Abstract
Staphylococcus aureus is a major cause of nosocomial infections. During the period from March 1992 to March 1994, the patients admitted to the intensive care unit of the University of Maryland Shock Trauma Center were monitored for the development of S. aureus infections. Among the 776 patients eligible for the study, 60 (7.7%) patients developed 65 incidents of nosocomial S. aureus infections. Of the clinical isolates, 43.1% possessed a polysaccharide type 5 capsule, 44.6% possessed a type 8 capsule, and the remaining 12.3% had capsules that were not typed by the type 5 or type 8 antibodies. Six antibiogram types were noted among the infection-related isolates, with the majority of the types being resistant only to penicillin and ampicillin. It was noted that the majority of cases of pneumonia were caused by relatively susceptible strains, while resistant strains were isolated from patients with bacteremia and other infections. Only 16 (6.3%) of the isolates were found to be methicillin-resistant S. aureus (MRSA). DNA fingerprinting by pulsed-field gel electrophoresis showed 36 different patterns, with characteristic patterns being found for MRSA strains and the strains with different capsular types. Clonal relationships were established, and the origins of the infection-related isolates in each patient were determined. We conclude that (i) nosocomial infection-related isolates from the shock trauma patients did not belong to a single clone, although the predominance of a methicillin-resistant genotype was noted, (ii) most infection-related S. aureus isolates were relatively susceptible to antibiotics, but a MRSA strain was endemic, and (iii) for practical purposes, the combination of the results of capsular and antibiogram typing can be used as a useful epidemiological marker.
Staphylococcus aureus is a major cause of nosocomial infections that uses numerous virulence factors such as extracellular toxins and enzymes (13, 41). Studies from the Centers for Disease Control and Prevention (18) showed that during the period from 1985 to 1988, S. aureus was an important cause of postoperative wound infections and nosocomial bacteremia and pneumonia. Current therapy for infections due to S. aureus consists of removal of infected devices, surgical drainage of abscesses, and the use of antibiotics effective against the pathogen.
In the preantibiotic era S. aureus bacteremia resulted in 80% mortality (37); however, with the advent of antibiotics, the organism was reported to be susceptible to the earliest antimicrobial agents, the sulfonamides and penicillin. The widespread use of these antibiotics in the 1950s induced the predominance of β-lactamase-producing resistant strains. To solve the problem, the β-lactamase-resistant penicillins were developed, but reports of resistance to this new group started to appear in the 1960s in Europe (20) and in the 1970s in the United States (30).
The emergence of antibiotic-resistant strains of S. aureus is now considered to be a major problem in most hospitals. Virtually, all nosocomial strains produce a β-lactamase(s) and thus are resistant to penicillins. Moreover, data from the Centers for Disease Control and Prevention indicate that throughout the United States there has been an increase in the frequency of methicillin-resistant S. aureus (MRSA) strains resistant to multiple antibiotics in both large and small hospitals (19). Thus far, all strains of MRSA have been susceptible to vancomycin, although certain strains have exhibited tolerance. It is possible, however, that vancomycin-resistant gram-positive cocci such as Enterococcus spp. (2, 40) and Staphylococcus haemolyticus (36) may transmit the gene(s) responsible for this phenotype to S. aureus, leaving few if any options for antimicrobial chemotherapy of infections caused by the organism.
Previous studies with different populations indicated that S. aureus ranks second only to Staphylococcus epidermidis as a cause of nosocomial bacteremia, accounting for 13% of all cases in 1988 and increasing to 19% of the total in 1990 (23). In this study we investigated the prevalence of MRSA among isolates causing infection in patients admitted to the intensive care unit (ICU) of the University of Maryland Shock Trauma Center (ST-ICU). Our goal was to determine the relatedness of the patient nasal isolates and infection-related isolates by studying their characteristics by phenotypic methods (capsular and antibiogram typing) and genotypic methods (pulsed-field gel electrophoresis [PFGE] of genomic DNA) and to appraise the ability of phenotypic markers to discriminate among strains whose identities have been confirmed by modern strain delineation techniques based on DNA content.
MATERIALS AND METHODS
Study design and data collection.
A total of 785 patients admitted to the Shock Trauma Center between March 1992 and March 1994 were screened for nasal carriage of S. aureus at admission. Carriage of S. aureus was determined by obtaining swab specimens from both anterior nares of patients at various times, as described below. The swabs were inoculated onto 5% sheep blood agar plates (BBL, Cockeysville, Md.), and the plates were incubated for 24 h at 35°C and observed for the growth of suspected S. aureus colonies. After the identity of the isolates was confirmed (6), they were stored at −70°C in freezer vials pending further analysis.
The patients enrolled in the study were monitored daily until discharge for the subsequent development of a nosocomial infection (bacteremia, pneumonia, wound infection, etc.). S. aureus infections were defined as follows. Bacteremia was defined as any blood sample growing only S. aureus on culture. Pneumonia was defined by a diagnosis of pneumonia by an attending infectious diseases physician based upon new parenchymal abnormalities on chest radiograph that failed to resolve with chest physical therapy; temperature, >101°F; total leukocyte count, >10,000/mm3; abnormal arterial-to-alveolar oxygen gradient; and an endotracheal aspirate with >10 polymorphonuclear leukocytes per oil immersion field and numerous gram-positive cocci in clusters; and a heavy growth of S. aureus as the predominant organism. Wound infection was defined as fever, local cellulitis, and a purulent discharge from a wound yielding a pure culture of S. aureus. For patients who developed an S. aureus infection, nasal samples were recultured at the time of diagnosis. For patients who were nasal culture positive for S. aureus on admission but who did not develop an infection, samples from the nares were routinely recultured on day 14 of hospitalization or hospital discharge, whichever was first. The total number of S. aureus isolates recovered from the patients described above and included in the study was 254.
