Abstract
Apelin (APLN) is recently demonstrated a direct association with many malignant diseases. However, its effects on cervical cancer remain unclear. This study therefore aims to evaluate the association between APLN expression and cervical cancer using publicly available data from The Cancer Genome Atlas (TCGA). The Pearson χ2 test and Fish exact test, as well as logistic regression, were used to evaluate the relationship between clinicopathological factors in cervical cancer and the expression of APLN. Additionally, the Cox regression and Kaplan-Meier methods were conducted to analyze the Overall Survival (OS) of cervical cancer patients in TCGA. Finally, gene set enrichment analysis (GSEA) was performed to establish its biological functions. High expression of APLN in cervical cancer was significantly associated with a more advanced clinical stage (OR = 1.91 (1.21–3.05) for Stage II, Stage III, and Stage IV vs Stage I, p = 0.006). Additionally, it was associated with poor outcome after primary therapy (OR = 2.14 (1.03–4.59) for Progressive Disease (PD), Stable Disease (SD), and Partial Response (PR) vs Complete Remission (CR), p = 0.045) and high histologic grade (OR = 1.67 (1.03–2.72) for G3 and G4 vs G1 and G2, p = 0.037). Moreover, multivariate analysis showed that high expression of APLN was associated with a shorter OS. GSEA demonstrated that six KEGG pathways, including PPAR signaling, ECM-receptor interaction, focal adhesion, MAPK signaling, TGF-beta signaling, and Gap junction pathways were differentially enriched in the high expression APLN phenotype. The recent study suggests that APLN plays an important role in the progression of cervical cancer and might be a promising prognostic biomarker of the disease.
Keywords: APLN, cervical cancer, prognosis, biomarker, the cancer genome atlas
Introduction
Cervical cancer is the fourth most common female malignancy worldwide. A total of 570,000 new cases were reported in 2018, accounting for about 6.9% of all female cancers. 1 The disease presents a significant threat to global health, especially in low- and middle-income countries. A notable decrease in the incidence of cervical cancer has been seen over the last few decades due to the introduction of mass screening programs at the community level. However, the mortality of the disease is still high, with 311,365 deaths occurred in 2018. 2 Cervical cancer is aggressive and timely diagnosis remains a challenge as it is often detected late. Therefore, it is urgent to better understand the underlying molecular mechanisms and identify novel biomarkers for effective diagnosis, treatment, and prognosis.
Apelin (APLN) was originally reported in 1998 by Tatemoto et al. 3 and is an endogenous ligand of the orphan G protein-coupled receptor APJ. Apelin/APJ mRNA and protein are widely expressed in various tissues and systems including the central nervous system, gastrointestinal tract, cardiomyocytes and vascular endothelial and smooth muscle cells, lung endothelial cells, liver, kidneys, and mammary glands. 4 Additionally, the Apelin/APJ system was reported to mediate several pathophysiological and physiological processes including metabolic disorders, modulation of the cardiovascular system, inflammatory response, hepatic, and kidney diseases. 5 Moreover, previous studies revealed that the Apelin/APJ system plays a significant role in tumor development. APLN was found to be highly expressed in Human Non-small Cell Lung Cancer (NSCLC) specimens compared to normal lung tissue samples. Moreover, it was reported to contribute to poor clinical outcomes. 6 Furthermore, APLN was identified in plasma samples and found to be associated with the systemic inflammatory response in gastroesophageal and colonic cancer. 7 A high level of APLN expression was also detected in Hepatocellular Carcinoma (HCC) and was involved in arteriogenesis in cancer. 8 Additionally, the up-regulation of APLN was associated with aggressive progression and short recurrence-free survival in patients with prostate cancer. 9 Besides, high expression levels of APLN in Oral Squamous Cell Carcinomas (OSCC) were significantly correlated with tumor recurrence and decreased disease-free survival rate. 10 In addition, high levels of circulating APLN presented a greater chance of developing endometrial cancer in obese patients. 11 However, few studies on the role of APLN in cervical cancer currently exist.
In this study, the expression levels of APLN in cervical cancer were analyzed. This was done to explore in detail the possible mechanisms underlying the expression of APLN in cervical cancer. To achieve this, a total of 304 cervical cancer cases were obtained from The Cancer Genome Altas (TCGA). The correlation between APLN and clinicopathological features as well as its clinical significance were then investigated to ascertain the potential prognostic value of the gene in cervical cancer. Furthermore, the Gene Set Enrichment Analysis (GSEA) was used to identify relevant functional pathways utilized by APLN in tumor development and progression in cervical cancer. Finally, the potential molecular mechanisms underlying cervical cancer were investigated by exploring the relationship between APLN and immune cell infiltration.
Materials and methods
RNA-sequencing data preprocessing
The RNA-seq data (Workflow Type: HTSeq-Counts) and corresponding clinical information were downloaded from the TCGA-Cervical Adenocarcinoma and Cervical Squamous Cell Carcinoma datasets. A total of 304 patients and 3 normal cervical tissue samples were included after excluding cervical cancer samples with incomplete or illegible clinical information. Thereafter, level 3 HTSeq-Counts data was transformed into Transcripts Per Million (TPM) reads for further analyses. Unavailable clinical information and unknown features were regarded as missing values. In addition, 10 normal cervical tissue samples from the GTEx dataset were included. Furthermore, the expression level of APLN in GSE7410 and GSE29570 were downloaded from NCBI-GEO (Gene Expression Omnibus). The two datasets contain 5 normal cervical tissues and 40 CESC tissues, 17 normal cervical tissues, and 45 CESC tissues, respectively. The study followed publication guidelines provided by TCGA (http://cancergenome.nih.gov/publications/publicationguidelines). Therefore, ethical approval was not required since all the data used in this study was acquired from TCGA.
