Figure Legend
In the original publication [1], there was a mistake in the legend for Figure 4, a log scale for the axis has been applied to each graph to better show the included data points.
The correct legend appears below:
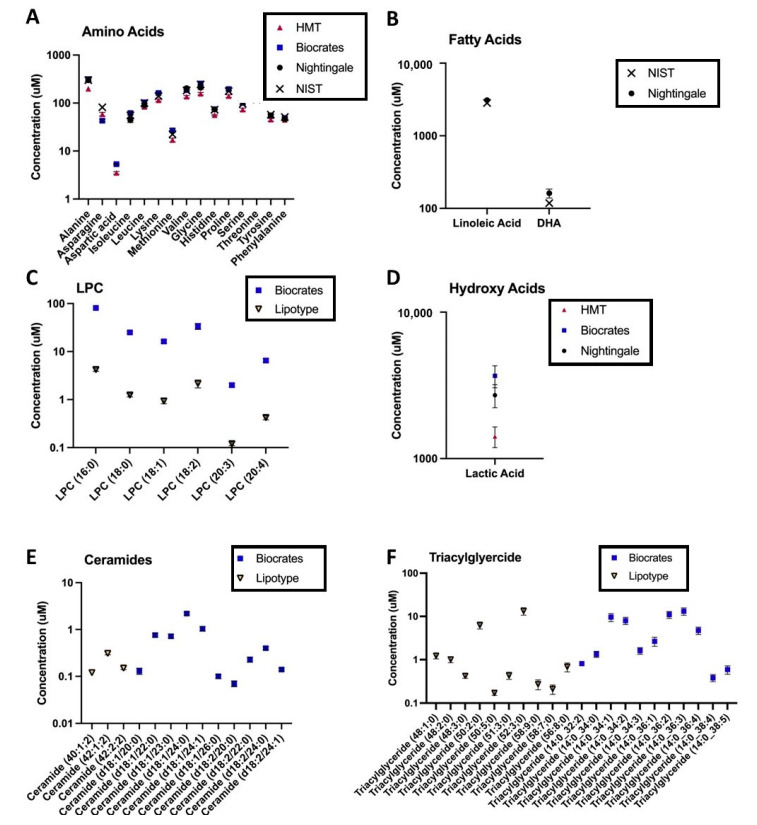
Figure 4. Platform-specific, log-transformed, average metabolite levels in control samples for vendors reporting absolute units; each point represents mean ± SEM for 11 control samples in total: 9 control samples from 6 individuals (with 3 technical replicates), and 2 NIST pooled reference plasma samples. Each panel depicts the range of covered metabolites, across all assays, for an exemplar metabolite class: (A) amino acids, (B) fatty acids, (C) lysophosphatidylcholines (LPC), (D) hydroxy acids, (E) ceramides, and (F) triglycerides. Depicted data are from the second sample shipment. NIST = concentrations reported in the National Institute of Standards and Technology (NIST) SRM 1950 Certificate of Analysis (COA, revised June 2020).
Error in Figure/Table
In the original publication, there was a mistake in:
1. Table 1, Table 2 and Table 3. The color coding in these tables does not match the footnote. This has been corrected.
2. Table 3. The amino acid, alanine, was omitted from the published manuscript. This is now included in the corrected table.
3. Table 3 as published. NIST standards were used for part of the study to compare the accuracy of the different measurements. In the measurement of fatty acids and cholesterol, the NIST standards reflect the total concentration of fatty acids and cholesterol. The vendors in the published manuscript were reporting free fatty acids and free cholesterol, and therefore the comparison represented in Table 3 is not valid. This discrepancy was brought to our attention after the paper was published. The fatty acid and cholesterol values and percent accuracy for Biocrates and Lipotype were removed.
4. Figure 4. Reflecting the updates from Table 3, Figure 4b has been edited and a log scale applied for all graphs in the figure to better show the included data points.
The corrected Table 1, Table 2 and Table 3 and Figure 4 appear below:
Table 1.
