Abstract
Background
Kaposi sarcoma, caused by the pathogen Kaposi sarcoma–associated herpesvirus (KSHV), is the most common neoplasm for patients with AIDS. Susceptibility to KSHV has been associated with several different genetic risk variants. The purpose of this study was to test whether variants of killer cell immunoglobulin-like receptors (KIRs) and their human leukocyte antigen (HLA-I) ligands influence the risk of KSHV infection.
Methods
A case-control study was performed in Xinjiang, a KSHV-endemic region of China. We recruited 299 individuals with HIV, including 123 KSHV-seropositive persons and 176 KSHV-seronegative controls. We used logistic regression and the MiDAS package to evaluate the association between KIR/HLA-I polymorphisms and KSHV infection.
Results
HLA-A*31:01, HLA-C*03:04, and HLA-C*12:03 were found to be associated with KSHV infection, with A*31:01 showing a protective effect under 3 different models (dominant: 0.30 [95% confidence interval {CI}, .08–.82], P = .031; additive: 0.30 [95% CI, .09–.80], P = .030; overdominant: 0.31 [95% CI, .09–.88], P = .042). The effect of A*31:01 might cause the variants of amino acid at HLA-A position 56, with individuals carrying an arginine having a lower KSHV infection risk. The increased homozygous KIR2DL3 was associated with a relatively high KSHV viral load (16.30% vs 41.94%, P = .010).
Conclusions
This study provides further insight into the link between HLA-I alleles and KIR genes and KSHV infection, highlighting KSHV-susceptible variants of HLA-I and KSHV replication caused by specific KIR genotype, and revealing a potential role of KIR-mediated natural killer cell activation in anti-KSHV infection.
Keywords: human leukocyte antigen, infection, Kaposi sarcoma–associated herpesvirus, killer cell immunoglobulin-like receptor, polymorphism
Kaposi sarcoma–associated herpesvirus (KSHV; also known as human herpesvirus 8) is the causative agent of Kaposi sarcoma (KS), the most common neoplasm and the major cause of death for patients with AIDS [1, 2]. Regardless of the wide application of antiretroviral therapy (ART), KS continues to affect people with human immunodeficiency virus (PWH), posing a great challenge to the 2022–2030 global health sector strategy of the World Health Organization (WHO) on human immunodeficiency virus (HIV) [3–5]. As reported in a newly published study [6], the overall infection rate of KSHV in Xinjiang was 25.60%, with a higher infection rate of 29.79% in the Uyghur population. Liu et al have reported that KSHV seroprevalence among PWH was 48.9% [7]. The disproportionately higher prevalence of KSHV infection and KS incidence in PWH compared to immunocompetent individuals indicates the importance of host immunity in KSHV infection and restricting viral pathogenesis [8, 9].
Human natural killer (NK) cells, an integral part of innate immunity, can rapidly provide defenses against herpesvirus infection [9]. Killer cell immunoglobulin-like receptors (KIRs) are a polymorphic family of receptors on the surface of NK cells that regulate NK-mediated cytotoxicity when ligated to requisite human leukocyte antigen (HLA) class I molecules [10, 11]. Both KIR and the specific HLA ligands are indispensable to regulating NK cell activity, such that one without the other is functionally inert [12]. When KSHV invades, the K3 and K5 proteins encoded by KSHV can downregulate HLA-I molecules, thereby disrupting the balance of KIRs and activating NK cells to recognize and clear infected cells [13].
Previous studies have shown that KSHV susceptibility is caused by virus-host-environment interactions [14]. The characteristics of KSHV infection in China are quite distinct from those in the Mediterranean region, suggesting that genetic background plays an important role [2, 7]. Thus, the genes encoding KIR and HLA-I are centrally involved in the immunological response to KSHV infection, and the KIR/HLA-I polymorphisms would be expected to affect the host susceptibility to KSHV infection. However, only tenuous support for this hypothesis has been reported [15–20], and most focused on Mediterranean populations and classic KS. For example, KIR2DS1 binding to HLA-C2 was prevalent in Italian patients with classical KS [16], whereas KIR3DS1-specific binding to HLA-B Bw4-80I and homozygous HLA-C1 might prevent KSHV infection [17, 18].
Considering that the extensive polymorphisms of KIR/HLA-I are closely related to the immune function against viral infections and the risk genes of KSHV infection vary substantially across different ethnic groups and individuals, the present study focused on AIDS-related KSHV infection among participants with HIV in Xinjiang, an endemic area for KSHV/KS in China [7], to examine the possible association between KIR/HLA-I polymorphisms and KSHV infection. Findings would have important implications for the development of targeted prevention and control measures in areas with a high prevalence of KSHV and the precision protection of KSHV-susceptible populations.
MATERIALS AND METHODS
Study Design and Sample Collection
This study was conducted in the Kazakh Autonomous Prefecture of Ili, Xinjiang, China. From May to December 2019, individuals from the Center for Disease Control and Prevention (CDC) in Ili were consecutively recruited. Participants were all aged 18 years or older and receiving regular ART for HIV infection. The HIV-positive status of all participants was confirmed by the CDC. Participants were interviewed in person by trained staff from local health facilities. A standard questionnaire was used to collect information on demographic characteristics and information related to HIV infection. Then, participants were stratified based on their ethnicity, sex, age, and KSHV infection status. We further performed random sampling from different subgroups to select individuals for this study to ensure the representativeness of the selected participants. All recruited individuals agreed to participation in the study with written informed consent and provided blood samples. This study was approved by the research ethics review committee of Fudan University, Shanghai, China (approval number IRB#03-0506).
Laboratory Testing
KSHV Serology
Antibodies against KSHV were detected using an enzyme-linked immunosorbent assay (ELISA) that employs the most immunogenic lytic antigens ORF65 and K8.1 and latent antigen ORF73. When used together, serological assays against KSHV lytic and latent antigens show the best combination of sensitivity (89.1%) and specificity (94.9%) [21–23]. The ORF65, K8.1, and ORF73 coding sequences were recombined into the pQE-80L vector to express the respective proteins in competent Escherichia coli cells, followed by the purification of the 3 proteins using a nickel column, as antigens. Three target proteins were used as antigens to coat the three 96-well plates, and alkaline phosphatase–labeled sheep antihuman immunoglobulin G was used as the secondary antibody. Detailed information has been previously described [6, 24]. Each serum to be tested was subjected to an ELISA for each of the 3 target proteins, with detection based on positive wells, negative wells, and blank wells. The absorbance value of each plasma sample was measured at 405 nm using a microplate reader, and the average optical density value of the negative control sample plus 2 times the standard deviation was used as the cutoff value. Participants who were positive for any of the 3 KSHV antibodies were considered KSHV seropositive.