Capsular polysaccharide typing.
Capsular serotyping of the coded isolates (no patient identifiers were used) was performed by direct cell agglutination and immunoprecipitation of cell extracts as described previously (21). Two different types of rabbit serum samples were used to identify the capsular type of each isolate: sera from animals vaccinated with a whole-cell vaccine (21) and monospecific sera against either type 5 or type 8 capsular polysaccharide conjugated vaccine prepared in rabbits (11, 12). Monoclonal antibodies to capsular types 5 and 8 produced and standardized by NABI (Rockville, Md.) were used to confirm the capsular serotypes of the isolates.
Antimicrobial susceptibility testing.
The susceptibilities of all isolates to different antimicrobial agents were tested by the disk-agar method as standardized by the National Committee for Clinical Laboratory Standards (29). The following antimicrobial disks and concentrations were used: ampicillin, 10 μg; cephalothin, 30 μg; clindamycin, 2 μg; erythromycin, 15 μg; oxacillin, 1 μg; penicillin, 10 U; tetracycline, 30 μg; and vancomycin, 30 μg (Becton Dickinson Microbiology Systems, BBL, Cockeysville, Md.). The results were recorded after 24 h of incubation at 35°C. A standard strain of S. aureus (ATCC 25923) was used as a control. MRSA isolates were detected by two standard methods: by the disk-agar diffusion susceptibility method with 1-μg oxacillin disks, incubation at 35°C for 24 h, and inspection of the plates for colonies after 48 h (29) and by the ability of the isolates to grow on Mueller-Hinton agar supplemented with 4% sodium chloride and 6 μg of oxacillin per ml (MRSA Screen Agar; BBL) after incubation at 35°C for 24 h (25, 43).
β-Lactamase production and inhibition tests.
The ability of the isolates to produce β-lactamase was determined as recommended by the National Committee for Clinical Laboratory Standards (29). Confirmatory tests were performed by the disk-agar diffusion method with amoxicillin-clavulanic acid (AMC 30) and ampicillin-sulbactam (SAM 20) disks (Difco Laboratories, Detroit, Mich.), as recommended previously (29).
Genomic DNA Analysis by PFGE.
The genomic DNA was extracted from logarithmic-phase S. aureus cultures grown in brain heart infusion broth (BBL) as described previously by Murray et al. (27) and modified by Wanger et al. (42). The DNA size standards used were a bacteriophage lambda ladder consisting of concatemers starting at 48.5 kbp and increasing to approximately 1,000 kbp (Bio-Rad Laboratories, Hercules, Calif.). The gels were stained with ethidium bromide, rinsed, and photographed under UV light.
Statistical analysis.
Tests of statistical significance were performed by the use of the statistical packages SAS for Windows (SAS Institute, Cary, N.C.) and Sigma-Stat for Windows (Jandel Scientific, San Rafael, Calif.). Means are reported as X ± 2 standard errors. Differences in proportions were tested for significance by the chi-square test with Yate’s correction or by Fisher’s exact test (for frequencies of less than 5). Differences in means were tested for significance by Student’s t test. All P values are two-sided.
RESULTS
Demographics.
A total of 785 patients were enrolled in the study, with the majority being males (73.3%). Most of the patients were Caucasians (60%), followed by blacks (36%) and people of other races (4.0%). The median age of all patients enrolled in the study was 32 years (age range, 12 to 92 years). The mean age of the patients who developed a nosocomial infection was 46.2 ± 19.6 years, while that of the patients who did not develop any infection was 37.2 ± 18.4 years. The difference between the mean ages was found to be significant by the t test (P = 0.001). Of the 776 patients with adequate nasal specimen cultures, 150 (19.4%) were positive for S. aureus. Fifty-six patients (7.2%) were reported to have acquired 65 nosocomial S. aureus infections, namely, bacteremia (36; 55.4%), pneumonia (21; 32.3%), and wound and other infections (8; 12.3%). Seven of the patients developed multiple infections.
Capsular and antibiogram typing.
Among nasal carriers of S. aureus at admission, the prevalent capsule type was type 8 (65.9%). Among the isolates causing infections, however, there was an almost equal prevalence of both capsular types 5 and 8 (43.1 and 44.6%, respectively). When the infections were classified by the capsular type of the organisms causing the infection, it was found that among the isolates causing pneumonia, S. aureus strains of type 8 were prevalent (61.9%), while among the isolates causing bacteremia and wound infections, type 5 isolates were more common. Some isolates noted not to express either the type 5 or the 8 capsule (12.3%) were isolated from patients with bacteremia and wound infections but not from patients with pneumonia (28).
Table 1 demonstrates the eight different antibiogram types identified among the isolates tested in the study. The data presented in Table 2 indicate the frequency of retrieval of each of the antibiogram types at different stages of the hospital stay, i.e., admission, infection, and discharge. Strains of antibiogram type I were predominant at all stages and comprised 81.5% of isolates at admission, 64% of infection-related isolates and 65% of isolates at discharge. In contrast, the multidrug-resistant type, type III (MRSA), comprised 0.8, 14.6, and 5.7% of isolates, respectively. Other multidrug-resistant isolates but not MRSA, such as type II, were isolated only from patients with infection. The retrieval of isolates with specific antibiogram types only during certain stages of the hospital stay was statistically valid (P = 0.002).
