurodegenerative diseases, influencing gene expression and cellular functions relevant to neuronal health [112]. Epigenetics refers to modifications that occur on the genome without altering the underlying DNA sequence, and these changes can be inherited or influenced by environmental factors. In neurodegenerative diseases, dysregulation of epigenetic mechanisms has been implicated in disrupting normal cellular processes and the progressive loss of neurons [16,113].
Chromatin remodeling is a fundamental epigenetic mechanism that regulates gene expression by modifying the chromatin structure, comprising DNA and histone proteins [114]. ATP-dependent chromatin remodeling complexes can lead to the misregulation of crucial genes involved in neuronal survival and function [115]. Moreover, histone modifications, such as acetylation and methylation, dynamically regulate gene expression in neurons, and their perturbations have been observed in various neurodegenerative conditions [116]. These epigenetic alterations in chromatin remodeling can impact the expression of genes associated with disease pathology, highlighting the significance of chromatin remodeling in the neurodegeneration [117]. Targeting chromatin remodeling factors may hold promise for developing epigenetic-based therapies to counteract neurodegenerative disease progression and promote neuroprotection. Further research is needed to elucidate the precise molecular mechanisms and potential therapeutic implications of chromatin remodeling in neurodegenerative diseases [28].
Three primary epigenetic mechanisms are particularly relevant to neurodegeneration: DNA methylation; histone modification; and non-coding RNAs [25].
3.2.1. DNA Methylation
DNA methylation involves adding a methyl group to specific cytosine residues in the DNA sequence, typically occurring at CpG sites (Cytosine-phosphate-Guanine) [118,119,120]. Methylation of CpG islands in the promoter regions of genes is associated with gene silencing, leading to reduced gene expression. In neurodegenerative diseases, aberrant DNA methylation patterns have been observed in genes that play crucial roles in neuronal function, such as synaptic plasticity, neuroinflammation, and oxidative stress response. These changes in DNA methylation can impact the expression of genes linked to disease pathogenesis, contributing to the dysfunction and death of neurons [120].
Emerging research has shed light on the dynamic nature of DNA methylation in neurodegenerative disorders and its impact on disease progression [121]. For instance, studies have shown altered DNA methylation patterns in genes associated with amyloid-beta processing and tau phosphorylation in Alzheimer’s disease (AD) [122]. Similarly, in Parkinson’s disease (PD) [123], dysregulated DNA methylation has been observed in genes linked to mitochondrial function, dopamine signaling, and neuroinflammation [124].
Moreover, DNA methylation changes have been implicated in regulating genes involved in response to oxidative stress, a process closely linked to neurodegeneration. Oxidative stress-induced DNA methylation alterations can affect the expression of antioxidant defense genes, exacerbating neuronal vulnerability to oxidative damage [125].
Advancements in epigenomic technologies, such as genome-wide DNA methylation profiling, have provided valuable insights into the specific genes and pathways affected by DNA methylation changes in neurodegenerative diseases [119].
3.2.2. Histone Modification
Histones are proteins around which DNA is wrapped to form chromatin, the complex structure that packages DNA within the cell nucleus. Histone modifications, such as acetylation, methylation, phosphorylation, and ubiquitination, can alter the accessibility of DNA to transcriptional machinery, affecting gene expression. Dysregulation of histone modifications has been involved in neurodegenerative diseases, leading to altered gene expression patterns that might contribute to disease progression. For example, histone deacetylases (HDACs), enzymes involved in histone deacetylation, have been shown to regulate gene expression in Alzheimer’s and Huntington’s disease [126].
In Alzheimer’s disease (AD), perturbations in histone acetylation and deacetylation processes have been linked to disease pathology. Histone deacetylases (HDACs), a class of enzymes responsible for histone deacetylation, play a crucial role in regulating gene expression in AD [117]. Studies have shown that dysregulation of specific HDACs, such as HDAC2, is associated with synaptic dysfunction and cognitive impairment in AD. Additionally, histone acetyltransferases (HATs), the enzymes responsible for histone acetylation, have been implicated in AD pathogenesis. HATs are involved in the acetylation of histones, leading to a relaxed chromatin structure and increased gene transcription. Notably, dysregulation of HATs may contribute to the altered expression of genes involved in neuroinflammation and amyloid-beta processing [116].
Moreover, histone modifications have been linked to other neurodegenerative diseases, such as Parkinson’s disease (PD) [127]. In PD, altered histone acetylation levels have been associated with mitochondrial dysfunction and oxidative stress, contributing to dopaminergic neuronal degeneration [128]. Additionally, histone methylation patterns have been reported to regulate alpha-synuclein expression, a protein implicated in PD pathology [60].
In Huntington’s disease (HD), an inherited neurodegenerative disorder, histone modifications have also been implicated in disease pathophysiology. For instance, aberrant histone methylation patterns have been observed in HD, leading to changes in gene expression associated with neuronal dysfunction. Furthermore, HDAC inhibitors have shown promising effects in preclinical models of HD, indicating the therapeutic potential of targeting histone modifications in this disorder [129].
3.2.3. Non-Coding RNAs
Non-coding RNAs (ncRNAs) are RNA molecules that do not code for proteins but have regulatory functions in the cell. Two major types of ncRNAs involved in epigenetic regulation are microRNAs (miRNAs) and long non-coding RNAs (lncRNAs).
MiRNAs are small RNA molecules that can bind to target messenger RNAs (mRNAs), leading to mRNA degradation or translational repression. The dysregulation of miRNAs has been linked to neurodegenerative diseases. Aberrant expression of specific miRNAs can disrupt key pathways related to neuroinflammation, synaptic plasticity, and mitochondrial function, contributing to the pathogenesis of neurodegenerative diseases.
LncRNAs, on the other hand, are a diverse group of transcripts that are longer than 200 nucleotides and do not encode proteins [130]. LncRNAs can interact with chromatin-modifying complexes, influencing gene expression by epigenetic mechanisms. Altered expression of lncRNAs has been associated with neurodegenerative disorders, contributing to the dysregulation of gene expression and cellular functions [131,132,133,134].
Accumulating evidence has revealed the pivotal roles of lncRNAs in epigenetic regulation, acting as scaffolds for chromatin-modifying complexes or interacting with various epigenetic regulators to influence gene expression [135].
In neurodegenerative diseases, altered expression of lncRNAs has been associated with dysregulated gene expression patterns and cellular dysfunctions [136]. For instance, some lncRNAs have been found to interact with histone-modifying enzymes, such as histone methyltransferases or demethylases, leading to changes in histone methylation patterns and subsequent transcriptional alterations [25].
Additionally, lncRNAs can function as competing endogenous RNAs (ceRNAs) by competitively binding to miRNAs, thereby modulating the availability of miRNAs for target mRNAs. This ceRNA crosstalk may play a critical role in fine-tuning gene expression networks in the context of neurodegeneration [137].
Dysregulated lncRNA-miRNA interactions have been reported in neurodegenerative diseases, and their effects on target gene expression may impact pathways involved in neuronal survival, neuroinflammation, and protein aggregation [138].
Moreover, recent studies have highlighted the involvement of circular RNAs (circRNAs) in neurodegenerative diseases. CircRNAs are a unique class of ncRNAs with covalently closed circular structures. They have been shown to regulate gene expression by interacting with miRNAs or RNA-binding proteins, and their dysregulation has been implicated in the pathogenesis of neurodegenerative disorders [139].
The dynamic nature of epigenetic modifications presents opportunities for therapeutic interventions, as these changes are potentially reversible. Targeting epigenetic mechanisms holds promise for developing novel therapies to modify disease progression and improve the outcomes for individuals affected by neurodegenerative diseases. However, a comprehensive understanding of the specific epigenetic changes and their functional consequences in different neurodegenerative disorders remains an area of active research. Unraveling the complexities of epigenetic regulation in these diseases may lead to identifying biomarkers and novel therapeutic targets, ultimately providing hope for more effective treatments in the future [121,140,141].
Given the complex nature of neurodegenerative diseases, including Alzheimer’s, Parkinson’s, and Huntington’s, it is evident that epigenetic regulation plays a significant role in disease progression. The intricate interplay between genetic, environmental, and epigenetic factors contributes to the loss of neurons observed in these disorders.
H2S has been found to interact with various epigenetic mechanisms, influencing gene expression and cellular functions relevant to neuronal health. By interacting with key epigenetic regulators such as DNA methylation, histone modifications, and non-coding RNAs, H2S can influence the expression of genes crucial for neuronal function and survival [8,142,143].
