Abstract
A seminested-PCR assay, based on the amplification of the pneumococcal penicillin-binding protein 2B gene (pbp2B), was developed for the detection of penicillin-resistant and -susceptible pneumococci in cerebrospinal fluid (CSF) specimens. Species-specific primers (P5 and P6) which amplified a 682-bp conserved region of the transpeptidase-encoding region of the pbp2B gene were used. Four “resistance” primers were designed to bind to altered areas of the pbp2B gene identified in penicillin-resistant South African wild-type strains. Together with the downstream primer P6, the upstream resistance primers amplified fragments which were used to detect the presence of penicillin resistance. This system identified all 35 of the S. pneumoniae isolates evaluated, including strains of 11 different serotypes and a range of penicillin-resistant and -susceptible strains. The specificity of the assay was demonstrated by its inability to amplify DNA from other bacterial species which commonly cause meningitis. It was possible to detect pneumococcal DNA from culture-negative CSF inoculated with 2.5 pg of purified DNA or 18 CFU. Analysis of 285 CSF specimens showed that PCR detected the pneumococcus in 18 samples positive by culture, including the identification of four penicillin-resistant isolates. The positive predictive value and the negative predictive value of the assay were each 100%.
Streptococcus pneumoniae (the pneumococcus) is the most common cause of acute community-acquired bacterial pneumonia and accounts for 30 to 40% of lower respiratory tract infections (35). It is the second most common cause of bacterial meningitis and is a prevalent cause of diseases such as sinusitis and otitis media (22). During the early 1940s, clinical isolates of pneumococci exhibited high degrees of susceptibility to antibiotics including penicillin, the antibiotic recommended for the treatment of suspected pneumococcal infections (20). The first appearance of clinically significant penicillin-resistant and multidrug-resistant pneumococci in South Africa occurred in 1977 and 1978. Penicillin-resistant isolates have since been reported worldwide (2). The prevalence of penicillin-resistant strains of pneumococci in South Africa is among the highest in the world, with penicillin MICs being up to a 1,000-fold greater than those for penicillin-susceptible strains (16). In Soweto, South Africa, near Johannesburg, the rate of resistance among pneumococcal strains isolated from meningitis patients is 41.7% (17).
Effective treatment of meningitis requires rapid detection of both the organism and the susceptibility pattern. Currently, the most sensitive method of diagnosis is based on the successful culture and identification of bacteria from cerebrospinal fluid (CSF). By standard culture methods, presumptive identification of S. pneumoniae takes 12 to 24 h, followed by biochemical tests for confirmation (7, 14, 34). Susceptibility testing requires a further 24 h, which means that a result is rarely available within less than 48 h. Empiric therapy must therefore have a wide spectrum to include coverage against penicillin-resistant pneumococci. The problem of having to use drugs such as broad-spectrum cephalosporins and vancomycin is that it encourages the development of drug resistance.
Alternate methods of diagnosing pneumococcal disease are based on the detection of bacterial antigens in body fluids. The detection of pneumococcal capsular antigen by counterimmunoelectrophoresis and latex agglutination has been proven to be useful; however, these techniques are not entirely satisfactory because of inadequate sensitivity and specificity (4, 9, 25). Reliable results are obtained only for samples containing more than 105 CFU per ml (3, 18), and since samples from approximately 45% of patients with meningitis have less than 105 CFU per ml (3, 18), the applicability of antigen testing is limited. In addition, these methods do not allow for susceptibility testing of the isolates.
Due to the development of molecular biology-based diagnostic techniques, such as the PCR, it is now possible to detect low numbers of pathogens in clinical specimens (28, 36). The PCR is a rapid and sensitive method, and since it does not depend on the presence of viable organisms, it may be more applicable in cases of prior antibiotic treatment. Previous studies have used PCR for the detection of bacterial pathogens in various specimens including blood, sputum, middle ear fluid, and CSF (11, 13, 27, 33).
