Abstract
The gene (omlA) coding for an outer membrane protein of Actinobacillus pleuropneumoniae serotypes 1 and 5 has been described earlier and has formed the basis for development of a specific PCR assay. The corresponding regions of all 12 A. pleuropneumoniae reference strains of biovar 1 were sequenced. Alignment of the sequences revealed conserved terminal and variable middle regions, which divided the reference strains into four distinct groups. Primers were selected from the conserved 5′ and 3′ termini of the gene. A 950-bp amplicon was obtained from each of 102 tested field isolates of A. pleuropneumoniae obtained from lungs. Their identity was verified by sequencing approximately 500 bp of the amplification product from 50 of the A. pleuropneumoniae isolates, which all showed the expected DNA sequence characteristic of the serotype. To test the specificity of the reaction, 23 other bacterial species related to A. pleuropneumoniae or isolated from pigs were assayed. They were all found negative in the PCR, as were tonsil cultures from 50 pigs of an A. pleuropneumoniae-negative herd. The sensitivity assessed by agarose gel analysis of the PCR product was 102 CFU/PCR test tube. The specificity and sensitivity of this PCR compared to those of culture suggest the use of this PCR for routine identification of A. pleuropneumoniae.
Infection with the contagious respiratory pathogen Actinobacillus pleuropneumoniae is the main cause of pleuropneumonia in pigs. Characteristic symptoms of the disease range from acute fibrinous pneumonia and pleuritis with high mortality to nearly asymptomatic colonization by the bacterium (18). After recovery some infected animals will suffer from chronic lung lesions, resulting in reduced weight gain. Pigs which survive an infection can still be carriers of the pathogen, so a herd once infected remains infected (7). Acute outbreak of the disease causes considerable economic losses for the pig industry (12).
A. pleuropneumoniae can be divided into two biovars (20), and in general biovar 1 strains are considered more virulent than those of biovar 2 (6, 14). Twelve different serotypes have been described (19), the prevalence and presence of which vary with geographic location. Serotypes 1 and 5 of biovar 1 are prevalent in North America, whereas serotypes 2 and 9 of biovar 1 have been isolated in many European countries (16). The prevalence of the infection seems to be increasing because of intensified modern swine production.
In vivo detection of the infection has until now mainly been performed by serological tests, such as enzyme-linked immunosorbent assays and complement fixation tests. However, antigenic variability among the serotypes of A. pleuropneumoniae has hampered attempts to produce species-specific diagnostic tests which include all serotypes. Conventional cultivation of the bacteria from healthy carrier pigs has been improved by development of A. pleuropneumoniae-selective media (15, 22). A PCR method for detection of A. pleuropneumoniae has also been developed (23) and evaluated and has been shown to be more sensitive than cultivation (10). Evaluation of this PCR assay showed that its specificity was not complete, as it also reacted with Actinobacillus lignieresii. Effective detection methods which are species specific rather than serotype specific are therefore necessary in order to control the spread of the disease.
The aim of this study was to develop a species-specific PCR for detection and identification of A. pleuropneumoniae for diagnosis of subclinical infections. A specific set of primers was designed from a previously described gene of an outer membrane protein from A. pleuropneumoniae serotype 1 and 5 isolates (3, 9, 13).
To test if the gene was generally present in A. pleuropneumoniae isolates, the corresponding regions of the omlA genes from all the A. pleuropneumoniae serotype reference strains of biovar 1 were sequenced. Four different but homologous omlA sequence regions divided the serotypes and lung isolates into four groups. These results open the possibility of developing a system of combined typing and detection of A. pleuropneumoniae.
MATERIALS AND METHODS
Bacterial strains.
A total of 102 Danish field isolates of A. pleuropneumoniae were obtained from the lungs of pigs with signs of pleuropneumonia. The predominant serotypes of A. pleuropneumoniae found in Denmark were represented in the collection, which comprised serotypes 1 (n = 5), 2 (n = 27), 5 (n = 26), 6 (n = 28), 7 (n = 3), 8 (n = 5), 10 (n = 5), and 12 (n = 3) (Table 1). Furthermore, a set of reference strains of A. pleuropneumoniae representing all serotypes of biovar 1 and two strains of biovar 2 were used (Table 1). The specificity of the PCR was tested on a collection of 48 strains representing 23 bacterial species other than A. pleuropneumoniae (Table 1). Strains of A. pleuropneumoniae and Haemophilus spp. were cultivated on PPLO agar (17). The rest of the tested strains were cultivated on Columbia agar base (Oxoid) supplemented with 5% bovine blood.
TABLE 1.