3.3. Hydrogen Sulfide and DNA Methylation
The modulation of DNA methylation by hydrogen sulfide (H2S) represents a fascinating interplay between this gasotransmitter and epigenetic regulation in the context of neurodegenerative diseases. Studies have revealed that H2S can modulate DNA methylation patterns by affecting the activity of enzymes involved in DNA methylation, such as DNA methyltransferases (DNMTs). For instance, H2S has been shown to inhibit DNMT activity, resulting in decreased DNA methylation at specific gene promoter regions. This reduced methylation can lead to altered gene expression, potentially impacting pathways crucial to neuronal survival, neuroinflammation, and oxidative stress response. Moreover, H2S has been found to influence the expression of genes’ expression in regulating H2S metabolism, creating a feedback loop that further impacts the epigenetic landscape. This intricate interplay between H2S and DNA methylation highlights the potential importance of epigenetic mechanisms in the pathogenesis of neurodegenerative disorders, offering new avenues for therapeutic interventions targeting H2S-mediated epigenetic dysregulation. Further research in this area may unveil the full extent of H2S’s role in shaping the epigenetic landscape and its implications for neurodegenerative disease progression and potential treatment strategies [51,144,145,146,147,148] (Table 1).
Table 1.
Data regarding Hydrogen Sulfide and DNA Methylation in Neurodegenerative diseases.
| Ref. | Title | Pathology | Extracted Data Regarding Epigenetics | Major Outcome |
|---|---|---|---|---|
| [51] | Therapeutic importance of hydrogen sulfide in age-associated neurodegenerative diseases |
Neurodegenerative diseases |
H2S, a crucial signaling molecule, regulates DNA methylation, an essential epigenetic modification impacting gene expression and cellular function. H2S influences DNA methylation in oxidative stress and aging conditions, safeguarding against DNA damage and preserving genomic integrity. | H2S modulates DNA methyltransferases. |
| [144] | Abnormal Homocysteine Metabolism: An Insight of Alzheimer’s Disease from DNA Methylation |
Alzheimer’s disease | DNA methylation involves adding methyl groups to cytosine-phosphate-guanine (CpG) sequences catalyzed by DNMT enzymes. DNMT1 maintains existing methylation during cell division, while DNMT3a and DNMT3b create new methylation patterns on unmethylated DNA strands. H2S interference with these processes opens the potential for novel therapeutic strategies against neurodegenerative diseases. | In Alzheimer’s disease (AD), changes in DNA methylation impact the production of amyloid-beta (Aβ) plaques and tau hyperphosphorylation, key factors in AD pathology. |
| [145] | Hydrogen sulfide signalling in the CNS—Comparison with NO | Schizophrenia | In C3H mice, DNA methylation levels at the MPST gene were significantly increased and positively correlated with MPST expression. In schizophrenia patients, MPST levels were positively associated with symptom severity scores. In MPST-transgenic mice, genes related to energy formation were downregulated, and mitochondrial energy metabolism was impaired. | H2S involvement in DNA methylation regulation of the MPST gene in schizophrenia may shed light on the molecular basis of energy metabolism dysregulation in the CNS. |
| [146] | Hydrogen Sulfide Improves Angiogenesis by Regulating the Transcription of pri-miR-126 in Diabetic Endothelial Cells |
Parkinson’s disease | DNA methylation, an essential epigenetic modification, regulates gene expression. In diabetic mice, DNMT1 overexpression reduces miR-126-3p levels, impairing endothelial cell function and blood flow recovery. Exogenous H2S reverses these effects by downregulating DNMT1 expression, enhancing miR-126-3p levels, and promoting angiogenesis. | The interplay between H2S and DNA methylation in the regulation of miR-126-3p expression. |
| [147] | Hydrogen sulfide in ageing, longevity, and disease | Alzheimer’s disease Parkinson’s disease |
Metformin interacts with H2S signaling and activates AMPK, inhibiting mTOR and IIS signaling pathways. It removes homocysteine-stimulated hypermethylation of the CSE promotor region, resulting in increased CSE expression and H2S production. | Metformin’s ability to increase H2S levels is linked to its role in remodeling DNA methylation patterns. |
| [148] | Cell Rearrangement and Oxidant/Antioxidant Imbalance in Huntington’s Disease |
Huntington’s disease | In HD, the toxic protein mHtt can interfere with the transcriptional machinery, altering histone modifications and DNA methylation and impairing gene expression and neuronal dysfunction. | HD is linked to accelerated epigenetic aging. Epigenetic clocks show a correlation between HD progression and epigenetic age. |
3.4. Hydrogen Sulfide and Histone Modifications
H2S exerts its effects on histones through interactions with histone-modifying enzymes, affecting histones’ acetylation, methylation, and phosphorylation. By influencing these histone modifications, H2S can modulate the accessibility of DNA to transcriptional machinery, leading to changes in gene expression patterns. Notably, H2S has been shown to impact the activity of histone acetyltransferases (HATs) and histone deacetylases (HDACs), enzymes involved in histone acetylation, which play crucial roles in regulating gene expression in neurodegenerative diseases. Dysregulation of histone modifications by H2S may contribute to altered expression of genes linked to neuroinflammation, neuroprotection, and other processes involved in neurodegeneration. Further investigations into the specific molecular interactions between H2S and histone-modifying enzymes will be crucial for unraveling the complex mechanisms underlying H2S-mediated epigenetic regulation in the context of neurodegeneration, potentially leading to the development of novel epigenetic-based interventions for neurodegenerative diseases [45,84,149,150,151,152] (Table 2).
Table 2.
Data regarding Hydrogen Sulfide and Histone Modifications in Neurodegenerative Diseases.
| Ref. | Title | Pathology | Extracted Data Regarding Epigenetics | Major Outcome |
|---|---|---|---|---|
| [45] | Hydrogen sulfide-induced post-translational modification as a potential drug target |
Neurodegenerative disease, Alzheimer’s, Parkinson’s, Huntington’s diseases |
H2S affects histones. S-sulfhydration of histones can regulate gene expression and epigenetic modifications. H2S-mediated S-sulfhydration of histone modifiers, such as Sirt1, affects aging, metabolism, and oxidative stress tolerance. | H2S-S-sulfhydration histone modifiers, such as Sirt1, influences aging, metabolism, and oxidative stress. |
| [84] | Exploring mitochondrial hydrogen sulfide signalling for therapeutic interventions in vascular diseases |
Neurodegenerative diseases, Parkinson’s disease |
H2S administration has been found to increase antioxidant proteins such as Trx-1 through the Nrf-2 pathway, leading to cardioprotection in ischemia-induced heart failure. H2S also regulates members of the SIRT family, such as SIRT1, SIRT3, and SIRT6, which play critical roles in histone and non-histone protein modifications. These findings highlight the importance of H2S-SIRT interactions in mediating cellular protection and physiological effects. | H2S-SIRT interactions mediate cellular protection, impacting histone modifications and cellular functions. |
| [149] | Hydrogen Sulfide Biology and Its Role in Cancer | Neurodegenerative diseases |
H2S can increase E-cadherin levels, inhibit histone deacetylase, and modulate NF-κB signaling, resulting in anti-metastatic and tumor-suppressive effects. However, the exact molecular targets underlying H2S’s diverse effects on biological processes, including cancer, require further investigation. Chronic exposure to H2S or its derivatives may have detrimental effects, including NF-κB inhibition and apoptosis. | H2S influences histone deacetylase and acetyltransferase activities, impacting gene expression and chromatin structure. |
| [150] | Protective effect of hydrogen sulfide is mediated by negative regulation of epigenetic histone acetylation in Parkinson’s disease |
Neurodegenerative diseases Parkinson’s disease |
Histone modifications and DNA methylation have been linked to the pathogenicity of Parkinson’s disease (PD). Histone deacetylase (HDAC) enzymes mediate chromatin condensation and inhibit gene transcription, while histone acetyltransferases (HAT) reverse these effects. Imbalances in HDAC and HAT activities are associated with neurodegenerative diseases, including PD. Inhibiting HDAC has shown promise in rescuing cells from degeneration in PD models. This study investigated the impact of HDAC inhibitor TSA on 6-hydroxydopamine-induced neurotoxicity in PD animal models. | H2S negatively regulates histone acetylation, impacting gene expression and neuronal survival in PD. |
| [151] | One-carbon epigenetics and redox biology of neurodegeneration |
Alzheimer’s disease Parkinson’s disease Amyotrophic lateral sclerosis |
Histone proteins form the histone octamer around which DNA is wrapped to create nucleosomes. Post-translational modifications (PTMs) of histone tails, including acetylation and methylation, regulate chromatin structure and gene expression. Histone acetyltransferases (HATs) add acetyl groups to lysine residues, promoting transcription, while histone deacetylases (HDACs) remove acetyl groups, leading to chromatin compaction and transcriptional inhibition. | H2S modulates histone acetyltransferases (HATs) and histone deacetylases (HDACs), influencing gene expression and chromatin remodeling. |
| [152] | Brain energy rescue: an emerging therapeutic concept for neurodegenerative disorders of ageing |
Neurodegenerative disorders | Histones, as crucial chromatin components, are subject to post-translational modifications, including acetylation, which influences gene expression. Hydrogen sulfide (H2S) plays a role in cellular energetics by regulating the availability of acetyl-CoA, a precursor of acetyl groups used in histone acetylation. H2S-related pathways, such as sirtuin 1 activation, mitochondrial function, and gut microbiota-produced short-chain fatty acids, also impact histone modifications, linking cellular energetics to epigenetic regulation. Targeting these mechanisms may hold therapeutic potential for neurodegenerative disorders. | H2S-related pathways influence histone modifications, linking cellular energetics to epigenetic regulation in neurodegenerative disorders. |
3.5. Hydrogen Sulfide and Non-Coding RNAs
H2S has been shown to modulate the expression of specific microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) that play regulatory roles in gene expression. By influencing the levels of these ncRNAs, H2S can affect the stability of mRNAs and protein translation, leading to changes in cellular functions. Dysregulation of miRNAs and lncRNAs has been observed in neurodegenerative diseases, and the interplay between H2S and these ncRNAs may contribute to disease pathogenesis [75,146,153,154] (Table 3).