Intra-assay Percent Coefficient of Variance (CV%) within Metabolite Classes and CV% Standard Deviation (SD) for Technical Replicates of PTSD and Control Samples in Shipment 1.
| Intra-Assay Precision: Shipment 1 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PTSD Average CV% | Control Average CV% | Count | Range | SD | PTSD Average CV% | Control Average CV% | Count | SD | PTSD Average CV% | Control Average CV% | Count | SD | PTSD Average CV% | Control Average CV% | Count | SD | PTSD Average CV% | Control Average CV% | Count | SD | |
| Metabolite Class | Biocrates | HMT | Nightingale | Lipotype | Metabolon | ||||||||||||||||
| Acylcarnitines | 10.33 | 9.88 | 14 | 2.07–76.09 | 15.67 | 5.66 | 5.81 | 35 | 7.43 | 9.03 | 8.39 | 21 | 7.70 | ||||||||
| Amino Acids | 3.53 | 8.12 | 40 | 0.87–19.01 | 3.14 | 6.85 | 6.93 | 22 | 2.56 | 2.78 | 6.01 | 9 | 3.76 | 8.84 | 9.56 | 52 | 8.22 | ||||
| Amino Acid Related | 3.44 | 8.71 | 1.73–12.88 | 3.21 | |||||||||||||||||
| Carboxylic Acids | 3.85 | 7.20 | 2 | 2.70–8.02 | 2.25 | 5.10 | 5.62 | 8 | 2.83 | 2.99 | 6.66 | 3 | 2.38 | 11.32 | 9.13 | 101 | 13.59 | ||||
| Cholesteryl ester | 8.46 | 14.69 | 18 | 3.42–48.13 | 8.44 | 4.23 | 13.30 | 13 | 6.84 | 4.63 | 4.00 | 26 | 2.20 | ||||||||
| Diglycerides | 8.46 | 12.28 | 14 | 1.95–36.34 | 7.53 | 4.84 | 9.80 | 2 | 3.38 | 4.29 | 4.00 | 19 | 2.73 | ||||||||
| Diazines | 21.00 | 18.45 | 3 | 21.16 | 15.39 | 13.46 | 5 | 12.60 | |||||||||||||
| Organonitrogen compounds | 7.29 | 9.37 | 13 | 5.02 | 15.55 | 11.3 | 14 | 12.61 | |||||||||||||
| Purine nucleotides | 9.69 | 16.16 | 5 | 9.32 | 14.18 | 12.48 | 10 | 8.09 | |||||||||||||
| Organooxygen compounds | 12.03 | 9.63 | 6 | 7.35 | 1.04 | 6.44 | 2 | 4.07 | 9.19 | 9.90 | 6 | 8.38 | |||||||||
| Hydroxy acids and derivatives | 4.04 | 6.79 | 5 | 2.47 | 0.88 | 3.05 | 1 | NA | 11.28 | 11.80 | 16 | 10.81 | |||||||||
| Keto acids and derivatives | 1.62 | 5.52 | 3 | 2.24 | 4.29 | 26.20 | 2 | 18.87 | 8.35 | 8.18 | 13 | 4.94 | |||||||||
| Ceramides | 6.53 | 8.60 | 25 | 0.59–29.33 | 5.33 | 11.36 | 10.43 | 5 | 4.62 | 5.03 | 6.77 | 2 | 3.06 | 7.22 | 7.10 | 11 | 5.55 | ||||
| Lactosylceramide | 19.98 | 15.25 | 13 | 13.62 | 10.15 | 14.99 | 12 | 10.36 | |||||||||||||
| Glucosylceramide | 21.84 | 19.55 | 13 | 11.15 | |||||||||||||||||
| Dihexosylceramides | 10.30 | 9.78 | 10 | 2.45–34.65 | 7.59 | ||||||||||||||||
| Trihexosylceramides | 14.16 | 17.94 | 6 | 4.69–43.82 | 11.60 | ||||||||||||||||
| Dihydroceramide | 19.07 | 17.15 | 12 | 14.73 | |||||||||||||||||
| Hexosylceramide | 9.56 | 11.32 | 19 | 1.33–27.20 | 5.81 | 6.28 | 8.33 | 12 | 4.34 | ||||||||||||