Strains used in this study and their reaction in PCR with the primers LPF and LPRa
| Strain(s) | No. PCR positive | No. PCR negative |
|---|---|---|
| Actinobacillus pleuropneumoniae biovar 1, field isolatesb | 102 | 0 |
| Actinobacillus pleuropneumoniae biovar 1, reference strainsc | 13 | 0 |
| Actinobacillus pleuropneumoniae biovar 2 (P597, P574) | 2 | 0 |
| Actinobacillus ureae (NCTC 10219, P1144) | 0 | 2 |
| Actinobacillus lignieresii (NCTC 4189, P155, P670, P671) | 0 | 4 |
| Actinobacillus capsulatus (NCTC 11408, P1364) | 0 | 2 |
| Actinobacillus hominis (NCTC 11529, P1336) | 0 | 2 |
| Actinobacillus rossii (NCTC 10801) | 0 | 1 |
| Actinobacillus equuli (NCTC 8529, P1284) | 0 | 2 |
| Actinobacillus suis (CCM 5586, P1143) | 0 | 2 |
| Escherichia coli (K31, 406, 203) | 0 | 3 |
| Bordetella bronchiseptica (1317) | 0 | 2 |
| Streptococcus suis (St. 735, St. 8074, St. 24636/74) | 0 | 3 |
| Pasteurella haemolytica (NCTC 9380, L78-94, K4017) | 0 | 4 |
| Pasteurella multocida (NCTC 10320, 2982-1, 272-3) | 0 | 3 |
| Pasteurella multocida subsp. multocida (NCTC 10322) | 0 | 1 |
| Pasteurella multocida subsp. septica (HIM 746-6) | 0 | 1 |
| Pasteurella aerogenes (ATCC 27883) | 0 | 1 |
| Pasteurella avium, biovar 2 (St. 5) | 0 | 1 |
| Pasteurella canis, biovar 2 (St. 25) | 0 | 1 |
| Pasteurella mairi (P 637) | 0 | 1 |
| Haemophilus parasuis (26489, 31637, 5015) | 0 | 3 |
| Haemophilus sp. Taxon 15 (EF 16500) | 0 | 1 |
| Haemophilus sp. Taxon Minor (305, 567, 558) | 0 | 3 |
| Haemophilus sp. Taxon D/E (319, 536, 595) | 0 | 3 |
| Haemophilus sp. Taxon F (46 KC2, 39C1) | 0 | 2 |
Five microliters of each strain was suspended in 200 μl of sterile distilled water. The bacteria were lysed and centrifuged, and 1 μl of the supernatant was used for PCR.
A. pleuropneumoniae biovar 1 field isolates from separate herds isolated from lungs of pigs with pleuropneumonia during 1993. The tested field strains of A. pleuropneumoniae consisted of serotypes 1, 2, 5, 6, 7, 8, 10, and 12.
A. pleuropneumoniae biovar 1 strains s 4074, s 1536, s 1421, M62, K17, L20, Femø, WF83, 405, 13261, D13039, 16153, and 1096.
Tonsil samples.
Tonsils from 101 pigs from nine conventional herds were obtained at slaughter. Samples from 50 tonsils from a specific-pathogen-free (SPF) herd served as negative controls. The SPF herd had been closed for the last 15 years, during which period no clinical, serological, or microbiological evidence of A. pleuropneumoniae infection had appeared.
Cultivation of samples.
Samples were cultured as previously described (15). In brief, tissue scrapings from seared cut surfaces of tonsils were suspended in phosphate-buffered saline and spread on four different agar media: two selective chocolate agar plates with added antibiotics and fungicide (300 μg of bacitracin/ml, 1 μg of lincomycin/ml, 1 μg of crystal violet/ml, 50 μg of nystatin/ml), one selective blood agar plate with added antibiotics and fungicide (100 μg of bacitracin/ml, 1 μg of lincomycin/ml, 1 μg of crystal violet/ml, 50 μg of nystatin/ml) and 0.07% NAD, one nonselective blood agar plate with a nurse strain, and one nonselective chocolate agar plate. The plates were incubated at 37°C for 48 h. A. pleuropneumoniae-like colonies were subcultivated from all media and biochemically confirmed as A. pleuropneumoniae (15). The second selective chocolate agar plate was used for PCR.
Sequencing the omlA gene.