Table 3.
Data regarding Hydrogen Sulfide and Non-Coding RNAs in Neurodegenerative diseases.
| Ref. | Title | Pathology | Extracted Data Regarding Epigenetics | Major Outcome |
|---|---|---|---|---|
| [146] | Hydrogen Sulfide Improves Angiogenesis by Regulating the Transcription of pri-miR-126 in Diabetic Endothelial Cells |
Parkinson’s disease |
MicroRNAs (miRNAs) are non-coding RNAs that modulate various cellular processes, including angiogenesis. Specific miRNAs, such as miR-126-3p, regulate angiogenesis in vascular endothelial cells. H2S is involved in miRNA transcription regulation, and the interplay between H2S and miRNAs is critical in cardiovascular disease pathophysiology. H2S has been shown to decrease cardiomyocyte apoptosis and impact Parkinson’s disease through miRNA regulation. | miRNAs such as miR-126-3p regulate angiogenesis—connected to PD |
| [75] | Regulating of LncRNA2264/ miR-20b-5p/IL17RD axis on hydrogen sulfide exposure-induced inflammation in broiler thymus by activating MYD88/NF-κB pathway |
Neurodegenerative disorders | lncRNA2264/miR-20b-5p/IL17RD axis was identified as part of the H2S-induced thymic inflammatory response. NcRNAs, including miRNAs and lncRNAs, can be potential biomarkers of environmental chemical exposure. In this study, lncRNA-sequencing revealed differentially expressed lncRNAs and miRNAs in the H2S-exposed group compared to the control group. Notably, lncRNA2264 showed significant downregulation, and it was identified as a molecular sponge for miR-20b-5p. MiR-20b-5p, which plays a role in immune cell function and inflammation, was significantly increased after H2S exposure. | NcRNAs, including miR-20b-5p and lncRNA2264, were identified as part of the H2S-induced thymic inflammatory response. |
| [153] | Overview on hydrogen sulfide-mediated suppression of vascular calcification and hemoglobin/heme-mediated vascular damage in atherosclerosis |
Neurodegenerative disorders | Epigenetic alterations, including DNA methylation and microRNAs (miRNAs), are implicated in atherosclerosis development and are linked to H2S pathways. H2S influences histone modifications, enhancing SIRT1 activity to reduce endothelial inflammation and foam cell formation, potentially reducing atherosclerotic plaque development. Targeting these epigenetic regulatory checkpoints holds promise for atherosclerosis therapy. | H2S-mediated epigenetic changes may alleviate atherosclerosis by modulating SIRT1 activity. |
| [154] | The emerging role of long non-coding RNAs and microRNAs in neurodegenerative diseases: A perspective of machine learning |
Neurodegenerative disease Alzheimer’s Parkinson’s Huntington’s diseases |
Neurodegenerative diseases (NDs) exhibit similar early symptoms, making their timely detection and differentiation crucial. Dysregulation of microRNAs and long non-coding RNAs is associated with NDs, highlighting their potential as diagnostic and therapeutic targets. Machine learning can effectively classify non-coding RNA expression profiles between healthy and affected individuals, aiding in accurate ND diagnosis with accuracy rates of 85% to 95%. Artificial intelligence offers a promising approach to enhance clinical diagnosis and early disease identification based on non-coding RNAs. | ncRNAs, potential diagnostic and therapeutic targets in neurodegenerative diseases, and machine learning improve ND diagnosis based on ncRNA expression. |
3.6. Future Directions
Future research efforts will be pivotal in advancing the potential of H2S-based therapies for neurodegenerative diseases, offering new hope to patients facing these devastating disorders. To achieve this objective, several crucial areas require investigation. Firstly, elucidating the precise molecular mechanisms through which H2S interacts with epigenetic regulation and cellular pathways in neurodegeneration is essential to fully understanding its neuroprotective effects and therapeutic applications. Secondly, comprehensive studies assessing long-term safety and efficacy are necessary before translating H2S-based therapies to clinical settings. Understanding potential side effects, dose-response relationships, and effects on cellular processes will ensure the therapies’ safety and effectiveness. Thirdly, it is crucial to identify optimal delivery methods for H2S-based therapies, considering their bioavailability and tissue distribution in different administration routes for varying disease stages and patient populations. Furthermore, targeted research is needed to determine the suitability of H2S-based therapies for specific neurodegenerative diseases, such as Alzheimer’s, Parkinson’s, and Huntington’s, and personalized medicine approaches should be explored to develop tailored therapies based on individual disease profiles and patient characteristics.
Investigating the potential synergistic effects of H2S-based therapies with existing treatments or emerging therapeutic agents may lead to innovative combination therapies that enhance neuroprotection and disease modification. Moving H2S-based therapies from preclinical research to clinical trials will require well-designed translational studies to establish safety, efficacy, and dosage recommendations. Neuroimaging techniques can provide valuable insights into the mechanisms of action and potential benefits of H2S-based therapies. Determining the optimal therapeutic window and identifying reliable biomarkers for monitoring treatment response are also crucial steps in advancing the field of H2S-based therapies for neurodegenerative diseases. Emphasizing research in these areas will pave the way for innovative and targeted therapies, bringing us closer to effective treatments for these debilitating conditions.
4. Conclusions
Epigenetic regulation has emerged as a critical determinant in the pathogenesis and progression of neurodegenerative diseases. The interaction between H2S and different epigenetic mechanisms, such as DNA methylation, histone modifications, and non-coding RNAs, suggests that H2S could influence gene expression and cellular functions relevant to neurodegenerative diseases. Understanding the precise molecular mechanisms underlying H2S’s interactions with epigenetic processes is essential to develop targeted and effective therapeutic strategies. Furthermore, investigating the long-term safety and efficacy of H2S-based therapies will be critical for their clinical translation.
Identifying optimal delivery methods, targeting disease-specific effects, and developing personalized medicine approaches will ensure the efficacy of H2S-based therapies for individual patients. Additionally, research on combination therapies and the development of reliable biomarkers for monitoring treatment response will further enhance the potential benefits of H2S interventions.