Gene set enrichment analysis
The GSEA is a computational analysis method used to judge whether a priori defined set of genes shows statistically significant differences between two biological states. 12 In this study, the R package clusterProfiler (3.14.3) 13 was used to perform GSEA between the high and low APLN expression groups. Functional or pathway terms with adjusted p-values <0.05 and False Discovery Rate (FDR) q-value <0.25 were considered statistically significant.
Immune infiltration analysis by ssGSEA
Immune infiltration analysis of cervical cancer was performed through the ssGSEA (single-sample Gene Set Enrichment Analysis) method using the GSVA package (http://www.bioconductor.org/packages/release/bioc/html/GSVA.html). 14 The signatures of immune infiltration included 509 genes encoding 24 types of immunocytes as reported by Bindea et al. 15 The relative enrichment score for each immunocyte was then obtained from the gene expression profile in each tumor sample. 15 Additionally, Spearman correlation was used to find the association between the expression of APLN and these immune cells.
ROC analysis
The Receiver Operating Characteristic (ROC) curve analysis was utilized to evaluate the value of APLN as a biomarker of cervical cancer, using the R package. This was based on the ability of APLN to distinguish cervical cancer from normal controls according to specificity and sensitivity, in the diagnosis of cervical cancer. An Area Under the Curve (AUC) value was then calculated and used to evaluate the ROC effect.
Statistical analysis
The R software (version 3.6.2) was used for statistical analysis. The expression levels of APLN in tumor and no-paired normal cervical tissue samples were compared using the Wilcoxon rank-sum test. Additionally, the correlation between clinicopathological factors in cervical cancer and expression of APLN was analyzed using Pearson χ2 test (or Fish exact test when needed) and univariate logistic regression. In addition to APLN expression, several other clinical factors may influence the prognosis of cervical cancer. Such include age, lymph node metastasis (N stage), distant metastasis (M stage), race, menopause status, radiation therapy, histological type, tumor grade, clinical stage, PIK3CA status, and primary therapy outcome. Therefore, the univariate and multivariate Cox regression analyses were used to identify independent variables, accordingly. Moreover, the Kaplan-Meier method was used to draw survival curves and the log-rank test was used to examine the difference in the survival curves. Median APLN expression was regarded as the cut-off value and p values of less than 0.05 were considered statistically significant. All hypothesis tests were two-sided.
Results
Comparison of APLN expression in CESC with those in normal cervical tissues
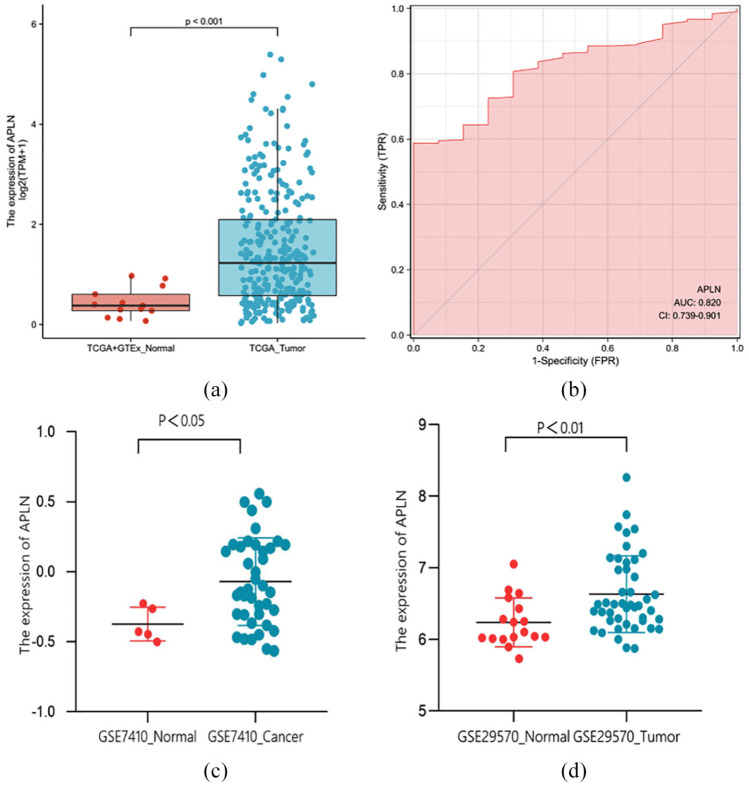
The results revealed that the expression levels of APLN in tumor tissues were remarkably higher than that in normal tissues (p < 0.001; Figure 1(a), (c), and (d)). Moreover, the ROC curve analysis was performed to further validate the potential diagnostic role of APLN in cervical cancer. The AUC was found to be 0.820 (95% Confidence Interval (CI): 0.739–0.901) as shown in Figure 1(b), indicating that APLN maybe a potential novel diagnostic biomarker of cervical cancer.
Figure 1.
APLN was highly expressed in tumors compared to normal cervical tissue samples in TCGA + GTEx dataset (a), GSE7410 dataset (c), and GSE29570 dataset (d). (b) ROC analysis showed that the AUC was 0.820 (95% CI: 0.739–0.901), indicating that APLN was a potential diagnostic biomarker of cervical cancer.