In total seven primers, designated LPF, LPF1, LPR, LPR1, LPR2, LPR3, and LPR4 (Table 2), were used for sequencing the omlA gene. For the production of amplification products from the A. pleuropneumoniae reference serotypes, PCR with primers LPF1 and LPR1 was performed under low-stringency conditions (denaturation at 94°C for 1 min, annealing at 40°C for 1 min, primer extension at 72°C for 2 min, 35 cycles). PCR with the rest of the primers was performed with annealing temperatures around 60°C, when used for production of amplification products for sequencing. The nucleotide sequences of the amplification products were determined by cycle sequencing (21) with an Amplitaq FS dye terminator kit and a 373A automatic sequencer (Applied Biosystems Division, Perkin-Elmer, Foster City, Calif.). Analysis of the sequence similarities was performed with the HIBIO DNASIS program for Windows, Higgins and Sharp algorithm (11) (CLUSTAL 4).
TABLE 2.
Positions and sequences of primers used for PCR amplification of the omlA genea
| Primer | Sequence | Position (bp) |
|---|---|---|
| LPF | 5′-AAGGTTGATATGTCCGCACC-3′ | 272–290 |
| LPF1 | 5′-ATTGTAAACTTTAGAGCTTTATATT-3′ | 34–58 |
| LPR | 5′-CACCGATTACGCCTTGCCA-3′ | 1223–1205 |
| LPR1 | 5′-ATTAAAAAGTAAAAAAGCTATCCC-3′ | 1312–1289 |
| LPR2 | 5′-ATCTTTTACCGATGCACTATT-3′ | 1340–1319 |
| LPR3 | 5′-TAGATGATTACGATTAATCTTATCC-3′ | 667–642 |
| LPR4 | 5′-AAAAGTAAAAAAGCTATCCCG-3′ | 1308–1287 |
Preparation of samples for PCR.
Samples of pure cultures were prepared by suspending approximately 10 μl of bacterial culture in 200 μl of sterile water. Mixed bacterial cultures from selective chocolate agar plates were harvested for PCR by washing them in 2 ml of distilled sterile water. All samples were stored frozen at −80°C. Bacterial cells of both pure and mixed cultures were lysed as described by Starnbach et al. (25). One microliter of the supernatant was used in the PCR as described below.
PCR amplification.
The LPF and LPR primers (Table 2) were used for amplification in the A. pleuropneumoniae-specific PCR. Before use the primers were purified by high-performance liquid chromatography (HPLC). The expected PCR amplification product was about 950 bp, and the sequences were analyzed for gene specificity by the FASTA program from the Genetics Computer Group package (8). The PCR was performed with an automated DNA thermal cycler. The PCR assay was performed with 0.5 U of Taq polymerase (Perkin-Elmer) in a total volume of 50 μl in a buffer containing 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 50 mM KCl, 0.005% Tween 20, 0.005% Nonidet P-40 detergent, 100 μM each deoxynucleoside triphosphate, and 0.2 μM each HPLC-purified primer. The PCR test tubes were subjected to an initial denaturation at 94°C for 3 min followed by 30 cycles of denaturation at 94°C for 30 s, annealing of primers at 63°C for 20 s, and primer extension by DNA polymerase at 72°C for 2 min. To ensure complete strand extension, the reaction mixture was incubated for 10 min at 72°C after the last cycle. Twelve-microliter samples of the final reaction mixture were analyzed by electrophoresis in a 1.5% agarose gel in 1× Tris-borate-EDTA (TBE) buffer. The PCR products were stained with ethidium bromide (10 μg/ml) and visualized under UV light.
For determination of PCR detection limits, serial 10-fold dilutions of A. pleuropneumoniae (104 to 10−1 CFU/PCR test tube) in distilled sterile water were mixed with Escherichia coli in a concentration of 108 CFU/ml.
Nucleotide sequence accession numbers.
The omlA genes of A. pleuropneumoniae reference serotypes 1 to 12 (including 5a and 5b) correspond to the following GenBank accession numbers: serotype 1, U86675; serotype 2, U86676; serotype 3, U86677; serotype 4, U86678; serotype 5a, U86679; serotype 5b, U86680; serotype 6, U86681; serotype 7, U86682; serotype 8, U86683; serotype 9, U86684; serotype 10, U86685; serotype 11, U86686; and serotype 12, U86687.
RESULTS
Sequencing the omlA gene.