| Triglycerides | 7.33 | 13.27 | 235 | 0.78–38.42 | 3.72 | 4.88 | 6.49 | 32 | 2.46 | 1.92 | 5.96 | 21 | 2.19 | ||||||||
| Hormones/Steroids | 2.53 | 6.43 | 3 | 1.48–7.15 | 2.34 | 5.88 | 6.28 | 9 | 4.77 | 8.65 | 6.18 | 26 | 3.79 | ||||||||
| Fatty Acids | 8.10 | 8.16 | 7 | 3.63–11.43 | 2.60 | 6.92 | 9.11 | 32 | 4.97 | 27.57 | 3.89 | 2 | 15.39 | 3.58 | 3.53 | 29 | 2.15 | ||||
| Fatty Acyls | 28.92 | 29.75 | 4 | 19.76 | 18.17 | 13.20 | 83 | 12.09 | |||||||||||||
| Biogenic Amines | 4.74 | 10.52 | 3 | 3.19–11.69 | 3.47 | ||||||||||||||||
| Bile Acids | 5.84 | 7.73 | 12 | 2.48–12.04 | 2.77 | 4.66 | 6.47 | 4 | 7.22 | 11.42 | 8.94 | 22 | 6.95 | ||||||||
| Indoles and Derivatives | 3.36 | 10.40 | 3 | 2.69–14.21 | 4.56 | 6.51 | 7.37 | 1 | NA | 7.45 | 5.82 | 9 | 3.97 | ||||||||
| Lysophosphatidyl-cholines (LPC) | 13.05 | 11.32 | 14 | 0.91–36.05 | 10.66 | 3.34 | 3.71 | 27 | 1.69 | 4.38 | 14.44 | 6 | 5.36 | 7.28 | 10.10 | 15 | 5.14 | ||||
| Glycerophosphocholines | 7.09 | 7.76 | 19 | 10.79 | |||||||||||||||||
| Phosphatidyl-cholines (PC) | 7.98 | 9.78 | 73 | 1.78–67.84 | 9.56 | 7.96 | 11.15 | 63 | 5.22 | 8.12 | 5.98 | 18 | 4.44 | ||||||||
| Sphingomyelins | 3.89 | 7.68 | 15 | 1.88–11.73 | 2.64 | 5.79 | 8.62 | 12 | 2.89 | 3.45 | 3.05 | 12 | 1.79 | ||||||||
| Sphingolipids | 14.28 | 13.47 | 12 | 10.86 | 6.13 | 7.79 | 2 | 2.11 | |||||||||||||
| Sphinganine | 6 | ||||||||||||||||||||
| Sphingosine | 11.76 | 12.89 | 6 | 9.48 | |||||||||||||||||
| Glycerophospholipids | 10.41 | 9.67 | 15 | 8.70 | 19.61 | 22.64 | 7 | 16.45 | |||||||||||||
| Glycerolipids (Monoacylglycerol) | 21.69 | 26.95 | 25 | 14.48 | |||||||||||||||||
| Carboximidic acids and derivatives | 4.93 | 4.33 | 1 | NA | 15.21 | 17.34 | 4 | 10.22 | |||||||||||||
| lyso-Phosphatidylethanolamine (LPE) | 10.36 | 8.40 | 22 | 7.95 | 8.83 | 15.24 | 9 | 5.24 | 5.18 | 6.38 | 8 | 4.88 | |||||||||
| Phosphatidylcholine (-ether) (LPC-O) | 13.02 | 16.40 | 41 | 8.64 | |||||||||||||||||
| Phosphatidylethanolamine (PE) | 9.22 | 15.06 | 15 | 6.79 | 4.38 | 8.11 | 12 | 6.46 | |||||||||||||
| Phosphatidylethanolamine (-ether) (LPE-O) | 11.54 | 15.20 | 16 | 7.76 | |||||||||||||||||
| Phosphatidylinositol (LPI) | 7.37 | 8.44 | 14 | 6.05 | 8.95 | 16.88 | 15 | 7.62 | 10.83 | 18.82 | 6 | 8.90 | |||||||||
| Lyso-Phosphatidylserine (LPS) | 11.21 | 15.84 | 7 | 8.97 | |||||||||||||||||
| Glycerophosphoglycerols (LPG) | 9.23 | 10.92 | 14 | 6.71 | |||||||||||||||||
| Vitamins and Cofactors | 1.09 | 10.44 | 1 | NA | 2.37 | 5.87 | 1 | NA | |||||||||||||
| Alkaloids | 4.69 | 13.74 | 1 | NA | 3.86 | 2.48 | 2 | 0.80 | |||||||||||||
