Abstract
The diagnosis of amebiasis by microscopic identification of the parasite in stool is insensitive and unable to distinguish the invasive parasite Entamoeba histolytica from the commensal parasite E. dispar. In this study, we have tested a PCR technique for the detection of E. histolytica and compared it with isoenzyme analysis and the TechLab E. histolytica-specific antigen detection test. The nested-PCR test we used is based on amplification of the small subunit rRNA gene of E. histolytica and E. dispar followed by restriction digest analysis of the PCR product. Single stool samples were obtained from 98 patients from Dhaka, Bangladesh, with diarrhea: 88 patients diagnosed by microscopy and/or culture with E. histolytica and/or E. dispar infection and 10 patients without infection. Isoenzyme analysis identified 53 of the infections as E. histolytica and 28 as E. dispar. PCR and isoenzyme identification of E. histolytica agreed in 96% (51 of 53) of amebic cultures. PCR for E. histolytica was negative in all 10 samples that were negative for E. histolytica by isoenzyme and antigen detection. PCR and antigen detection had comparable sensitivities when performed directly on fresh stool specimens, identifying 87% (46 of 53) and 85% (45 of 53), respectively, of E. histolytica infections identified by isoenzyme analysis. The correlation of results by antigen detection and PCR for identification of E. histolytica in stool was 93% (45 of 48 cases). Mixed infections with E. histolytica and E. dispar were detected by PCR in 14% (12 of 88) of cases. In conclusion, all three techniques for specific identification of E. histolytica in fresh stool showed excellent correlation. Only the TechLab E. histolytica antigen detection test was both rapid and technically simple.
It has long been known that although about 500 million people each year have amebiasis, only about 10% experience symptomatic disease (20, 21). After much research and argument, it is now generally accepted that what was earlier known as Entamoeba histolytica actually comprises two genetically distinct but morphologically indistinguishable species—a pathogenic one, for which it has been suggested that the name E. histolytica be retained, and a nonpathogenic one, for which the name Entamoeba dispar has been revived (4, 21). E. histolytica can cause invasive intestinal and extraintestinal disease, while E. dispar cannot. A WHO-Pan American Health Organization-United Nations Educational, Scientific, and Cultural Organization Expert Panel recently recommended the development of improved methods, using technologies appropriate for developing countries, for the specific diagnosis of E. histolytica infection (21).
Identification and differentiation of E. dispar and E. histolytica in stool sample by microscopy is imprecise. While E. histolytica trophozoites are more likely than E. dispar to contain ingested erythrocytes, the organisms are identical in appearance (4–6). Not only is microscopy unable to differentiate E. histolytica from E. dispar; it is at best only 60% sensitive and can be confounded by false-positive results due to misidentification of macrophages and nonpathogenic species of Entamoeba (6, 9, 11, 17). Culture is more sensitive than microscopy, and isoenzyme analysis of cultured amebae enables the differentiation of E. histolytica from E. dispar. However, amebic cultures and isoenzyme analysis require a week to complete and are negative in many microscopy-positive samples, in some cases due to delays in sample processing or due to the institution of antiamebic therapy prior to stool collection (6, 9, 17).
New approaches to the detection of E. histolytica and E. dispar are based on antigen detection in stool (1, 7–9, 17) and detection of E. histolytica-specific DNA by PCR amplification (2, 3, 10, 13, 16, 18). Antigenic differences in the lectin of E. histolytica and E. dispar amebae enable specific identification of the disease-causing amebae E. histolytica (14). Antigen detection tests have proven to be more sensitive and specific than microscopy (7–9). In this study, we have used a PCR technique for detection of E. histolytica and compared it with antigen detection and isoenzyme analysis.
MATERIALS AND METHODS
Stool specimens.
Stool samples were collected from patients with diarrhea seen in 1995 and 1996 at the International Center for Diarrheal Disease Research, Bangladesh (ICDDR,B). Analyzed were single stool samples from 88 patients diagnosed with E. histolytica and/or E. dispar infection (by microscopy and/or culture) and from 10 patients whose stool samples were negative by microscopy and culture for E. histolytica and E. dispar infection.
Antigen detection, culture, and isoenzyme analyses.