TABLE 1.
S. aureus antibiogram types retrieved in this studya
| Type | Susceptibility to the following antibiotic:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Tet | Ox | P | E | Am | Va | Cc | Cf | |
| I | s | s | r | s | r | s | s | s |
| II | r | s | r | r | r | s | s | s |
| III | s | r | r | r | r | s | r | r |
| IV | r | s | r | s | r | s | s | s |
| V | s | s | s | s | s | s | s | s |
| VI | s | s | r | r | r | s | s | s |
| VII | s | s | r | r | r | s | r | s |
| VIII | s | s | s | r | s | s | s | s |
Abbreviations: Tet, tetracycline; Ox, oxacillin; P, penicillin G; E, erythromycin; Am, ampicillin; Va, vancomycin; Cc, clindamycin; Cf, cephalothin; s, susceptible; r, resistant.
TABLE 2.
Antibiogram types of isolates obtained from patients included in the study at different stages of hospitalizationa
| Stage | No. (%) of isolates with the following antibiogram type:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | V | VI | VII | VIII | Total | |
| NCb at admission | 106 | 0 | 1 | 2 | 9 | 9 | 3 | 0 | 130 |
| Infection | 44 | 2 | 8 | 1 | 5 | 5 | 0 | 0 | 65 |
| NC at infection | 13 | 2 | 5 | 1 | 1 | 2 | 0 | 0 | 24 |
| NC at discharge | 23 | 0 | 2 | 2 | 4 | 3 | 0 | 1 | 35 |
| Total | 186 (73.2) | 4 (1.6) | 16 (6.3) | 6 (2.3) | 19 (7.5) | 19 (7.5) | 3 (1.2) | 1 (0.4) | 254 (100.0) |
P = 0.002.
NC, nasal carriage of S. aureus.
The S. aureus infection-related isolates were found to have any one of six antibiogram types (Table 2). The antibiogram types of the infection-related isolates distributed by type of infection and capsular type are presented in Table 3. Isolates of antibiogram type I continued to be the most prevalent in all infections (66 to 76%). The multidrug-resistant strains of antibiogram type III were mainly isolated from patients with bacteremia and wound infections (16.7 and 40.0%, respectively).
TABLE 3.
Summary of the antibiogram types of the infection-related S. aureus isolates distributed by type of infection and capsular type
| Type of infection | Capsular typea | No. of isolates with the following antibiogram type:
|
||||||
|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | V | VI | Total | ||
| Bacteremia | T5 | 9 | 1 | 6 | 0 | 0 | 1 | 17 |
| T8 | 12 | 1 | 0 | 0 | 0 | 1 | 14 | |
| NT | 2 | 0 | 0 | 0 | 1 | 2 | 5 | |
| Pneumonia | T5 | 7 | 0 | 0 | 0 | 0 | 0 | 7 |
| T8 | 8 | 0 | 0 | 1 | 3 | 1 | 13 | |
| NT | 1 | 0 | 0 | 0 | 0 | 0 | 1 | |
| Wound | T5 | 0 | 0 | 2 | 0 | 0 | 0 | 2 |
| T8 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | |
| NT | 1 | 0 | 0 | 0 | 0 | 0 | 1 | |
| Otherb | T5 | 1 | 0 | 1 | 0 | 0 | 0 | 2 |
| T8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| NT | 1 | 0 | 0 | 0 | 0 | 0 | 1 | |
| Subtotal | T5 | 17 | 1 | 9 | 0 | 0 | 1 | 28 |
| T8 | 22 | 1 | 0 | 1 | 3 | 2 | 29 | |
| NT | 5 | 0 | 0 | 0 | 1 | 2 | 8 | |
| Total | 44 | 2 | 9 | 1 | 4 | 5 | 65 | |
Abbreviations: T5, capsular type 5; T8, capsular type 8; NT, nontypeable with the monoclonal capsular type 5 and capsular type 8 antisera.
Pleural fluid, gall bladder, and sputum.
Analysis of the relationship between a specific antibiogram type and expression of a specific capsular type for all the study isolates (Table 4) indicated that this relationship was statistically valid (P < 0.0001) and that there was a tendency for strains expressing a capsular type to have only specific antibiogram patterns. It must be noted that the number of isolates in some of the antibiograms was too small for inclusion of the data for those isolates in the analysis.
TABLE 4.
Association of capsular types with specific antibiogram types of the S. aureus isolates in the study
| Antibiogram type | No. of isolates with the following capsular typea:
|
|||
|---|---|---|---|---|
| T5 | T8 | NT | Total | |
| I | 53 | 119 | 14 | 186 |
| II | 2 | 2 | 0 | 4 |
| III | 16 | 0 | 0 | 16 |
| IV | 0 | 4 | 2 | 6 |
| V | 7 | 10 | 2 | 19 |
| VI | 7 | 9 | 3 | 19 |
| VII | 0 | 2 | 1 | 3 |
| VIII | 1 | 0 | 0 | 1 |
| Total | 86 | 146 | 22 | 254 |
Abbreviations: T5, capsular type 5; T8, capsular type 8; NT, nontypeable with the monoclonal capsular type 5 and capsular type 8 antisera.
Production of β-lactamase and methicillin resistance.
