Abstract
The internal areas and the position of integration of the glycopeptide resistance element Tn1546 were characterized by using PCR fragment length polymorphism, sequencing, and DNA hybridization techniques with 38 high-level vancomycin-resistant Enterococcus faecium isolates of human and animal origins from Europe and the United States. Only minor variations in the coding regions within Tn1546 were found, suggesting high genetic stability. The isolates originated from broilers (n = 5), a chicken (n = 1), a duck (n = 1), a turkey (n = 1), pigs (n = 8), a pony (n = 1), and humans (n = 23). A total of 13 different types were defined based on a single-nucleotide difference in the vanX gene, the presence of insertion sequences, and hybridization patterns. For some types more than one isolate were found. For type 1, 10 isolates of both human and animal origins were 1found. All were indistinguishable from the reference strain, BM4147. For type 2, 11 isolates of human and animal origins were found. Six human isolates from England were all of type 3. Two human isolates from the United States, indistinguishable from each other, were type 9. These results showed that vancomycin-resistant E. faecium of animal and human origins can contain indistinguishable genetic elements coding for vancomycin resistance, indicating either horizontal gene transfer between E. faecium organisms of human and animal origins or the existence of a common reservoir for glycopeptide resistance.
The enterococci are a dominant bacterial group in the intestinal flora of human and animals (14). Over the last two decades, the number of infections in hospitalized patients due to enterococci has increased (26–29). Until recently most of these infections have been successfully treated by combinations of vancomycin and other antibiotics, but since the emergence of vancomycin-resistant Enterococcus faecium in 1986 (23), isolates resistant to all known antibiotics can be found. Vancomycin-resistant E. faecium has been found increasingly not only in hospitalized patients (10, 13, 15, 18), but also in the healthy human population (20, 34), in animals (1, 8, 22), and in sewage plants (8, 21, 32). In Europe a glycopeptide, avoparcin, has been used as a growth promoter in animal feed, and its use has been shown to create a reservoir for vancomycin-resistant E. faecium in animals (7). Avoparcin is not used as a growth promoter in the United States, and no VanA-positive isolates have been found in animals (31) or in healthy volunteers (11) in the United States.
Studies of vancomycin-resistant enterococci have shown high clonal diversity, indicating that horizontal gene transfer to some extent plays a part in the dissemination of vancomycin resistance (13, 15, 25). The VanA gene cluster, encoding high-level glycopeptide resistance, is located on the mobile DNA element Tn1546 (3, 6). By investigation of selected vancomycin-resistant E. faecium isolates, variations in this element have been found (16, 25). Molecular characterization of the VanA gene cluster could therefore provide additional information regarding the variation or identities of isolates of different origins and could allow for epidemiological studies of the dissemination of vancomycin resistance due to horizontal gene transfer.
In this study we have characterized isolates of human and animal origins in order to determine if identical Tn1546 could be found in nonidentical strains, and results obtained from 38 vancomycin-resistant E. faecium isolates of human and animal origins in Europe and the United States are described.
MATERIALS AND METHODS
Strains.
A total of 40 vancomycin-resistant E. faecium isolates were included (Table 1). The Danish isolates were selected from a previously described collection (1). The English isolates have been described previously (8, 19). Seven clinical isolates of vancomycin-resistant E. faecium from the United States, collected from patients in Columbus, Ohio (EFM 10-601), Philadelphia, Pa. (EFM 11-803), Bronx, N.Y. (EFM 12-901), New York, N.Y. (EFM 15-1201), Richmond, Va. (EFM 17-2302), New Haven, Conn. (EFM 19-2501), and Chicago, Ill. (EFM 26-4507), were included (Table 1).
TABLE 1.
Vancomycin-resistant E. faecium isolates of animal and human origins
| No. | Source | Country | Size of vanA-positive fragmenta (kb) | Designation |
|---|---|---|---|---|
| 1 | Pony | England | <50 | C15VF9 |
| 2 | Duck | England | <50 | C13VF7 |
| 3 | Chicken | England | <50 | C14VF8 |
| 4 | Turkey | England | <50 | C12VF4 |
| 5 | Human | England | 85 | 63910 |
| 6 | Human | England | <48 | 67668 |
| 7 | Human | England | 100 | GP3 61741 |
| 8 | Pig | England | 45 | A10 Pig 2.19 |
| 9 | Pig | England | 160 | A6 Pig22 |
| 10 | Pig | England | 160 | A1 VF1 |
| 11 | Human | England | 60 | 60761(1) |
| 12 | Human | United States | Negative | EFM 10-601 |
| 13 | Human | United States | 55 | EFM 11-803 |
| 14 | Human | United States | 200 | EFM 12-901 |
| 15 | Human | United States | 38 | EFM 15-1201 |
| 16 | Human | United States | 250 | EFM 17-2302 |
| 17 | Human | United States | 200 | EFM 19-2501 |
| 18 | Human | United States | Negative | EFM 26-4507 |
| 19 | Human | England | 60 | 72801 |
| 20 | Human | England | 60 | 59479 |
| 21 | Human | England | 60 | 62899 |
| 22 | Human | England | 60 | 68521 |
| 23 | Human | England | 60 | 80103 |
| 24 | Human | England | 50 | 89407 |
| 25 | Human | England | 35 | 58538 |
| 26 | Human | England | 60 | 58155 |
| 27 | Human | France | 35 | BM4147 |
| 28 | Human | Denmark | 180 | H17575 |
| 29 | Broiler | Denmark | 50 | 73343-2-1 |
| 30 | Broiler | Denmark | 45 | 73281-7-2 |
| 31 | Pig | Denmark | 160 | A17-SV1 |
| 32 | Pig | Denmark | 160 | D1-SV1 |
| 33 | Broiler | Denmark | 45 | 73583-8-1 |
| 34 | Broiler | Denmark | 45 | 73449-1-2 |
| 35 | Pig | Denmark | 160 | S3-SV1 |
| 36 | Pig | Denmark | 160 | 014-S2 |
| 37 | Human | Denmark | 29 | 5979 |
| 38 | Broiler | Denmark | 50 | 73452-4-2 |
| 39 | Pig | Denmark | 160 | E8-SV3 |
| 40 | Human | Denmark | 145 | H17243 |
The collected strains were tested for the position of integration of Tn1546-like sequences by PFGE of total DNA digested with SmaI. This enzyme has no internal recognition site in the VanA gene cluster. Positive bands vary in size because, in addition to Tn1546, these fragments contain the flanking regions from the point of integration to the nearest SmaI sites. These areas vary as to the physical position of the target site on the chromosome.