Penicillin-resistant pneumococci produce altered penicillin-binding proteins (PBPs) which have reduced affinities for β-lactam antibiotics. Alterations in pbp2B genes are highly divergent, particularly in the transpeptidase-encoding region (6, 12). Within this divergent region, changes which are common to all penicillin-resistant pneumococcal strains tested in South Africa have been identified (31). These changes are universal in that they are present in pbp2B genes from pneumococci found in other parts of the world. Our study describes a seminested-PCR strategy used to detect the presence of penicillin-susceptible and -resistant S. pneumoniae organisms in CSF specimens. The presence of pneumococcal DNA was detected with species-specific primers which amplify a conserved region of the transpeptidase-encoding region of the pbp2B gene. Four different “resistance” primers were designed to bind to altered areas of the pbp2B gene identified in penicillin-resistant South African wild-type strains of pneumococci (31). These altered areas occur internal to the species-specific primer binding sites. Together with the downstream primer, the upstream resistance primers amplify resistance products, which are used to detect the existence of penicillin resistance in the pneumococcus.
MATERIALS AND METHODS
Strains.
All organisms used in this study were obtained from the South African Institute for Medical Research (SAIMR; Johannesburg, South Africa). Thirty-three S. pneumoniae isolates of various serotypes and with various degrees of susceptibility to penicillin were used. Isolates were serotyped by latex agglutination and were confirmed by the Quellung method with specific antisera from the Staten Seruminstitut (Copenhagen, Denmark) (22). Penicillin MICs were determined by the agar dilution method in Mueller-Hinton agar (Difco Laboratories, Detroit, Mich.) supplemented with 3% lysed horse blood (24). The properties of these isolates are listed in Table 1. Two reference strains, namely, strain R6, an unencapsulated laboratory strain, and S. pneumoniae ATCC 49619 were also included in the study. Twenty nonpneumococcal organisms were tested in the study. These included coagulase-negative staphylococcus, Staphylococcus aureus, Escherichia coli, Pseudomonas aeruginosa, Enterococcus faecalis, Streptococcus pyogenes, Streptococcus sanguis, Streptococcus faecium, Streptococcus agalactiae, Streptococcus milleri, Streptococcus mutans, Streptococcus bovis, viridans group streptococcus Streptococcus mitior, Haemophilus influenzae, Mycobacterium tuberculosis, Listeria monocytogenes, Cryptococcus neoformans, Moraxella catarrhalis, and Neisseria meningitidis. These organisms were isolated from clinical specimens submitted to the routine microbiology laboratory and were identified by standard laboratory methods.
TABLE 1.
Properties of pneumococcal isolatesa
| Isolate | Hospital | Specimen | Serotype | Yr of isolation | Penicillin MIC (μg/ml) |
|---|---|---|---|---|---|
| 69602 | RXH | T/Asp | 6 | 1987 | 2 |
| 50800 | RXH | B/C | 6 | 1990 | 2 |
| 45525 | BH | B/C | 14 | 1992 | 2 |
| 47937 | RXH | Spt | 19 | 1992 | 0.125 |
| 47921 | RXH | Spt | 14 | 1992 | 2 |
| 47926 | RXH | E/S | 6 | 1992 | 0.125 |
| 14309 | RXH | E/S | 14 | 1987 | 0.03 |
| 24984 | RXH | E/S | 23 | 1990 | 2 |
| 20396 | RXH | B/C | 6 | 1996 | 0.25 |
| 20482 | BH | B/C | 19 | 1996 | 0.125 |
| 20713 | BH | B/C | 23 | 1996 | 2 |
| 20402 | RXH | B/C | 6 | 1996 | 0.25 |
| 20685 | NJH | B/C | 14 | 1996 | 0.125 |
| 12569 | BH | B/C | 23 | 1991 | 2 |
| 22453 | BH | CSF | 14 | 1989 | 4 |
| 85697 | BH | B/C | 19 | 1988 | 2 |
| 48478 | BH | CSF | 19 | 1989 | 4 |
| 4916 | TH | G/juice | 23 | 1988 | 2 |
| 17283 | MCT | Spt | 14 | 1987 | 1 |
| 65372 | TH | G/juice | 19 | 1987 | 2 |
| 23252 | BH | B/C | 1 | 1996 | 0.03 |
| 23253 | BH | B/C | 1 | 1996 | 0.03 |
| 23255 | BH | B/C | 1 | 1996 | 0.03 |
| 23254 | BH | B/C | 1 | 1996 | 0.03 |
| 23241 | HH | B/C | 10 | 1996 | 0.03 |
| 23312 | BBH | E/S | 19 | 1996 | 0.125 |
| 23474 | NJH | B/C | 15 | 1996 | 0.03 |
| 23475 | NJH | B/C | 12 | 1996 | 0.03 |
| 23380 | NJH | B/C | 5 | 1996 | 0.03 |
| 23328 | BH | B/C | 3 | 1996 | 0.03 |
| 9833 | Ermelo | B/C | 19 | 1995 | 4 |
| 8303 | VD | Spt | 23 | 1995 | 4 |
| 10036 | BH | CSF | 23 | 1995 | 0.5 |
Abbreviations: B/C, blood culture; T/Asp, tracheal aspirate; G/juice, gastric juice; E/S, ear swab; Spt, sputum; BH, Baragwanath Hospital; HH, Hillbrow Hospital; NJH, New Johannesburg Hospital; MCT, Medical School Observatory, Cape Town; BBH, Boksburg Benoni Hospital; VD, Van Drimmelen Laboratories; RXH, Red Cross Hospital; TH, Tshepong Hospital.