HIV and KSHV Viral Load
The HIV viral load was quantified by Cobas AmpliPrep Cobas TaqMan HIV-1 Test, version 2.0 (Roche). HIV type 1 (HIV-1) RNA was extracted from plasma and reverse transcribed into complementary DNA for detection. Using real-time polymerase chain reaction (PCR) to amplify the gag region of HIV-1, samples were quantified using the internal standard method, with a linear range of 20–107 copies/mL.
KSHV viral loads were detected using real-time fluorescence quantitative PCR (qPCR) among KSHV-seropositive individuals. Primer pairs used were KS-1 and KS-2 (forward, 5′-AGCCGAAAGGATTCCACCAT-3′; reverse, 5′-TCCGTGTTGTCTACGTCCAG-3′), amplifying the KS330Bam233 region of ORF26 (a viral capsid protein). Each 20-μL PCR reaction contained 10 μL SYBR 2× mix (NO ROX), 2 μL DNA template, 7 μL double-distilled water, and 0.5 μL of each primer. The PCR conditions were 95°C for 3 minutes, followed by 40 cycles at 95°C for 5 seconds, 55°C for 30 seconds, 72°C for 30 seconds, and a final step at 72°C for 5 minutes. A cutoff value of ≥1000 copies/μL was used to identify participants positive for KSHV DNA, which also indicated a high KSHV viral load.
KIR-HLA Genotyping
DNA was extracted using the TIANGEN blood genomic DNA extraction kit DP318-03 (centrifugal column type). DNA samples were stored at −80°C before genotyping.
KIR Typing
Genotyping for the 11 KIR genes (excluding the framework genes KIR3DL3, 2DL4, and 3DL2 and pseudogenes 2DP1 and 3DP1), including 6 activating loci (KIR2DS2, 3DS1, 2DS3, 2DS5, 2DS1, and 2DS4) and 5 inhibitory loci (KIR2DL2, 2DL3, 2DL1, 3DL1, and 2DL5), was conducted by PCR sequence-specific primer [25]. The primer sequences of these 11 KIR genes are shown in Supplementary Table 1. PCR conditions were 95°C for 2 minutes, 30 cycles at 94°C for 20 seconds, 63°C for 30 seconds, 68°C (2DS3-2, 2DS5, 3DS1, 3DL1, and 2DL3 for 1.5 minutes; 2DS2-1, 2DL1-1, 2DL2-1, 2DS1, 2DS4, and 2DL5 for 2.5 minutes; 2DS2-2, 2DL2-2, 2DS3-1, and 2DL1-2 for 10 minutes), and 72°C for 10 minutes. The amplification products were analyzed by stained agarose gel electrophoresis. The genotypes were classified based on the criteria adopted by Middleton (Allele Frequency Net Database: http://www.allelefrequencies.net/kir6001a.asp) [26], and 2 broad haplotypes, termed A and B, were defined.
HLA-I Typing
HLA-I alleles were detected via multiplex PCR amplification using the FastTarget system. Sequencing libraries were generated using the FastTarget Custom Panel following the manufacturer's recommendations, with index codes being added to each sample. The fragment length distribution of the library was verified using Agilent 2100 Bioanalyzer. Following the accurate quantification of the molar concentration of the library, FastQ data were obtained using the Illumina HiSeq platform for high-throughput sequencing using the 2 × 250 bp double-terminal sequencing mode. Consequently, 4-digit HLA-A/B/C allele typing results were acquired.
KIR/HLA-I Complex Genotypes
KIR2DL1 and 2DS1 bind the HLA-C2 epitope (asparagine at position 77, lysine at position 80), and KIR2DL2, 2DL3, and 2DS2 bind the C1 epitope (serine at position 77, asparagine at position 80) [27]. HLA-B*46 allotype also was a ligand of 2DL2 and 2DL3 [28]. HLA-Bw4*80I (isoleucine at position 80) subset was considered the ligand for KIR3DL1 and 3DS1 [29, 30]. In addition, HLA-A*23, -*24, and -*32 molecules (all carrying Bw4 epitope) were included among the ligands of 3DL1 [31, 32].
Statistical Analysis
All data were double-entered using EpiData version 3.1 and subsequently transferred to R version 3.5.2 for further statistical analysis. Comparisons between different groups was performed by the Mann–Whitney U test for continuous variables and χ2 analysis or Fisher exact test (each when adequate) for categorical variables. The detection rate (F) of each KIR gene was determined as F = number of gene detections/number of individuals; and the gene frequency (GF) of each KIR gene was determined as GF = 1–. Logistic regression was used to calculate odds ratios (ORs) and 95% confidence intervals (CIs) under different genetic patterns (dominant, additive, and overdominant models). The association analysis of KIR/HLA-I polymorphisms with KSHV infection was performed using the MiDAS package [33], in which Hardy-Weinberg equilibrium (HWE) testing for HLA-I alleles can be performed. MiDAS is based on the Immuno Polymorphism Database-ImMunoGeneTics (IPD-IMGT)/HLA database (http://www.allelefrequencies.net/hla.asp) to perform HLA sequence alignment and can obtain the variable amino acid residues of individuals from HLA allele data and then determine the variable amino acid residues associated with KSHV infection in the HLA protein by the likelihood ratio test. ORs and their 95% CIs were calculated for all of the coefficients of regression. Differences were considered statistically significant at P < .05, and P values were calculated for Bonferroni method corrected for multiple testing.