The TechLab (Blacksburg, Va.) Entamoeba test (designed to detect but not differentiate E. histolytica and E. dispar antigen in stool specimens) and TechLab E. histolytica test (designed to detect specifically E. histolytica in stool specimens) were performed on the stool specimens according to the manufacturer’s instructions. Fresh stool samples were examined microscopically in a 0.9% saline smear for the presence of E. histolytica-E. dispar complex cysts and trophozoites. Stools were cultured for Entamoeba species in Robinson’s medium within 6 h of collection, and isoenzyme analysis was performed as described previously (7, 15).
Extraction of DNA from stool samples and cultures.
Trophozoites and cysts present in the stool and cultured amebae from the stool samples were the source of target DNA, which was purified by a modified version of the method of Katzwinkel-Wladarsch et al. (10). All procedures were performed with sterile, disposable plastic tubes and pipette tips. Aliquots (0.2 g) of stool samples or cultures in Robinson’s medium were taken to 1.5 ml in microcentrifuge tubes, and 33.3 μl of 1 M KOH and 9.3 μl of 1 M dithiothreitol were added. The samples were mixed thoroughly by stirring with a pipette tip, followed by brief shaking. After incubation at 65°C for 15 min, the samples were neutralized with 4.3 μl of 25% HCl and buffered with 80 μl of 2 M Tris-HCl (pH 8.3) and the suspension was mixed again. The DNA was extracted by shaking with 250 μl of phenol-chloroform-isoamyl alcohol (25:24:1) (PCI) saturated with 10 mM Tris (pH 8.0) and 1 mM EDTA. The phases were separated by a 4-min spin in a microcentrifuge. The aqueous phase was transferred to a new tube, and the DNA was further purified by adsorption to 5 μl of Glassmax matrix suspension (Gibco BRL). The DNA was eluted in 39 μl of deionized water.
PCR method.
Primer construction was based on sequences from the small subunit rRNA gene of E. histolytica and E. dispar (10). For the first PCR, the primer pair E-1 (TTT GTA TTA GTA CAA A) and E-2 (GTA [A/G]TA TTG ATA TAC T), which amplified a 0.9 kb fragment of the rRNA gene, was used. The primer pair E-1 and E-2 is complementary to both E. histolytica and E. dispar sequences, with the E-2 primer constructed twofold degenerately, i.e., as a mixture with half corresponding to the E. histolytica sequence and the other half corresponding to the E. dispar sequence. The first PCR amplification with E-1 and E-2 was followed by two additional PCRs, each of which was specific for either the E. histolytica or the E. dispar sequence. The primers used for these reactions were located downstream of E-1 and E-2, making this a nested PCR. For the second (nested) PCR, two different primer pairs specific for E. histolytica (EH-1, AAT GGC CAA TTC ATT CAA TG, and EH-2, TTT AGA AAC AAT GCT TCT CT) or E. dispar (ED-1, AGT GGC CAA TTT ATG TAA GT, and ED-2, TTT AGA AAC AAT GTT TCT TC) were used. All primers were obtained from Oswel DNA, University of Southampton, Southampton, United Kingdom.
Both of the PCRs used a hot-start technique. In the first PCR, 18.4 μl of the DNA extracts was denatured at 96°C for 2 min after the addition of 0.6 μl each of 40 μM solutions of the primers (E-1 and E-2) and 1 drop of mineral oil. After cooling to 80°C, 5.4 μl of freshly prepared “mastermix” (2.5 μl of 10× PCR buffer [catalog no. 18038; Gibco BRL], 2 μl of 50 mM MgCl2, 0.64 μl of deoxynucleoside triphosphate mix [10 mM each; Perkin-Elmer, Norwalk, Conn.], and 0.25 μl (5 IU/μl) of Taq polymerase [Gibco BRL]) was added. Fifty cycles with denaturation at 92°C for 60 s, annealing at 43°C for 60 s, and extension at 72°C for 90 s were performed. In the second (nested) PCR, 3 μl of the first PCR product was taken in 26 μl of water and denatured at 96°C for 2 min after the addition of 1 μl each of 40 μM solutions of the primers (EH-1 and EH-2 for E. histolytica; ED-1 and ED-2 for E. dispar) and 2 drops of mineral oil. After cooling to 80°C, 8.6 μl of freshly prepared mastermix (10× PCR buffer, 3.2 μl of 50 mM MgCl2, 1 μl of deoxynucleoside triphosphate mix, and 0.4 μl of Taq polymerase) was added and PCR was performed as described above, except that the annealing temperature was 62°C. PCR amplifications were performed with a Bio-Rad gene cycler. Products were visualized on a 1.3% agarose gel containing ethidium bromide (0.2 μg/ml; Sigma).