PCR primers.
The selection of primers was based on the published gene sequences of the pbp2B transpeptidase-encoding region of South African penicillin-resistant wild-type S. pneumoniae strains (31). The properties of the primers are listed in Table 2.
TABLE 2.
Sequences of oligonucleotide primers
| Primera | Sequence (5′→3′) | Position in pbp2B geneb | Product length (bp) following PCR with downstream primer P6 |
|---|---|---|---|
| R1 | GCCTTTTCTAGGCCAATGCCGATTAC | 697–722 | 331 |
| R2 | GCCTACGATTCATTCCCGATT | 700–720 | 328 |
| R3 | AAATTGGCATATGGATCTTTTCCT | 694–717 | 334 |
| R4 | GTTTTAACTAACAATTTAGAATCC | 814–837 | 214 |
| P5 | CTGACCATTGATTTGGCTTTCCAA | 346–369 | 682 |
| P6 | TTTGCAATAGTTGCTACATACTG | 1006–1028 |
Primers P5 and P6 are specific for pneumococci. Primers R1 to R4 are specific for penicillin-resistant pneumococci only.
According to the published sequences of Smith and Klugman (31).
PCR sample preparation.
(i) Bacterial chromosomal DNA was purified from freshly cultured bacteria grown on Columbia agar supplemented with 5% horse blood. N. meningitidis and H. influenzae were cultured on chocolate agar (SAIMR). The DNA was extracted by standard phenol-chloroform extraction methods (29).
(ii) A small loopful of bacterial cells was inoculated into a culture-negative CSF specimen, and DNA was released from the cells by boiling for 10 min. This CSF specimen consisted of pooled, culture-negative CSF specimens with various macroscopic properties, which would include any potential PCR inhibitors.
PCR.
A seminested-PCR strategy was used. Each assay required two reactions containing primers R1, R3, P5, and P6 and primers R2, R4, P5, and P6, respectively. Amplification was carried out with a Hybaid Omnigene Thermal Cycler (Middlesex, United Kingdom). The 50-μl reaction mixture contained 0.5 μg of chromosomal DNA or 5 μl of boiled cells, 2 mM MgCl2, 200 μM deoxynucleoside triphosphates (Boehringer Mannheim, Mannheim, Germany), 50 mM KCl, 10 mM Tris-HCl (pH 8), 1.0 μM (each) primer, and 2.5 U of Taq DNA polymerase (Promega Corp., Madison, Wis.). The PCR process included an initial 3 min of incubation at 93°C to denature the target DNA. This was followed by 30 cycles of 93°C for 1 min, 55°C for 1 min and 30 s, and 72°C for 1 min and 30 s. A 5-min extension at 72°C was included at the end of the final cycle. The PCR products were analyzed by electrophoresis through 2% agarose gels containing ethidium bromide and were visualized with a UV transilluminator.
Specificity of the assay.
Representative strains of the currently recognized species of streptococci and other pathogens colonizing humans were studied. We ensured that all bacterial species commonly known to cause meningitis were included.
Sensitivity of the assay. (i) Bacterial detection.
An isolate of S. pneumoniae was grown overnight on Columbia agar supplemented with 5% horse blood (SAIMR). A single colony was picked off and inoculated into 50 μl of culture-negative CSF. A series of 10-fold dilutions was prepared from this bacterial suspension by using culture-negative CSF as the diluent. For each dilution, 25 μl was plated onto Columbia blood agar plates and the plates were incubated overnight at 37°C in 5% CO2. The samples were also boiled for 10 min and analyzed by PCR for 30 and 40 cycles by the protocol mentioned above.