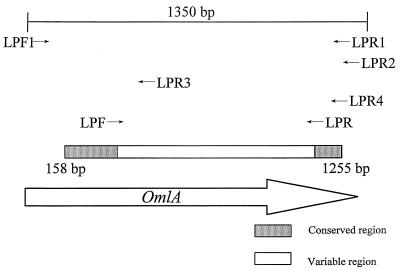
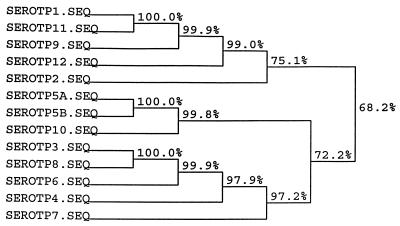
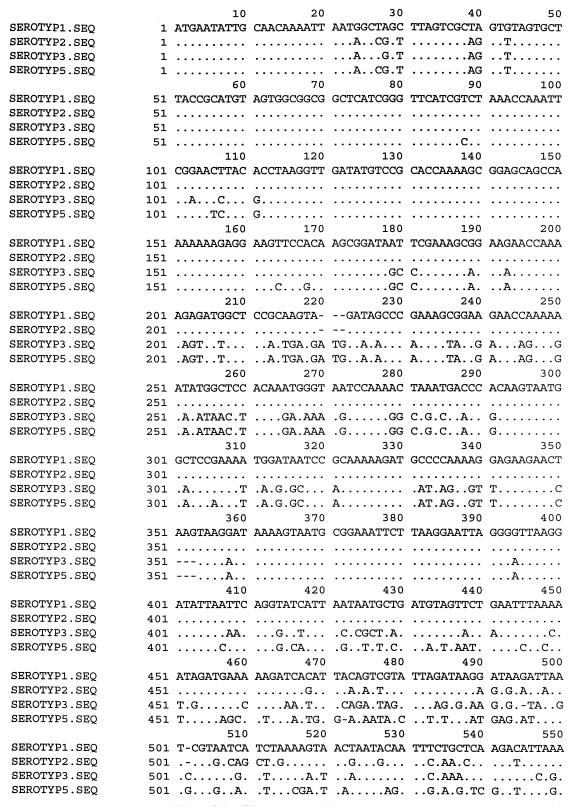
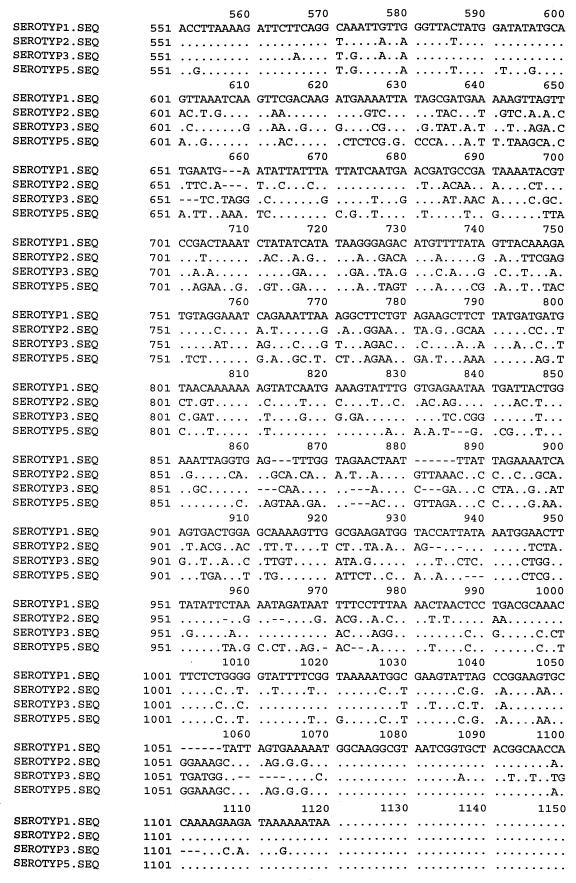
Construction of the first set of primers (LPF1 and LPR1) for sequencing was based on the alignments of the published omlA genes of serotypes 1 and 5 (3, 9). The primers were designed from the 5′ and 3′ termini of the genes. To locate identical regions of the omlA genes, the corresponding sequences of all reference strains of the A. pleuropneumoniae biovar 1 serotypes were sequenced. Alignment of the PCR amplification products from primers LPF1 and LPR1 formed the basis for construction of the A. pleuropneumoniae-specific primers designated LPF and LPR. Additionally, three reverse primers were designed for sequencing the omlA gene. The LPR2 primer (Table 2) was used for production of PCR products from A. pleuropneumoniae reference serotypes 2 and 5. For sequencing the omlA gene of reference serotype 1, it was necessary to construct two reverse LPR3 and LPR4 primers (Table 2), with one placed in the variable middle region of the gene. All seven primers were used in different combinations for amplification of PCR products for sequencing. The schematic positions of the primers are shown in Fig. 1. An alignment of the nucleotide sequences of all the reference serotypes revealed common 5′ and 3′ termini of the genes. Differences in the middle regions of the sequences divided the A. pleuropneumoniae reference strains into four groups. The group similarities of the omlA genes of the A. pleuropneumoniae reference serotypes are shown in Fig. 2. Serotypes 1, 9, 11, and 12 had overall sequence identities between 99 and 100%. Serotype 2 constituted a separate group, showing an overall sequence identity of 75% with this cluster. Serotypes 3, 4, 6, 7, and 8 all belonged to the same group of omlA-related strains, having overall sequence identities between 97 and 100%. The last of the four clusters included serotypes 10, 5a, and 5b, with sequence identities of nearly 100%. An alignment of one representative of each group-specific sequence is shown in Fig. 3. The common 5′ termini of the omlA sequences consisted of approximately 200 bp (Fig. 1 and 3). The variable middle region of approximately 700 bp showed large differences among the sequences of the four groups (Fig. 1 and 3). The last 100 bp of the 3′ termini of the genes characteristic of the serotypes showed a high degree of homology (Fig. 1 and 3).