Overall, exploring H2S’s role in epigenetic regulation and neurodegeneration represents a promising avenue for future research. Advancements in this field have the potential to revolutionize the treatment landscape for neurodegenerative diseases, offering new hope to patients and their families facing these currently incurable conditions.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Tello J.A., Williams H.E., Eppler R.M., Steinhilb M.L., Khanna M. Animal Models of Neurodegenerative Disease: Recent Advances in Fly Highlight Innovative Approaches to Drug Discovery. Front. Mol. Neurosci. 2022;15:883358. doi: 10.3389/fnmol.2022.883358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akhtar A., Andleeb A., Waris T.S., Bazzar M., Moradi A.-R., Awan N.R., Yar M. Neurodegenerative diseases and effective drug delivery: A review of challenges and novel therapeutics. J. Control. Release. 2021;330:1152–1167. doi: 10.1016/j.jconrel.2020.11.021. [DOI] [PubMed] [Google Scholar]
- 3.Wareham L.K., Liddelow S.A., Temple S., Benowitz L.I., Di Polo A., Wellington C., Goldberg J.L., He Z., Duan X., Bu G., et al. Solving neurodegeneration: Common mechanisms and strategies for new treatments. Mol. Neurodegener. 2022;17:23. doi: 10.1186/s13024-022-00524-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo J., Huang X., Dou L., Yan M., Shen T., Tang W., Li J. Aging and aging-related diseases: From molecular mechanisms to interventions and treatments. Signal Transduct. Target. Ther. 2022;7:391. doi: 10.1038/s41392-022-01251-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.VandeVrede L., Boxer A.L., Polydoro M. Targeting tau: Clinical trials and novel therapeutic approaches. Neurosci. Lett. 2020;731:134919. doi: 10.1016/j.neulet.2020.134919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hammond T.R., Marsh S.E., Stevens B. Immune Signaling in Neurodegeneration. Immunity. 2019;50:955–974. doi: 10.1016/j.immuni.2019.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cecil C.A.M., Nigg J.T. Epigenetics and ADHD: Reflections on Current Knowledge, Research Priorities and Translational Potential. Mol. Diagn. Ther. 2022;26:581–606. doi: 10.1007/s40291-022-00609-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olufunmilayo E.O., Gerke-Duncan M.B., Holsinger R.M.D. Oxidative Stress and Antioxidants in Neurodegenerative Disorders. Antioxidants. 2023;12:517. doi: 10.3390/antiox12020517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ortega M.A., Fraile-Martínez Ó., García-Montero C., Alvarez-Mon M.A., Lahera G., Monserrat J., Llavero-Valero M., Mora F., Rodríguez-Jiménez R., Fernandez-Rojo S., et al. Nutrition, Epigenetics, and Major Depressive Disorder: Understanding the Connection. Front. Nutr. 2022;9:867150. doi: 10.3389/fnut.2022.867150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zanin M., Aitya N.A., Basilio J., Baumbach J., Benis A., Behera C.K., Bucholc M., Castiglione F., Chouvarda I., Comte B., et al. An Early Stage Researcher’s Primer on Systems Medicine Terminology. Netw. Syst. Med. 2021;4:2–50. doi: 10.1089/nsm.2020.0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McManus C. Cerebral Polymorphisms for Lateralisation: Modelling the Genetic and Phenotypic Architectures of Multiple Functional Modules. Symmetry. 2022;14:814. doi: 10.3390/sym14040814. [DOI] [Google Scholar]
- 12.Kular L., Jagodic M. Epigenetic insights into multiple sclerosis disease progression. J. Intern. Med. 2020;288:82–102. doi: 10.1111/joim.13045. [DOI] [PubMed] [Google Scholar]
- 13.Millan M.J. The epigenetic dimension of Alzheimer’s disease: Causal, consequence, or curiosity? Dialog.-Clin. Neurosci. 2014;16:373–393. doi: 10.31887/DCNS.2014.16.3/mmillan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akhtar A., Gupta S.M., Dwivedi S., Kumar D., Shaikh M.F., Negi A. Preclinical Models for Alzheimer’s Disease: Past, Present, and Future Approaches. ACS Omega. 2022;7:47504–47517. doi: 10.1021/acsomega.2c05609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y. Modern epigenetics methods in biological research. Methods. 2021;187:104–113. doi: 10.1016/j.ymeth.2020.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sibuh B.Z., Quazi S., Panday H., Parashar R., Jha N.K., Mathur R., Jha S.K., Taneja P., Jha A.K. The Emerging Role of Epigenetics in Metabolism and Endocrinology. Biology. 2023;12:256. doi: 10.3390/biology12020256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang K., Liu H., Hu Q., Wang L., Liu J., Zheng Z., Zhang W., Ren J., Zhu F., Liu G.-H. Epigenetic regulation of aging: Implications for interventions of aging and diseases. Signal Transduct. Target. Ther. 2022;7:374. doi: 10.1038/s41392-022-01211-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qureshi I.A., Mehler M.F. Epigenetic mechanisms underlying nervous system diseases. Handb. Clin. Neurol. 2018;147:43–58. doi: 10.1016/b978-0-444-63233-3.00005-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kan R.L., Chen J., Sallam T. Crosstalk between epitranscriptomic and epigenetic mechanisms in gene regulation. Trends Genet. 2022;38:182–193. doi: 10.1016/j.tig.2021.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fitz-James M.H., Cavalli G. Molecular mechanisms of transgenerational epigenetic inheritance. Nat. Rev. Genet. 2022;23:325–341. doi: 10.1038/s41576-021-00438-5. [DOI] [PubMed] [Google Scholar]
- 21.Li S., Yang D., Gao L., Wang Y., Peng Q. Epigenetic regulation and mechanobiology. Biophys. Rep. 2020;6:33–48. doi: 10.1007/s41048-020-00106-x. [DOI] [Google Scholar]
- 22.Policarpi C., Dabin J., Hackett J.A. Epigenetic editing: Dissecting chromatin function in context. Bioessays. 2021;43:e2000316. doi: 10.1002/bies.202000316. [DOI] [PubMed] [Google Scholar]
- 23.Fallet M., Blanc M., Di Criscio M., Antczak P., Engwall M., Bosagna C.G., Rüegg J., Keiter S.H. Present and future challenges for the investigation of transgenerational epigenetic inheritance. Environ. Int. 2023;172:107776. doi: 10.1016/j.envint.2023.107776. [DOI] [PubMed] [Google Scholar]
- 24.Lee H.-T., Oh S., Ro D.H., Yoo H., Kwon Y.-W. The Key Role of DNA Methylation and Histone Acetylation in Epigenetics of Atherosclerosis. J. Lipid Atheroscler. 2020;9:419–434. doi: 10.12997/jla.2020.9.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bure I.V., Nemtsova M.V., Kuznetsova E.B. Histone Modifications and Non-Coding RNAs: Mutual Epigenetic Regulation and Role in Pathogenesis. Int. J. Mol. Sci. 2022;23:5801. doi: 10.3390/ijms23105801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J., Zhang Y.-Z., Jiang J., Duan C.-G. The Crosstalk Between Epigenetic Mechanisms and Alternative RNA Processing Regulation. Front. Genet. 2020;11:998. doi: 10.3389/fgene.2020.00998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobow K., Reid C.A., van Vliet E.A., Becker A.J., Carvill G.L., Goldman A.M., Hirose S., Lopes-Cendes I., Khiari H.M., Poduri A., et al. Epigenetics explained: A topic “primer” for the epilepsy community by the ILAE Genetics/Epigenetics Task Force. Epileptic Disord. 2020;22:127–141. doi: 10.1684/epd.2020.1143. [DOI] [PubMed] [Google Scholar]
- 28.Rasmi Y., Shokati A., Hassan A., Aziz S.G.-G., Bastani S., Jalali L., Moradi F., Alipour S. The role of DNA methylation in progression of neurological disorders and neurodegenerative diseases as well as the prospect of using DNA methylation inhibitors as therapeutic agents for such disorders. IBRO Neurosci. Rep. 2022;14:28–37. doi: 10.1016/j.ibneur.2022.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nikolac Perkovic M., Videtic Paska A., Konjevod M., Kouter K., Svob Strac D., Nedic Erjavec G., Pivac N. Epigenetics of Alzheimer’s Disease. Biomolecules. 2021;11:195. doi: 10.3390/biom11020195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gil N., Ulitsky I. Regulation of gene expression by cis-acting long non-coding RNAs. Nat. Rev. Genet. 2019;21:102–117. doi: 10.1038/s41576-019-0184-5. [DOI] [PubMed] [Google Scholar]