(ii) Chromosomal DNA detection.
A total of 0.25 μg of S. pneumoniae chromosomal DNA per μl was diluted 10-fold, and 1 μl of each dilution was used per PCR.
CSF samples.
A total of 285 CSF samples from patients suspected of having meningitis were analyzed. In most cases the specimens had been briefly centrifuged and the supernatant was used for the assay. Previous experiments (data not shown) showed that the PCR was able to detect the presence of the pneumococcus in both the supernatants and the deposit fractions of the CSF specimens. For each specimen, 15 μl was aliquoted and boiled for 10 min and 5 μl was used per PCR. The number of cycles was extended to 40 to increase the sensitivity of the assay. The study was conducted in a blinded fashion such that the culture results were not known prior to the PCR assay.
RESULTS
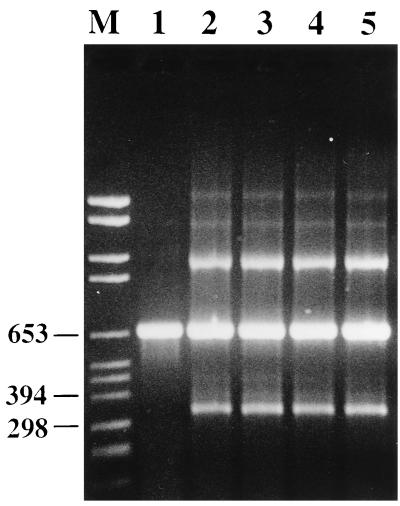
The PCR assay detected S. pneumoniae DNA from all 35 pneumococcal isolates tested. The properties of these isolates are listed in Table 1. The resistance primers R1 to R4 amplified the DNAs from isolates for which the penicillin MICs were 0.125 μg/ml and higher (Fig. 1). All of the nonpneumococcal organisms tested except S. sanguis were PCR negative.
FIG. 1.
Agarose gel electrophoresis of PCR-amplified DNA fragments of the pbp2B gene from S. pneumoniae. Lane M, molecular size markers (in base pairs). The penicillin MICs for the isolates are as follows: 0.03 μg/ml (lane 1), 0.125 μg/ml (lane 2), 0.5 μg/ml (lane 3), 1 μg/ml (lane 4), and 2 μg/ml (lane 5).
Increasing the number of cycles in the PCR from 30 to 40 resulted in a 10-fold increase in the sensitivity of the assay. The sensitivity limit of the assay with serial dilutions of chromosomal DNA from a pneumococcal isolate was approximately 2.5 pg of DNA, while the lowest number of bacterial cells that gave a positive result was 18 CFU.
Analysis of 285 CSF specimens showed that the PCR was able to detect the pneumococcus in all samples positive by culture (18 of 285), including the identification of four penicillin-resistant isolates. For these four resistant isolates penicillin MICs were 0.125 μg/ml. No false-positive results were found among the culture-negative CSF specimens. One specimen was culture negative but latex agglutination positive for S. pneumoniae, and this specimen gave a negative PCR result.