Phenotypic test.
A phenotypic test for vancomycin and teicoplanin resistance was performed with the Sensititre system (Accumed International Limited, East Grinstead, West Sussex, England). All isolates were tested for resistance to concentrations of vancomycin and teicoplanin in twofold dilutions from 64 to 0.5 μg/ml. Avoparcin resistance was determined on Mueller-Hinton II agar (Becton Dickinson Microbiology Systems, Cockeysville, Md.) according to the description provided by Aarestrup et al. (1). All isolates were tested for resistance to concentrations of avoparcin in twofold dilutions from 256 to 4 μg/ml.
Total DNA extraction.
Total DNA was extracted from bacteria grown on Mueller-Hinton plates supplemented with 5% (bovine) blood. A tablet (Rosco Diagnostica, Taastrup, Denmark) containing 70 μg of vancomycin was placed on each plate. A 1-ml suspension of the resistant bacteria in phosphate-buffered saline was transferred to an Eppendorf tube, centrifuged at 20,000 rpm for 5 min, and then resuspended in 100 μl of Tris-EDTA buffer. Cells were lysed by boiling (for 10 min), and 2 μl was used for PCR amplification.
Pulsed-field gel electrophoresis (PFGE).
For preparations of DNA embedded in agarose plugs, whole-cell DNA was prepared as previously described (28), with the following modifications: 3 ml of a solution containing 1 M NaCl, 10 mM Tris-HCl, 200 mM EDTA, 0.5% sodium lauryl sarcosine, 1.7 mg of lysozyme/ml, and 3.3 μg of Rnase A/ml was used as lysis buffer, and proteins were digested with 1 mg of proteinase K/ml and removed by a wash in 2 ml of Tris-EDTA buffer (10:1) containing 0.25 mg of phenylmethylsulfonyl fluoride/ml.
A small slice of the agarose plug was digested with 20 U of SmaI (Amersham Life Science) for a minimum of 4 h. Digested DNA was electrophoresed in a 1% agarose gel. Electrophoresis was performed in a Pharmacia LKB Gene Navigator unit by using the following settings: 2 s, 5 h, 5 s, 6 h, 9 s, 6 h, and 12 s, 5 h in 0.5× Tris-borate-EDTA buffer at 12°C at 180 V. DNA was transferred onto a nylon membrane (Hybond-N; Amersham International, Little Chalfont, Buckinghamshire, United Kingdom) by capillary transfer and fixed by UV fixation. Hybridizations were performed by using a minihybridization oven (Hybaid Ltd., Teddington, Middlesex, England). A digoxigenin-labelled DNA vanA probe (535 bp) was used for hybridization.
PCR amplification of internal regions of Tn1546.
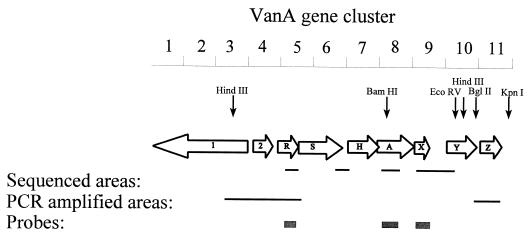
DNA extractions and PCR amplification were performed according to the method of Aarestrup et al. (1). Primers were designed according to the published sequence of Tn1546 (3, 6). All primers used are listed in Table 2. Both coding and noncoding areas were amplified. The positions and sizes of the PCR-amplified regions are indicated in Fig. 1. The melting temperatures for the individual primers were calculated by using the Tm DETERMINATION software (9) available on the Internet (http: //alces.med.umn.edu/rawtm.html). All PCR amplifications were run with a MgCl2 concentration of 1 mM.
TABLE 2.