| Amine (Oxides) | 6.92 | 8.23 | 1 | NA | 45.46 | 12.49 | 1 | NA | |||||||||||||
| Carbohydrates and Related | 2.50 | 6.33 | 1 | NA | 10.81 | 14.37 | 34 | 13.62 | |||||||||||||
| Cresols | 2.07 | 7.14 | 1 | NA | |||||||||||||||||
| Imidazopyrimidines | 11.35 | 5.44 | 1 | NA | 10.32 | 7.60 | 17 | 8.68 | |||||||||||||
| 5′-deoxyribonucleosides | 19.82 | 18.62 | 1 | NA | 7.84 | 8.63 | 1 | NA | |||||||||||||
| Nucleoside and nucleotide analogues | 13.05 | 3.4 | 1 | NA | |||||||||||||||||
| Pyrimidine nucleosides | 1.33 | 7.94 | 1 | NA | 9.24 | 9.59 | 7 | 7.33 | |||||||||||||
| Pyridines and derivatives | 6.69 | 7.90 | 10 | 5.76 | |||||||||||||||||
| Quinolines and derivatives | 11.85 | 13.64 | 3 | 7.42 | |||||||||||||||||
| Phenols | 9.22 | 4.79 | 3 | 5.40 | |||||||||||||||||
| Prenol lipids | 17.87 | 8.77 | 7 | 8.13 | |||||||||||||||||
| Imidazole ribonucleosides and ribonucleotides | 12.17 | 5.5 | 1 | NA | |||||||||||||||||
| Benzene and substituted derivatives | 12.72 | 9.49 | 14 | 10.36 | |||||||||||||||||
| Phenylpropanoic acids | 6.52 | 5.82 | 9 | 8.63 | |||||||||||||||||
| Tetrapyrroles and derivatives | 3.45 | 11.6 | 2 | 5.43 | |||||||||||||||||
| Cholesterol and derivatives | 7.53 | 9.5 | 2 | 4.40 | |||||||||||||||||
| Non-metal oxoanionic compounds | 2.94 | 3.17 | 2 | 1.81 | |||||||||||||||||
| Organic sulfuric acids and derivatives | 6.1 | 4.03 | 22 | 3.49 | |||||||||||||||||
| Organic sulfonic acids and derivatives | 2.47 | 5.59 | 2 | 3.48 | |||||||||||||||||
| Organic carbonic acids and derivatives | 6.33 | 9.48 | 2 | 3.88 | |||||||||||||||||
| Organic phosphoric acids and derivatives | 9.53 | 7.87 | 1 | NA | |||||||||||||||||
| Benzothiazepines | 2.29 | 6.17 | 2 | 5.83 | |||||||||||||||||
| Bilirubins | 9.03 | 5.41 | 2 | 6.66 | |||||||||||||||||
| Dihydrofurans | 6.94 | 3.07 | 2 | 3.68 | |||||||||||||||||
| Alkyl halides | 2.9 | 3.98 | 2 | 1.49 | |||||||||||||||||
| Sulfinic acids and derivatives | 11.13 | 16.04 | 1 | NA | |||||||||||||||||
| Azoles | 11.11 | 10.6 | 7 | 5.90 | |||||||||||||||||
| Azolidines | 4.34 | 6.92 | 1 | NA | |||||||||||||||||
| Cinnamic acids and derivatives | 4.07 | 13.92 | 1 | NA | |||||||||||||||||
| Peptidomimetics | 20.02 | 14.52 | 1 | NA | |||||||||||||||||
| Piperidines | 14.81 | 16.27 | 1 | NA | |||||||||||||||||
| Pyrrolidines | 4.09 | 5.48 | 1 | NA | |||||||||||||||||
| Coumarins and derivatives | 0 | 14.1 | 1 | NA | |||||||||||||||||
Notes: “High” precision is shown in green (≤10%), “moderate” in yellow (10% < x < 20%), and “low” in red (≥20%).
Table 2.