DISCUSSION
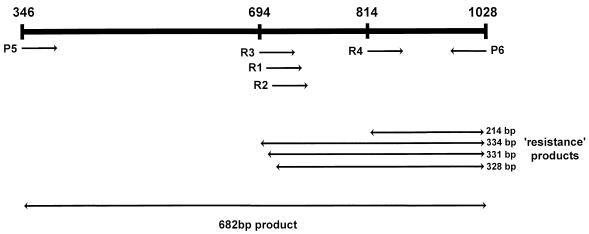
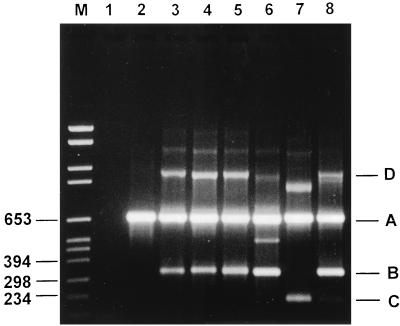
The PCR assay was able to detect S. pneumoniae DNA from all 35 pneumococcal isolates tested. The resistance primers R1 to R4 amplified the DNAs from isolates for which penicillin MICs were 0.125 μg/ml and higher (Fig. 1), which is the usual definition of clinically relevant penicillin resistance. All primers used in this study were designed from the S. pneumoniae pbp2B gene, which encodes a PBP binding protein that is unique to the pneumococcus (5, 19). The positions of primer binding to the pbp2B gene are indicated in Fig. 2. Alterations in the structural gene of PBP 2B, together with alterations in other high-molecular-weight PBP genes, lead to a remodeling of the active sites of these enzymes in such a way that their reactivities with β-lactam antibiotics are greatly decreased (10, 12). Two species-specific primers (primers P5 and P6) homologous to conserved areas of the pbp2B gene were designed and were used for the diagnosis of pneumococcal infection. Nucleotide sequence analysis of the pbp2B gene from penicillin-resistant strains has shown extensive alterations in the gene compared with the sequence of the pbp2B gene from susceptible strains (8). Smith and Klugman (31) showed that all penicillin-resistant pneumococci tested in their study had nucleotide sequence divergence within a ±300-bp area at the center of the pbp2B transpeptidase-encoding region. They also revealed that the amino acid substitutions occurring within this area could be grouped into five different profiles. For the PCR diagnosis of penicillin resistance, we therefore designed four resistance primers which encompassed the multiple mutational pathways which seem to exist for PBP 2B to remodel itself in order to inhibit the binding of penicillin. The resistance primers R1, R2, R3, and R4 identify resistance profiles 1 + 2, 3, 4, and 5, respectively. Therefore, in the PCRs we decided to combine primers R1 and R3 and primers R2 and R4. Together with the downstream primer P6, primers R1 and R3 amplify DNA fragments of 331 and 334 bp, respectively, while primers R2 and R4 amplify fragments of 328 and 214 bp, respectively (Fig. 3). We found that by combining the species-specific primers (primers P5 and P6) and two of the resistance primers in a PCR, we were able to both detect the presence of the organism and determine whether it was penicillin resistant or susceptible. Therefore, two PCRs were set up for each specimen. The first PCR contained primers R1, R3, P5, and P6, and the second PCR contained primers R2, R4, P5, and P6.
FIG. 2.
Primer binding sites in the S. pneumoniae pbp2B gene. P5 and P6 represent species-specific primers. R1 to R4 represent the four resistance primers.
FIG. 3.
Agarose gel electrophoresis of PCR-amplified DNA fragments of the pbp2B gene from S. pneumoniae. Lane M, molecular size marker (in base pairs). Lane 1, negative control; lane 2, penicillin-susceptible S. pneumoniae. Primer combinations are as follows: R1 + P5 + P6 (lane 3), R3 + P5 + P6 (lane 4), R1 + R3 + P5 + P6 (lane 5), R2 + P5 + P6 (lane 6), R4 + P5 + P6 (lane 7), and R2 + R4 + P5 + P6 (band C is poorly visible) (lane 8). (A) A 682-bp species-specific product arising from amplification with primers P5 and P6. (B) A 328- to 334-bp products arising from amplification with primers R1 to R3 and P6. (C) A 214-bp product arising from amplification with primers R4 and P6. (D) Amplification products produced as a result of annealing between a resistance product(s) and the 682-bp product and which are subsequently extended by Taq DNA polymerase to produce a larger product (±900 to 1,000 bp).
Ubukata and coworkers (32) have designed a similar system whereby they detected penicillin resistance with DNA extracted from clinical isolates of S. pneumoniae. Their system is based on three sets of primers designed for amplification of the pbp2B gene from penicillin-susceptible S. pneumoniae, as well as two classes of mutations of the pbp2B gene which are present in penicillin-resistant pneumococci in Japan. The primer used to detect penicillin-susceptible strains in their study most likely also amplifies DNA from resistant strains (on the basis of pbp2B sequence data of Smith and Klugman [31]), since the primer sequence covers an area of the pbp2B gene which is not unique only to penicillin-susceptible isolates. In fact, it is identical to sequences which are also found in penicillin-resistant pneumococcal isolates. In our study we describe four resistance primers which expand the genetic variabilities of resistance detected in the pbp2B gene.