- 31.Ashe A., Colot V., Oldroyd B.P. How does epigenetics influence the course of evolution? Philos. Trans. R. Soc. B Biol. Sci. 2021;376:20200111. doi: 10.1098/rstb.2020.0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mandrioli M. From Environmental Epigenetics to the Inheritance of Acquired Traits: A Historian and Molecular Perspective on an Unnecessary Lamarckian Explanation. Biomolecules. 2023;13:1077. doi: 10.3390/biom13071077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guerrero-Bosagna C. From epigenotype to new genotypes: Relevance of epigenetic mechanisms in the emergence of genomic evolutionary novelty. Semin. Cell Dev. Biol. 2019;97:86–92. doi: 10.1016/j.semcdb.2019.07.006. [DOI] [PubMed] [Google Scholar]
- 34.Jaenisch R., Bird A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33((Suppl. S3)):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 35.Holland M.L. Epigenetic Regulation of the Protein Translation Machinery. Ebiomedicine. 2017;17:3–4. doi: 10.1016/j.ebiom.2017.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stephens K.E., Miaskowski C.A., Levine J.D., Pullinger C.R., Aouizerat B.E. Epigenetic Regulation and Measurement of Epigenetic Changes. Biol. Res. Nurs. 2013;15:373–381. doi: 10.1177/1099800412444785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun W., Xie G., Jiang X., Khaitovich P., Han D., Liu X. Epigenetic regulation of human-specific gene expression in the prefrontal cortex. BMC Biol. 2023;21:123. doi: 10.1186/s12915-023-01612-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Colicchio J.M., Herman J. Empirical patterns of environmental variation favor adaptive transgenerational plasticity. Ecol. Evol. 2020;10:1648–1665. doi: 10.1002/ece3.6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kogenaru M., Nghe P., Poelwijk F.J., Tans S.J. Predicting Evolutionary Constraints by Identifying Conflicting Demands in Regulatory Networks. Cell Syst. 2020;10:526–534.e3. doi: 10.1016/j.cels.2020.05.004. [DOI] [PubMed] [Google Scholar]
- 40.Dornburg A., Mallik R., Wang Z., Bernal M.A., Thompson B., Bruford E.A., Nebert D.W., Vasiliou V., Yohe L.R., Yoder J.A., et al. Placing human gene families into their evolutionary context. Hum. Genom. 2022;16:56. doi: 10.1186/s40246-022-00429-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsankova N., Renthal W., Kumar A., Nestler E.J. Epigenetic regulation in psychiatric disorders. Nat. Rev. Neurosci. 2007;8:355–367. doi: 10.1038/nrn2132. Erratum in Nat. Rev. Neurosci. 2019, 20, 187–188. [DOI] [PubMed] [Google Scholar]
- 42.Flöttmann M., Scharp T., Klipp E. A Stochastic Model of Epigenetic Dynamics in Somatic Cell Reprogramming. Front. Physiol. 2012;3:216. doi: 10.3389/fphys.2012.00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sen N. Functional and Molecular Insights of Hydrogen Sulfide Signaling and Protein Sulfhydration. J. Mol. Biol. 2017;429:543–561. doi: 10.1016/j.jmb.2016.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Citi V., Martelli A., Gorica E., Brogi S., Testai L., Calderone V. Role of hydrogen sulfide in endothelial dysfunction: Pathophysiology and therapeutic approaches. J. Adv. Res. 2020;27:99–113. doi: 10.1016/j.jare.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen H.-J., Qian L., Li K., Qin Y.-Z., Zhou J.-J., Ji X.-Y., Wu D.-D. Hydrogen sulfide-induced post-translational modification as a potential drug target. Genes Dis. 2022;10:1870–1882. doi: 10.1016/j.gendis.2022.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu L.-F., Lu M., Hon Wong P.T., Bian J.-S. Hydrogen Sulfide: Neurophysiology and Neuropathology. Antioxid. Redox Signal. 2011;15:405–419. doi: 10.1089/ars.2010.3517. [DOI] [PubMed] [Google Scholar]
- 47.Perridon B.W., Leuvenink H.G., Hillebrands J.-L., Van Goor H., Bos E.M. The role of hydrogen sulfide in aging and age-related pathologies. Aging. 2016;8:2264–2289. doi: 10.18632/aging.101026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Munteanu C., Rotariu M., Turnea M., Dogaru G., Popescu C., Spînu A., Andone I., Postoiu R., Ionescu E.V., Oprea C., et al. Recent Advances in Molecular Research on Hydrogen Sulfide (H2S) Role in Diabetes Mellitus (DM)—A Systematic Review. Int. J. Mol. Sci. 2022;23:6720. doi: 10.3390/ijms23126720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodkin S., Nwosu C., Sannikov A., Tyurin A., Chulkov V.S., Raevskaya M., Ermakov A., Kirichenko E., Gasanov M. The Role of Gasotransmitter-Dependent Signaling Mechanisms in Apoptotic Cell Death in Cardiovascular, Rheumatic, Kidney, and Neurodegenerative Diseases and Mental Disorders. Int. J. Mol. Sci. 2023;24:6014. doi: 10.3390/ijms24076014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qiao P., Zhao F., Liu M., Gao D., Zhang H., Yan Y. Hydrogen sulfide inhibits mitochondrial fission in neuroblastoma N2a cells through the Drp1/ERK1/2 signaling pathway. Mol. Med. Rep. 2017;16:971–977. doi: 10.3892/mmr.2017.6627. [DOI] [PubMed] [Google Scholar]
- 51.Jeong N.Y., Jung J., Tabassum R. Therapeutic importance of hydrogen sulfide in age-associated neurodegenerative diseases. Neural Regen. Res. 2020;15:653–662. doi: 10.4103/1673-5374.266911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rao S.P., Dobariya P., Bellamkonda H., More S.S. Role of 3-Mercaptopyruvate Sulfurtransferase (3-MST) in Physiology and Disease. Antioxidants. 2023;12:603. doi: 10.3390/antiox12030603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aschner M., Skalny A.V., Ke T., da Rocha J.B., Paoliello M.M., Santamaria A., Bornhorst J., Rongzhu L., Svistunov A.A., Djordevic A.B., et al. Hydrogen Sulfide (H2S) Signaling as a Protective Mechanism against Endogenous and Exogenous Neurotoxicants. Curr. Neuropharmacol. 2022;20:1908–1924. doi: 10.2174/1570159X20666220302101854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou L., Wang Q. Advances of H2S in Regulating Neurodegenerative Diseases by Preserving Mitochondria Function. Antioxidants. 2023;12:652. doi: 10.3390/antiox12030652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen Z., Li S., Subramaniam S., Shyy J.Y.-J., Chien S. Epigenetic Regulation: A New Frontier for Biomedical Engineers. Annu. Rev. Biomed. Eng. 2017;19:195–219. doi: 10.1146/annurev-bioeng-071516-044720. [DOI] [PubMed] [Google Scholar]
- 56.Berson A., Nativio R., Berger S.L., Bonini N.M. Epigenetic Regulation in Neurodegenerative Diseases. Trends Neurosci. 2018;41:587–598. doi: 10.1016/j.tins.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Minarovits J., Banati F., Szenthe K., Niller H.H. Epigenetic Regulation. 2015, 879, 1–25. Adv. Exp. Med. Biol. 2015;879:1–25. doi: 10.1007/978-3-319-24738-0_1. [DOI] [PubMed] [Google Scholar]
- 58.Wang R. Physiological Implications of Hydrogen Sulfide: A Whiff Exploration That Blossomed. Physiol. Rev. 2012;92:791–896. doi: 10.1152/physrev.00017.2011. [DOI] [PubMed] [Google Scholar]
- 59.Panthi S., Manandhar S., Gautam K. Hydrogen sulfide, nitric oxide, and neurodegenerative disorders. Transl. Neurodegener. 2018;7:3. doi: 10.1186/s40035-018-0108-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ghosh P., Saadat A. Neurodegeneration and epigenetics: A review. Neurologia. 2021;38:e62–e68. doi: 10.1016/j.nrl.2021.01.016. [DOI] [PubMed] [Google Scholar]
- 61.Calabrese V., Cornelius C., Dinkova-Kostova A.T., Calabrese E.J., Mattson M.P., Catani M.V., Gasperi V., Bisogno T., Maccarrone M., Depp C., et al. Cellular Stress Responses, The Hormesis Paradigm, and Vitagenes: Novel Targets for Therapeutic Intervention in Neurodegenerative Disorders. Antioxid. Redox Signal. 2010;13:1763–1811. doi: 10.1089/ars.2009.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pomierny B., Krzyżanowska W., Skórkowska A., Jurczyk J., Przejczowska-Pomierny K., Szafarz M., Marcinkowska M., Torregrossa R., Whiteman M., Pera J., et al. Neuroprotection by Post-Stroke Administration of the Slow-Releasing Hydrogen Sulfide (H2S) Delivery Molecule AP39: Novel Insight into Stroke Therapy. Res. Sq. 2023 doi: 10.21203/rs.3.rs-3100208/v1. preprint . [DOI] [Google Scholar]