Inter-Assay percent Coefficient of Variance (CV%) within Metabolite Classes for Technical Replicates of PTSD and Control Samples across Shipment 1 and Shipment 2.
| Inter-assay Precision: Shipment 1 vs. Shipment 2 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PTSD Average CV% | Control Average CV% | Standard deviation (SD) | PTSD Average CV% | Control Average CV% | Standard deviation (SD) | PTSD Average CV% | Control Average CV% | Standard deviation (SD) | PTSD Average CV% | Control Average CV% | Standard deviation (SD) | PTSD Average CV% | Control Average CV% | Standard deviation (SD) | |
| Metabolite Class | Biocrates | HMT | Nightingale | Lipotype | Metabolon | ||||||||||
| Acylcarnitines | 7.21 | 11.48 | 2.78 | 12.21 | 12.10 | 8.64 | 15.27 | 13.52 | 9.93 | ||||||
| Amino Acids | 5.88 | 11.95 | 3.57 | 9.10 | 9.79 | 4.02 | 4.80 | 7.37 | 3.48 | 12.84 | 14.21 | 8.03 | |||
| Amino Acid Related | 7.63 | 13.90 | 6.86 | ||||||||||||
| Carboxylic Acids | 7.54 | 13.71 | 4.05 | 10.60 | 8.90 | 5.83 | 5.92 | 7.90 | 1.51 | 15.38 | 13.91 | 12.75 | |||
| Cholesteryl ester | 11.71 | 14.47 | 2.58 | 8.07 | 8.93 | 5.25 | 10.16 | 8.68 | 7.77 | ||||||
| Diglycerides | 17.42 | 22.27 | 10.62 | 9.13 | 13.25 | 6.27 | 10.97 | 10.06 | 4.53 | ||||||
| Diazines | 16.42 | 21.57 | 14.04 | 33.22 | 29.05 | 24.92 | |||||||||
| Organonitrogen compounds | 13.31 | 11.50 | 8.24 | 14.25 | 13.66 | 7.67 | |||||||||
| Purine nucleotides | 30.06 | 38.64 | 18.99 | 11.49 | 11.84 | 5.98 | |||||||||
| Organooxygen compounds | 31.07 | 37.08 | 13.17 | 4.89 | 6.94 | 2.62 | 31.34 | 26.85 | 28.72 | ||||||
| Hydroxy acids and derivatives | 10.88 | 11.10 | 6.05 | 1.88 | 3.57 | NA | 17.76 | 17.90 | 12.03 | ||||||
| Keto acids and derivatives | 14.10 | 16.66 | 2.83 | 10.58 | 19.49 | 9.58 | 15.64 | 13.55 | 8.45 | ||||||
| Ceramides | 16.04 | 19.50 | 8.65 | 11.34 | 15.68 | 4.34 | 6.58 | 6.47 | 0.78 | 10.10 | 7.88 | 4.44 | |||
| Lactosylceramide | 15.32 | 16.79 | 8.06 | 16.61 | 17.51 | 13.28 | |||||||||
| Glucosylceramide | 19.30 | 23.43 | 9.77 | ||||||||||||
| Dihexosylceramides | 11.99 | 17.82 | 5.18 | ||||||||||||
| Trihexosylceramides | 17.08 | 23.39 | 4.52 | ||||||||||||
| Dihydroceramide | 18.63 | 21.21 | 12.20 | ||||||||||||
| Hexosylceramide | 14.23 | 17.72 | 6.01 | 11.44 | 10.57 | 3.49 | |||||||||
| Triglycerides | 15.42 | 19.35 | 7.56 | 8.34 | 6.67 | 2.20 | 9.3 | 7.65 | 2.61 | ||||||
| Hormones/Steroids | 15.59 | 18.54 | 9.04 | 8.08 | 10.35 | 5.86 | 15.39 | 13.53 | 5.84 | ||||||
| Fatty Acids | 13.80 | 18.43 | 7.40 | 24.26 | 27.33 | 33.29 | 53.25 | 9.39 | 26.15 | 6.89 | 6.60 | 3.39 | |||
| Fatty Acyls | 4.21 | 4.99 | NA | 20.94 | 17.94 | 9.44 | |||||||||
| Biogenic Amines | 6.37 | 18.78 | 10.04 | ||||||||||||