S. aureus strains capable of producing β-lactamase were, as expected, prevalent (92.1%). Only 3 of the 20 isolates found not to be β-lactamase producers were infection-related isolates, constituting only 4.6% of the infection-related isolates.
Table 5 indicates the frequency of MRSA isolates among the different isolates included in the study. Only 16 (6.3%) of the study isolates were MRSA strains, as detected by standard methods (25, 29, 43) and confirmed by performing the β-lactamase production and inhibition tests to ensure the exclusion of the false-positive hyper-β-lactamase-producing strains of S. aureus (26). The majority (81.3%) of the MRSA isolates were found to be infection-related isolates (infection and postinfection nasal swabs). All the MRSA strains were significantly found to have multidrug-resistant antibiogram type III and to possess the type 5 capsule.
TABLE 5.
Frequency of methicillin resistance among the different isolates obtained from patients during the studya
| Source of isolate | No. (%) of isolates
|
||
|---|---|---|---|
| Methicillin resistant | Methicillin susceptible | Total | |
| NC at admission | 1 (0.8) | 129 (99.2) | 130 |
| Infection | 8 (12.3) | 57 (87.7) | 65 |
| NC at infection | 5 (20.8) | 19 (79.2) | 24 |
| NC at discharge | 2 (5.7) | 33 (94.3) | 35 |
| Total | 16 (6.3) | 238 (93.7) | 254 |
Abbreviations: NC, nasal carriage of S. aureus.
DNA fingerprinting.
A total of 74 S. aureus isolates obtained from the 60 patients who suffered from infections were fingerprinted by PFGE. In addition to the infection-related isolates, the total represents nasal S. aureus isolates from positive cultures of nasal specimens taken from infected patients upon admission, infection, or discharge, when available. Of these, 32 (43.2%) expressed a type 5 capsule, 33 (44.6%) expressed a type 8 capsule, and 9 (12.2%) were not typed by the monoclonal capsular type 5 and capsular type 8 antibodies. It was notable that the percentages of retrieval of the different capsular serotypes were not changed after adding the data for the types of the few nasal isolates from infected patients.
The DNA fragment patterns were stable features of the isolates and were consistent in different DNA isolation procedures. Upon repeat subculture, the DNA restriction patterns of the isolates did not change. In this study a genotype was defined as any fragment pattern which varied from another pattern with regard to the number and size of the DNA fragments. Two or more isolates were defined as identical if they showed analogous patterns. Types with small differences in the fragment patterns were considered variants belonging to the same major type (16, 35), because it was demonstrated that a single molecular event (deletion, substitution, etc.) can potentially give rise to mobility alterations in one to three restriction fragments (isolates that showed patterns that differed in one to three bands were considered genetically related but not identical [16]). Strains having differences in more than three bands were considered unrelated.
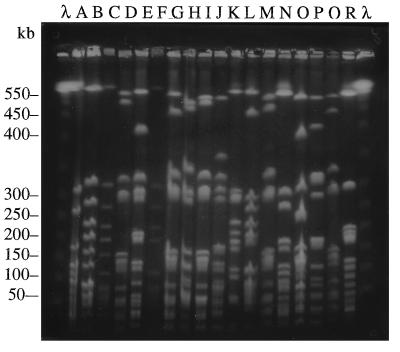
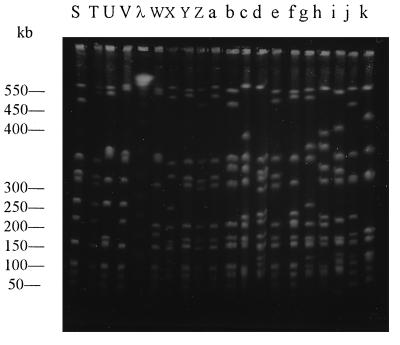
In total, 36 different genotypes were obtained. Figure 1 illustrates the 18 different PFGE patterns obtained for the S. aureus isolates that expressed the type 8 capsule. Among these the most frequent pattern was pattern A, which was characteristic of 11 (33.3%) isolates. Figure 2 demonstrates the 16 patterns obtained for the isolates that expressed the type 5 capsule and the 3 patterns recovered for the non-type 5, non-type 8 isolates. Among the S. aureus type 5 isolates, pattern X was the most common pattern and was retrieved for 5 (15.6%) isolates, followed by pattern S for 4 (12.5%) isolates and pattern Y for 3 (9.4%) isolates. Patterns E, k, and j were restricted to the non-type 5, non-type 8 S. aureus isolates. The first four patterns shown in Fig. 2 (patterns S, T, U, and V) are the patterns obtained solely for the MRSA strains. There was minimal overlapping of the patterns between capsular type groups; only three patterns (patterns D, F, and i) appeared in more than one capsular type. Two patterns (patterns D and F) overlapped between the type 5 isolates (one isolate with pattern D and one isolates with pattern F) and the type 8 isolates (two isolates with pattern D and one isolate with pattern F), while the third pattern (pattern i) overlapped between the type 5 (two isolates) and the non-type 5, non-type 8 isolates (six isolates). It was also noted that 66.7% of the non-type 5, non-type 8 isolates demonstrated pattern i and that none of the patterns were shared by the type 5 and non-type 5, non-type 8 isolates.
FIG. 1.
Contour-clamped homogeneous electric field characterization of isolates by PFGE. Chromosomal DNA was digested with the endonuclease SmaI and separated by PFGE and was then photographed under UV light. The different patterns shown were obtained for the isolates that expressed the type 8 capsular polysaccharide. Lanes λ, size markers.