FIG. 1.
Schematic representation of the omlA gene and the positions of the primers. The sequenced regions are represented by the box.
FIG. 2.
Similarities among sequences of the omlA genes from the reference serotypes of A. pleuropneumoniae biovar 1. Analysis of the sequence similarities was performed with the HIBIO DNASIS program for Windows, Higgins and Sharp algorithm (11) (CLUSTAL 4). The program is based on a dendrogram produced by applying the UPGMA method (24) to a matrix of similarity scores for all the aligned sequences. SEROTP, serotype; SEQ, sequence.
FIG. 3.
Alignment of one representative of each group-specific sequence of the omlA genes of the reference serotypes of A. pleuropneumoniae biovar 1. Serotype (SEROTYP) 1 represents the homologous omlA genes of A. pleuropneumoniae serotypes 1, 9, 11, and 12. Serotype 2 represents a separate group of its own. Serotype 3 represents serotypes 3, 4, 6, 7, and 8. Serotypes 5a, 5b, and 10 are represented by the sequence of serotype 5a. The dots represent sequence identity with the sequence of serotype 1; the dashes represent deletions. The positions of the sequences were calculated from the published omlA gene region (9) of an A. pleuropneumoniae strain of serotype 1. The schematic positions of the sequenced regions are shown in Fig. 1. Nucleotide 1 corresponds to nucleotide 158, and nucleotide 1120 corresponds to nucleotide 1255 of the published omlA gene sequence (9).
To test if A. pleuropneumoniae lung isolates generally had omlA sequence compositions characteristic of their serotypes, about 500 bp of 50 A. pleuropneumoniae lung isolates of serotypes 1 (n = 4), 2 (n = 9), 5 (n = 10), 6 (n = 10), 7 (n = 3), 8 (n = 4), 10 (n = 5), and 12 (n = 5) were sequenced with the LPR primer. Alignment of these 500-bp regions with those of the representatives of the group-specific sequences revealed that they each possessed the DNA sequence characteristic of their serotype.
PCR with pure cultures of bacterial strains.
The PCR assay amplified a product of approximately 950 bp with all 102 tested Danish isolates of A. pleuropneumoniae and with the A. pleuropneumoniae reference strains. All other Haemophilus, Actinobacillus, and Pasteurella species and other respiratory tract strains were negative in the test (Table 1).
To evaluate the detection limit of the PCR assay, dilution series of A. pleuropneumoniae suspensions representing 104 to 10−1 CFU/PCR test tube with the addition of E. coli at 108 CFU/ml were tested. Following agarose gel electrophoresis and ethidium bromide staining of the products from these PCRs, the detection limit of the assay was found to be approximately 102 CFU/PCR test tube.
Tonsil samples.
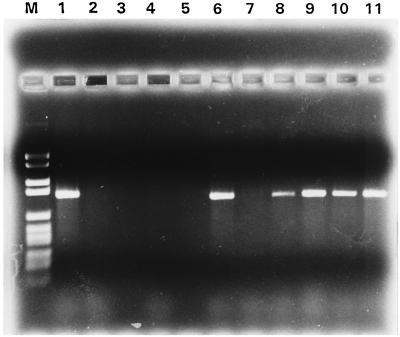
After PCR testing, 57 of the 101 mixed tonsil cultures were positive, producing an A. pleuropneumoniae-specific product of the expected size (Fig. 4). In comparison, only 23 of the 101 tonsils were found to be A. pleuropneumoniae positive by conventional culture. These 23 tonsils were all found to be PCR positive as well. No PCR amplification products were observed when we tested 50 mixed bacterial cultures from tonsils originating from an SPF herd, which served as a negative control.
FIG. 4.