RESULTS
A total of 299 PWH were finally recruited into the present study, including 123 KSHV-seropositive persons and 176 KSHV-seronegative controls. There were 162 (54.2%) males and 201 (67.2%) Uyghurs, with a mean age of 44.35 years (range, 19–89 years). Sex, age, and ethnic distribution did not differ between the infected and uninfected groups. We found that compared with KSHV-seronegative participants, individuals infected with KSHV had significantly lower CD4+ T-cell counts (593.28 ± 303.55 cells/μL vs 505.54 ± 234.13 cells/μL, P = .006). However, no significant difference between the 2 groups was detected in terms of HIV stage, HIV viral load, or duration of HIV infection (Supplementary Table 2). Moreover, among the KSHV-seropositive individuals, there were 92 persons with KSHV viral load <1000 copies/μL and 31 persons with high KSHV viral load (≥1000 copies/μL).
HLA Alleles and KIR Genes With KSHV Seroprevalence
HLA alleles and KIR genes were successfully determined in all 299 participants. We compared each HLA-A, HLA-B, and HLA-C allele and KIR gene in different KSHV infection groups to explore their possible associations with KSHV infection.
HLA-I alleles with frequencies >2.5% were screened, and alleles strongly deviating from the HWE equilibrium were filtered out (removing alleles with PHWE <3.5 × 10−4). We obtained 40 alleles, including 13 HLA-A, 13 HLA-B, and 14 HLA-C alleles. Since the inheritance pattern of HLA alleles was not determined, each allele was discussed separately under common genetic models (recessive model was not applicable here). The association between HLA-A/-B/-C and KSHV infection was analyzed under dominant, additive, and overdominant models. Three HLA-I alleles (HLA-A*31:01, HLA-C*03:04, and HLA-C*12:03) were nominally associated with KSHV infection (Table 1). Among them, HLA-A*31:01 showed a protective effect against KSHV infection under all 3 models (dominant: OR, 0.30 [95% CI, .08–.82], P = .031; additive: OR, 0.30 [95% CI, .09–.80], P = .030; overdominant: OR, 0.31 [95% CI, .09–.88], P = .042), and HLA-C*03:04 showed protection against KSHV infection only in the additive model (OR, 0.45 [95% CI, .20–.95], P = .047). In contrast, individuals carrying HLA-C*12:03 were more prevalent in the KSHV-positive group, suggesting that it might be a risk allele for KSHV infection under the dominant and overdominant models (ORs, 2.50 [95% CI, 1.18–5.49], P = .019 and 3.03 [95% CI, 1.38–7.03], P = .007, respectively). However, none of these 3 alleles was statistically different after Bonferroni correction (P = 0.05/40/3 = 4.2 × 10−4).
Table 1.
Associations Between HLA-A, HLA-B, and HLA-C Alleles and Kaposi Sarcoma–Associated Herpesvirus Infection Under the Dominant, Additive, and Overdominant Models
| Allele | Dominant Modela | Additive Modelb | Overdominant Modelc | |||
|---|---|---|---|---|---|---|
| OR (95% CI) | P Value | OR (95% CI) | P Value | OR (95% CI) | P Value | |
| A*01:01 | 1.05 (.52–2.08) | .897 | 1.12 (.57–2.15) | .746 | 0.97 (.47–1.95) | .937 |
| A*02:01 | 0.91 (.51–1.61) | .750 | 1.00 (.61–1.63) | .995 | 0.81 (.44–1.48) | .499 |
| A*02:06 | 1.23 (.52–2.85) | .626 | 1.32 (.59–2.94) | .487 | 1.11 (.46–2.61) | .812 |
| A*02:07 | 0.95 (.43–2.03) | .894 | 0.90 (.42–1.86) | .779 | 1.01 (.45–2.19) | .978 |
| A*03:01 | 1.51 (.77–2.97) | .223 | 1.51 (.77–2.97) | .223 | 1.51 (.77–2.97) | .223 |
| A*11:01 | 1.35 (.77–2.35) | .294 | 1.18 (.72–1.95) | .504 | 1.50 (.85–2.67) | .164 |
| A*24:02 | 0.92 (.54–1.56) | .771 | 1.03 (.65–1.62) | .886 | 0.80 (.46–1.39) | .438 |
| A*26:01 | 0.79 (.32–1.82) | .586 | 0.76 (.32–1.66) | .499 | 0.85 (.35–1.97) | .706 |
| A*30:01 | 0.81 (.36–1.75) | .600 | 0.83 (.40–1.62) | .591 | 0.83 (.35–1.85) | .650 |
| A*31:01 | 0.30 (.08–.82) | .031 | 0.30 (.09–.80) | .030 | 0.31 (.09–.88) | .042 |
| A*32:01 | 0.85 (.28–2.36) | .761 | 0.85 (.28–2.36) | .761 | 0.85 (.28–2.36) | .761 |
| A*33:03 | 1.63 (.67–4.04) | .281 | 1.50 (.68–3.39) | .310 | 1.64 (.64–4.26) | .297 |
| A*68:01 | 1.29 (.47–3.47) | .610 | 1.41 (.56–3.56) | .459 | 1.12 (.39–3.09) | .827 |
| B*07:02 | 0.95 (.36–2.37) | .915 | 0.95 (.36–2.37) | .915 | 0.95 (.36–2.37) | .915 |
| B*13:02 | 0.79 (.40–1.52) | .487 | 0.81 (.43–1.46) | .483 | 0.80 (.40–1.57) | .524 |