FIG. 2.
Contour-clamped homogenous electric field characterization of isolates by PFGE. Chromosomal DNA was digested with endonuclease SmaI and separated by PFGE and was then photographed under UV light. The different patterns were obtained from the isolates that expressed the type 5 capsular polysaccharide (patterns S to h) and the patterns obtained from the isolates denoted as being non-type 5, non-type 8 strains (patterns i to k). Patterns S to V represent the patterns obtained for the MRSA strains. Lane λ, size markers.
The isolates available for each S. aureus infection episode were studied collectively, clonal relationships were established (Fig. 3), and the origin of the infection-related isolate from each patient was determined. Although in most patients the profiles indicated either a hospital-related transmission or autoinfection (due to nasal carriage), no outbreaks were detected during the period of the study.
FIG. 3.
Contour-clamped homogeneous electric field characterization of isolates from bacteremic patients by PFGE. Chromosomal DNA was digested with endonuclease SmaI and separated by PFGE and was then photographed under UV light. The different patterns are shown in groups for different patients to determine strain delineation. Patterns designated a and b are those for isolates from blood (pattern a) and nasal swab at infection (pattern b) from patient 1; patterns designated 1, 2, 3, and 4 refer to S. aureus isolates from patients with nasal carriage at admission (pattern 1), infection (bacteremia) (pattern 2), infection (gall bladder) (pattern 3), and nasal carriage at discharge (pattern 4), respectively. Lanes A, B, C, and three isolates from blood, nasal swab at infection, and nasal swab at discharge, respectively, from patient 3; Lane λ, size markers.
DISCUSSION
This study was designed to determine the prevalence of MRSA isolates among infection-related isolates from patients admitted to the ST-ICU, characterize the infection-related isolates phenotypically and genotypically to determine the genetic relatedness of the organisms involved, and evaluate whether the different typing methods are efficient tools in routine epidemiological investigations.
Most of the S. aureus isolates retrieved in this study (91.0%) expressed either capsular polysaccharide type 5 or capsular polysaccharide type 8 (Table 3), in compliance with previous reports (38, 42). The relationship between nasal carriage of S. aureus and subsequent infection has been well established by many investigators (7, 28, 33, 41, 44). Notably, the type 8 capsules were expressed more in the isolates from the nares of patients at admission to the hospital, while no significant prevalence of any of the types was noted among the infection-related isolates. When the infections were classified by the capsular type of the organism causing the infection, the type 5 capsular type was noted to predominate in patients with bacteremia, while capsular type 8 was the prevalent type in patients with pneumonia (Table 3).
The antibiogram typing of the isolates revealed that antibiogram type I (resistant only to penicillin G and ampicillin) was predominant for 73.2% of all the nasal and infection-related isolates (Table 2). This result reflects the predominance of relatively susceptible strains in our setting. The retrieval of more resistant strains among the infection-related isolates was expected (Table 3). An interesting finding was not only that some isolates with specific antibiogram types expressed a specific type of capsule (all type III isolates had a type 5 capsule) but also that there existed a significant relationship (P < 0.0001) between the recovery of certain antibiogram types from isolates with a specific capsular type (Table 4).
Of the isolates tested, 6.3% were found to be resistant to methicillin. The results presented in Table 5 indicate that most of the MRSA isolates are infection-related isolates, implying that MRSA is present in a reservoir in the hospital environment. The finding that only 16.7% of the bacteremias in this study were caused by MRSA reflected a proper hospital management plan for MRSA, because MRSA comprised 20% of all isolates causing bacteremia at the University of Maryland Hospital in 1988, a proportion that increased dramatically to 50% in 1990 (23).
Our study disclosed a significant relationship between the property of methicillin resistance and the possession of a specific type of capsule. All our MRSA isolates (100%) possessed a type 5 capsule, in agreement with previous reports noting the prevalence of type 5 capsules in MRSA isolates (5, 14, 42).
The finding that most infection-related S. aureus isolates belonged to capsular polysaccharide type 5 or 8 did not signify whether these strains were derived from resident (patient), commensal isolates or were the result of the nosocomial spread of a single endemic strain. To perform proper epidemiological typing and delineation of genetic relatedness, a number of methods were introduced and used effectively by other investigators. These methods included bacteriophage typing (22, 44), plasmid pattern analysis (4, 8), whole-cell protein profile analysis (9), immunoblotting profile analysis (15), ribotyping (10, 39), restriction endonuclease analysis of plasmid DNA (17, 34), and PFGE of restriction enzyme-digested genomic DNA (1, 24, 32). We chose to use PFGE for our study because the exceptional properties of PFGE have, with time, turned it into a favored epidemiological tool (3, 27, 31). PFGE detects the distribution of restriction sites throughout the chromosome, giving fingerprints that reflect the structural organization of the bacterial chromosome (31). This distribution of restriction sites, which varies because of ancestral strain-to-strain mutational differences and because of the variations in the gene content of the staphylococcal chromosome, give the method its acknowledged discriminatory ability.