Association between the expression of APLN and clinicopathologic features
A total of 304 patients with detailed clinical information were divided into two groups based on the median APLN expression level. Correlation analysis showed that high level of APLN expression was significantly associated with advanced M stage (p = 0.049), clinical stage (p = 0.006), and a higher possibility of radiation therapy (p = 0.007). However, no correlation was found between the expression of APLN and other clinicopathologic features (Table 1). Additionally, a univariate logistic regression analysis of APLN expression (as a categorical dependent variable based on median expression value) indicated that higher expression levels were significantly associated with poor prognostic features (Table 2). These included more advanced clinical stage (OR = 1.91 (1.21–3.05) for Stage II, Stage III, and Stage IV vs Stage I, p = 0.006), poor primary therapy outcome (OR = 0.47 (0.22–0.97) for Complete Remission (CR) vs Progressive Disease (PD), Stable Disease (SD) and Partial Response (PD), p = 0.045), and high histologic grade (OR = 1.67 (1.03–2.72) for G3 and G4 vs G1 and G2, p = 0.037). These results indicated that tumors with high expression of APLN were associated with poor clinicopathologic factors.
Table 1.
Association between APLN expression and clinicopathological features in the TCGA cohort.
| Characters | Level | Low expression of APLN | High expression of APLN | p Value |
|---|---|---|---|---|
| n | 152 | 152 | ||
| N stage (%) | N0 | 75 (68.2%) | 58 (69.9%) | 0.924 |
| N1 | 35 (31.8%) | 25 (30.1%) | ||
| M stage (%) | M0 | 63 (96.9%) | 53 (86.9%) | 0.049a,b |
| M1 | 2 (3.1%) | 8 (13.1%) | ||
| Clinical stage (%) | Stage I | 92 (62.6%) | 70 (46.7%) | 0.006 a |
| Stage II–IV | 55 (37.4%) | 80 (53.3%) | ||
| Radiation therapy (%) | No | 73 (48.0%) | 49 (32.2%) | 0.007 a |
| Yes | 79 (52.0%) | 103 (67.8%) | ||
| Primary therapy outcome (%) | CR | 99 (88.4%) | 82 (78.1%) | 0.129 b |
| PD | 8 (7.1%) | 14 (13.3%) | ||
| PR | 4 (3.6%) | 4 (3.8%) | ||
| SD | 1 (0.9%) | 5 (4.8%) | ||
| Race (%) | Asian | 15 (11.2%) | 5 (4.0%) | 0.054 b |
| Black or African American | 12 (9.0%) | 18 (14.4%) | ||
| White | 107 (79.9%) | 102 (81.6%) | ||
| Histological type (%) | Adenosquamous | 29 (19.1%) | 23 (15.1%) | 0.446 |
| Squamous cell carcinoma | 123 (80.9%) | 129 (84.9%) | ||
| Histologic grade (%) | G1 | 11 (7.7%) | 7 (5.4%) | 0.120 b |
| G2 | 78 (54.5%) | 57 (44.2%) | ||
| G3 | 54 (37.8%) | 64 (49.6%) | ||
| G4 | 0 (0.0%) | 1 (0.8%) | ||
| Menopause status (%) | Peri | 15 (12.9%) | 10 (8.7%) | 0.230 |
| Post | 45 (38.8%) | 37 (32.2%) | ||
| Pre | 56 (48.3%) | 68 (59.1%) | ||
| PIK3CA status (%) | Mut | 40 (27.8%) | 42 (29.6%) | 0.837 |
| WT | 104 (72.2%) | 100 (70.4%) | ||
| Age (%) | ≤50 | 91 (59.9%) | 95 (62.5%) | 0.724 |
| >50 | 61 (40.1%) | 57 (37.5%) |
CR: complete response; Mut: mutant type; PD: progressive disease; PR: partial response; SD: stable disease; WT: wild type.
Statistically significant.
Fisher’s exact test.
Table 2.
APLN expression a associated with clinical pathological characteristic (logistic regression).
| Characteristics | Odds ratio in APLN expression | Odds ratio (OR) | p Value |
|---|---|---|---|
| N stage (N1 vs N0) | 193 | 0.92 (0.50–1.71) | 0.801 |
| M stage (M1 vs M0) | 126 | 4.75 (1.13–32.42) | 0.055 |
| Clinical stage (stage II and stage III and stage IV vs stage I) | 297 | 1.91 (1.21–3.05) | 0.006 b |
| Histological type (squamous vs adenosquamous cell carcinoma) | 304 | 1.32 (0.73–2.43) | 0.362 |
| Histologic grade (G3 and G4 vs G1 and G2) | 272 | 1.67 (1.03–2.72) | 0.037 b |
| PIK3CA status (Mut vs WT) | 286 | 1.09 (0.65–1.83) | 0.737 |
| Primary therapy outcome (PD, SD, and PR vs CR) | 217 | 2.14 (1.03–4.59) | 0.045 b |
CR: complete response; PD: progressive disease; PR: partial response; SD: stable disease.
Categorical dependent variable, greater or less than the median expression level.
Statistically significant.