Restriction endonuclease digests.
Bands excised from the agarose gel were silica gel purified as described above, eluted in 9.6 μl of buffer, and digested with 0.8 μl (10 U/μl) of DraI (Gibco BRL) for 60 min at 37°C, followed by the addition of 0.4 μl (10 U/μl) of Sau96I (Amersham) and further incubation at the same temperature for another 90 min, by the method of Katzwinkel-Wladarsch et al. (10).
RESULTS
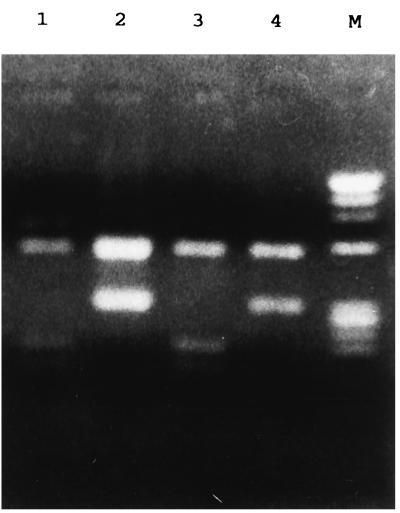
The nested-PCR amplifications, performed by a modification of the method of Katzwinkel-Wladarsch et al., yielded a 0.9-kb band with E. histolytica and E. dispar DNA. Restriction enzyme digestion of the DNA amplified with the E. histolytica-specific primers exhibited characteristic bands of 0.55 and 0.35 kb, usually with some of the undigested 0.9-kb band remaining. The E. dispar-amplified DNA yielded a band of 0.55 kb and a confluent band of 0.15 to 0.2 kb, often with a partial digestion product of 0.7 kb (Fig. 1).
FIG. 1.
Restriction endonuclease digestion of the products of nested PCR. The restriction fragments of DNA amplified with the E. dispar-specific nested primers (lanes 1 and 3) and E. histolytica-specific nested primers (lanes 2 and 4) are shown. The starting materials for the PCRs were stool samples from patients with culture-confirmed infections with E. dispar (lane 1), E. histolytica (lane 2), and E. histolytica (lanes 3 and 4 [a mixed infection based on PCR]). The marker (lane M) is φX174 DNA digested with HaeIII.
Nested PCR, antigen detection tests, and culture and isoenzyme analyses were performed on 98 stool specimens from patients with diarrhea in Dhaka. Stool specimens from 68 patients were positive for E. histolytica and/or E. dispar infection by microscopy and culture, 13 were positive only by culture, 7 were positive only by microscopy, and 10 were negative by both microscopy and culture (data not shown). Isoenzyme analysis identified 53 E. histolytica and 28 E. dispar infections from the 81 culture-positive isolates. Nested PCR using cultured amebae as the source of DNA demonstrated a 96% correlation with isoenzyme analysis in detecting E. histolytica infection: 51 of 53 isolates identified as E. histolytica by isoenzyme analysis demonstrated E. histolytica-specific DNA amplification products (data not shown).
Results of nested PCR using stool were compared with those of culture and isoenzyme analysis (Table 1). For the 53 stool specimens that were positive for E. histolytica by isoenzyme analysis, the nested PCR performed directly on stool was positive for 46 specimens (including 9 specimens positive for both E. histolytica and E. dispar), for a correlation of 87% (Table 1). The PCR and antigen detection tests had similar sensitivities when performed directly on stool specimens, identifying 87% (46 of 53) and 85% (45 of 53), respectively, of E. histolytica infections identified by culture and isoenzyme analyses (data not shown).
TABLE 1.