The DNA components of all of the isolates from infected patients were fingerprinted by PFGE. Each of the genotypes obtained by PFGE of DNA for the S. aureus isolates tested was given a different designation, without consideration of band differences and relatedness, in order to present large numbers of patterns, as was previously done by others (31, 35). Except for the few exceptions mentioned previously, the 36 patterns obtained were clearly separated on the basis of capsular type. The isolates that expressed the type 8 capsular polysaccharide were found to have to a specific set of patterns (Fig. 1), while those that expressed the type 5 capsular polysaccharide had a different set of patterns (Fig. 2). The isolates that did not produce either the type 5 or the type 8 capsule were found to have one of three patterns (patterns i, j, and k in Fig. 2). An interesting result was the finding that the MRSA isolates that were found to produce a type 5 capsule had one of only four possible patterns that were not shared by any of the other isolates that produced type 5 patterns (patterns S to V in Fig. 2), with pattern S being the most prevalent (56.3%). This finding confirmed the clonal relatedness of the MRSA strains isolated in the study.
Figure 3 presents the PFGE patterns for MRSA isolates from three different bacteremic patients. The data demonstrate the clonal relationships of these isolates. Isolates from the same patient had identical DNA patterns. Furthermore, isolates from the different patients had similar patterns (a difference of only one or two bands). These data establish the presence of a resident MRSA population endemic in our setting, probably as nosocomial colonization of hospital personnel. This finding is a reasonable incentive for initiating an infection control study that aims at detecting and treating MRSA carriers in shock trauma units.
PFGE demonstrated typeability, reproducibility, and an ability to discriminate between strains. These properties demarcate an excellent tool for epidemiological studies; however, the only problem with the routine use of this method is the difficulty in performing the technique and the need for expensive instruments and chemicals.
The trend toward developing precise molecular biology-based methods has diverted attention from the development of simpler and more common typing methods (like capsular and antibiogram typing techniques). Attempts were made to use a combination of antibiogram and capsular typing methods, both of which are simple techniques that are routinely performed in many hospitals, to assess the possibility of using them as discriminatory tools for epidemiological studies. For practical purposes, the epidemiological conclusions based on the results of the tests based on the use of the previous combination of methods correlated well with those based on the use of the DNA fingerprinting by PFGE, especially with regard to the MRSA isolates.
The previous results indicated the possibility of using the two typing methods in combination in order to provide a more effective tool for epidemiological studies. Because antibiogram typing has been used as an effective tool in associating isolates to outbreaks in hospitals (42) and because both capsular typing and antibiogram typing are easy and reproducible techniques, we suggest that a combination of these simple typing methods be used for the routine processing of clinical specimens in hospitals, because the results obtained may provide valuable epidemiological information. However, we suggest increasing the number of antibiotics used in antibiogram typing, because this will increase the technique’s potential for discrimination.
We conclude that (i) the nosocomial infection isolates from the shock trauma patients were not found to belong to a single clone, although the predominance of one methicillin-resistant genotype was noted. (ii) Most S. aureus infections were caused by organisms relatively susceptible to antibiotics, although a well-defined multidrug resistant hospital strain of MRSA was present in our setting. (iii) For practical purposes, we suggest the use of a combination of the results of capsular and antibiogram typing as an epidemiological marker, based on the fact that in our study the combined results correlated well with those based on the use of the DNA fingerprinting by PFGE.
ACKNOWLEDGMENT
T. Na’was was a visiting researcher from the Faculty of Medicine of the Jordan University of Science and Technology supported by a Fulbright grant sponsored by the United States Information Agency.
REFERENCES
- 1.Allardet-Servent A, Bouziges N, Carles-Nurit M J, Bourg G, Gouby A, Ramuz M. Use of low-frequency-cleavage restriction endonucleases for DNA analysis in epidemiological investigations of nosocomial bacterial infections. J Clin Microbiol. 1989;27:2057–2061. doi: 10.1128/jcm.27.9.2057-2061.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Obeid S, Collatz E, Gutman L. Mechanism of resistance to vancomycin in Enterococcus faecium D366 and Enterococcus fecalis A256. Antimicrob Agents Chemother. 1990;34:252–256. doi: 10.1128/aac.34.2.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arbeit R D, Mathur M, Dunn R, Kim C, Selander R K, Goldstein R. Resolution of recent evolutionary divergence among Escherichia coli from related lineages: the application of pulsed-field electrophoresis to molecular epidemiology. J Infect Dis. 1990;161:230–235. doi: 10.1093/infdis/161.2.230. [DOI] [PubMed] [Google Scholar]