Univariate and multivariate Cox analyses of survival
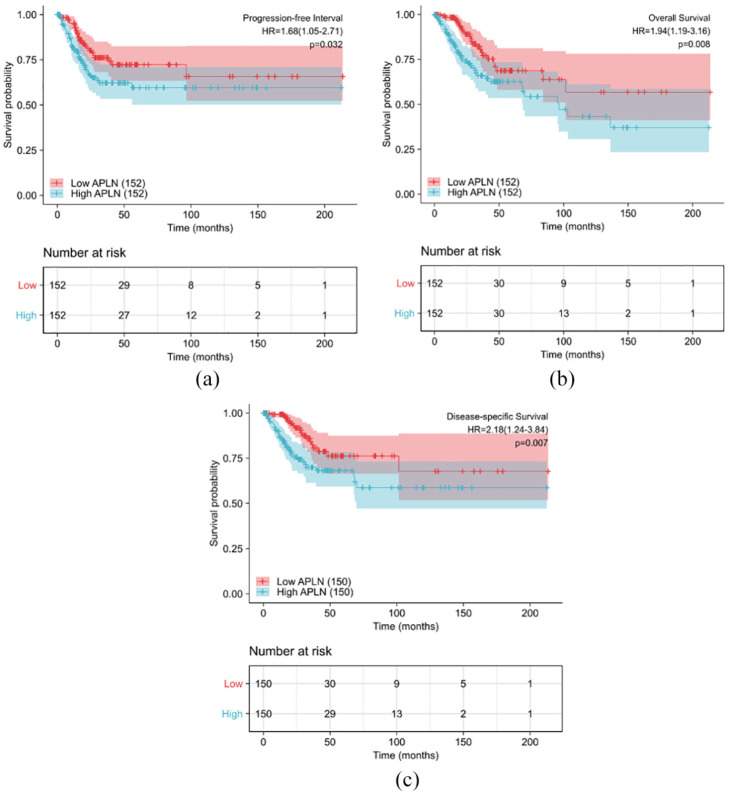
Patients with high expression of APLN had a significantly shorter OS, progression free survival (PFS), and disease specific survival (DSS) compared to those with low expression as shown in Figure 2(a) to (c) (both log-rank and p < 0.05). Univariate analysis revealed that N stage (HR 2.029; 95% CI: 1.358, 5.349; p = 0.005), primary therapy outcome (HR 13.514; 95% CI: 7.250, 25.189; p < 0.001), and APLN expression (HR 1.942; 95% CI: 1.192, 3.164; p = 0.008) were associated with OS. Additionally, Age (HR 1.612; 95% CI: 1.012, 2.568; p = 0.044), primary therapy outcome (HR 7.097; 95% CI: 4.631, 13.500; p < 0.001), and APLN expression (HR 1.683; 95% CI: 1.045, 2.709; p = 0.032) were associated with PFS. Finally, N stage (HR 3.303; 95% CI: 1.446–7.541; p = 0.005), Primary therapy outcome (HR 16.934; 95% CI: 8.629, 33.235; p < 0.001), and APLN expression (HR 2.178; 95% CI: 1.236, 3.838; p = 0.007) were associated with DSS. Moreover, multivariate analysis showed that APLN was an independent prognostic risk factor associated with OS, PFS, and DSS. The HR values were 3.428 (CI: 1.390–8.456; p = 0.007) for OS, 1.741 (CI: 1.010–2.999; p = 0.046) for PFS, and 5.134 (CI: 1.854–14.216; p = 0.002) for DSS along with primary therapy outcome (Tables 3–5).
Figure 2.
Impact of APLN expression on progression free interval (a), overall survival (b), and disease specific survival (c) in cervical cancer patients in the TCGA cohort.
Table 3.
Univariate and multivariate Cox proportional hazard analysis of APLN expression and OS for patients with cervical cancer in the TCGA cohort.
| Characteristics | Total (N) | HR (95% CI) univariate analysis | p Value univariate analysis | HR (95% CI) multivariate analysis | p Value multivariate analysis |
|---|---|---|---|---|---|
| N stage (N1 vs N0) | 193 | 2.695 (1.358–5.349) | 0.005 a | 2.887 (1.206–6.912) | 0.017 a |
| Clinical stage (stage II and stage III and stage IV vs stage I) | 297 | 1.429 (0.896–2.280) | 0.134 | ||
| Radiation therapy (yes vs no) | 304 | 1.153 (0.681–1.951) | 0.596 | ||
| Histological type (squamous vs adenosquamous cell carcinoma) | 304 | 1.010 (0.530–1.926) | 0.976 | ||
| Menopause status (post vs pre and peri) | 231 | 1.275 (0.744–2.185) | 0.376 | ||
| Histologic grade (G3 and G4 vs G1 and G2) | 272 | 0.889 (0.527–1.502) | 0.661 | ||
| Age (>50 vs ≤50) | 304 | 1.317 (0.825–2.101) | 0.248 | ||
| PIK3CA status (Mut vs WT) | 286 | 1.011 (0.599–1.707) | 0.967 | ||
| Primary therapy outcome (PD, SD, and PR vs CR) | 217 | 13.514 (7.250–25.189) | <0.001 a | 7.877 (2.792–22.222) | <0.001 a |
| Race (Asian and Black or African American vs White) | 259 | 0.841 (0.427–1.658) | 0.618 | ||
| APLN (high vs low) | 304 | 1.942 (1.192–3.164) | 0.008 a | 3.428 (1.390–8.456) | 0.007 a |
CI: confidence interval; CR: complete response; HR: hazard ratio; PD: progressive disease; PR: partial response; SD: stable disease.
Statistically significant.
Table 4.