Results of PCR and culture and isoenzyme analysis using stool specimens
| PCR result | Culture and isoenzyme analysis result (no. of specimens positive)
|
Total no. of specimens | ||
|---|---|---|---|---|
| E. histolytica | E. dispar | Culture negative | ||
| E. histolytica | 37 | 7 | 2 | 46 |
| E. dispar | 3 | 16 | 3 | 22 |
| E. histolytica-E. dispar | 9 | 3 | 0 | 12 |
| Negative | 4 | 2 | 12 | 18 |
| Total | 53 | 28 | 17 | 98 |
Results of the nested PCR using stool specimens were also compared with those of the antigen detection tests (Table 2). The antigen detection tests revealed that there were 50 samples positive for E. histolytica, 29 positive for E. dispar, and 19 negative for E. histolytica and E. dispar antigens. For the 50 specimens that were positive for E. histolytica by the antigen detection tests, the nested PCR performed directly on stool specimen was positive for 47 specimens (including 8 specimens positive for both E. histolytica and E. dispar), for a correlation of 94%. Out of 12 mixed infections of E. histolytica and E. dispar detected by the nested PCR performed directly on stool, 8 specimens were identified as E. histolytica by the antigen detection test (Table 2).
TABLE 2.
Results of PCR and antigen detection using stool specimens
| PCR result | Antigen detection result (no. of specimens positive)
|
Total no. of specimens | ||
|---|---|---|---|---|
| E. histolytica | E. dispar | Antigen negative | ||
| E. histolytica | 39 | 5 | 2 | 46 |
| E. dispar | 1 | 19 | 2 | 22 |
| E. histolytica-E. dispar | 8 | 4 | 0 | 12 |
| Negative | 2 | 1 | 15 | 18 |
| Total | 50 | 29 | 19 | 98 |
DISCUSSION
Microscopy is an outmoded technique that should not be used to diagnose amebic colitis: it is insensitive, incapable of differentiating pathogenic E. histolytica from nonpathogenic E. dispar, and prone to giving false-positive results. Our experience in Bangladesh has highlighted the danger of relying on microscopy. Of all children with diarrhea diagnosed with amebiasis by microscopy, only 40% were proven to have E. histolytica infection when specific methods (antigen detection and culture-isoenzyme analysis) were used. And of all children diagnosed with E. histolytica infection by specific methods, the majority were missed by microscopy (9).
For this reason the WHO, the Pan American Health Organization, and UNESCO issued in 1997 the joint recommendation that “E. histolytica should be specifically identified” (21). The only E. histolytica-specific test that is approved for in vitro diagnostic use is the TechLab E. histolytica antigen detection kit. The Alexon and Merlin Optimum S antigen detection kit cross-react with E. dispar (12).
This study represents the first time that three independent techniques for species-specific identification of E. histolytica have been compared by using the same stool samples. The nested-PCR test for detection of E. histolytica (based on amplification of the rRNA gene) correlated well with antigen detection and isoenzyme techniques for detection of E. histolytica in diarrheal stool specimens. The overall correlation between the nested-PCR results from stool specimens and those of antigen detection tests for detecting E. histolytica infection was greater than 90%. This agreement between techniques provides confidence that any one of the techniques may be used alone to yield an accurate assessment of the presence of E. histolytica in a stool specimen. However, of the three techniques, the TechLab antigen detection test was by far the most rapid and simple, providing an answer within 2 to 3 h. In contrast, it took several days to complete the PCR test, and 1 to 2 weeks were needed to culture the amebae and perform isoenzyme analysis.
PCR amplification of rRNA genes has been shown to be more sensitive than antigen detection when cultured parasites are used as the source of DNA and antigen (12). However, in the real-world situation where the parasite has to be identified in stool specimens, our results indicate that the two techniques are approximately equal in sensitivity. Thus, there is no disadvantage to the use of antigen detection and there is a decided advantage to its use due to its speed and technical simplicity.
Dual infection with E. dispar and E. histolytica in the same stool specimen and occult infection (PCR-detected infection in stool specimens negative for E. histolytica by other techniques) were both uncommon, similar to the observations of other investigators studying smaller numbers of samples (3, 19). This is a marked contrast to studies of Mexican children by Samuelson and colleagues (13, 16), in which PCR detected dual and occult infections in the majority of stool specimens tested. This could reflect real differences in the epidemiology of E. histolytica in Mexico and Bangladesh. However, it is also possible that the PCR test used in the Mexican studies (which was based on highly repetitive sequences in the noncoding region of the rRNA gene) was prone to a high level of false-positive results. Unfortunately, in the studies from Mexico the PCR results were not confirmed by either culture or antigen detection.