- 4.Archer G L, Mayhall C G. Comparison of epidemiological markers used in the investigation of an outbreak of methicillin-resistant Staphylococcus aureus infections. J Clin Microbiol. 1983;18:395–399. doi: 10.1128/jcm.18.2.395-399.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boutonnier A, Nato F, Bouvet A, Lebrun L, Audurier A, Mazie J C, Fournier J M. Direct testing of blood cultures for detection of the serotype 5 and 8 capsular polysaccharides of Staphylococcus aureus. J Clin Microbiol. 1989;27:989–993. doi: 10.1128/jcm.27.5.989-993.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chambers H F, Hackbarth C J. Effect of NaCl and nafcillin on penicillin-binding protein 2a and heterogeneous expression of methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1987;31:1982–1988. doi: 10.1128/aac.31.12.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chow J W, Yu V L. Staphylococcus aureus nasal carriage in hemodialysis patients. Arch Intern Med. 1989;149:1258–1262. [PubMed] [Google Scholar]
- 8.Collins J K, Smith J S, Kelly M T. Comparison of phage typing, plasmid mapping, and antibiotic resistance patterns as epidemiologic markers in a nosocomial outbreak of methicillin-resistant Staphylococcus aureus infections. Diagn Microbiol Infect Dis. 1984;2:233–245. doi: 10.1016/0732-8893(84)90036-1. [DOI] [PubMed] [Google Scholar]
- 9.Costas M, Cookson B D, Talsania H G, Owen R J. Numerical analysis of electrophoresis protein patterns of methicillin-resistant strains of Staphylococcus aureus. J Clin Microbiol. 1989;27:2574–2581. doi: 10.1128/jcm.27.11.2574-2581.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Buyser M L, Morvan A, Grimont F, El-Solh N. Characterization of Staphylococcus aureus species by ribosomal gene restriction patterns. J Gen Microbiol. 1989;135:989–999. doi: 10.1099/00221287-135-4-989. [DOI] [PubMed] [Google Scholar]
- 11.Fattom A, Schneerson R, Szu S C, Vann W F, Shiloach J, Karakawa W W, Robbins J B. Synthesis and immunologic properties in mice of vaccines composed of Staphylococcus aureus type 5 and type 8 capsular polysaccharides conjugated to Pseudomonas aeruginosa exotoxin A. Infect Immun. 1990;58:2367–2374. doi: 10.1128/iai.58.7.2367-2374.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fattom A, Schneerson R, Watson D C, Karakawa W W, Fitzgerald D, Pastan I, Li X, Shiloach J, Bryla D A, Robbins J B. Laboratory and clinical evaluation of conjugate vaccines composed of Staphylococcus aureus type 5 and type 8 capsular polysaccharides bound to Pseudomonas aeruginosa recombinant exoprotein A. Infect Immun. 1993;61:1023–1032. doi: 10.1128/iai.61.3.1023-1032.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster T J. Potential for vaccination against infections caused by Staphylococcus aureus. Vaccine. 1991;9:221–227. doi: 10.1016/0264-410x(91)90103-d. [DOI] [PubMed] [Google Scholar]
- 14.Fournier J M, Bouvet A, Boutonnier A, Audurier A, Goldstein F, Pierre J, Bure A, Lebrun L, Hochkeppel H K. Predominance of capsular polysaccharide type 5 among oxacillin-resistant Staphylococcus aureus. J Clin Microbiol. 1987;25:1932–1933. doi: 10.1128/jcm.25.10.1932-1933.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaston M S, Duff P S, Naidoo J, Ellis K, Roberts J I S, Richardson J F, Marples R R, Cooke E M. Evaluation of electrophoretic methods for typing methicillin-resistant Staphylococcus aureus. J Med Microbiol. 1988;26:189–197. doi: 10.1099/00222615-26-3-189. [DOI] [PubMed] [Google Scholar]
- 16.Goering R V the Molecular Epidemiological Study Group. Third International Meeting of Bacterial Epidemiological Markers, Cambridge, England. 1994. Guidelines for evaluating pulsed-field restriction fragment patterns in the epidemiological analysis of nosocomial infections, abstr. F19. [Google Scholar]
- 17.Hartsein A I, Morthland V H, Eng S, Archer G L, Schoenknecht F D, Rashad A L. Restriction enzyme analysis of plasmid DNA and bacteriophage typing of paired Staphylococcus aureus blood culture isolates. J Clin Microbiol. 1989;27:1874–1879. doi: 10.1128/jcm.27.8.1874-1879.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horan T, Culver D, Jarvis W, et al. Pathogens causing nosocomial infections: preliminary data from the national nosocomial infections surveillance system. Antimicrob Newsl. 1988;5:65–67. [Google Scholar]
- 19.Hughes J M. Setting priorities: nationwide nosocomial infection prevention and control programs in the USA. Eur J Clin Microbiol. 1987;6:348–351. doi: 10.1007/BF02017638. [DOI] [PubMed] [Google Scholar]
- 20.Jessen O, Rosendal K, Bülow P, Faber V, Eriksen K R. Changing staphylococci and staphylococcal infections: a ten-year study of bacteria and cases of bacteremia. N Engl J Med. 1969;281:627–635. doi: 10.1056/NEJM196909182811201. [DOI] [PubMed] [Google Scholar]
- 21.Karakawa W W, Fournier J M, Vann W F, Arbeit R, Schneerson R S, Robbins J B. Method for the serological typing of the capsular polysaccharides of Staphylococcus aureus. J Clin Microbiol. 1985;22:445–447. doi: 10.1128/jcm.22.3.445-447.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luzar M A, Coles G A, Faller B, Slingeneyer A, DahDah G, Briat C, Wone C, Knefati Y, Kessler M, Peluso F. Staphylococcus aureus nasal carriage and infection in patients on continuous ambulatory peritoneal dialysis. N Engl J Med. 1990;322:505–509. doi: 10.1056/NEJM199002223220804. [DOI] [PubMed] [Google Scholar]
- 23.Martin, M. A. Unpublished data.