Univariate and multivariate Cox proportional hazard analysis of APLN expression and PFS for patients with cervical cancer in the TCGA cohort.
| Characteristics | Total (N) | HR (95% CI) univariate analysis | p Value univariate analysis | HR (95% CI) multivariate analysis | p Value multivariate analysis |
|---|---|---|---|---|---|
| N stage (N1 vs N0) | 193 | 1.983 (0.986–3.990) | 0.055 | ||
| Clinical stage (stage II, stage III and stage IV vs stage I) | 297 | 1.308 (0.821–2.084) | 0.258 | ||
| Radiation therapy (yes vs no) | 304 | 1.288 (0.754–2.200) | 0.354 | ||
| Histological type (squamous vs adenosquamous cell carcinoma) | 304 | 0.778 (0.433–1.397) | 0.400 | ||
| Menopause status (post vs pre and peri) | 231 | 1.091 (0.640–1.861) | 0.749 | ||
| Histologic grade (G3 and G4 vs G1 and G2) | 272 | 1.594 (0.967–2.627) | 0.067 | ||
| Age (>50 vs ≤50) | 304 | 1.612 (1.012–2.568) | 0.044 a | 1.174 (0.688–2.002) | 0.557 |
| PIK3CA status (Mut vs WT) | 286 | 0.994 (0.589–1.679) | 0.983 | ||
| Primary therapy outcome (PD, SD, and PR vs CR) | 217 | 7.907 (4.631–13.500) | <0.001 a | 7.461 (4.342–12.821) | <0.001 a |
| Race (Asian and Black or African American vs White) | 259 | 1.066 (0.553–2.057) | 0.848 | ||
| APLN (high vs low) | 304 | 1.683 (1.045–2.709) | 0.032 a | 1.741 (1.010–2.999) | 0.046 a |
CI: confidence interval; CR: complete response; HR: hazard ratio; PD: progressive disease; PR: partial response; SD: stable disease.
Statistically significant.
Table 5.
Univariate and multivariate Cox proportional hazard analysis of APLN expression and DSS for patients with cervical cancer in the TCGA cohort.
| Characteristics | Total (N) | HR (95% CI) univariate analysis | p Value univariate analysis | HR (95% CI) multivariate analysis | p Value multivariate analysis |
|---|---|---|---|---|---|
| N stage (N1 vs N0) | 191 | 3.303 (1.446–7.541) | 0.005 a | 2.970 (1.187–7.430) | 0.02 a |
| Clinical stage (stage II, stage III and stage IV vs stage I) | 293 | 1.608 (0.941–2.749) | 0.082 | ||
| Radiation therapy (yes vs no) | 300 | 1.772 (0.891–3.522) | 0.103 | ||
| Histological type (squamous vs adenosquamous cell carcinoma) | 300 | 0.956 (0.467–1.958) | 0.901 | ||
| Menopause status (post vs pre and peri) | 228 | 1.233 (0.678–2.243) | 0.493 | ||
| Histologic grade (G3 and G4 vs G1 and G2) | 269 | 0.955 (0.530–1.720) | 0.877 | ||
| Age (>50 vs ≤50) | 300 | 1.333 (0.780–2.278) | 0.292 | ||
| PIK3CA status (Mut vs WT) | 282 | 1.097 (0.606–1.989) | 0.759 | ||
| Primary therapy outcome (PD, SD, and PR vs CR) | 217 | 16.934 (8.629–33.235) | <0.001 a | 10.497 (3.470–31.753) | <0.001 a |
| Race (Asian and Black or African American vs White) | 257 | 0.770 (0.344–1.724) | 0.526 | ||
| APLN (high vs low) | 300 | 2.178 (1.236–3.838) | 0.007 a | 5.134 (1.854–14.216) | 0.002 a |
CI: confidence interval; CR: complete response; HR: hazard ratio; PD: progressive disease; PR: partial response; SD: stable disease.
Statistically significant.
APLN-related signaling pathways based on gene set enrichment analysis (GSEA)
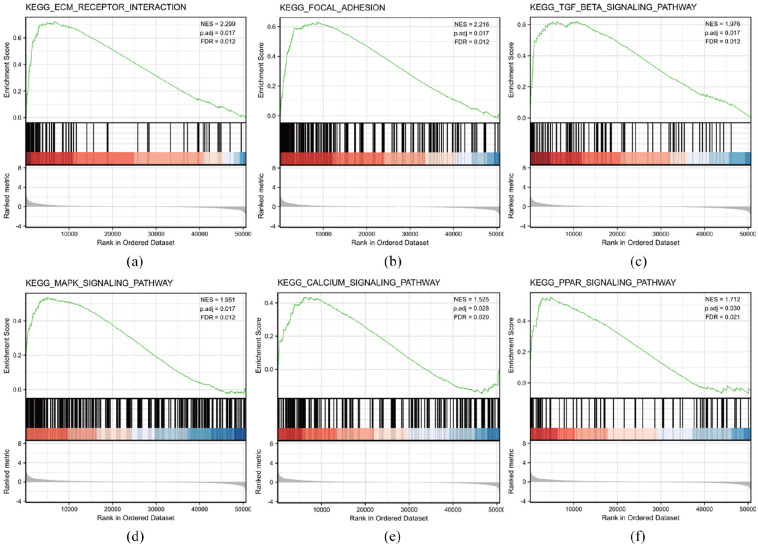
GSEA of differences between low and high APLN expression data sets was performed to identify signaling pathways involved in cervical cancer. The results demonstrated significant differences (FDR <0.05, adjusted p < 0.05) in enrichment of the MSigDB Collection (c2.all.v7.0.symbols.gmt (Curated)). Additionally, Six KEGG pathways, including the PPAR signaling pathway, ECM-receptor interaction pathway, focal adhesion pathway, MAPK-Signaling pathway, TGF-beta signaling pathway, and Calcium signaling pathway significantly showed differential enrichment in the APLN high expression phenotype based on adjusted p-values, Normalized Enrichment Scores (NES), and FDR values (Figure 3(a)–(f) and Table 6). This indicated the potential role of APLN in the development of cervical cancer.
Figure 3.