PCR products from 10 of the 101 tested tonsils. A suspension from the cut surfaces of the tonsils was cultivated on A. pleuropneumoniae-selective chocolate agar and incubated at 37°C. Bacterial growths from the plates were harvested after 48 h, the bacterial suspension was lysed and centrifuged, and 1 μl of the supernatant was used for PCR. Lane M, molecular weight markers (DNA molecular weight marker VI; Boehringer Mannheim); lanes 1 to 10, tonsil cultures; lane 11, A. pleuropneumoniae (s 4074).
DISCUSSION
The main objective of this study was to develop a species-specific PCR for detection of A. pleuropneumoniae. Although PCR assays possess many advantages, such as specificity, sensitivity, and rapidity, the presence or absence of an amplification product of an expected size does not necessarily reflect the presence or absence of a pathogenic agent. Designing primers from a coding region instead of a functionally unknown sequence will probably minimize the risk of persistent point mutations, which could hinder amplification. We chose the coding region of an outer membrane protein previously described for A. pleuropneumoniae isolates of serotypes 1 and 5 (3, 9, 13). In the earlier publications the presence of homologous genes in most A. pleuropneumoniae reference serotype strains had been suggested. To investigate the presence of genes homologous to omlA in A. pleuropneumoniae reference strains, the DNA regions of the omlA genes from all the A. pleuropneumoniae reference serotypes of biovar 1 were sequenced. Differences in the nucleotide sequences of the approximately 700 bp from the middle regions of the omlA genes allowed a division of the A. pleuropneumoniae serotypes into four groups (Fig. 2 and 3). A homology search showed that serotypes 1, 9, 11, and 12 were nearly identical at the DNA level. This is generally in agreement with previous results, where an omlA gene of serotype 1 was used as a probe in a Southern blot analysis (9). In this investigation DNA from serotypes 1, 2, 8, 9, 11, and 12 hybridized even under high-stringency washing conditions. The same study revealed that sera from pigs immunized with a recombinant omlA protein from serotype 1 reacted more strongly in Western blotting with serotypes 1, 9, and 11 than with serotypes 2, 8, and 12. We found that serotype 2 constituted a separate group at the DNA level, which resembled the cluster of serotypes 1, 9, 11, and 12. Sequence homology of serotype 8 placed it in another group, consisting of serotypes 3, 4, 6, 7, and 8. The DNA sequences of the omlA genes from serotypes 5a, 5b, and 10 were nearly identical. This is in complete agreement with previously published results, where hybridization experiments showed the presence of genes highly homologous to the omlA5a gene of serotypes 5b and 10 (3, 13). The presence of specific omlA genes in different groups of the A. pleuropneumoniae serotypes resembles the serotype-specific distribution of the Apx genes (1). Serotypes 2, 3, 4, 6, and 8 have been shown to have identical Apx toxin patterns, with the exception of serotype 3, which lacks the transport genes ApxBD of ApxI (1). The omlA genes of these serotypes are also similar, except for that of serotype 2. The common Apx gene pattern of serotypes 1, 5, 9, and 11 (1) closely agrees with the presence of the same omlA gene in serotypes 1, 9, 11, and 12. However, further investigations of the presence of different genes in the A. pleuropneumoniae serotypes are necessary to reveal the genetic background of the differences among the serotypes. The described DNA variations of the omlA genes among A. pleuropneumoniae serotypes indicate the possibility of developing a combined DNA typing and detection system for A. pleuropneumoniae based on omlA gene homologies. Examination of such a system is in progress.
After optimization of the PCR assay, the LPF and LPR primers did not react with any of the A. pleuropneumoniae-related species or with any of the other tested species (Table 1) nor did the PCR produce any amplification products when we examined 50 mixed bacterial cultures from tonsils originating from an SPF herd, indicating a specificity for the PCR of 100%. The detection assay evaluated in the present study did not react with A. lignieresii. This species is related to A. pleuropneumoniae at the DNA level (2, 5) and, in a previously published A. pleuropneumoniae PCR test, gave rise to an amplification product (10, 23).
To mimic the presence of bacterial cultures other than A. pleuropneumoniae, evaluation of the detection limit of the PCR was performed with dilution series of A. pleuropneumoniae mixed with suspensions of E. coli at a concentration of 108 CFU/ml. The lower detection limit of the PCR assay of 102 CFU/PCR test tube is comparable to the detection limits of various published PCR tests (4, 26), and it was more sensitive than a previously published A. pleuropneumoniae PCR (10, 22).