The specificity of the assay was demonstrated by the inability of the PCR to amplify DNAs from 19 nonpneumococcal organisms. Three S. sanguis isolates were tested, and amplification products identical to the 682-bp species-specific fragment were detected. The closest relatives of pneumococci are the viridans group streptococci which, along with pneumococci, coinhabit the human oropharynx. Studies have demonstrated that viridans group streptococci have the potential to transfer resistance genes to pneumococci. In particular, it has been demonstrated that the transfer of penicillin resistance between S. pneumoniae and S. sanguis or S. mitis via transformation occurs at high frequencies (5, 26). However, it has also been shown that the movement of DNA can occur from pneumococci into the viridans group streptococci (23). None of the bacterial species which commonly cause meningitis gave amplification products which interfered with the interpretation of our results. The reactivity of S. sanguis in this assay, it should be noted, is unlikely to cause significant problems in the diagnosis of the etiology of meningitis (since it is unlikely to be present in a CSF specimen), and we found no false-positive results in our study of 285 CSF specimens.
Analysis of 285 CSF specimens again demonstrated the high degrees of specificity and sensitivity of the assay by detecting the pneumococcus in all samples positive by culture (18 of 285), including the identification of four penicillin-resistant isolates. No false-positive results were recorded among the culture-negative CSF specimens. Only one specimen was culture negative and latex agglutination positive for S. pneumoniae, and this specimen gave a negative PCR result. Previous work has demonstrated the limited specificity and sensitivity of antigen testing (1, 9, 25). Evaluations of bacterial latex agglutination kits have found them to range from having high degrees of specificity and sensitivity to having extremely low degrees of specificity and sensitivity, depending on the kit used (4, 15, 30). Perkins and coworkers (25) have reported a high incidence of false-positive results (54%) by latex agglutination testing. It has been suggested that latex agglutination only be used in cases in which the Gram staining result is negative or when the patients have received a lumbar puncture late in their therapy because of the severity of their illness (21). In addition, antigen testing gives no indication of the antibiotic susceptibility pattern of the organism.
The objective of this study was to develop a seminested-PCR assay which could potentially be used for the simultaneous diagnosis of pneumococcal meningitis and identification of penicillin-resistant isolates of S. pneumoniae in CSF specimens. The specificity and sensitivity for CSF specimens were each 100%, and the predictive values of both a positive and a negative result were each 100%; therefore, these values make the technique attractive as a diagnostic method. Currently, at least 48 h is required in order to culture the causative organism and carry out the susceptibility testing. By this PCR assay, a result can be achieved within a few hours. Unlike other methods, CSF only has to be boiled, and therefore laborious DNA extraction methods are eliminated. Since there is often insufficient specimen available for numerous laboratory tests, this method is convenient in that it requires only 15 μl of CSF. The results presented here are of sufficient value to merit further clinical development and potentially extend the spectrum of the assay for the detection of resistance to other β-lactam antibiotics such as broad-spectrum cephalosporins. This system could also be developed to include other meningitis-causing pathogens.
ACKNOWLEDGMENTS
We thank Mani Khoosal and Shabir Madhi, Baragwanath Hospital; Debbie Lethman, New Johannesburg Hospital; and Jay Patel and Susana Gomes, SAIMR Central, for supplying CSF specimens.
REFERENCES
- 1.Ajello G W, Bolan G A, Hayes P S, Lehman D, Mongomery J, Feeley J C, Perlino C A, Broome C V. Commercial latex agglutination tests for detection of Haemophilus influenzae type B and Streptococcus pneumoniae antigens in patients with bacteremic pneumonia. J Clin Microbiol. 1987;25:1388–1391. doi: 10.1128/jcm.25.8.1388-1391.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bacquero F. Pneumococcal resistance to β-lactam antibiotics: a global geographic overview. Microb Drug Resist. 1995;1:115–120. doi: 10.1089/mdr.1995.1.115. [DOI] [PubMed] [Google Scholar]