KEGG pathway enrichment analysis of APLN: (a) enrichment of genes in the KEGG ECM-receptor interaction pathway by GSEA, (b) enrichment of genes in the KEGG focal adhesion pathway by GSEA, (c) enrichment of gens in the KEGG TGF-beta signaling pathway by GSEA, (d) enrichment of KEGG MAPK-signaling pathway by GSEA, (e) enrichment of genes in the KEGG Calcium signaling pathway by GSEA, and (f) enrichment of genes in the KEGG PPAR signaling pathway by GSEA.
Table 6.
KEGG pathways enriched in the high APLN expression group using GSEA.
| Name | NES | p. adjust | FDR |
|---|---|---|---|
| KEGG_ECM_RECEPTOR_INTERACTION | 2.299 | 0.017 | 0.012 |
| KEGG_FOCAL_ADHESION | 2.216 | 0.017 | 0.012 |
| KEGG_TGF_BETA_SIGNALING | 1.976 | 0.017 | 0.012 |
| KEGG_MAPK_SIGNALING | 1.951 | 0.017 | 0.012 |
| KEGG_PPAR_SIGNALING | 1.712 | 0.03 | 0.021 |
| KEGG_CALCIUM_SIGNALING | 1.525 | 0.028 | 0.020 |
FDR: false discovery rate; GSEA: gene set enrichment analysis; NES: normalized enrichment score.
The correlation between APLN expression and immune infiltration
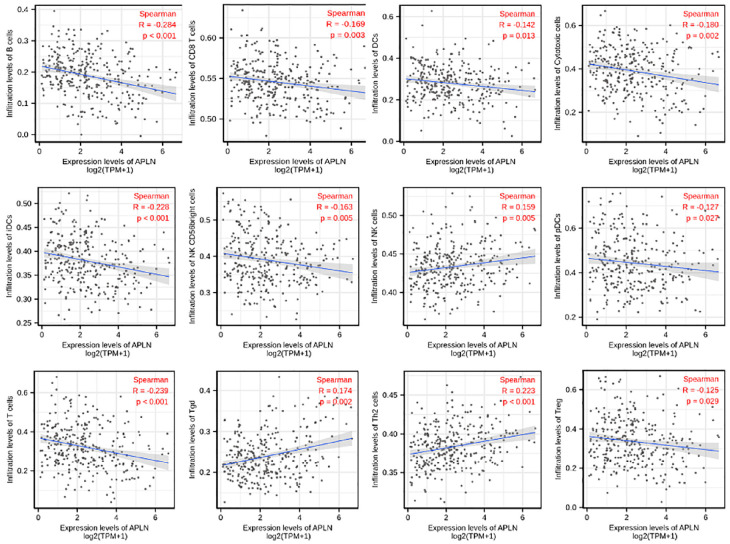
Spearman correlation was used to analyze the relationship between the expression level (TPM) of APLN and the extent of immune cell enrichment (generated by ssGSEA). The results showed that there was a negative correlation between the expression of APLN and infiltration levels of B cells(r = −0.284, p < 0.001), T cells(r = −0.239, p < 0.001), iDCs (r = −0.228, p < 0.001), Cytotoxic cells (r = −0.180, p = 0.002), CD8 T cells (r = −0.169, p = 0.003), DCs (r = −0.142, p = 0.013), NK CD56bright cells (r = −0.163, p = 0.005), pDCs (r = −0.127, p = 0.027), Treg (r = −0.125, p = 0.029) in CESC. However, APLN expression was positively correlated with the abundance of Th2 cells (r = 0.223, p < 0.001), Tgd (r = 0.174, p = 0.002), NK cells (r = 0.159, p = 0.005) in CESC (Figure 4, p < 0.05).
Figure 4.
The expression level of APLN was associated with immune infiltration in the tumor microenvironment. APLN expression was negatively correlated with the abundance of adaptive immunocytes (B cells, T cells, iDCs, Cytotoxic cells, CD8 T cells, DCs, NK CD56bright cells, pDCs, Treg). In contrast, expression of APLN was positively correlated with the abundance of innate immune cells (Th2 cells, Tgd, NK cells).
p < 0.05.
Discussion
Cervical cancer is one of the most common cancers that affect women’s health globally, especially in developing counties. Recently, screening is used as the main method for early diagnosis of cervical cancer. However, the accuracy of screening needs further improvement. Nonetheless, the prognosis of the disease remains poor. Notably, the prognosis and treatment of cervical cancer are based on the FIGO stage. This can especially be challenging since patients at the same FIGO stage may have different prognoses. Therefore, new biomarkers are urgently needed for early diagnosis and prognostic assessment.
APLN, a regulatory peptide, exhibits its effects by binding to the G protein-coupled receptor APJ. Upon binding to APJ, a series of different downstream signaling pathways are activated leading to many pathophysiological processes. 5 In recent years, the role of the APLN/APJ system in angiogenesis has been widely reported.16–18 Angiogenesis is an extremely important process during tumorigenesis. Furthermore, anti-angiogenesis drugs were shown to be effective inhibitors of tumor growth. 19 APLN has widely been reported to be involved in the oncogenicity and development of multiple tumor types. However, the correlation between APLN and cervical carcinoma has not been illustrated. Therefore, this study aimed to explore the potential role and value of APLN in cervical cancer.