In conclusion, the nested PCR described in the present work is comparable to isoenzyme analysis and the TechLab E. histolytica antigen detection test for identifying E. histolytica infection in stool. However, the PCR technique described is time-consuming, cumbersome, and expensive and therefore not well suited for use in developing countries where amebiasis is prevalent. The antigen detection test, which is rapid and simple and does not require any special equipment, is presently the only practical means for diagnosis of E. histolytica infection.
ACKNOWLEDGMENTS
This research was supported by National Institutes of Health grant AI-26649 and the ICDDR,B. TechLab, Inc., provided the E. histolytica antigen detection kits. The ICDDR,B is supported by countries and agencies which share its concern for the health problems of developing countries. Current donors include the aid agencies of the governments of Australia, Bangladesh, Belgium, Canada, China, Germany, Japan, The Netherlands, Norway, Republic of Korea, Saudi Arabia, Sweden, Switzerland, the United Kingdom, and the United States; international organizations, including the Arab Gulf Fund, the Asian Development Bank, the International Atomic Energy Center, the United Nations Children’s Fund (UNICEF), the United Nations Development Program (UNDP), the United Nations Population Fund (UNFPA), and the WHO; private foundations, including the Child Health Foundation, the Ford Foundation, the Population Council, the Rockefeller Foundation, and the Sasakawa Foundation; and private organizations, including American Express Bank, Bayer AG, CARE, Family Health International, Helen Keller International, the Johns Hopkins University, Procter Gamble, RAND, SANDOZ, Swiss Red Cross, the University of California, Davis, and others.
REFERENCES
- 1.Abd-Alla M D, Jackson T F H G, Gathiram V, El-Hawey A M, Ravdin J I. Differentiation of pathogenic Entamoeba histolytica infections from nonpathogenic infections by detection of galactose-inhibitable adherence protein antigen in sera and feces. J Clin Microbiol. 1993;31:2845–2850. doi: 10.1128/jcm.31.11.2845-2850.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Acuna-Soto R, Samuelson J, De Girolami P, et al. Application of the polymerase chain reaction to the epidemiology of pathogenic and nonpathogenic Entamoeba histolytica. Am J Trop Med Hyg. 1994;48:58–70. doi: 10.4269/ajtmh.1993.48.58. [DOI] [PubMed] [Google Scholar]
- 3.Britten D, Wilson S M, McNerney R, Moody A H, Chiodini P L, Ackers J P. An improved colorimetric PCR-based method for detection and differentiation of Entamoeba histolytica and Entamoeba dispar in feces. J Clin Microbiol. 1997;35:1108–1111. doi: 10.1128/jcm.35.5.1108-1111.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diamond L S, Clark C G. A redescription of Entamoeba histolytica Shaudinn 1903 (amended Walker 1911) separating it from Entamoeba dispar (Brumpt 1925) J Eukaryot Microbiol. 1993;40:340–344. doi: 10.1111/j.1550-7408.1993.tb04926.x. [DOI] [PubMed] [Google Scholar]
- 5.Gathiram V, Jackson T F H G. Frequency distribution of Entamoeba histolytica zymodemes in a rural South African population. Lancet. 1985;i:719–721. doi: 10.1016/s0140-6736(85)91263-2. [DOI] [PubMed] [Google Scholar]
- 6.Gonzalez-Ruiz A, Haque R, Aguirre A, et al. Value of microscopy in the diagnosis of dysentery associated with invasive Entamoeba histolytica. J Clin Pathol. 1994;47:236–239. doi: 10.1136/jcp.47.3.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haque R, Kress K, Wood S, et al. Diagnosis of pathogenic Entamoeba histolytica infection using a stool ELISA based on monoclonal antibodies to the galactose specific adhesin. J Infect Dis. 1993;167:247–249. doi: 10.1093/infdis/167.1.247. [DOI] [PubMed] [Google Scholar]