- 3.Bingen E, Lambert-Zechovsky N, Mariani-Kurkdjian P, Doit C, Aujard Y, Fournerie F, Mathieu H. Bacterial counts in cerebrospinal fluid of children with meningitis. Eur J Clin Microbiol Infect Dis. 1990;9:278–281. doi: 10.1007/BF01968060. [DOI] [PubMed] [Google Scholar]
- 4.Cerosaletti K M, Roghman M C, Bentley D W. Comparison of latex agglutinatination and counterimmunoelectrophoresis for the detection of pneumococcal antigen in elderly pneumonia patients. J Clin Microbiol. 1985;22:553–557. doi: 10.1128/jcm.22.4.553-557.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chalkley L J, Koornhof H J. Intra- and interspecific transformation of S. pneumoniae to penicillin resistance. J Antimicrob Chemother. 1990;26:21–28. doi: 10.1093/jac/26.1.21. [DOI] [PubMed] [Google Scholar]
- 6.Coffey T J, Dowson C G, Daniels M, Spratt B G. Genetics and molecular biology of β-lactam-resistant pneumococci. Microb Drug Resist. 1995;1:29–34. doi: 10.1089/mdr.1995.1.29. [DOI] [PubMed] [Google Scholar]
- 7.Davis T E, Fuller D A, Aeschleman E C. Rapid, direct identification of Staphylococcus aureus and Streptococcus pneumoniae from blood cultures using commercial immunologic kits and modified conventional tests. Diag Microbiol Infect Dis. 1992;15:295–300. doi: 10.1016/0732-8893(92)90014-k. [DOI] [PubMed] [Google Scholar]
- 8.Dowson C G, Hutchinson A, Brannigan J A, George R C, Hansman D, Linares J, Tomasz A, Maynard Smith J, Spratt B G. Horizontal transfer of penicillin-binding protein genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1989;86:8842–8846. doi: 10.1073/pnas.86.22.8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forward K R. Prospective evaluation of bacterial antigen detection in cerebrospinal fluid in the diagnosis of bacterial meningitis in a predominantly adult hospital. Diagn Microbiol Infect Dis. 1988;11:61–63. doi: 10.1016/0732-8893(88)90074-0. [DOI] [PubMed] [Google Scholar]
- 10.Frere J M, Joris B. Penicillin sensitive enzymes in peptidoglycan synthesis. Crit Rev Microbiol. 1985;11:299–396. doi: 10.3109/10408418409105906. [DOI] [PubMed] [Google Scholar]
- 11.Gillespie S H, Ullman C, Smith M D, Emery V. Detection of Streptococcus pneumoniae in sputum samples by PCR. J Clin Microbiol. 1994;32:1308–1311. doi: 10.1128/jcm.32.5.1308-1311.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hakenbeck R, Tarpay M, Tomasz A. Multiple changes in penicillin-binding proteins in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1980;17:364–371. doi: 10.1128/aac.17.3.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hassan-King M, Baldeh I, Secka O, Falade A, Greenwood B. Detection of Streptococcus pneumoniae DNA in blood cultures by PCR. J Clin Microbiol. 1994;32:1721–1724. doi: 10.1128/jcm.32.7.1721-1724.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heitzman-Rowley A, Wald E R. The incubation period necessary for detection of bacteremia in immunocompetent children with fever. Clin Pediatr. 1986;25:485–489. doi: 10.1177/000992288602501001. [DOI] [PubMed] [Google Scholar]
- 15.Hoban D J, Witwicki E, Hammond G W. Bacterial antigen detection in cerebrospinal fluid of patients with meningitis. Diagn Microbiol Infect Dis. 1985;3:373–379. doi: 10.1016/0732-8893(85)90075-6. [DOI] [PubMed] [Google Scholar]
- 16.Klugman K P, Koornhof H J, Wasas A, Storey K, Gilbertson I. Carriage of penicillin-resistant pneumococci. Arch Dis Child. 1986;61:377–381. doi: 10.1136/adc.61.4.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koornhof H J, Wasas A, Klugman K P. Antimicrobial resistance in Streptococcus pneumoniae: a South African perspective. Clin Infect Dis. 1992;15:84–94. doi: 10.1093/clinids/15.1.84. [DOI] [PubMed] [Google Scholar]
- 18.La Scolea L, Jr, Dryja D. Quantitation of bacteria in cerebrospinal fluid and blood of children with meningitis and its diagnostic significance. J Clin Microbiol. 1984;19:187–190. doi: 10.1128/jcm.19.2.187-190.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Markowicz Z, Tomasz A. Variation in penicillin-binding protein patterns of penicillin-resistant clinical isolates of pneumococci. J Clin Microbiol. 1989;27:405–410. doi: 10.1128/jcm.27.3.405-410.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marton A, Gulyas M, Munoz R, Tomasz A. Extremely high incidence of antibiotic resistance in clinical isolates of Streptococcus pneumoniae in Hungary. J Infect Dis. 1991;163:542–548. doi: 10.1093/infdis/163.3.542. [DOI] [PubMed] [Google Scholar]