- 63.Ide M., Ohnishi T., Toyoshima M., Balan S., Maekawa M., Shimamoto-Mitsuyama C., Iwayama Y., Ohba H., Watanabe A., Ishii T., et al. Excess hydrogen sulfide and polysulfides production underlies a schizophrenia pathophysiology. EMBO Mol. Med. 2019;11:e10695. doi: 10.15252/emmm.201910695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu D., Li M., Tian W., Wang S., Cui L., Li H., Wang H., Ji A., Li Y. Hydrogen sulfide acts as a double-edged sword in human hepatocellular carcinoma cells through EGFR/ERK/MMP-2 and PTEN/AKT signaling pathways. Sci. Rep. 2017;7:5134. doi: 10.1038/s41598-017-05457-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mateus I. Importance of Mitochondrial Hydrogen Sulfide Oxidation in Liver Pathophysiology. Tissues and Organs [q-bio.TO] Université Paris Cité; Paris, France: 2021. NNT:2021UNIP5049. (In English) [Google Scholar]
- 66.Yang Y., Liu Y., Wang Y., Chao Y., Zhang J., Jia Y., Tie J., Hu D. Regulation of SIRT1 and Its Roles in Inflammation. Front. Immunol. 2022;13:831168. doi: 10.3389/fimmu.2022.831168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pozzi G., Gobbi G., Masselli E., Carubbi C., Presta V., Ambrosini L., Vitale M., Mirandola P. Buffering Adaptive Immunity by Hydrogen Sulfide. Cells. 2022;11:325. doi: 10.3390/cells11030325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Inuzuka H., Liu J., Wei W., Rezaeian A.-H. PROTAC technology for the treatment of Alzheimer’s disease: Advances and perspectives. Acta Mater. Med. 2022;1:24–41. doi: 10.15212/AMM-2021-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shi Y., Zhang H., Huang S., Yin L., Wang F., Luo P., Huang H. Epigenetic regulation in cardiovascular disease: Mechanisms and advances in clinical trials. Signal Transduct. Target. Ther. 2022;7:200. doi: 10.1038/s41392-022-01055-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Effendi W.I., Nagano T. Epigenetics Approaches toward Precision Medicine for Idiopathic Pulmonary Fibrosis: Focus on DNA Methylation. Biomedicines. 2023;11:1047. doi: 10.3390/biomedicines11041047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang S., Duan S., Xie Z., Bao W., Xu B., Yang W., Zhou L. Epigenetic Therapeutics Targeting NRF2/KEAP1 Signaling in Cancer Oxidative Stress. Front. Pharmacol. 2022;13:924817. doi: 10.3389/fphar.2022.924817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zatterale F., Raciti G.A., Prevenzano I., Leone A., Campitelli M., De Rosa V., Beguinot F., Parrillo L. Epigenetic Reprogramming of the Inflammatory Response in Obesity and Type 2 Diabetes. Biomolecules. 2022;12:982. doi: 10.3390/biom12070982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Linowiecka K., Slominski A.T., Reiter R.J., Böhm M., Steinbrink K., Paus R., Kleszczyński K. Melatonin: A Potential Regulator of DNA Methylation. Antioxidants. 2023;12:1155. doi: 10.3390/antiox12061155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lettieri-Barbato D., Aquilano K., Punziano C., Minopoli G., Faraonio R. MicroRNAs, Long Non-Coding RNAs, and Circular RNAs in the Redox Control of Cell Senescence. Antioxidants. 2022;11:480. doi: 10.3390/antiox11030480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang Y., He Y., Hu X., Chi Q., Zhao B., Ye J., Li S. Regulating of LncRNA2264/miR-20b-5p/IL17RD axis on hydrogen sulfide exposure-induced inflammation in broiler thymus by activating MYD88/NF-κB pathway. Toxicology. 2022;467:153086. doi: 10.1016/j.tox.2021.153086. [DOI] [PubMed] [Google Scholar]
- 76.Zeng M., Zhang T., Lin Y., Lin Y., Wu Z. The Common LncRNAs of Neuroinflammation-Related Diseases. Mol. Pharmacol. 2022;103:113–131. doi: 10.1124/molpharm.122.000530. Erratum in Mol. Pharmacol. 2023, 103, 298. [DOI] [PubMed] [Google Scholar]
- 77.Jayasuriya R., Ganesan K., Xu B., Ramkumar K.M. Emerging role of long non-coding RNAs in endothelial dysfunction and their molecular mechanisms. Biomed. Pharmacother. 2022;145:112421. doi: 10.1016/j.biopha.2021.112421. [DOI] [PubMed] [Google Scholar]
- 78.Hu Q., Zhang X., Sun M., Jiang B., Zhang Z., Sun D. Potential epigenetic molecular regulatory networks in ocular neovascularization. Front. Genet. 2022;13:970224. doi: 10.3389/fgene.2022.970224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kaziród K., Myszka M., Dulak J., Łoboda A. Hydrogen sulfide as a therapeutic option for the treatment of Duchenne muscular dystrophy and other muscle-related diseases. Cell. Mol. Life Sci. 2022;79:608. doi: 10.1007/s00018-022-04636-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu J., Mesfin F.M., Hunter C.E., Olson K.R., Shelley W.C., Brokaw J.P., Manohar K., Markel T.A. Recent Development of the Molecular and Cellular Mechanisms of Hydrogen Sulfide Gasotransmitter. Antioxidants. 2022;11:1788. doi: 10.3390/antiox11091788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhao H., Liu H., Yang Y., Lan T., Wang H., Wu D. Hydrogen Sulfide Plays an Important Role by Regulating Endoplasmic Reticulum Stress in Diabetes-Related Diseases. Int. J. Mol. Sci. 2022;23:7170. doi: 10.3390/ijms23137170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Le Corre L., Padovani D. Mechanism-based and computational modeling of hydrogen sulfide biogenesis inhibition: Interfacial inhibition. Sci. Rep. 2023;13:7287. doi: 10.1038/s41598-023-34405-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sharif A.H., Iqbal M., Manhoosh B., Gholampoor N., Ma D., Marwah M., Sanchez-Aranguren L. Hydrogen Sulphide-Based Therapeutics for Neurological Conditions: Perspectives and Challenges. Neurochem. Res. 2023;48:1981–1996. doi: 10.1007/s11064-023-03887-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sanchez L.D., Sanchez-Aranguren L., Marwah M., Wang K., Spickett C.M., Griffiths H.R., Dias I.H. Exploring mitochondrial hydrogen sulfide signalling for therapeutic interventions in vascular diseases. Adv. Redox Res. 2022;4:100030. doi: 10.1016/j.arres.2022.100030. [DOI] [Google Scholar]
- 85.Piragine E., Malanima M.A., Lucenteforte E., Martelli A., Calderone V. Circulating Levels of Hydrogen Sulfide (H2S) in Patients with Age-Related Diseases: A Systematic Review and Meta-Analysis. Biomolecules. 2023;13:1023. doi: 10.3390/biom13071023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fang H., Yu Z., Xing K., Zhou L., Shao Y., Zhang X., Pei Y., Zhang L. Transcriptomic analysis reveals the functions of H2S as a gasotransmitter independently of Cys in Arabidopsis. Front. Plant Sci. 2023;14:1184991. doi: 10.3389/fpls.2023.1184991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lin Q., Geng B. The Role of Hydrogen Sulfide in Plaque Stability. Antioxidants. 2022;11:2356. doi: 10.3390/antiox11122356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lu D., Wang L., Liu G., Wang S., Wang Y., Wu Y., Wang J., Sun X. Role of hydrogen sulfide in subarachnoid hemorrhage. CNS Neurosci. Ther. 2022;28:805–817. doi: 10.1111/cns.13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huang Y., Omorou M., Gao M., Mu C., Xu W., Xu H. Hydrogen sulfide and its donors for the treatment of cerebral ischaemia-reperfusion injury: A comprehensive review. Biomed. Pharmacother. 2023;161:114506. doi: 10.1016/j.biopha.2023.114506. [DOI] [PubMed] [Google Scholar]
- 90.Tu Z., Zhong Y., Hu H., Shao D., Haag R., Schirner M., Lee J., Sullenger B., Leong K.W. Design of therapeutic biomaterials to control inflammation. Nat. Rev. Mater. 2022;7:557–574. doi: 10.1038/s41578-022-00426-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu Y., Chen Q., Li Y., Bi L., Lin S., Ji H., Sun D., Jin L., Peng R. Hydrogen sulfide-induced oxidative stress mediated apoptosis via mitochondria pathway in embryo-larval stages of zebrafish. Ecotoxicol. Environ. Saf. 2022;239:113666. doi: 10.1016/j.ecoenv.2022.113666. [DOI] [PubMed] [Google Scholar]