| B*14:02 | 0.87 (.34–2.13) | .769 | 0.82 (.33–1.90) | .653 | 0.95 (.36–2.37) | .915 |
| B*35:01 | 0.50 (.14–1.51) | .251 | 0.50 (.14–1.51) | .251 | 0.50 (.14–1.51) | .251 |
| B*35:03 | 2.06 (.81–5.48) | .132 | 2.06 (.81–5.48) | .132 | 2.06 (.81–5.48) | .132 |
| B*40:01 | 0.61 (.24–1.41) | .265 | 0.69 (.30–1.46) | .349 | 0.56 (.21–1.35) | .219 |
| B*40:06 | 1.46 (.52–4.08) | .461 | 1.46 (.52–4.08) | .461 | 1.46 (.52–4.08) | .461 |
| B*44:02 | 1.27 (.43–3.62) | .656 | 1.40 (.53–3.73) | .490 | … | … |
| B*46:01 | 1.16 (.51–2.57) | .714 | 0.96 (.46–1.91) | .904 | 1.48 (.63–3.44) | .360 |
| B*50:01 | 0.68 (.32–1.40) | .312 | 0.67 (.32–1.33) | .268 | 0.72 (.33–1.48) | .382 |
| B*51:01 | 0.97 (.49–1.87) | .925 | 0.97 (.49–1.87) | .925 | 0.97 (.49–1.87) | .925 |
| B*52:01 | 2.14 (.80–6.04) | .135 | 2.14 (.80–6.04) | .135 | 2.14 (.80–6.04) | .135 |
| B*58:01 | 1.47 (.61–3.55) | .383 | 1.56 (.68–3.61) | .289 | 1.33 (.54–3.25) | .532 |
| C*01:02 | 1.25 (.70–2.24) | .446 | 1.25 (.70–2.24) | .446 | 1.25 (.70–2.24) | .446 |
| C*03:02 | 1.33 (.54–3.25) | .532 | 1.28 (.58–2.84) | .535 | 1.31 (.51–3.35) | .569 |
| C*03:03 | 1.11 (.46–2.61) | .812 | 1.11 (.46–2.61) | .812 | 1.11 (.46–2.61) | .812 |
| C*03:04 | 0.46 (.20–.98) | .053 | 0.45 (.20–.95) | .047 | 0.48 (.20–1.03) | .070 |
| C*04:01 | 0.95 (.50–1.75) | .858 | 0.95 (.50–1.75) | .858 | 0.95 (.50–1.75) | .858 |
| C*06:02 | 0.86 (.52–1.42) | .567 | 0.78 (.50–1.21) | .276 | 1.03 (.62–1.72) | .907 |
| C*07:01 | 0.49 (.16–1.32) | .183 | 0.49 (.16–1.32) | .183 | 0.49 (.16–1.32) | .183 |
| C*07:02 | 0.96 (.52–1.76) | .903 | 0.96 (.52–1.76) | .903 | 0.96 (.52–1.76) | .903 |
| C*08:01 | 1.79 (.75–4.39) | .189 | 1.79 (.75–4.39) | .189 | 1.79 (.75–4.39) | .189 |
| C*08:02 | 0.87 (.34–2.14) | .769 | 0.87 (.34–2.14) | .769 | 0.87 (.34–2.14) | .769 |
| C*12:02 | 2.14 (.80–6.04) | .135 | 2.14 (.80–6.04) | .135 | 2.14 (.80–6.04) | .135 |
| C*12:03 | 2.50 (1.18–5.49) | .019 | 1.94 (.98–3.99) | .062 | 3.03 (1.38–7.03) | .007 |
| C*14:02 | 1.18 (.46–2.95) | .717 | 1.30 (.54–3.06) | .552 | 1.04 (.39–2.66) | .929 |
| C*15:02 | 0.95 (.40–2.17) | .904 | 0.95 (.40–2.17) | .904 | 0.95 (.40–2.17) | .904 |
Statistically significant associations are marked in bold. P values were calculated for uncorrected.
Abbreviations: CI, confidence interval; OR, odds ratio.
Dominant model: comparing individuals who carry 1 allele against individuals who do not carry it (CA + AA vs CC).
Additive model: comparing pure individuals carrying 1 allele and pure sum individuals not carrying that allele (AA vs CC).
Overdominant model: comparing individuals carrying pure alleles and individuals carrying heterozygous alleles (CC + AA vs CA).
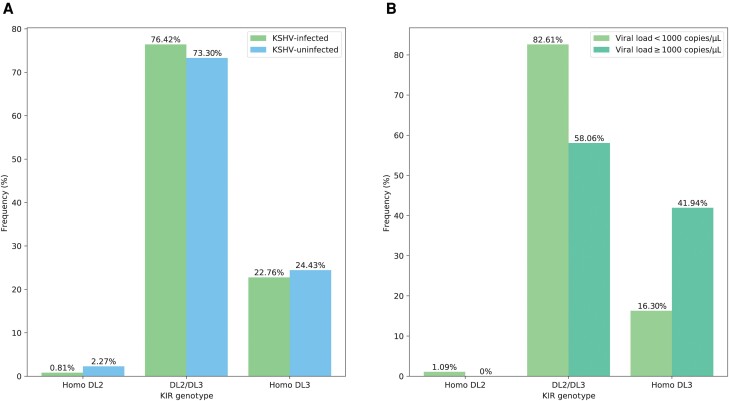
Genotyping for 11 KIR genes showed that the frequency of KIR genes was similar in the KSHV-infected group versus controls (Supplementary Table 3). Consequently, we classified the KIR genotypes and found a total of 28 genotypes, which were all the Bx (AB or BB genotypes) (Supplementary Table 4). Previous research has highlighted that KIR2DL2/DS2 (considered together because of their strong linkage disequilibrium) and KIR2DL3 genotypes are the most significant KIR variations for the risk of developing herpesvirus-associated diseases [34, 35]. In our study, we observed differences in KIR2DL2/DS2 and KIR2DL3 genotypes between the KSHV infection status groups. Specifically, there was a slight increase in KIR2DL2/2DL3 heterozygosity (76.42% vs 73.30%) in the KSHV-infected group, although this difference was not statistically significant (P = .636). Intriguingly, subdividing the KSHV-infected group accordingly to KSHV viral load, the results evidenced a significantly different distribution of the KIR2DL2/DS2 and KIR2DL3 genotypes in these 2 groups. Individuals with KSHV carrying the homozygous KIR2DL3 had a relatively high viral load (16.30% vs 41.94%, P = .010) (Figure 1).
Figure 1.
A, Frequency of KIR genotypes in different Kaposi sarcoma–associated herpesvirus (KSHV) serum infection groups. B, Frequency of KIR genotypes in different KSHV viral load groups. Abbreviations: Homo DL2, KIR2DL2/DS2 homozygosity; DL2/DL3, KIR2DL2/KIR2DL3 heterozygosity; Homo DL3, KIR2DL3 homozygosity.