Primers used to characterize Tn1546 from vancomycin-resistant enterococcia
| Primer | Sequence (5′→3′) | Position |
|---|---|---|
| VanA gene cluster | ||
| Van A1 | AAA TGT GCG AAA AAC CTT GC | 7127–7146 |
| Van A2 | AAC AAC TAA CGC GGC ACT | 7662–7645 |
| VanS1 | ATT GTT CAG CAT GGA GGG C | 5696–5714 |
| VanH2 | GAG CAT GGA ATG CAT CTG CC | 6081–6063 |
| VanR1 | AAA TAA GGG ACA AGC ACA CC | 4125–4194 |
| VanR2 | CCC ATA TCT CAT GAA ATA GC | 4534–4515 |
| VanX1 | ACT TGG GAT AAT TTC ACC GG | 8082–8101 |
| VanX2 | TGC GAT TTT GCG CTT CAT TG | 8505–8486 |
| VanX3 | CTC ATC ATG CGG CAA ATG G | 8458–8476 |
| VanY1 | TGG GTA TTT TCA GAA GTC CC | 9212–9193 |
| VanY2-1 | GTT TCC CGG ATC AAC ACA TAC TA | 9927–9949 |
| VanZ2-2 | CCC AGT AGC AGT AAA TGG AGT CA | 10262–10240 |
| VanZ1 | CTG GGA ATT TCA GAG AGA TG | 10258–10277 |
| VanZ2 | AAT GGG TAC GGT AAA CGA GC | 10581–10562 |
| ORF12 | CCA TTC CTC GTA TGT ATT CG | 2318–2337 |
| VanR2 | CCC ATA TCT CAT GAA ATA GC | 4534–4515 |
| ORF14-2 | GCC CTT AGG TTG GGA ACA TA | 174–155 |
| VanSma-2 | ATA AAA TGA CTA ACG CCA CC | 9446–9427 |
| IS1216V– IS3-like | ||
| IS1216V-1 | AAA GCA ATT TCA GCA GGA TG | 256–275 |
| IS1216V-2 | GTA CGA TGT TCT GTC CCT TG | 711–692 |
| IS3-like-1 | ACT GGG TAT CGC CAA ATC CA | 1251–1270 |
| IS3-like-2 | TTT GTC CCA TTG GTC AAC CG | 1619–1600 |
| IS3-like-2-1 | CCA CAC TGA TTC ATA GCG AC | 1748–1767 |
| IS1251 | ||
| IS1251-3 | GCA TCC ACT GTA AAC ACC AG | 273–292 |
| IS1251-4 | CGC TGT GTT TGA CCA TCC AT | 699–680 |
| IS1476, IS1476-2-1 | CTT TCG GGC ACG GAT CTA TT | 1366–1385 |
All primers were used for PCR amplification of internal areas of Tn1546. Positions refer to sequences published in GenBank; access numbers are M97297 for Tn1546, L40841 for IS1216V, L34675 for IS1251, and U63997 for IS1476. Correct positions of the IS sequences were determined by using one primer inside the IS sequence and one in the flanking region of Tn1546. For the position of IS1251, primers VanH2 and IS1251-4 were used; for linking of IS1216V to the IS3-like element, primers IS1216V-1 and IS3-like-2 were used; and IS3-like-2-1 and ORF14-2 were used to position this complex in the left end of Tn1546. Primers IS1476-2-1 and VanSma-2 were used to determine whether IS1476 was present.
FIG. 1.
Map of Tn1546. Numbers at the top represent kilobases. Open arrows represent coding sequences, and letters inside them stand for genes (e.g., R, vanR). Positions of recognition sites for selected restriction enzymes are marked by vertical arrows. Only essential sizes for the hybridizations are indicated.
Presence of IS sequences.
The presence of published insertion element (IS) sequences in the VanA gene cluster was tested by designing one primer in the published sequence of Tn1546 and one in the published IS sequence. All primers used are listed in Table 2. Only if the IS sequence is present at the published position in the VanA gene cluster can a positive PCR amplicon of the bridging area between the VanA gene cluster and the IS sequence be obtained. Table 3 lists positive and negative PCR amplification results as the numbers 1 and 0, respectively.
TABLE 3.
Genetic characterization of Tn1546-like elements from vancomycin-resistant enterococci by PCR and DNA hybridization
| Isolate typea | Isolate no. | Source | PCR result
|
DNA hybridization signald (kb)
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| M vanXb | IS1216V–IS3-likec | IS1251c | VanR (H3) | VanX
|
|||||
| BHI-KI | BHI-BII | BHI-EV | |||||||
| 1 | 1 | Pony | G | 0 | 0 | 7 | 3.5 | 2.5 | |
| 1 | 2 | Duck | G | 0 | 0 | 7 | 3.5 | 2.5 | |
| 1 | 3 | Chicken | G | 0 | 0 | 7 | 3.5 | 2.5 | |
| 1 | 4 | Turkey | G | 0 | 0 | 7 | 3.5 | 2.5 | |
| 1 | 13 | Human | G | 0 | 0 | 7 | 3.5 | 2.5 | |
| 1 | 27 | Human | G | 0 | 0 | 7 | 3.5 | 2.5 | |
| 1 | 30 | Broiler | G | 0 | 0 | 7 | 3.5 | 2.5 | 2.1 |
| 1 | 33 | Broiler | G | 0 | 0 | 7 | 3.5 | 2.5 | |
| 1 | 34 | Broiler | G | 0 | 0 | 7 | 3.5 | 2.5 | |
| 1 | 38 | Broiler | G | 0 | 0 | 7 | 3.5 | 2.5 | |
| 2 | 8 | Pig | T | 1 | 0 | 7 | 3.5 | 2.5 | |
| 2 | 9 | Pig | T | 1 | 0 | 7 | 3.5 | 2.5 | |