- 92.Cornwell A., Badiei A. From Gasotransmitter to Immunomodulator: The Emerging Role of Hydrogen Sulfide in Macrophage Biology. Antioxidants. 2023;12:935. doi: 10.3390/antiox12040935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Khan M.S.S., Islam F., Ye Y., Ashline M., Wang D., Zhao B., Fu Z.Q., Chen J. The Interplay between Hydrogen Sulfide and Phytohormone Signaling Pathways under Challenging Environments. Int. J. Mol. Sci. 2022;23:4272. doi: 10.3390/ijms23084272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vinton A.C., Gascoigne S.J., Sepil I., Salguero-Gómez R. Plasticity’s role in adaptive evolution depends on environmental change components. Trends Ecol. Evol. 2022;37:1067–1078. doi: 10.1016/j.tree.2022.08.008. [DOI] [PubMed] [Google Scholar]
- 95.Paul B.D., Pieper A.A. Protective Roles of Hydrogen Sulfide in Alzheimer’s Disease and Traumatic Brain Injury. Antioxidants. 2023;12:1095. doi: 10.3390/antiox12051095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Singh S., Yang F., Sivils A., Cegielski V., Chu X.-P. Amylin and Secretases in the Pathology and Treatment of Alzheimer’s Disease. Biomolecules. 2022;12:996. doi: 10.3390/biom12070996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cheng G., Xie A., Yan Z., Zhu X., Song Y., Chen T. Nanomedicines for Alzheimer’s disease: Therapies based on pathological mechanisms. Brain-x. 2023;1:e27. doi: 10.1002/brx2.27. [DOI] [Google Scholar]
- 98.Fišar Z. Linking the Amyloid, Tau, and Mitochondrial Hypotheses of Alzheimer’s Disease and Identifying Promising Drug Targets. Biomolecules. 2022;12:1676. doi: 10.3390/biom12111676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gupta R., Sahu M., Tripathi R., Ambasta R.K., Kumar P. Protein S-sulfhydration: Unraveling the prospective of hydrogen sulfide in the brain, vasculature and neurological manifestations. Ageing Res. Rev. 2022;76:101579. doi: 10.1016/j.arr.2022.101579. [DOI] [PubMed] [Google Scholar]
- 100.Calabresi P., Mechelli A., Natale G., Volpicelli-Daley L., Di Lazzaro G., Ghiglieri V. Alpha-synuclein in Parkinson’s disease and other synucleinopathies: From overt neurodegeneration back to early synaptic dysfunction. Cell Death Dis. 2023;14:176. doi: 10.1038/s41419-023-05672-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vidović M., Rikalovic M.G. Alpha-Synuclein Aggregation Pathway in Parkinson’s Disease: Current Status and Novel Therapeutic Approaches. Cells. 2022;11:1732. doi: 10.3390/cells11111732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ravenhill S.M., Evans A.H., Crewther S.G. Escalating Bi-Directional Feedback Loops between Proinflammatory Microglia and Mitochondria in Ageing and Post-Diagnosis of Parkinson’s Disease. Antioxidants. 2023;12:1117. doi: 10.3390/antiox12051117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fan H., Sheng S., Zhang F. New hope for Parkinson’s disease treatment: Targeting gut microbiota. CNS Neurosci. Ther. 2022;28:1675–1688. doi: 10.1111/cns.13916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Murros K.E. Hydrogen Sulfide Produced by Gut Bacteria May Induce Parkinson’s Disease. Cells. 2022;11:978. doi: 10.3390/cells11060978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Arning L., Nguyen H.P. Huntington disease update: New insights into the role of repeat instability in disease pathogenesis. Med. Genet. 2021;33:293–300. doi: 10.1515/medgen-2021-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Paul B.D. Cysteine metabolism and hydrogen sulfide signaling in Huntington’s disease. Free. Radic. Biol. Med. 2022;186:93–98. doi: 10.1016/j.freeradbiomed.2022.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kim D.-S., Pessah I.N., Santana C.M., Purnell B.S., Li R., Buchanan G.F., Rumbeiha W.K. Investigations into hydrogen sulfide-induced suppression of neuronal activity in vivo and calcium dysregulation in vitro. Toxicol. Sci. 2023;192:247–264. doi: 10.1093/toxsci/kfad022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Aroca A., Gotor C. Hydrogen Sulfide: A Key Role in Autophagy Regulation from Plants to Mammalians. Antioxidants. 2022;11:327. doi: 10.3390/antiox11020327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhao H., Yang Y., Liu H., Wang H. The Role of Hydrogen Sulfide Targeting Autophagy in the Pathological Processes of the Nervous System. Metabolites. 2022;12:879. doi: 10.3390/metabo12090879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhu L., Duan W., Wu G., Zhang D., Wang L., Chen D., Chen Z., Yang B. Protective effect of hydrogen sulfide on endothelial cells through Sirt1-FoxO1-mediated autophagy. Ann. Transl. Med. 2020;8:1586. doi: 10.21037/atm-20-3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li J., Li M., Li L., Ma J., Yao C., Yao S. Hydrogen sulfide attenuates ferroptosis and stimulates autophagy by blocking mTOR signaling in sepsis-induced acute lung injury. Mol. Immunol. 2021;141:318–327. doi: 10.1016/j.molimm.2021.12.003. Erratum in Mol. Immunol. 2023, 156, 156. [DOI] [PubMed] [Google Scholar]
- 112.Giallongo S., Longhitano L., Denaro S., D’aprile S., Torrisi F., La Spina E., Giallongo C., Mannino G., Furno D.L., Zappalà A., et al. The Role of Epigenetics in Neuroinflammatory-Driven Diseases. Int. J. Mol. Sci. 2022;23:15218. doi: 10.3390/ijms232315218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chen Z., Wu M., Lai Q., Zhou W., Wen X., Yin X. Epigenetic regulation of synaptic disorder in Alzheimer’s disease. Front. Neurosci. 2022;16:888014. doi: 10.3389/fnins.2022.888014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gopinathan G., Diekwisch T.G.H. Epigenetics and Early Development. J. Dev. Biol. 2022;10:26. doi: 10.3390/jdb10020026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jiang D., Li T., Guo C., Tang T.-S., Liu H. Small molecule modulators of chromatin remodeling: From neurodevelopment to neurodegeneration. Cell Biosci. 2023;13:10. doi: 10.1186/s13578-023-00953-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Park J., Lee K., Kim K., Yi S.-J. The role of histone modifications: From neurodevelopment to neurodiseases. Signal Transduct. Target. Ther. 2022;7:217. doi: 10.1038/s41392-022-01078-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Santana D.A., Smith M.d.A.C., Chen E.S. Histone Modifications in Alzheimer’s Disease. Genes. 2023;14:347. doi: 10.3390/genes14020347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mahana Y., Ohki I., Walinda E., Morimoto D., Sugase K., Shirakawa M. Structural Insights into Methylated DNA Recognition by the Methyl-CpG Binding Domain of MBD6 from Arabidopsis thaliana. ACS Omega. 2022;7:3212–3221. doi: 10.1021/acsomega.1c04917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kaur G., Rathod S.S.S., Ghoneim M.M., Alshehri S., Ahmad J., Mishra A., Alhakamy N.A. DNA Methylation: A Promising Approach in Management of Alzheimer’s Disease and Other Neurodegenerative Disorders. Biology. 2022;11:90. doi: 10.3390/biology11010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mattei A.L., Bailly N., Meissner A. DNA methylation: A historical perspective. Trends Genet. 2022;38:676–707. doi: 10.1016/j.tig.2022.03.010. [DOI] [PubMed] [Google Scholar]