- 21.Maxson S, Lewno M J, Schultze G E. Clinical usefulness of cerebrospinal bacterial antigen studies. J Pediatr. 1994;125:235–238. doi: 10.1016/s0022-3476(94)70201-2. [DOI] [PubMed] [Google Scholar]
- 22.Mufson M A. Streptococcus pneumoniae. In: Mandell G L, Douglas R G Jr, Bennett J E, editors. Principles and practice of infectious diseases. New York, N.Y: Churchill Livingstone; 1990. pp. 1539–1580. [Google Scholar]
- 23.Munoz R, Musser J M, Crain M, Briles D E, Marton A, Parkinson A J, Sorenson U, Tomasz A. Geographic distribution of penicillin-resistant clones of Streptococcus pneumoniae: characterization by penicillin-binding protein profile, surface protein A typing and multilocus enzyme electrophoresis. Clin Infect Dis. 1992;15:112–118. doi: 10.1093/clinids/15.1.112. [DOI] [PubMed] [Google Scholar]
- 24.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A3. 3rd ed. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1993. [Google Scholar]
- 25.Perkins M D, Mirrett S, Barth Reller L. Rapid bacterial antigen detection is not clinically useful. J Clin Microbiol. 1995;33:1486–1491. doi: 10.1128/jcm.33.6.1486-1491.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Potgieter E, Chalkley L J. Reciprocal transfer of penicillin resistance genes between Streptococcus pneumoniae, Streptococcus mitior and Streptococcus sanguis. J Antimicrob Chemother. 1991;28:463–465. doi: 10.1093/jac/28.3.463. [DOI] [PubMed] [Google Scholar]
- 27.Rådström P, Backman A, Qian N, Kragsbjerg P, Pahlson C, Olcen P. Detection of bacterial DNA in cerebrospinal fluid by an assay for simultaneous detection of Neisseria meningitidis, Haemophilus influenzae and streptococci using a seminested PCR strategy. J Clin Microbiol. 1994;32:2738–2744. doi: 10.1128/jcm.32.11.2738-2744.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rudolph K M, Parkinson A J, Black C M, Mayer L W. Evaluation of polymerase chain reaction for diagnosis of pneumococcal pneumonia. J Clin Microbiol. 1993;31:2661–2666. doi: 10.1128/jcm.31.10.2661-2666.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Singhal A, Lalitha M K, Jacob John T, Thomas K, Raghupathy P, Jacob S, Steinhoff M C. Modified latex agglutination test for rapid detection of Streptococcus pneumoniae and Haemophilus influenzae in cerebrospinal fluid and direct serotyping of Streptococcus pneumoniae. Eur J Clin Microbiol Infect Dis. 1996;15:472–477. doi: 10.1007/BF01691314. [DOI] [PubMed] [Google Scholar]
- 31.Smith A M, Klugman K P. Alterations in penicillin-binding protein 2B from penicillin-resistant wild-type strains of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1995;39:859–867. doi: 10.1128/aac.39.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ubukata K, Asahi Y, Yamane A, Konno M. Combinational detection of autolysin and penicillin-binding protein 2B genes of Streptococcus pneumoniae by PCR. J Clin Microbiol. 1996;34:592–596. doi: 10.1128/jcm.34.3.592-596.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Virolainen A, Salo P, Jero J, Karma P, Eskola J, Leinonen M. Comparison of PCR assay with bacterial culture for detecting Streptococcus pneumoniae in middle ear fluid of children with acute otitis media. J Clin Microbiol. 1994;32:2667–2670. doi: 10.1128/jcm.32.11.2667-2670.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson M L, Weinstein M P, Reimer L G, Mirret S, Barth Reller L. Controlled comparison of the BacT/Alert and BACTEC 660/730 nonradiometric blood culture systems. J Clin Microbiol. 1992;30:323–329. doi: 10.1128/jcm.30.2.323-329.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woodhead, M. 1994. Pneumonia in the elderly. J. Antimicrob. Chemother. 34(Suppl. A):85–92. [DOI] [PubMed]
- 36.Zhang Y, Isaacman D J, Wadowsky R M, Rydquist-White J, Post J C, Ehrlich G D. Detection of Streptococcus pneumoniae in whole blood by PCR. J Clin Microbiol. 1995;33:596–601. doi: 10.1128/jcm.33.3.596-601.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]