| 2 | 10 | Pig | T | 1 | 0 | 7 | 3.5 | 2.5 | 2.1 |
| 2 | 14 | Human | T | 1 | 0 | 7 | 3.5 | 2.5 | |
| 2 | 28 | Human | T | 1 | 0 | 7 | 3.5 | 2.5 | 2.1 |
| 2 | 31 | Pig | T | 1 | 0 | 7 | 3.5 | 2.5 | |
| 2 | 32 | Pig | T | 1 | 0 | 7 | 3.5 | 2.5 | |
| 2 | 35 | Pig | T | 1 | 0 | 7 | 3.5 | 2.5 | |
| 2 | 36 | Pig | T | 1 | 0 | 7 | 3.5 | 2.5 | |
| 2 | 39 | Pig | T | 1 | 0 | 7 | 3.5 | 2.5 | |
| 2 | 40 | Human | T | 1 | 0 | 7 | 3.5 | 2.5 | |
| 3 | 19 | Human | G | 0 | 0 | 8 | 3.0 | 10.5 | 10.5 |
| 3 | 20 | Human | G | 0 | 0 | 8 | 3.0 | 10.5 | 10.5 |
| 3 | 21 | Human | G | 0 | 0 | 8 | 3.0 | 10.5 | 10.5 |
| 3 | 22 | Human | G | 0 | 0 | 8 | 3.0 | 10.5 | 10.5 |
| 3 | 23 | Human | G | 0 | 0 | 8 | 3.0 | 10.5 | 10.5 |
| 3 | 26 | Human | G | 0 | 0 | 8 | 3.0 | 10.5 | 10.5 |
| 4 | 5 | Human | G | 1 | 0 | 7 | 3.5 | 2.5 | |
| 5 | 6 | Human | G | 1 | 0 | 7, 9 | 3.5, 4.4 | 2.5, 3.5 | 2.1, 2.9 |
| 6 | 7 | Human | G | 1 | 0 | 7 | 3.5 | 2.5 | |
| 7 | 11 | Human | G | 0 | 0 | 8 | 3.0 | 10.5 | 2.1 |
| 8 | 15 | Human | T | 0 | 1 | 4, 5 | 3.5 | 2.5 | |
| 9 | 16 | Human | T | 1 | 1 | 4 | 3.5 | 2.5 | |
| 9 | 17 | Human | T | 1 | 1 | 4 | 3.5 | 2.5 | 2.1 |
| 10 | 24 | Human | G | 0 | 0 | 8, 11 | 3.0, 7.0 | 5.5, 10.5 | 6.8, 10.5 |
| 11 | 25 | Human | G | 0 | 0 | 15 | 4.4 | 3.5 | 2.9 |
| 12 | 29 | Broiler | G | 0 | 0 | 8 | 4.4 | 3.5 | 2.9 |
| 13 | 37 | Human | T | 0 | 0 | 7 | 3.5 | 2.5 | |
Isolates are divided into 13 types according to the results obtained.
Sequence result of PCR-amplified region of the vanX gene. T, sequence homologous to published sequence of Tn1546; G, sequence with a single-nucleotide variation (T to G) at position 8234.
Presence of the IS1216V–IS3-like element and of IS1251 at the published positions in the left end of Tn1546 and in the vanSH intergenic region, respectively. Symbols: 1, correct position; 0, absence of a positive PCR amplification.
Where two values are given, two bands were detected. Abbreviations: H3, HindIII; BHI, BamHI; KI, KpnI; BII, BglII; EV, EcoRV.
Sequencing.
The nucleotide sequences of the amplification products were determined by cycle sequencing (30) using the Amplitaq FS dye terminator kit and a 373A automatic sequencer (Applied Biosystems, Perkin-Elmer, Foster City, Calif.). DNASIS software (Hitachi Software Engineering Co., Ltd.) was used for sequence analysis.
Hybridizations.
Total DNA was digested with the restriction enzymes HindIII, BamHI-BglII, BamHI-EcoRV, and BamHI-KpnI. All enzymes were purchased from Amersham International. Fragments were separated in a 0.8% agarose gel. DNA was transferred to Hybond-N membranes (Amersham International).
Digoxigenin-labelled DNA probes for vanR (409 bp) and vanX (423 bp) were prepared by PCR amplification using the primers described in Table 2 and were subsequently labelled with the Boehringer Mannheim DNA labelling and detection kit. The obtained PCR products were purified by using Qiagen (Hilden, Germany) spin columns. The vanR probe was used for HindIII-digested total DNA, and the vanX probe was used for BamHI-BglII-, BamHI-EcoRV-, and BamHI-KpnI-digested total DNA.
RESULTS
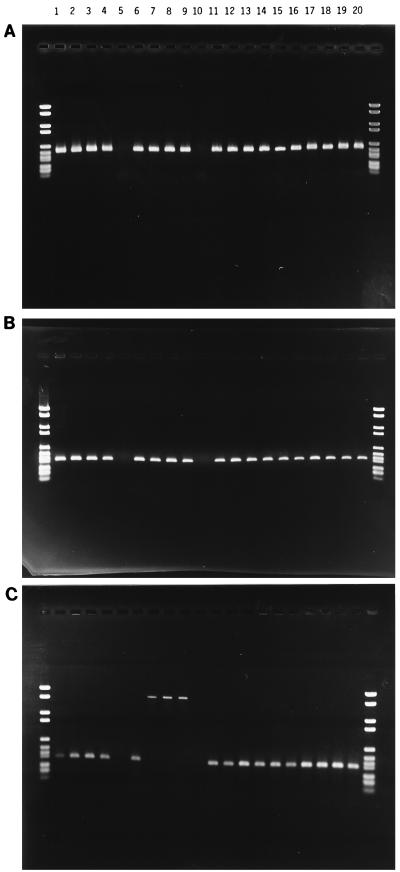
All isolates were resistant to vancomycin (>64 μg/ml) and avoparcin (>16 μg/ml). Thirty-eight were resistant to teicoplanin (>16 μg/ml). Two isolates (no. 12 and 18) were sensitive to teicoplanin, indicating that these strains contain vanB. Of the 40 isolates, 38 hybridized to the vanA probe and gave positive results in the PCR amplifications. The two negative isolates were identical to the teicoplanin-sensitive isolates (no. 12 and 18). Positive hybridization signals of different sizes were obtained (Table 1), indicating different positions of integration for Tn1546 on the genome. This is an indication that the tested strains are not identical. The position of integration (target) could be identical for all strains, since several identical targets could exist at different locations on the chromosome. Isolates 12 and 18 gave no positive amplicons for vanA (Fig. 2A), vanR, or vanX (Fig. 2B). These isolates were later defined as vanB (28a).