- 121.Fodder K., de Silva R., Warner T.T., Bettencourt C. The contribution of DNA methylation to the (dys)function of oligodendroglia in neurodegeneration. Acta Neuropathol. Commun. 2023;11:106. doi: 10.1186/s40478-023-01607-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gao X., Chen Q., Yao H., Tan J., Liu Z., Zhou Y., Zou Z. Epigenetics in Alzheimer’s Disease. Front. Aging Neurosci. 2022;14:911635. doi: 10.3389/fnagi.2022.911635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yazar V., Dawson V.L., Dawson T.M., Kang S.-U. DNA Methylation Signature of Aging: Potential Impact on the Pathogenesis of Parkinson’s Disease. J. Park. Dis. 2023;13:145–164. doi: 10.3233/JPD-223517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Walton E., Baltramonaityte V., Calhoun V., Heijmans B.T., Thompson P.M., Cecil C.A.M. A systematic review of neuroimaging epigenetic research: Calling for an increased focus on development. Mol. Psychiatry. 2023 doi: 10.1038/s41380-023-02067-2. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Xie J., Xie L., Wei H., Li X.-J., Lin L. Dynamic Regulation of DNA Methylation and Brain Functions. Biology. 2023;12:152. doi: 10.3390/biology12020152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Aricthota S., Rana P.P., Haldar D. Histone acetylation dynamics in repair of DNA double-strand breaks. Front. Genet. 2022;13:926577. doi: 10.3389/fgene.2022.926577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Angelopoulou E., Paudel Y.N., Papageorgiou S.G., Piperi C. Environmental Impact on the Epigenetic Mechanisms Underlying Parkinson’s Disease Pathogenesis: A Narrative Review. Brain Sci. 2022;12:175. doi: 10.3390/brainsci12020175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Paccosi E., Proietti-De-Santis L. Parkinson’s Disease: From Genetics and Epigenetics to Treatment, a miRNA-Based Strategy. Int. J. Mol. Sci. 2023;24:9547. doi: 10.3390/ijms24119547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jurcau A. Molecular Pathophysiological Mechanisms in Huntington’s Disease. Biomedicines. 2022;10:1432. doi: 10.3390/biomedicines10061432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Saha P., Verma S., Pathak R.U., Mishra R.K. Long Noncoding RNAs in Mammalian Development and Diseases. Adv. Exp. Med. Biol. 2017;1008:155–198. doi: 10.1007/978-981-10-5203-3_6. [DOI] [PubMed] [Google Scholar]
- 131.Lee Y.S. Are We Studying Non-Coding RNAs Correctly? Lessons from nc886. Int. J. Mol. Sci. 2022;23:4251. doi: 10.3390/ijms23084251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Fort R.S., Chavez S., Barnech J.M.T., Oliveira-Rizzo C., Smircich P., Sotelo-Silveira J.R., Duhagon M.A. Current Status of Regulatory Non-Coding RNAs Research in the Tritryp. Non-Coding RNA. 2022;8:54. doi: 10.3390/ncrna8040054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mattick J.S., Amaral P.P., Carninci P., Carpenter S., Chang H.Y., Chen L.-L., Chen R., Dean C., Dinger M.E., Fitzgerald K.A., et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023;24:430–447. doi: 10.1038/s41580-022-00566-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nukala S.B., Jousma J., Cho Y., Lee W.H., Ong S.-G. Long non-coding RNAs and microRNAs as crucial regulators in cardio-oncology. Cell Biosci. 2022;12:24. doi: 10.1186/s13578-022-00757-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Conforti F., Ruffo P., De Amicis F., Giardina E. Long-noncoding RNAs as epigenetic regulators in neurodegenerative diseases. Neural Regen. Res. 2023;18:1243–1248. doi: 10.4103/1673-5374.358615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Srinivas T., Mathias C., Oliveira-Mateos C., Guil S. Roles of lncRNAs in brain development and pathogenesis: Emerging therapeutic opportunities. Mol. Ther. 2023;31:1550–1561. doi: 10.1016/j.ymthe.2023.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhang M., He P., Bian Z. Long Noncoding RNAs in Neurodegenerative Diseases: Pathogenesis and Potential Implications as Clinical Biomarkers. Front. Mol. Neurosci. 2021;14:685143. doi: 10.3389/fnmol.2021.685143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gentile G., Morello G., La Cognata V., Guarnaccia M., Conforti F.L., Cavallaro S. Dysregulated miRNAs as Biomarkers and Therapeutical Targets in Neurodegenerative Diseases. J. Pers. Med. 2022;12:770. doi: 10.3390/jpm12050770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ren S., Lin P., Wang J., Yu H., Lv T., Sun L., Du G. Circular RNAs: Promising Molecular Biomarkers of Human Aging-Related Diseases via Functioning as an miRNA Sponge. Mol. Ther.-Methods Clin. Dev. 2020;18:215–229. doi: 10.1016/j.omtm.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Martínez-Iglesias O., Naidoo V., Carrera I., Corzo L., Cacabelos R. Natural Bioactive Products as Epigenetic Modulators for Treating Neurodegenerative Disorders. Pharmaceuticals. 2023;16:216. doi: 10.3390/ph16020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sundaramoorthy T.H., Castanho I. The Neuroepigenetic Landscape of Vertebrate and Invertebrate Models of Neurodegenerative Diseases. Epigenetics Insights. 2022;15:25168657221135848. doi: 10.1177/25168657221135848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Louit A., Galbraith T., Berthod F. In Vitro 3D Modeling of Neurodegenerative Diseases. Bioengineering. 2023;10:93. doi: 10.3390/bioengineering10010093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Morozova A., Zorkina Y., Abramova O., Pavlova O., Pavlov K., Soloveva K., Volkova M., Alekseeva P., Andryshchenko A., Kostyuk G., et al. Neurobiological Highlights of Cognitive Impairment in Psychiatric Disorders. Int. J. Mol. Sci. 2022;23:1217. doi: 10.3390/ijms23031217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pi T., Liu B., Shi J. Abnormal Homocysteine Metabolism: An Insight of Alzheimer’s Disease from DNA Methylation. Behav. Neurol. 2020;2020:8438602. doi: 10.1155/2020/8438602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kimura H. Hydrogen sulfide signalling in the CNS-Comparison with NO. Br. J. Pharmacol. 2020;177:5031–5045. doi: 10.1111/bph.15246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Xue W., Zhang Q., Chen Y., Zhu Y. Hydrogen Sulfide Improves Angiogenesis by Regulating the Transcription of pri-miR-126 in Diabetic Endothelial Cells. Cells. 2022;11:2651. doi: 10.3390/cells11172651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wilkie S.E., Borland G., Carter R.N., Morton N.M., Selman C. Hydrogen sulfide in ageing, longevity and disease. Biochem. J. 2021;478:3485–3504. doi: 10.1042/BCJ20210517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.D’egidio F., Castelli V., Cimini A., D’Angelo M. Cell Rearrangement and Oxidant/Antioxidant Imbalance in Huntington’s Disease. Antioxidants. 2023;12:571. doi: 10.3390/antiox12030571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Khattak S., Rauf M.A., Khan N.H., Zhang Q.-Q., Chen H.-J., Muhammad P., Ansari M.A., Alomary M.N., Jahangir M., Zhang C.-Y., et al. Hydrogen Sulfide Biology and Its Role in Cancer. Molecules. 2022;27:3389. doi: 10.3390/molecules27113389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sun Y., Li D., Su Y., Zhao H., Pang W., Zhao W., Wu S. Protective effect of hydrogen sulfide is mediated by negative regulation of epigenetic histone acetylation in Parkinson’s disease. Arch. Med. Sci. 2020;16:1124–1135. doi: 10.5114/aoms.2020.93121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Coppedè F. One-carbon epigenetics and redox biology of neurodegeneration. Free. Radic. Biol. Med. 2021;170:19–33. doi: 10.1016/j.freeradbiomed.2020.12.002. [DOI] [PubMed] [Google Scholar]
- 152.Cunnane S.C., Trushina E., Morland C., Prigione A., Casadesus G., Andrews Z.B., Beal M.F., Bergersen L.H., Brinton R.D., de la Monte S., et al. Brain energy rescue: An emerging therapeutic concept for neurodegenerative disorders of ageing. Nat. Rev. Drug Discov. 2020;19:609–633. doi: 10.1038/s41573-020-0072-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gáll T., Nagy P., Garai D., Potor L., Balla G.J., Balla J. Overview on hydrogen sulfide-mediated suppression of vascular calcification and hemoglobin/heme-mediated vascular damage in atherosclerosis. Redox Biol. 2022;57:102504. doi: 10.1016/j.redox.2022.102504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.García-Fonseca Á., Martin-Jimenez C., Barreto G.E., Pachón A.F.A., González J. The Emerging Role of Long Non-Coding RNAs and MicroRNAs in Neurodegenerative Diseases: A Perspective of Machine Learning. Biomolecules. 2021;11:1132. doi: 10.3390/biom11081132. [DOI] [PMC free article] [PubMed] [Google Scholar]