RNA-seq data and corresponding clinical information were downloaded from the TCGA-Cervical Adenocarcinoma and Cervical Squamous Cell Carcinoma datasets. The results demonstrated that the expression level of APLN was significantly higher in cervical cancer compared to normal cervical tissues. Additionally, increased APLN was positively associated with advanced clinicopathologic features, poor primary therapy outcome, and short survival time in cervical cancer tissues. Moreover, the ROC curve analysis used for distinguishing cervical cancer from normal cervical tissues demonstrated that APLN may be a promising biomarker for the diagnosis of the malignancy. Furthermore, through GSEA, six KEGG pathways including the PPAR signaling pathway, ECM-receptor interaction pathway, focal adhesion pathway, MAPK-Signaling pathway, TGF-beta signaling pathway, and Gap junction pathway, were enriched differently in the APLN high expression phenotype of cervical cancer. All of these pathways have previously been reported to be involved in cancer cell proliferation, invasion, and metastasis.20–25 In addition, analysis of immune cell infiltration showed that high expression levels of APLN were positively associated with the abundance of Th2 cells, while negatively associated with infiltration levels of B cells. It has been reported that immune cell infiltration affects tumor prognosis and response to therapy, different tumor-infiltrating immune cells have different effect. 26 Th2 cells were associated with poor prognosis in various types of cancer. 27 In contrast, B cells were found to have positive prognostic effect on many cancer types. 28 Therefore, these results indicate that APLN may potentially drive the progression of cervical cancer.
The APLN/APJ system plays important role in vascular physiology. 29 However, increasing reports have highlighted its importance in cancer. APLN was reported to stimulate the proliferation of colon adenocarcinomas 30 as well as their migration and invasion. 31 Additionally, inhibition of the APLN/APJ system could lead to decreased proliferation and promote apoptosis in hepatocellular carcinoma. 32 Furthermore, increased expression of APLN was observed in muscle-invasive bladder cancer and was associated with poor clinical outcomes. 33 Our findings were consistent with these reports. The gene could therefore be considered as a potential druggable target.
To the best of our knowledge, this study is the first one to report on the correlation between APLN and cervical cancer. The findings provide a potentially novel biomarker for the prognosis of cervical carcinoma. However, this research had a few limitations. First, the study was performed based on data mining from the TCGA database. Therefore, results need to be verified by both in vitro and in vivo experiments. Second, the number of normal cervical tissues used as controls was not proportional to that of the cervical cancer tissues. This might have led to a misrepresentation of the results. Therefore, further studies with an equal number of cases and controls are required. Third, the study only illustrated the differences in expression of APLN in cervical cancer at the mRNA level. Consequently, differences in protein levels are unknown. Therefore, further research should be conducted to uncover the direct role of APLN in the development of cervical cancer.
Conclusions
In conclusion, elevated expression of APLN was significantly associated with progression and poor survival in cervical cancer. Therefore, APLN might be a potential prognostic molecular maker as well as a treatment target for cervical cancer. However, further experiments are needed to validate these results and ascertain the underlying mechanisms and clinical applications for cervical cancer patients.
Acknowledgments
Thanks are due to Xin for helpful suggestions.
Author biographies
Yusha Chen graduated from Fujian Medical University in 2011, then worked in Fujian Maternity and Child Health Hospital. Now her research focuses on cervical disease diagnosis and treatment.
Xiaoqian Lin received her PhD in Obstetrics in 2016 and currently she is an attending obstetrician and gynecologist in Fujian Maternity and Child Health Hospital.
Jinwen Zheng is the chief of the Cervical Disease Diagnosis and Treatment Health Center, Cervical Disease Diagnosis and Treatment Health Center. She has been engaged in the diagnosis and treatment of cervical diseases for more than twenty years.
Jiancui Chen is a chief physician of the Cervical Disease Diagnosis and Treatment Health Center, Cervical Disease Diagnosis and Treatment Health Center. She is committed to the prevention and treatment of cervical cancer.
Huifeng Xue is an excellent colposcope physician and dedicated to the diagnosis and treatment of cervical disease.
Xiangqin Zheng is a Professor of Obstetrics and Gynecology, Director of Gynecologic Surgery, he is dedicated to research and treatment of gynecological tumors.
Footnotes
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
Ethics approval: Ethical approval was not sought for the present study because the study didn’t contain any studies with human participants or animals performed by any of the authors.
Informed consent: Informed consent was not sought for the present study because the study didn’t contain any studies with human participants or animals performed by any of the authors and our study was in accordance with the publication guidelines provided by TCGA.
Trial registration: The study was not registered because it was not randomized clinical trial.