FIG. 2.
PCR amplification of the internal region of the VanA gene cluster. (A) The vanA gene of selected isolates, from position 7266 to 7623. Isolates 12 and 18 (lanes 5 and 10) gave no positive product. (B) The vanX gene of selected isolates, from position 8129 to 8483. Isolates 12 and 18 (lanes 5 and 10) gave no positive product. (C) The vanSH intergenic region of selected isolates, from position 5715 to 6031. Isolates 12 and 18 (lanes 5 and 10) gave no positive product. Isolates 15 through 17 (lanes 7 through 9) gave a positive product of approximately 1,900 bp. The Boehringer IV (Boehringer Mannheim) was used for size determination. Sizes of markers are 2,176, 1,766, 1,230, 1,033, 653, 517, 453, 394, 298, 234, 220, and 154 bp. Results for isolates are shown as follows: lane 1, 5; lane 2, 6; lane 3, 7; lane 4, 11; lane 5, 12; lane 6, 14; lane 7, 15; lane 8, 16; lane 9, 17; lane 10, 18; lane 11, 22; lane 12, 24; lane 13, 25; lane 14, 27; lane 15, 28; lane 16, 29; lane 17, 30; lane 18, 34; lane 19, 37; lane 20, 40.
Coding regions of Tn1546.
PCR amplification products of vanA (457 bp) (Fig. 2A), vanR (303 bp), and vanX (354 bp) (Fig. 2B) from Tn1546 were obtained for 38 isolates. All PCR products from the three replicons were of the expected sizes. These were sequenced, and only a single-nucleotide variation (T to G) was found in the vanX gene at position 8234. For vanZ, a PCR product of the expected size (323 bp) was obtained from all isolates. No further investigations of these PCR products were performed.
Intergenic area.
PCR products of the expected size for the vanXY intergenic region (554 bp) were obtained from 38 isolates. No sequence or size variation was observed in the amplicons of the vanXY intergenic region (data not shown). PCR amplification of the vanSH intergenic region produced an amplicon of 318 bp for 35 isolates. For three isolates, an amplicon of approximately 1,900 bp was obtained. These three isolates (no. 15 through 17) (Fig. 2C) were all of human origin and all from the United States. For the region containing open reading frame 2 (ORF2), the ORF2-vanR intergenic region, and vanR (position 2318 to 4534), PCR products (2,216 bp) were obtained from 38 isolates. BglII digestion of the amplicons resulted in fragments of 1,085, 1,027, and 104 bp, respectively (data not shown). The vanYZ intergenic region was PCR amplified for 38 isolates, and no size variation in the PCR products was observed (data not shown).
IS sequences.
The presence of the IS1216V (17) sequence was detected in all 40 isolates, but only in 16 isolates was it linked to the IS3-like sequence at the published position inside the VanA gene cluster (Table 3) (17). In the remaining 24 isolates, the IS3-like sequence was positioned outside Tn1546. IS1251 (16) was detected in three human isolates from hospitals in the United States (no. 15 through 17) for which a larger DNA fragment was obtained from PCR amplification of the intergenic area (Fig. 2C), and its presence was confirmed by an amplicon positive for the presence of IS1251 at this position (Table 3). IS1476 (24) was not found in any of the isolates.
DNA hybridizations.
A probe specific for the vanR gene hybridized to one band ranging from 4 to 11 kb of HindIII (positions 2455 and 9571)-digested total DNA (Table 3) in 35 isolates. In three of the isolates (no. 6, 15, and 24), two bands were detected (Table 3). A probe specific for the vanX gene hybridized to one band ranging from 3 to 7 kb of BamHI (position 7295)-KpnI (position 10849)-digested total DNA (Table 3) in 36 isolates. Isolates 6 and 24 each gave two bands. With the vanX-specific probe, bands ranging from 2.5 to 10.5 kb of BamHI (position 7295)-BglII (position 9825)-digested total DNA were detected (Table 3); isolates 6 and 24 each gave two bands. BamHI (position 7295)-EcoRV (position 9304)-digested total DNA of selected strains was hybridized with the vanX-specific probe. Positive bands ranging from 2.1 to 10.5 kb were detected (Table 3); isolates 6 and 24 each gave two bands.
DISCUSSION
Since the emergence of vancomycin-resistant E. faecium, several investigations have been performed to study the dissemination of glycopeptide in these strains. Analyses of bacteria isolated from hospitalized patients have indicated both clonal spread of vancomycin-resistant E. faecium and horizontal gene transfer of Tn1546. After the detection of vancomycin-resistant E. faecium in sewage plants, animals, healthy humans, and pet animals (12, 33), it was hypothesized that the hospital environment was creating vancomycin-resistant E. faecium, which subsequently spread to the environment. After the discovery that the glycopeptide avoparcin, commonly used as a growth promoter in agriculture, selected for vancomycin-resistant E. faecium in production animals (2, 22), the hypothesis that vancomycin-resistant E. faecium could spread from animals to humans via the food chain was raised (8). To test the transfer between animals and humans, several isolates of human and animal origins have been typed (8, 19, 20). These experiments indicated high diversity among the strains of vancomycin-resistant E. faecium defined by ribotypes, and the focus was turned to the dissemination of the genetic element for vancomycin resistance, Tn1546.