KIR/HLA-I Complex With KSHV Seroprevalence
In contrast to single HLA alleles and KIR genes, the joint classification of participants according to their KIR and HLA-I ligand pairs was more beneficial to understand the pathogenesis of KIR-mediated NK cytotoxicity in KSHV infection. The analysis of the KIR/HLA-I complex genotypes, summarized in Table 2, suggested that although statistical significance was not reached, 5 complex genotypes appeared to be protective against KSHV infection, while others were expressed as risk genotypes for infection. Specifically, participants with C1-KIR2DL3 (OR, 1.85 [95% CI, .98–3.65], P = .064) and C1_KIR2DS2 (OR, 1.86 [95% CI, .97–3.75], P = .069) genotypes showed a marginal increase in the risk of KSHV infection.
Table 2.
Associations Between KIR/HLA-I Complex Genotypes and Kaposi Sarcoma–Associated Herpesvirus infection
| KIR/HLA-I | KSHV Positive (n = 123) |
KSHV Negative (n = 176) |
OR (95% CI) | P Value | ||
|---|---|---|---|---|---|---|
| No. | (%) | No. | (%) | |||
| A*03_KIR3DL2 | 25 | (20.33) | 25 | (14.20) | 1.54 (.84–2.84) | .165 |
| A*11:01_KIR2DS2 | 30 | (24.39) | 34 | (19.32) | 1.35 (.77–2.35) | .294 |
| A*11_KIR3DL2 | 32 | (26.02) | 36 | (20.45) | 1.37 (.79–2.36) | .260 |
| A*11_KIR2DS4 | 24 | (19.51) | 29 | (16.48) | 1.23 (.67–2.23) | .499 |
| A*23_KIR3DL1 | 2 | (1.63) | 9 | (5.11) | 0.31 (.04–1.22) | .135 |
| A*24_KIR3DL1 | 33 | (26.83) | 47 | (26.70) | 1.01 (.60–1.69) | .981 |
| A*32_KIR3DL1 | 6 | (4.88) | 10 | (5.68) | 0.85 (.28–2.36) | .761 |
| Bw4 80I_KIR3DL1 | 76 | (61.79) | 99 | (56.25) | 1.26 (.79–2.02) | .339 |
| B*46:01_KIR2DL2 | 9 | (7.32) | 10 | (5.68) | 1.31 (.51–3.35) | .569 |
| B*46:01_KIR2DL3 | 12 | (9.76) | 15 | (8.52) | 1.16 (.51–2.57) | .714 |
| B*51_KIR3DS1 | 19 | (15.45) | 25 | (14.20) | 1.10 (.57–2.10) | .765 |
| C1_KIR2DL3 | 108 | (87.80) | 140 | (79.55) | 1.85 (.98–3.65) | .064 |
| C1_KIR2DS2 | 109 | (88.62) | 142 | (80.68) | 1.86 (.97–3.75) | .069 |
| C1_KIR2DL2 | 83 | (67.48) | 107 | (60.80) | 1.34 (.83–2.18) | .238 |
| C2_KIR2DL2 | 65 | (52.85) | 83 | (47.16) | 1.26 (.79–2.00) | .334 |
| C2_KIR2DS1 | 66 | (53.66) | 88 | (50.00) | 1.16 (.73–1.84) | .534 |
| C2_KIR2DL3 | 75 | (60.98) | 104 | (59.09) | 1.08 (.68–1.74) | .744 |
| C2_KIR2DL1 | 75 | (60.98) | 108 | (61.36) | 0.98 (.61–1.58) | .946 |
| C*01:02_KIR2DS4 | 20 | (16.26) | 30 | (17.05) | 0.95 (.50–1.75) | .858 |
| C*04:01_KIR2DS4 | 17 | (13.82) | 20 | (11.36) | 1.25 (.62–2.50) | .526 |
| C*05:01_KIR2DS4 | 4 | (3.25) | 5 | (2.84) | 1.15 (.28–4.43) | .838 |
| C*14:02_KIR2DS4 | 8 | (6.50) | 10 | (5.68) | 1.15 (.43–3.02) | .769 |
C1 denotes HLA-C*01, C*03, C*07, and C*08, and C2 denotes HLA-C*02, C*04, C*05, and C*06 [27]. HLA-B*46 allotype was a ligand of 2DL2 and 2DL3 [28]. HLA-B Bw4 80I denotes HLA-B*27:02, B*38:01, B*49:01, B*51, B*57, and B*58 [29, 30]. HLA-A*23, -*24, and -*32 molecules (all carrying Bw4 epitope) are included among the ligands of 3DL1 [31, 32].
Abbreviations: CI, confidence interval; KSHV, Kaposi sarcoma–associated herpesvirus; OR, odds ratio.
HLA Association Fine Mapping on the Amino Acid Level
Corresponding to the results of our allele-level associations, we found the strongest associated amino acid positions in the HLA-I region, which can help to finely map the relevant variants to peptide binding regions or other functionally distinct parts of the protein. As shown in Table 3, a variety of amino acids exhibited an association with KSHV seropositivity, with position 56 of HLA-A having a significant association under all models. Individuals carrying an arginine had a lower risk of KSHV infection, whereas individuals carrying a glycine at the same position had an elevated risk of infection.
Table 3.
Associations Between Amino Acids of HLA-A and Kaposi Sarcoma–Associated Herpesvirus Infection Under the Dominant, Additive, and Overdominants Model
| Position of Amino Acid | Amino Acid Residues | KSHV Infection Group | OR (95% CI) | P Value | |
|---|---|---|---|---|---|
| KSHV Positive (n = 123) |
KSHV Negative (n = 176) |
||||
| Dominant model | |||||
| A_56 | Glycine (Gly, G) | 120 (48.78%) | 169 (48.01%) | 1.66 (.45–7.81) | .471 |
| Arginine (Arg, R) | 14 (5.69%) | 40 (11.36%) | 0.44 (.22–.83) | .014 | |
| A_149 | Alanine (Ala, A) | 123 (50.00%) | 173 (49.15%) | … | .986 |
| Threonine (Thr, T) | 10 (4.07%) | 26 (7.39%) | 0.51 (.23–1.07) | .087 | |
| B_41 | Alanine (Ala, A) | 111 (45.12%) | 142 (40.34%) | 2.21 (1.12–4.64) | .027 |
| Threonine (Thr, T) | 60 (24.39%) | 100 (28.41%) | 0.72 (.46–1.15) | .171 | |
| Additive model | |||||
| A_56 | Glycine (Gly, G) | 229 (93.09%) | 303 (86.08%) | 1.93 (1.15–3.42) | .017 |
| Arginine (Arg, R) | 17 (6.91%) | 45 (12.78%) | 0.55 (.31–.94) | .036 | |
| A_144 | Lysine (Lys, K) | 191 (77.64%) | 239 (67.90%) | 1.58 (1.11–2.30) | .013 |
| Glutamine (Gln, Q) | 55 (22.36%) | 109 (30.97%) | 0.66 (.45–.95) | .026 | |
| Overdominant model | |||||
| A_56 | Glycine (Gly, G) | 11 (4.47%) | 35 (9.94%) | 0.37 (.18–.79) | .012 |
| Arginine (Arg, R) | 11 (4.47%) | 35 (9.94%) | 0.37 (.18–.79) | .012 | |
| C_94 | Isoleucine (Ile, I) | 31 (12.60%) | 64 (18.18%) | 0.59 (.35–.98) | .042 |
| Threonine (Thr, T) | 31 (12.60%) | 64 (18.18%) | 0.59 (.35–.98) | .042 | |
| C_95 | Isoleucine (Ile, I) | 27 (10.98%) | 62 (17.61%) | 0.52 (.31–.87) | .014 |
| Leucine (Leu, L) | 30 (12.20%) | 67 (19.03%) | 0.52 (.31–.87) | .014 | |
Only statistically significant amino acids are shown.