- 8.Haque R, Neville L M, Hahn P, Petri W A., Jr Rapid diagnosis of Entamoeba infection by using Entamoeba and Entamoeba histolytica stool antigen detection kits. J Clin Microbiol. 1995;33:2558–2561. doi: 10.1128/jcm.33.10.2558-2561.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haque R, Faruque A S G, Hahn P, Lyerly D M, Petri W A., Jr Entamoeba histolytica and Entamoeba dispar infection in children in Bangladesh. J Infect Dis. 1997;175:734–736. doi: 10.1093/infdis/175.3.734. [DOI] [PubMed] [Google Scholar]
- 10.Katzwinkel-Wladarsch S, Loscher T, Rinder H. Direct amplification and differentiation of pathogenic and nonpathogenic Entamoeba histolytica DNA from stool specimens. Am J Trop Med Hyg. 1994;51:115–118. doi: 10.4269/ajtmh.1994.51.115. [DOI] [PubMed] [Google Scholar]
- 11.Krogstad D J, Spencer H C, Healy G R, et al. Amebiasis: epidemiologic studies in the United States, 1971–1974. Ann Intern Med. 1978;88:89–97. doi: 10.7326/0003-4819-88-1-89. [DOI] [PubMed] [Google Scholar]
- 12.Mirelman D, Nuchamowitz Y, Stolarsky T. Comparison of use of enzyme-linked immunosorbent assay-based kits and PCR amplification of rRNA genes for simultaneous detection of Entamoeba histolytica and E. dispar. J Clin Microbiol. 1997;35:2405–2407. doi: 10.1128/jcm.35.9.2405-2407.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newton-Sanchez O A, Sturm-Ramirez K, Romero-Zamora J L, Santos-Preciado J I, Samuelson J. High rate of occult infection with Entamoeba histolytica among non-dysenteric Mexican children. Arch Med Res. 1997;28:S311–S313. [PubMed] [Google Scholar]
- 14.Petri W A, Jr, Jackson T F H G, Gathiram V, Kress K, Saffer L D, Snodgrass T L, Chapman M D, Keren Z, Mirelman D. Pathogenic and nonpathogenic strains of Entamoeba histolytica can be differentiated by monoclonal antibodies to the galactose-specific adherence lectin. Infect Immun. 1990;58:1802–1806. doi: 10.1128/iai.58.6.1802-1806.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinson G L. The laboratory diagnosis of human parasitic amoebae. Trans R Soc Trop Med Hyg. 1968;62:285–294. doi: 10.1016/0035-9203(68)90170-3. [DOI] [PubMed] [Google Scholar]
- 16.Romero J L, Descoteaux S, Reed S, Orozco E, Santos J, Samuelson J. Use of polymerase chain reaction and nonradioactive DNA probes to diagnose Entamoeba histolytica in clinical samples. Arch Med Res. 1992;23:277–279. [PubMed] [Google Scholar]
- 17.Strachan W D, Spice W M, Chiodini P L, Moody A H, Ackers J P. Immunological differentiation of pathogenic and nonpathogenic isolates of E. histolytica. Lancet. 1988;i:561–563. doi: 10.1016/s0140-6736(88)91355-4. [DOI] [PubMed] [Google Scholar]
- 18.Tannich E, Burchard G D. Differentiation of pathogenic from nonpathogenic Entamoeba histolytica by restriction fragment analysis of a single gene amplified in vitro. J Clin Microbiol. 1991;29:250–255. doi: 10.1128/jcm.29.2.250-255.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Troll H, Marti H, Weiss N. Simple differential detection of Entamoeba histolytica and Entamoeba dispar in fresh stool specimens by sodium acetate-acetic acid-formalin concentration and PCR. J Clin Microbiol. 1997;35:1701–1705. doi: 10.1128/jcm.35.7.1701-1705.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walsh J A. Problems in recognition and diagnosis of amebiasis: estimation of the global magnitude of morbidity and mortality. Rev Infect Dis. 1986;8:228–238. doi: 10.1093/clinids/8.2.228. [DOI] [PubMed] [Google Scholar]
- 21.World Health Organization. Amoebiasis. W H O Weekly Epidemiol Rec. 1997;72:97–100. [Google Scholar]