In the present study, the genetic elements for vancomycin resistance in 40 high-level vancomycin-resistant E. faecium isolates of animal and human origins from Europe and the United States were studied. For 38 isolates, an amplicon positive for vanA was obtained (Table 1). For two isolates (no. 12 and 18), a PCR amplicon positive for vanB was obtained (Table 1).
A vanA probe gave specific bands of different sizes when hybridized to DNA fragments from PFGE of SmaI-digested total DNA from 38 isolates. The variations in size of positive bands (Table 1) indicated different positions of integration of Tn1546, suggesting that the isolates were nonidentical. For 7 of the 12 English human isolates and 7 of 8 porcine isolates, hybridization to fragments of similar sizes (Table 1) was detected.
PCR amplification and sequencing of internal areas of Tn1546 in the 38 vanA-positive isolates confirmed high homology to the published sequence of Tn1546 in coding regions. In total, 19% of Tn1546 was sequenced, and only a single-base-pair difference was detected in the vanX gene (Table 3). The detected difference at position 8234 in the vanX gene changed the amino acid at this position from a lysine to an asparagine but apparently did not affect the phenotype. No sequence or size variation was seen in the vanXY intergenic region, contrary to published results (25). Since some of the isolates in the previous investigation (25) were not vancomycin-resistant E. faecium, and others originated from a hospital in England different from the ones from which isolates for this survey were taken, the result is not surprising. This just indicates that more isolates need to be tested in order to define all types.
Two previously published IS sequences were found to have integrated into Tn1546. IS1216V was present in all 40 strains, but only in 16 of the 38 vanA-positive isolates could the position of this IS sequence combined with the IS3-like element (17) be confirmed in the left end of Tn1546 (Table 3). In the rest of the positive isolates, the IS1216V-like elements probably are positioned outside Tn1546. The presence of IS1251 was detected in three human isolates from the United States (no. 15 through 17), positioned in the vanSH intergenic region (Table 3). Handwerger and Skoble (17) presumed that the presence of the IS3-like sequence positioned Tn1546 on the chromosome. In the present work, no efforts were made to localize Tn1546 on the chromosome or on a plasmid, but sizes of positive fragments from PFGE of SmaI-digested total DNA (Table 1) of IS3-like-positive clones were all large (>85 kb) except for isolate 6 (<48 kb) and isolate 8 (48 kb). This could indicate a chromosomal position.
Based solely on Southern blot analysis of total DNA digested with several restriction enzymes, creating fragments covering most of Tn1546, 38 isolates could be divided into nine unique types (Table 3). Type 1 consists of 24 isolates. These gave fragment sizes corresponding to Tn1546 according to previous published sequences. Type 2 consists of two human clinical isolates from the United States (no. 16 and 17). These gave a smaller fragment. Type 3 consists of six human clinical isolates from England. These gave a larger fragment. These six isolates (no. 19 through 23 and no. 26) and also isolate 11 are believed to contain two uncharacterized size variations of approximately 8 kb. This is indicated by larger bands obtained from DNA hybridizations (Table 3). For isolate 11, a variation has been mapped between positions 9304 and 9825. For the rest, the variation has been mapped between positions 7295 and 9304 (Fig. 1 and Table 3). Since these areas contain the essential vanA and vanX genes, the vanXY intergenic area, and vanY (Fig. 1), and since no sequence variation was found in the vanA gene, the vanR gene, or the vanXY intergenic region, the insertions are believed to be in the vanY gene. Attempts to identify the insertions as the recently published IS1476 (24) residing in the vanY gene (position 9333) by PCR amplification have failed. The vanY gene codes for a d,d-carboxypeptidase, which contributes to the hydrolysis of soluble peptidoglycan precursors, complementary to vanX dipeptidase hydrolysis, and is for that reason not essential for the phenotype of the VanA gene cluster (4, 5).
For three isolates (no. 15 through 17), the size of HindIII-digested total DNA obtained by using a vanR-specific probe can be explained by the presence of IS1251. This IS sequence contains an additional HindIII recognition site, thus creating a 4-kb band instead of the 7-kb band (Table 3). The additional 5-kb band obtained with isolate 15 cannot be explained. Isolates 6 and 24 each contain two nonidentical copies of Tn1546. This is, to our knowledge, the first time this has been described.
At least five additional size variations have been mapped in seven unique types to the area between positions 7295 and 9304. Most of this area has been sequenced (Fig. 1), and the variations detected by hybridizations could be located in vanY and might be explained by an IS sequence “hot spot” in this nonessential gene. This is at present being investigated.
Adding together the results obtained, no significant variations were found in the Tn1546 elements. Only the presence or absence of IS sequences and a single difference in the vanX gene seem to differentiate the Tn1546 elements observed in this study from the published sequence (3, 6). However, some variations were found; thus, a genetic characterization of Tn1546 can provide information on dissemination of vancomycin resistance due to horizontal gene transfer.
In this study, the 38 vanA-positive isolates could be divided into 13 types based on hybridizations, obtained sequences, and the presence of IS sequences (Table 3). Types 1 and 2 consist of isolates of human and animal origins, suggesting that indistinguishable genetic elements for vancomycin resistance can be present in bacteria isolated from animals and humans. Type 1 includes the reference strain, BM4147 (isolate 27). Type 2 consists of isolates with the single-base-pair difference and the presence of the IS1216V–IS3-like sequence. Type 3 consists of clinical isolates of human origin from England and indicates an epidemiological relationship among the isolates. Type 9 consists of two human isolates from the United States. The last 10 unique types differ from the defined genetic subgroups by the presence of IS1251 and detected size variations of unknown origin.