The 102 Danish isolates of A. pleuropneumoniae all produced a product of approximately 950 bp when tested in the PCR assay. Homology comparisons of partial DNA sequences with sequences representative of the four omlA gene groups revealed that the isolates all possessed the expected DNA sequences characteristic of their serotypes. These results imply that the gene generally is present in A. pleuropneumoniae in a serotype group-characteristic form, indicating that the variations of the omlA gene are serotype group specific.
The ability of the PCR assay to detect A. pleuropneumoniae was compared to that of conventional culture by testing 101 mixed bacterial cultures from tonsils. A product of the expected size was produced from 57 of the tonsil cultures. In comparison, only 23 tonsil cultures were found to be A. pleuropneumoniae positive, suggesting that the sensitivity of the PCR test is superior to that of subcultivation. This difference was probably due to difficulties in the visual identification of A. pleuropneumoniae among the microbial flora of the tonsils during subcultivation. All culture-positive tonsils were found to be A. pleuropneumoniae positive in the PCR assay.
The absence of nonspecific products in this PCR assay, the relatively low detection level, and the complete specificity make it a promising diagnostic tool for detection of A. pleuropneumoniae from mixed cultures. If the PCR assay is used for routine detection of A. pleuropneumoniae from mixed bacterial cultures, more extensive testing of herds with known A. pleuropneumoniae infection status should be performed. However, when performed accurately the PCR assay is at present the most promising tool for routine DNA-based identification of A. pleuropneumoniae.
ACKNOWLEDGMENTS
We thank Lene Gertman for expert technical assistance. We also thank Magne Bisgaard, The Royal Veterinary and Agricultural University, Department of Veterinary Microbiology, Copenhagen, Denmark, for the kind donation of strains.
This study was supported by a grant from the Danish Ministry of Food, Agriculture and Fisheries.
ADDENDUM IN PROOF
After the submission of this work, we became aware of a work by a Japanese group using the omlA gene for molecular typing of A. pleuropneumoniae (M. Osaki, Y. Sato, H. Tomura, H. Ito, and T. Sekizaki, J. Vet. Med. Sci. 59:213–215, 1997).
REFERENCES
- 1.Beck M, Van Den Bosch J F, Jongenelelen I M C A, Loeffen P L W, Nielsen R, Nicolet J, Frey J. RTX toxin genotypes and phenotypes in Actinobacillus pleuropneumoniae field strains. J Clin Microbiol. 1994;32:2749–2754. doi: 10.1128/jcm.32.11.2749-2754.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borr J D, Ryan D A J, MacInnes J I. Analysis of Actinobacillus pleuropneumoniae and related organisms by DNA-DNA hybridization and restriction endonuclease fingerprinting. Int J Syst Bacteriol. 1991;41:121–129. doi: 10.1099/00207713-41-1-121. [DOI] [PubMed] [Google Scholar]
- 3.Bunka S, Christensen C, Potter A A, Willson P J, Gerlach G F. Cloning and characterization of a protective outer membrane lipoprotein of Actinobacillus pleuropneumoniae serotype 5. Infect Immun. 1995;63:2797–2800. doi: 10.1128/iai.63.7.2797-2800.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dedieu L, Mady V, Lefevre P C. Development of a selective polymerase chain reaction assay for the detection of Mycoplasma mycoides subsp. mycoides S.C. (contagious bovine pleuropneumonia agent) Vet Microbiol. 1994;42:327–339. doi: 10.1016/0378-1135(94)90064-7. [DOI] [PubMed] [Google Scholar]
- 5.Dewhirst F E, Paster B J, Olsen I, Fraser G J. Phylogeny of 54 representative strains of species in the family Pasteurellaceae as determined by comparison of 16S rRNA sequences. J Bacteriol. 1992;174:2002–2013. doi: 10.1128/jb.174.6.2002-2013.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dom P, Haesebrouck F. Comparative virulence of NAD-dependent and NAD-independent Actinobacillus pleuropneumoniae strains. Zentralbl Veterinaermed. 1992;39:303–306. doi: 10.1111/j.1439-0450.1992.tb01173.x. [DOI] [PubMed] [Google Scholar]
- 7.Fenwick B, Henry S. Porcine pleuropneumonia. J Am Vet Med Assoc. 1994;204:1334–1340. [PubMed] [Google Scholar]
- 8.Genetics Computer Group. Program manual for the Wisconsin package version 8. Madison, Wis: Genetics Computer Group; 1994. [Google Scholar]