ORCID iD: Yusha Chen  https://orcid.org/0000-0002-7706-9976
https://orcid.org/0000-0002-7706-9976
References
- 1.Arbyn M, Weiderpass E, Bruni L, et al. Estimates of incidence and mortality of cervical cancer in 2018: a worldwide analysis. Lancet Glob Health 2020; 8: e191–e203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68: 394–424. [DOI] [PubMed] [Google Scholar]
- 3.Tatemoto K, Hosoya M, Habata Y, et al. Isolation and characterization of a novel endogenous peptide ligand for the human APJ receptor. Biochem Biophys Res Commun 1998; 251: 471–476. [DOI] [PubMed] [Google Scholar]
- 4.Pitkin SL, Maguire JJ, Bonner TI, et al. International Union of Basic and Clinical Pharmacology. LXXIV. Apelin receptor nomenclature, distribution, pharmacology, and function. Pharmacol Rev 2010; 62: 331–342. [DOI] [PubMed] [Google Scholar]
- 5.Antushevich H, Wójcik M.Review: apelin in disease. Clin Chim Acta 2018; 483: 241–248. [DOI] [PubMed] [Google Scholar]
- 6.Berta J, Kenessey I, Dobos J, et al. Apelin expression in human non-small cell lung cancer: role in angiogenesis and prognosis. J Thorac Oncol 2010; 5: 1120–1129. [DOI] [PubMed] [Google Scholar]
- 7.Diakowska D, Markocka-Mączka K, Szelachowski P, et al. Serum levels of resistin, adiponectin, and apelin in gastroesophageal cancer patients. Dis Markers 2014; 2014: 619649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muto J, Shirabe K, Yoshizumi T, et al. The apelin-APJ system induces tumor arteriogenesis in hepatocellular carcinoma. Anticancer Res 2014; 34: 5313–5320. [PubMed] [Google Scholar]
- 9.Wan Y, Zeng ZC, Xi M, et al. Dysregulated microRNA-224/apelin axis associated with aggressive progression and poor prognosis in patients with prostate cancer. Hum Pathol 2015; 46: 295–303. [DOI] [PubMed] [Google Scholar]
- 10.Heo K, Kim YH, Sung HJ, et al. Hypoxia-induced up-regulation of apelin is associated with a poor prognosis in oral squamous cell carcinoma patients. Oral Oncol 2012; 48: 500–506. [DOI] [PubMed] [Google Scholar]
- 11.Altinkaya SO, Nergiz S, Küçük M, et al. Apelin levels are higher in obese patients with endometrial cancer. J Obstet Gynaecol Res 2015; 41: 294–300. [DOI] [PubMed] [Google Scholar]
- 12.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005; 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu G, Wang LG, Han Y, et al. ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012; 16: 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hänzelmann S, Castelo R, Guinney J.GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013; 14: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013; 39: 782–795. [DOI] [PubMed] [Google Scholar]
- 16.Kunduzova O, Alet N, Delesque-Touchard N, et al. Apelin/APJ signaling system: a potential link between adipose tissue and endothelial angiogenic processes. FASEB J 2008; 22: 4146–4153. [DOI] [PubMed] [Google Scholar]
- 17.Kälin RE, Kretz MP, Meyer AM, et al. Paracrine and autocrine mechanisms of apelin signaling govern embryonic and tumor angiogenesis. Dev Biol 2007; 305: 599–614. 2007/04/07. DOI: 10.1016/j.ydbio.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 18.Chen D, Lee J, Gu X, et al. Intranasal delivery of apelin-13 Is neuroprotective and promotes angiogenesis after ischemic stroke in mice. ASN Neuro 2015; 7: 1759091415605114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Viallard C, Larrivée B.Tumor angiogenesis and vascular normalization: alternative therapeutic targets. Angiogenesis 2017; 20: 409–426. [DOI] [PubMed] [Google Scholar]
- 20.Borland MG, Kehres EM, Lee C, et al. Inhibition of tumorigenesis by peroxisome proliferator-activated receptor (PPAR)-dependent cell cycle blocks in human skin carcinoma cells. Toxicology 2018; 404–405: 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andersen MK, Rise K, Giskeødegård GF, et al. Integrative metabolic and transcriptomic profiling of prostate cancer tissue containing reactive stroma. Sci Rep 2018; 8: 14269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gong J, Lu X, Xu J, et al. Coexpression of UCA1 and ITGA2 in pancreatic cancer cells target the expression of miR-107 through focal adhesion pathway. J Cell Physiol 2019; 234: 12884–12896. [DOI] [PubMed] [Google Scholar]
- 23.Sheng W, Chen C, Dong M, et al. Calreticulin promotes EGF-induced EMT in pancreatic cancer cells via Integrin/EGFR-ERK/MAPK signaling pathway. Cell Death Dis 2017; 8: e3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seoane J, Gomis RR.TGF-β Family signaling in tumor suppression and cancer progression. Cold Spring Harb Perspect Biol 2017; 9: a022277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crottès D, Lin YT, Peters CJ, et al. TMEM16A controls EGF-induced calcium signaling implicated in pancreatic cancer prognosis. Proc Natl Acad Sci U S A 2019; 116: 13026–13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thorsson V, Gibbs DL, Brown SD, et al. The immune landscape of cancer. Immunity 2018; 48: 812–830.e814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fridman WH, Pagès F, Sautès-Fridman C, et al. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer 2012; 12: 298–306. [DOI] [PubMed] [Google Scholar]
- 28.Wouters MCA, Nelson BH. Prognostic significance of tumor-infiltrating B Cells and plasma cells in human cancer. Clin Cancer Res 2018; 24: 6125–6135. [DOI] [PubMed] [Google Scholar]
- 29.Strohbach A, Pennewitz M, Glaubitz M, et al. The apelin receptor influences biomechanical and morphological properties of endothelial cells. J Cell Physiol 2018; 233: 6250–6261. [DOI] [PubMed] [Google Scholar]
- 30.Picault FX, Chaves-Almagro C, Projetti F, et al. Tumour co-expression of apelin and its receptor is the basis of an autocrine loop involved in the growth of colon adenocarcinomas. Eur J Cancer 2014; 50: 663–674. [DOI] [PubMed] [Google Scholar]
- 31.Podgórska M, Pietraszek-Gremplewicz K, Nowak D.Apelin effects migration and invasion abilities of colon cancer cells. Cells 2018; 7: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen H, Wong CC, Liu D, et al. APLN promotes hepatocellular carcinoma through activating PI3K/Akt pathway and is a druggable target. Theranostics 2019; 9: 5246–5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang L, Li YL, Li XQ, et al. High apelin level indicates a poor prognostic factor in muscle-invasive bladder cancer. Dis Markers 2019; 2019: 4586405. [DOI] [PMC free article] [PubMed] [Google Scholar]