Abbreviations: CI, confidence interval; KSHV, Kaposi sarcoma–associated herpesvirus; OR, odds ratio.
Mapping the significant amino acid residues back to the respective HLA alleles, the protective effect of arginine in position 56 of HLA-A may be due to the different HLA-A alleles, which include the protective allele HLA-A*31:01 that we previously found.
DISCUSSION
Although the cause of KSHV susceptibility is still unclear, the previous studies showed that in addition to differences in exposure chance, genetic background also plays an important impact [14, 36]. Furthermore, KIRs present in the surface of NK cells and their specific HLA-I ligands have been shown to predispose both to susceptibilities to herpesvirus infection [37] and to the development of herpesvirus-associated diseases, such as multiple sclerosis [35]. In addition, the invasion of KSHV downregulates the expression of HLA-I antigens, which can be recognized by KIR and then delivers activation signals to NK cells [13]. Thus, the polymorphisms of KIR/HLA-I influence the response of NK cells against herpesvirus infection [12, 38]. We conducted the current study based on the population of PWH in Xinjiang, which avoids confounding by HLA polymorphisms on the HIV infection, to elucidate the association between KIR/HLA-I polymorphisms and the risk of KSHV infection.
We found that 3 alleles (HLA-A*31:01, HLA-C*03:04, and HLA-C*12:03) in HLA-I were associated with KSHV infection, in which HLA-A*31:01 causes the variants of amino acid in position 56 of HLA-A, with individuals carrying arginine having a lower risk of KSHV infection. We also found homozygous KIR2DL3 may affect KSHV viral load in the infected hosts. These results reveal a potential role of KIR-mediated NK cell activation in anti-KSHV infection.
In our study, the risk of KSHV infection was reduced in people with HLA-A*31:01 and HLA-C*03:04, and it was increased for those with HLA-C*12:03. Among them, the protective effect of A*31:01 was shown significantly under all 3 models (dominant: OR, 0.30 [95% CI, .08–.82], P = .031; additive: OR, 0.30 [95% CI, .09–.80], P = .030; overdominant: OR, 0.31 [95% CI, .09–.88], P = .042). In addition, the analysis of the molecular basis revealed that arginine at position 56 of HLA-A also reduces the risk of KSHV infection. It is logical to hypothesize that the protective effect of the arginine at position 56 of HLA-A against KSHV infection was due to the association basis of HLA-A*31:01. Similar to other research, this rare allele was also shown to be a protective allele against nasopharyngeal carcinoma caused by Epstein-Barr virus [39], which, like KSHV, is a γ-herpesvirus. This allele was also found to be associated with macular papular erythema caused by human herpesvirus reactivation in the Chinese population [40]. This implies that A*31:01 may have the same manner of effective control of the herpesvirus, especially γ-herpesvirus infection.
The significant effects of the inhibitory KIR receptors on KSHV viral load also merit special attention. The KIR genotype of KSHV-seropositive individuals with different viral loads was characterized for the frequency of the KIR2DL2 and KIR2DL3 genes, to evidence any potential influence of NK receptor allotype in the risk of KSHV replication. The observed increased homozygous KIR2DL3 and a relatively high KSHV viral load in participants with KSHV suggest higher inhibitory capabilities of KIR2DL2 in comparison with the KIR2DL3 receptor. This result was consistent with a study in Italy [15], which found that this specific KIR genotype may impair NK cell function in controlling KSHV infection and proliferation, leading the virus to exert its angioproliferative activity. There is clinical evidence to suggest that lytic KSHV replication is important for the progression to KS [41]. Thus, it can be hypothesized that the modulation of NK cell–mediated cytotoxicity by KIR may be beneficial against infectious agents.
However, no statistically significant KIR/HLA-I complex genotypes associated with KSHV infection were found in our study, which was consistent with some results of other studies in the Mediterranean region [16]. KIR/HLA-I polymorphisms may have more impact on the development of KS. The significance of the HLA single allele and nonsignificance of the KIR/HLA-I complex genotypes suggest that there are other inflammatory effects against KSHV infection in addition to KIR-mediated NK cell cytotoxicity. HLA could present herpesvirus-related antigenic epitopes to cytotoxic T lymphocytes, resulting in effective control of the virus in the lytic phase [42]. Furthermore, the extensive polymorphisms in the KIR/HLA-I and the relatively small sample size included in the study may also influence the results.
Certain limitations also should be considered in this study. First, there are extensive polymorphisms in KIR/HLA-I, and the relatively small sample size of the study might have reduced the statistical power of the findings. Second, the association between the identified alleles and genes and KSHV infection was not significant after Bonferroni correction. Given the conservative results generated by the multiple correction methods and because this was a preliminary study, multiple comparisons could not be considered in the association analysis. Finally, KSHV infection was measured by serology, which may be confounded by exposure. Despite the inclusion of relevant covariates, it remains challenging to fully control for these exposure-related confounders, which may affect the interpretation of the results. The participants included were all HIV-infected with no KS occurring after ART, and there was a lack of association studies between KIR/HLA-I polymorphisms and KS progression. The role of NK cells and KIR in the development of KS has not been studied in depth. Further functional studies are warranted to clarify the findings described herein and to understand their role in the development of KS.