- 24.McClelland M, Jones R Y, Nelson M. Restriction endonucleases for pulsed field mapping of bacterial genomes. Nucleic Acids Res. 1987;15:5985–6005. doi: 10.1093/nar/15.15.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDougal L K, Thornsberry C. New recommendations for disk diffusion antimicrobial susceptibility tests for methicillin-resistant (heteroresistant) staphylococci. J Clin Microbiol. 1984;19:482–488. doi: 10.1128/jcm.19.4.482-488.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDougal L K, Thornsberry C. The role of β-lactamase in staphylococcal resistance to penicillinase-resistant penicillins and cephalosporins. J Clin Microbiol. 1986;23:832–839. doi: 10.1128/jcm.23.5.832-839.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murray B E, Singh K V, Heath J D, Sharma B R, Weinstock G M. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J Clin Microbiol. 1990;28:2059–2063. doi: 10.1128/jcm.28.9.2059-2063.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Na’was, T., A. Hawwari, E. Hendrix, J. Hebden, R. Edelman, M. Martin, W. Campbell, R. Naso, R. Schwalbe, and A. Fattom. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 29.National Committee for Clinical Laboratory Standards. Approved standard M2-A5. Performance standards for antimicrobial disk susceptibility tests. 5th ed. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1993. [Google Scholar]
- 30.Peacock J E, Jr, Marsik F J, Wenzel R P. Methicillin-resistant Staphylococcus aureus: introduction and spread within a hospital. Ann Intern Med. 1980;93:526–532. doi: 10.7326/0003-4819-93-4-526. [DOI] [PubMed] [Google Scholar]
- 31.Prevost G, Jaulhac B, Piemont Y. DNA fingerprinting by pulsed-field gel electrophoresis is more effective than ribotyping in distinguishing among methicillin-resistant Staphylococcus aureus isolates. J Clin Microbiol. 1992;30:967–973. doi: 10.1128/jcm.30.4.967-973.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prevost G, Pottecher B, Dahlet M, Bientz M, Mantz J M, Piemont Y. Pulsed-field gel electrophoresis as a new epidemiological tool for monitoring methicillin-resistant Staphylococcus aureus in an intensive care unit. J Hosp Infect. 1991;17:255–269. doi: 10.1016/0195-6701(91)90270-i. [DOI] [PubMed] [Google Scholar]
- 33.Raviglione M C, Mariuz P, Pablos-Mendez A, Battan R, Ottuso P, Taranta A. High Staphylococcus aureus nasal carriage rate in patients with acquired immunodeficiency syndrome or AIDS related complex. Am J Infect Control. 1990;18:64–69. doi: 10.1016/0196-6553(90)90083-5. [DOI] [PubMed] [Google Scholar]
- 34.Rhinehart E, Shlaes D M, Keys T F, Serkey J, Kirkley B, Kim C, Currie-McCumber C A, Hall G. Nosocomial clonal dissemination of methicillin-resistant Staphylococcus aureus. Arch Intern Med. 1987;92:7–13. [PubMed] [Google Scholar]
- 35.Schlichting C, Branger C, Fournier J-M, Witte W, Boutonnier A, Wolz C, Goullet P, Döring G. Typing of Staphylococcus aureus by pulsed-field gel electrophoresis, zymotyping, capsular typing, and phage typing: resolution of clonal relationships. J Clin Microbiol. 1993;31:227–232. doi: 10.1128/jcm.31.2.227-232.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwalbe R S, Stapleton J S, Gilligan P H. Emergence of vancomycin resistance in coagulase-negative staphylococci. N Engl J Med. 1987;316:927–931. doi: 10.1056/NEJM198704093161507. [DOI] [PubMed] [Google Scholar]
- 37.Smith I M, Vickers A B. Natural history of 338 treated and untreated patients with staphylococcal septicaemia (1936–1955) Lancet. 1960;i:1318–1322. [PubMed] [Google Scholar]
- 38.Sompolonsky D, Samra Z, Karakawa W W, Vann W F, Schneerson R, Malik Z. Encapsulation and capsular types in isolates of Staphylococcus aureus from different sources and relationship to phage types. J Clin Microbiol. 1985;22:828–834. doi: 10.1128/jcm.22.5.828-834.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomson-Carter F M, Carter P E, Pennington T H. Differentiation of Staphylococcus aureus species and strains by ribosomal RNA gene patterns. J Gen Microbiol. 1989;135:2093–2097. doi: 10.1099/00221287-135-7-2093. [DOI] [PubMed] [Google Scholar]
- 40.Uttley A H, George R C, Naidoo J, Woodford N, Johnson A P, Collins C H, Morrison D, Gilfillan A J, Fitch L E, Heptonstall J. High level vancomycin-resistant enterococci causing hospital infections. Epidemiol Infect. 1989;103:173–181. doi: 10.1017/s0950268800030478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waldvogel F A. Staphylococcus aureus. In G. L. Mandell, R. G. Douglas, and J. E. Bennett (ed.), Principles and practices of infectious disease. 3rd ed. New York, N.Y: Churchill Livingstone; 1990. [Google Scholar]
- 42.Wanger A R, Morris S L, Ericsson C, Singh K V, LaRocco M T. Latex agglutination-negative methicillin-resistant Staphylococcus aureus recovered from neonates: epidemiologic features and comparison of typing methods. J Clin Microbiol. 1992;30:2583–2588. doi: 10.1128/jcm.30.10.2583-2588.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Working Party of the Hospital Infection Society and British Society for Antimicrobial Chemotherapy. Guidelines for the control of epidemic methicillin resistant Staphylococcus aureus. J Hosp Infect. 1986;7:193–201. [PubMed] [Google Scholar]
- 44.Yu V L, Goetz A, Wagener M, Smith P B, Rihs J D, Hanchett J, Zuraleff J J. Staphylococcus aureus nasal carriage and infection in patients on hemodialysis: efficacy of antibiotic prophylaxis. N Engl J Med. 1986;315:91–96. doi: 10.1056/NEJM198607103150204. [DOI] [PubMed] [Google Scholar]