By hybridization with the vanA-specific probe to DNA fragments from PFGE of SmaI-digested total DNA, different integration positions of Tn1546 were detected. This indicates that the strain does not originate from a clonal spread. Therefore, the existence of two genetic subgroups containing isolates of both human and animal origins indicates two possibilities. Either the isolates have obtained Tn1546 from a common reservoir or a horizontal gene transfer has occurred. These two possibilities cannot be distinguished, but both show that bacteria of human and animal origins can harbor similar resistance genes.
The work described in this article can be used to study the epidemiological spread of vancomycin-resistant E. faecium where horizontal transfer of Tn1546 dominates the dissemination of vancomycin resistance. For this purpose it is essential to characterize all variations in Tn1546 by investigation of a broad variety of strains. This work is being performed at present.
ACKNOWLEDGMENTS
We acknowledge the following persons for their technical assistance: René Hendriksen, Mette Juul, Lissie Kjær Jensen, Karina Kristensen, Inge Hansen, Rikke Bruun, Jonas Michelsen. We also thank Rikke Lykke Poulsen, Statens Serum Institut, for performing PCR detection of vanB. Special thanks are due to J. Zoe Jordens, Public Health Laboratory Service, John Radcliffe Hospital, Oxford, England, for providing the English strains and to Knud Børge Pedersen and Henrik C. Wegener for helpful comments in the preparation of the manuscript.
REFERENCES
- 1.Aarestrup F M, Ahrens P, Madsen M, Pallesen L V, Poulsen R L, Westh H. Glycopeptide susceptibility among Danish Enterococcus faecium and Enterococcus faecalis isolates of animal and human origin and PCR identification of genes within the VanA cluster. Antimicrob Agents Chemother. 1996;40:1938–1940. doi: 10.1128/aac.40.8.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aarestrup F M. Occurrence of glycopeptide resistance among Enterococcus faecium isolates from conventional and ecological poultry farms. Microb Drug Resist. 1995;1:255–257. doi: 10.1089/mdr.1995.1.255. [DOI] [PubMed] [Google Scholar]
- 3.Arthur M, Molinas C, Depardieu F, Courvalin P. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1993;175:117–127. doi: 10.1128/jb.175.1.117-127.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arthur M, Depardieu F, Snaith H A, Reynolds P E, Courvalin P. Contribution of VanY d,d-carboxypeptidase to glycopeptide resistance in Enterococcus faecalis by hydrolysis of peptidoglycan precursors. Antimicrob Agents Chemother. 1994;38:1899–1903. doi: 10.1128/aac.38.9.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arthur M, Courvalin P. Genetics and mechanisms of glycopeptide resistance in enterococci. Antimicrob Agents Chemother. 1993;37:1563–1571. doi: 10.1128/aac.37.8.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arthur M, Depardieu F, Molinas C, Reynolds P, Courvalin P. The vanZ gene of Tn1546 from Enterococcus faecium BM4147 confers resistance to teicoplanin. Gene. 1995;154:87–92. doi: 10.1016/0378-1119(94)00851-i. [DOI] [PubMed] [Google Scholar]
- 7.Bager F, Madsen M, Christensen J, Aarestrup F M. Occurrence of vancomycin resistant Enterococcus faecium in pig and poultry farms using avoparcin as a growth promotant. Prev Vet Med. 1997;31:95–112. doi: 10.1016/s0167-5877(96)01119-1. [DOI] [PubMed] [Google Scholar]
- 8.Bates J, Jordens J Z, Griffiths D T. Farm animals as a putative reservoir for vancomycin-resistant enterococcal infection in man. J Antimicrob Chemother. 1994;34:507–516. doi: 10.1093/jac/34.4.507. [DOI] [PubMed] [Google Scholar]
- 9.Breslauer K J, Frank R, Blöcket H, Marky L A. Predicting DNA duplex stability from the base sequence. Proc Natl Acad Sci USA. 1986;83:3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention. Nosocomial enterococci resistant to vancomycin: United States, 1989–1993. National Nosocomial Infection Surveillance. Morbid Mortal Weekly Rep. 1993;42:597–599. [PubMed] [Google Scholar]
- 11.Coque T M, Tomayko J F, Ricke S C, Okhyusen P C, Murray B E. Vancomycin-resistant enterococci from nosocomial, community, and animal sources in the United States. Antimicrob Agents Chemother. 1996;40:2605–2609. doi: 10.1128/aac.40.11.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devriese L A, Ieven M, Goossens H, Vandamme P, Pot B, Hommez J, Haesebrouck F. Presence of vancomycin-resistant enterococci in farm and pet animals. Antimicrob Agents Chemother. 1996;40:2285–2287. doi: 10.1128/aac.40.10.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dutka-Malen S, Leclercq R, Coutant V, Duval J, Courvalin P. Phenotypic and genotypic heterogeneity of glycopeptide resistance determinants in gram-positive bacteria. Antimicrob Agents Chemother. 1990;34:1875–1879. doi: 10.1128/aac.34.10.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Facklam R R, Sahm D F. Enterococcus. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C: American Society for Microbiology; 1995. pp. 308–314. [Google Scholar]