In conclusion, even considering the exploratory characteristics of the study, our results provide further insight into the link between HLA-I alleles and KIR genes and KSHV infection, highlighting the potential role of KIR-mediated NK cells in anti-KSHV infection. Further studies should aim at completing the picture of the interplay among KSHV, genetic background, and immunosuppression.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Xin Zhang, Department of Epidemiology, School of Public Health, Fudan University, Shanghai, China; Key Laboratory of Public Health Safety, Fudan University, Ministry of Education, Shanghai, China.
Yi Li, Department of Epidemiology, School of Public Health, Fudan University, Shanghai, China; Key Laboratory of Public Health Safety, Fudan University, Ministry of Education, Shanghai, China.
Xinyu Han, Department of Epidemiology, School of Public Health, Fudan University, Shanghai, China; Key Laboratory of Public Health Safety, Fudan University, Ministry of Education, Shanghai, China.
Yiyun Xu, Department of Epidemiology, School of Public Health, Fudan University, Shanghai, China; Key Laboratory of Public Health Safety, Fudan University, Ministry of Education, Shanghai, China.
Haili Wang, Department of Epidemiology, School of Public Health, Fudan University, Shanghai, China; Key Laboratory of Public Health Safety, Fudan University, Ministry of Education, Shanghai, China.
Tianye Wang, Department of Epidemiology, School of Public Health, Fudan University, Shanghai, China; Key Laboratory of Public Health Safety, Fudan University, Ministry of Education, Shanghai, China.
Tiejun Zhang, Department of Epidemiology, School of Public Health, Fudan University, Shanghai, China; Key Laboratory of Public Health Safety, Fudan University, Ministry of Education, Shanghai, China; Shanghai Institute of Infectious Disease and Biosecurity, School of Public Health, Fudan University, Shanghai, China; Yiwu Research Institute, Fudan University, Yiwu, China.
Notes
Author contributions. All authors contributed to the study's conception and design. X. Z., Y. L., and X. H. were responsible for data collection. Y. X. and H. W. were responsible for sorting and cleaning the data. X. Z. and T. W. performed data analysis. X. Z. was responsible for interpreting the results and drafting the article. T. Z. was responsible for revising the manuscript critically for important intellectual content and final approval.
Disclaimer. The funders did not play a role in the design, conduct, or analysis of the study, nor in the drafting of this manuscript.
Financial support. This work was supported by National Natural Science Foundation of China (grant number 81772170) and by the Special Project on National Science and Technology Basic Resources Investigation (project number 2019FY101103).
References
- 1. Bouvard V, Baan R, Straif K, et al. A review of human carcinogens—part B: biological agents. Lancet Oncol 2009; 10:321–2. [DOI] [PubMed] [Google Scholar]
- 2. Cesarman E, Damania B, Krown SE, Martin J, Bower M, Whitby D. Kaposi sarcoma. Nat Rev Dis Primers 2019; 5:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Silverberg MJ, Chao C, Leyden WA, et al. HIV infection, immunodeficiency, viral replication, and the risk of cancer. Cancer Epidemiol Biomarkers Prev 2011; 20:2551–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Robey RC, Bower M. Facing up to the ongoing challenge of Kaposi's sarcoma. Curr Opin Infect Dis 2015; 28:31–40. [DOI] [PubMed] [Google Scholar]
- 5. World Health Organization . Global health sector strategies on, respectively, HIV, viral hepatitis and sexually transmitted infections for the period 2022–2030. 2022. Available at: https://www.who.int/publications/i/item/9789240053779. Accessed 1 May 2023.
- 6. Wen Z, Li W, Fang Y, et al. Prevalence and risk factors of Kaposi's sarcoma–associated herpesvirus infection among Han and Uygur populations in Xinjiang, China. Can J Infect Dis Med Microbiol 2021; 2021:2555865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu Z, Fang Q, Zhou S, et al. Seroprevalence of Kaposi's sarcoma–associated herpesvirus among HIV-infected Uygurs in Xinjiang, China. J Med Virol 2017; 89:1629–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mesri EA, Cesarman E, Boshoff C. Kaposi's sarcoma and its associated herpesvirus. Nat Rev Cancer 2010; 10:707–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Della Chiesa M, De Maria A, Muccio L, Bozzano F, Sivori S, Moretta L. Human NK cells and herpesviruses: mechanisms of recognition, response and adaptation. Front Microbiol 2019; 10:2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ahlenstiel G, Martin MP, Gao X, Carrington M, Rehermann B. Distinct KIR/HLA compound genotypes affect the kinetics of human antiviral natural killer cell responses. J Clin Invest 2008; 118:1017–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fang Q, Liu Z, Zhang T. Human leukocyte antigen polymorphisms and Kaposi's sarcoma-associated herpesvirus infection outcomes: a call for deeper exploration. J Med Virol 2019; 91:541–8. [DOI] [PubMed] [Google Scholar]
- 12. Vivier E, Nunès JA, Vély F. Natural killer cell signaling pathways. Science 2004; 306:1517–9. [DOI] [PubMed] [Google Scholar]
- 13. Ishido S, Wang C, Lee BS, Cohen GB, Jung JU. Downregulation of major histocompatibility complex class I molecules by Kaposi's sarcoma–associated herpesvirus K3 and K5 proteins. J Virol 2000; 74:5300–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sallah N, Miley W, Labo N, et al. Distinct genetic architectures and environmental factors associate with host response to the gamma2-herpesvirus infections. Nat Commun 2020; 11:3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Borghi A, D'Accolti M, Rizzo R, et al. High prevalence of specific KIR types in patients with HHV-8 positive cutaneous vascular lesions: a possible predisposing factor? Arch Dermatol Res 2016; 308:373–7. [DOI] [PubMed] [Google Scholar]