- 9.Gerlach G-F, Anderson C, Klashinsky S, Rossi-Campos A, Potter A A, Willson P J. Molecular characterization of a protective outer membrane lipoprotein (OmlA) from Actinobacillus pleuropneumoniae serotype 1. Infect Immun. 1993;61:565–572. doi: 10.1128/iai.61.2.565-572.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gram T, Ahrens P, Nielsen J P. Evaluation of a PCR for detection of Actinobacillus pleuropneumoniae in mixed bacterial cultures from tonsils. Vet Microbiol. 1996;51:95–104. doi: 10.1016/0378-1135(96)00013-2. [DOI] [PubMed] [Google Scholar]
- 11.Higgins D G, Sharp P M. CLUSTAL: a package for performing multiple sequence alignments on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 12.Hunneman W A. Incidence, economic effects and control of Haemophilus pleuropneumoniae infections in pigs. Vet Q. 1986;8:83–87. doi: 10.1080/01652176.1986.9694024. [DOI] [PubMed] [Google Scholar]
- 13.Ito H, Uchida I, Sekizaki T, Ooishi E, Kawai T, Okabe T, Taneno A, Terakado N. Molecular cloning of an Actinobacillus pleuropneumoniae outer membrane lipoprotein (OmlA) from serotype 5a. Microb Pathog. 1995;18:29–36. [PubMed] [Google Scholar]
- 14.Jacobsen M J, Nielsen J P, Nielsen R. Comparison of virulence of different Actinobacillus pleuropneumoniae serotypes and biotypes using an aerosol infection model. Vet Microbiol. 1996;9:159–168. doi: 10.1016/0378-1135(95)00184-0. [DOI] [PubMed] [Google Scholar]
- 15.Jacobsen M J, Nielsen J P. Development and evaluation of a selective and indicative medium for isolation of Actinobacillus pleuropneumoniae. Vet Microbiol. 1995;47:191–197. doi: 10.1016/0378-1135(95)00062-f. [DOI] [PubMed] [Google Scholar]
- 16.Macinnes J I, Smart N L. Actinobacillus and Haemophilus. In: Gyles C L, Thoen C O, editors. Pathogenesis of bacterial infections in animals. Ames: Iowa State University Press; 1993. pp. 188–200. [Google Scholar]
- 17.Nicolet J. Sur l’hemophilose du porc. III. Differenciation serologic de Haemophilus parahaemolyticus. Zentralbl Bakteriol Abt 1 Orig B. 1971;216:487–495. [PubMed] [Google Scholar]
- 18.Nicolet J. Actinobacillus pleuropneumoniae. In: Leman A D, Straw B, Mengeling W L, D’Allaire S, Taylor D J, editors. Diseases of swine. Ames: Iowa State University Press; 1992. pp. 401–408. [Google Scholar]
- 19.Nielsen R. New diagnostic techniques: a review of the HAP group of bacteria. Can J Vet Res. 1990;54:68–92. [PubMed] [Google Scholar]
- 20.Pohl S, Bertschinger H U, Frederiksen W, Mannheim W. Transfer of Haemophilus pleuropneumoniae and the Pasteurella haemolytica-like organism causing porcine necrotic pleuropneumonia to the genus Actinobacillus (Actinobacillus pleuropneumoniae comb. nov.) on the basis of phenotypic and deoxyribonucleic acid relatedness. Int J Syst Bacteriol. 1983;33:510–514. [Google Scholar]
- 21.Sears L E, Moran L S, Kissinger C, Slatko B E. Thermal cycle sequencing and alternative sequencing protocols using the highly thermostable VentR™(exo) DNA polymerase. BioTechniques. 1992;13:626–633. [PubMed] [Google Scholar]
- 22.Sidibé M, Messier S, Lariviére S, Gottschalk M, Mittal K R. Detection of Actinobacillus pleuropneumoniae in the porcine upper respiratory tract as a complement to serological tests. Can J Vet Res. 1993;57:204–208. [PMC free article] [PubMed] [Google Scholar]
- 23.Sirois M, Lemire E G, Levesque R C. Construction of a DNA probe and detection of Actinobacillus pleuropneumoniae by using polymerase chain reaction. J Clin Microbiol. 1991;29:1183–1187. doi: 10.1128/jcm.29.6.1183-1187.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sneath P H A, Sokay R R. Numerical taxonomy. W. H. San Francisco, Calif: Freeman; 1973. [Google Scholar]
- 25.Starnbach M N, Falkow S, Tompkins L S. Species-specific detection of Legionella pneumophila in water by DNA amplification and hybridization. J Clin Microbiol. 1989;27:1257–1261. doi: 10.1128/jcm.27.6.1257-1261.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stone G G, Oberst R D, Hays M P, McVey S, Chengappa M M. Detection of Salmonella serovars from clinical samples by enrichment broth cultivation-PCR procedure. J Clin Microbiol. 1994;32:1742–1749. doi: 10.1128/jcm.32.7.1742-1749.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]