- 15.Frieden T R, Munsiff S S, Low D E, Willey B M, Williams G, Faur Y, Eisner W, Warren S, Kreiswirth B. Emergence of vancomycin-resistant enterococci in New York City. Lancet. 1993;342:76–79. doi: 10.1016/0140-6736(93)91285-t. [DOI] [PubMed] [Google Scholar]
- 16.Handwerger S, Skoble J, Discotto L F, Pucci M J. Heterogeneity of the vanA gene cluster in clinical isolates of enterococci from the northeastern United States. Antimicrob Agents Chemother. 1995;39:362–368. doi: 10.1128/aac.39.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Handwerger S, Skoble J. Identification of chromosomal mobile element conferring high-level vancomycin resistance in Enterococcus faecium. Antimicrob Agents Chemother. 1995;39:2446–2453. doi: 10.1128/aac.39.11.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones R N, Sader H S, Erwin M E, Anderson S C the Enterococcus Study Group. Emerging multiple resistant enterococci among clinical isolates. I. Prevalence data from 97 medical center surveillance study in the United States. Diagn Microbiol Infect Dis. 1995;21:85–93. doi: 10.1016/0732-8893(94)00147-o. [DOI] [PubMed] [Google Scholar]
- 19.Jordens J Z, Bates J, Griffiths D T. Faecal carriage and nosocomial spread of vancomycin-resistant Enterococcus faecium. J Antimicrob Chemother. 1994;34:515–528. doi: 10.1093/jac/34.4.515. [DOI] [PubMed] [Google Scholar]
- 20.Klare I, Heier H, Claus H, Böhme G, Marin S, Seltmann G, Hakenbeck R, Antanassova V, Witte W. Enterococcus faecium strains with vanA-mediated high-level glycopeptide resistance isolated from animal foodstuffs and fecal samples of humans in the community. Microb Drug Resist. 1995;1:265–272. doi: 10.1089/mdr.1995.1.265. [DOI] [PubMed] [Google Scholar]
- 21.Klare I, Heier H, Claus H, Witte W. Environmental strains of Enterococcus faecium with inducible high-level resistance to glycopeptides. FEMS Microbiol Lett. 1993;106:23–30. doi: 10.1111/j.1574-6968.1993.tb05930.x. [DOI] [PubMed] [Google Scholar]
- 22.Klare I, Heier H, Claus H, Reissbrodt R, Witte W. vanA-mediated high-level glycopeptide resistance in Enterococcus faecium from animal husbandry. FEMS Microbiol Lett. 1995;125:165–172. doi: 10.1111/j.1574-6968.1995.tb07353.x. [DOI] [PubMed] [Google Scholar]
- 23.Leclercq R, Derlot E, Duval J, Courvalin P. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N Engl J Med. 1988;319:157–161. doi: 10.1056/NEJM198807213190307. [DOI] [PubMed] [Google Scholar]
- 24.MacKinnon M G, Drebot M A, Tyrrell G J. Identification and characterization of IS1476, an insertion sequence-like element that disrupts VanY function in a vancomycin-resistant Enterococcus faecium strain. Antimicrob Agents Chemother. 1997;41:1805–1807. doi: 10.1128/aac.41.8.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miele A, Bandera M, Goldstein B P. Use of primers selective for vancomycin resistance genes to determine van genotype in enterococci and to study gene organization in VanA isolates. Antimicrob Agents Chemother. 1995;39:1772–1778. doi: 10.1128/aac.39.8.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moellering R C. The enterococci: an enigma and a continuing therapeutic challenge. Eur J Clin Microbiol Infect Dis. 1990;9:73–74. doi: 10.1007/BF01963629. [DOI] [PubMed] [Google Scholar]
- 27.Murray B E. The life and times of Enterococcus. Clin Microbiol Rev. 1990;3:46–65. doi: 10.1128/cmr.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murray B E, Singh K V, Heath J D, Sharma B R, Weinstock G M. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J Clin Microbiol. 1990;28:2059–2063. doi: 10.1128/jcm.28.9.2059-2063.1990. . (Erratum, 29:418, 1991.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.Poulsen, R. L. Personal communication.
- 29.Schalberg D R, Culver D H, Raynes R P. Major trends in the microbial etiology of nosocomial infections. Am J Med. 1991;91:72–76. doi: 10.1016/0002-9343(91)90346-y. [DOI] [PubMed] [Google Scholar]
- 30.Sears L E, Moran L S, Kissinger C, Slatko B E. Thermal cycle sequencing and alternative sequencing protocols using the highly thermostable VentR (exo) DNA polymerase. BioTechniques. 1992;13:626–683. [PubMed] [Google Scholar]
- 31.Thal L A, Chow J W, Mahayni R, Bonilla H, Perri M B, Donabedian S A, Silverman J, Taber S, Zervos M J. Characterization of antimicrobial resistance in enterococci of animal origin. Antimicrob Agents Chemother. 1995;39:2112–2115. doi: 10.1128/aac.39.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torres C, Reguera J A, Sanmartin M J, Pérez-Díaz J C, Baquero F. vanA-mediated vancomycin-resistant Enterococcus spp. in sewage. J Antimicrob Chemother. 1994;33:553–561. doi: 10.1093/jac/33.3.553. [DOI] [PubMed] [Google Scholar]
- 33.van Belkum A, van den Braak N, Thomassen R, Verbrugh H, Endtz H. Vancomycin-resistant enterococci in cats and dogs. Lancet. 1996;348:1038–1039. doi: 10.1016/s0140-6736(05)64973-2. [DOI] [PubMed] [Google Scholar]
- 34.Van der Auwera P, Pensart N, Korten V, Murray B E, Leclercq R. Influence of oral glycopeptides on the fecal flora of human volunteers: selection of highly glycopeptide-resistant enterococci. J Infect Dis. 1996;173:1129–1136. doi: 10.1093/infdis/173.5.1129. [DOI] [PubMed] [Google Scholar]