- 16. Guerini FR, Mancuso R, Agostini S, et al. Activating KIR/HLA complexes in classic Kaposi's sarcoma. Infect Agents Cancer 2012; 7:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Agostini S, Guerini FR, Bandera A, et al. A role for activatory KIR/HLA complexes in HIV-associated Kaposi's sarcoma development. Retrovirology 2011; 8(Supp 1):A209. [Google Scholar]
- 18. Goedert JJ, Martin MP, Vitale F, et al. Risk of classic Kaposi sarcoma with combinations of killer immunoglobulin-like receptor and human leukocyte antigen loci: a population-based case-control study. J Infect Dis 2016; 213:432–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guerini FR, Agliardi C, Mancuso R, et al. Association of HLA-DRB1 and -DQB1 with classic Kaposi's sarcoma in mainland Italy. Cancer Genomics Proteomics 2006; 3(3–4):191–6. [PubMed] [Google Scholar]
- 20. Guech-Ongey M, Verboom M, Pfeiffer RM, et al. HLA polymorphisms and detection of Kaposi sarcoma–associated herpesvirus DNA in saliva and peripheral blood among children and their mothers in the Uganda sickle cell anemia KSHV study. Infect Agents Cancer 2010; 5:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tedeschi R, Dillner J, De Paoli P. Laboratory diagnosis of human herpesvirus 8 infection in humans. Eur J Clin Microbiol Infect Dis 2002; 21:831–44. [DOI] [PubMed] [Google Scholar]
- 22. Laney AS, Peters JS, Manzi SM, Kingsley LA, Chang Y, Moore PS. Use of a multiantigen detection algorithm for diagnosis of Kaposi's sarcoma–associated herpesvirus infection. J Clin Microbiol 2006; 44:3734–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mbisa GL, Miley W, Gamache CJ, et al. Detection of antibodies to Kaposi's sarcoma–associated herpesvirus: a new approach using K8.1 ELISA and a newly developed recombinant LANA ELISA. J Immunol Methods 2010; 356(1–2):39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fu B, Sun F, Li B, et al. Seroprevalence of Kaposi's sarcoma–associated herpesvirus and risk factors in Xinjiang, China. J Med Virol 2009; 81:1422–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Martin MP, Carrington M. KIR locus polymorphisms: genotyping and disease association analysis. Methods Mol Biol 2008; 415:49–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Middleton D, Gonzelez F. The extensive polymorphism of KIR genes. Immunology 2010; 129:8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Biassoni R, Falco M, Cambiaggi A, et al. Amino acid substitutions can influence the natural killer (NK)-mediated recognition of HLA-C molecules. Role of serine-77 and lysine-80 in the target cell protection from lysis mediated by “group 2” or “group 1” NK clones. J Exp Med 1995; 182:605–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gendzekhadze K, Norman PJ, Abi-Rached L, et al. Co-evolution of KIR2DL3 with HLA-C in a human population retaining minimal essential diversity of KIR and HLA class I ligands. Proc Natl Acad Sci U S A 2009; 106:18692–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Middleton D, Williams F, Halfpenny IA. KIR genes. Transpl Immunol 2005; 14(3–4):135–42. [DOI] [PubMed] [Google Scholar]
- 30. Gumperz JE, Litwin V, Phillips JH, Lanier LL, Parham P. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J Exp Med 1995; 181:1133–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Saunders PM, Pymm P, Pietra G, et al. Killer cell immunoglobulin-like receptor 3DL1 polymorphism defines distinct hierarchies of HLA class I recognition. J Exp Med 2016; 213:791–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stern M, Ruggeri L, Capanni M, Mancusi A, Velardi A. Human leukocyte antigens A23, A24, and A32 but not A25 are ligands for KIR3DL1. Blood 2008; 112:708–10. [DOI] [PubMed] [Google Scholar]
- 33. Migdal M, Ruan DF, Forrest WF, Horowitz A, Hammer C. MiDAS-meaningful immunogenetic data at scale. PLoS Comput Biol 2021; 17:e1009131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Caselli E, Rizzo R, Ingianni A, Contini P, Pompei R, Di Luca D. High prevalence of HHV8 infection and specific killer cell immunoglobulin-like receptors allotypes in Sardinian patients with type 2 diabetes mellitus. J Med Virol 2014; 86:1745–51. [DOI] [PubMed] [Google Scholar]
- 35. Rizzo R, Gentili V, Casetta I, et al. Altered natural killer cells’ response to herpes virus infection in multiple sclerosis involves KIR2DL2 expression. J Neuroimmunol 2012; 251(1–2):55–64. [DOI] [PubMed] [Google Scholar]
- 36. Gao Y, Jiang J, Yang S, et al. Genome-wide association study of Mycobacterium avium subspecies paratuberculosis infection in Chinese Holstein. BMC Genomics 2018; 19:972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moraru M, Cisneros E, Gomez-Lozano N, et al. Host genetic factors in susceptibility to herpes simplex type 1 virus infection: contribution of polymorphic genes at the interface of innate and adaptive immunity. J Immunol 2012; 188:4412–20. [DOI] [PubMed] [Google Scholar]
- 38. Campbell KS, Purdy AK. Structure/function of human killer cell immunoglobulin-like receptors: lessons from polymorphisms, evolution, crystal structures and mutations. Immunology 2011; 132:315–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tian W, Zhu FM, Wang WY, et al. Sequence-based typing of HLA-A gene in 930 patients with nasopharyngeal carcinoma in Hunan province, southern China. Tissue Antigens 2015; 86:15–20. [DOI] [PubMed] [Google Scholar]
- 40. Calligaris L, Stocco G, De Iudicibus S, et al. Carbamazepine hypersensitivity syndrome triggered by a human herpes virus reactivation in a genetically predisposed patient. Int Arch Allergy Immunol 2009; 149:173–7. [DOI] [PubMed] [Google Scholar]
- 41. Martin DF, Kuppermann BD, Wolitz RA, Palestine AG, Li H, Robinson CA. Oral ganciclovir for patients with cytomegalovirus retinitis treated with a ganciclovir implant. Roche Ganciclovir Study Group. N Engl J Med 1999; 340:1063–70. [DOI] [PubMed] [Google Scholar]
- 42. Chin YM, Mushiroda T, Takahashi A, et al. HLA-A SNPs and amino acid variants are associated with nasopharyngeal carcinoma in Malaysian Chinese. Int J Cancer 2015; 136:678–87. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.