| Bile Acids | 21.36 | 23.51 | 28.05 | 5.99 | 10.49 | 10.09 | 21.36 | 20.83 | 11.05 | ||||||
| Indoles and Derivatives | 8.60 | 15.22 | 4.59 | 13.11 | 12.47 | 5.93 | |||||||||
| Lysophosphatidyl-cholines (LPC) | 13.24 | 17.78 | 8.17 | 6.45 | 8.75 | 3.38 | 9.13 | 8.85 | 0.77 | 17.64 | 17.23 | 12.98 | |||
| Glycerophosphocholines | 6.56 | 7.21 | 2.76 | ||||||||||||
| Phosphatidyl-cholines (PC) | 10.11 | 14.43 | 6.97 | 10.24 | 13.18 | 10.69 | 13.65 | 13.98 | 13.68 | ||||||
| Sphingomyelins | 9.34 | 12.99 | 4.28 | 6.16 | 7.19 | 2.09 | 6.51 | 7.58 | 4.35 | ||||||
| Sphingolipids | 20.21 | 20.23 | 10.83 | 9.90 | 12.50 | 1.60 | |||||||||
| Sphinganine | |||||||||||||||
| Sphingosine | 19.34 | 19.49 | 10.01 | ||||||||||||
| Glycerophospholipids | 19.51 | 22.85 | 25.01 | 22.79 | 26.91 | 17.68 | |||||||||
| Glycerolipids (Monoacylglycerol) | 35.32 | 34.69 | 15.53 | ||||||||||||
| Carboximidic acids and derivatives | 23.26 | 3.55 | NA | 12.92 | 17.33 | 5.22 | |||||||||
| lyso-Phosphatidylethanolamine (LPE) | 11.88 | 13.87 | 17.60 | 9.92 | 9.85 | 1.89 | 22.80 | 19.85 | 12.54 | ||||||
| Phosphatidylcholine (-ether) (LPC-O) | 14.35 | 15.00 | 6.39 | ||||||||||||
| Phosphatidylethanolamine (PE) | 12.50 | 14.70 | 6.08 | 12.16 | 12.16 | 8.38 | |||||||||
| Phosphatidylethanolamine (-ether) (LPE-O) | 10.14 | 11.68 | 3.76 | ||||||||||||
| Phosphatidylinositol (LPI) | 9.81 | 10.24 | 6.62 | 13.27 | 13.27 | 7.59 | 39.03 | 49.61 | 11.64 | ||||||
| Lyso-Phosphatidylserine (LPS) | 19.00 | 20.64 | 7.74 | ||||||||||||
| Glycerophosphoglycerols (LPG) | 12.34 | 15.52 | 7.60 | ||||||||||||
| Vitamins and Cofactors | 7.59 | 13.77 | NA | 6.44 | 13.52 | NA | |||||||||
| Alkaloids | 17.05 | 46.83 | 21.81 | ||||||||||||
| Amine (Oxides) | 6.55 | 11.15 | NA | 28.84 | 24.42 | NA | |||||||||
| Carbohydrates and Related | 6.61 | 13.25 | NA | 23.31 | 22.25 | 22.84 | |||||||||
| Cresols | 4.39 | 11.88 | NA | ||||||||||||
| Imidazopyrimidines | 19.81 | 26.41 | 14.25 | ||||||||||||
| 5′-deoxyribonucleosides | 12.33 | 9.07 | 5.31 | 10.04 | 13.25 | NA | |||||||||
| Nucleoside and nucleotide analogues | 44.29 | 15.88 | NA | ||||||||||||
| Pyrimidine nucleosides | 21.49 | 19.06 | 15.04 | ||||||||||||
| Pyridines and derivatives | 11.51 | 11.75 | 5.71 | ||||||||||||
| Quinolines and derivatives | 20.42 | 20.85 | 13.19 | ||||||||||||
| Phenols | 22.06 | 21.35 | 6.87 | ||||||||||||
| Prenol lipids | 16.43 | 13.54 | 6.28 | ||||||||||||
| Imidazole ribonucleosides and ribonucleotides | 7.24 | 7.26 | NA | ||||||||||||
| Benzene and substituted derivatives | 23.37 | 23.26 | 13.29 | ||||||||||||
| Phenylpropanoic acids | 17.47 | 23.77 | 12.32 | ||||||||||||
| Tetrapyrroles and derivatives | 9.73 | 17.57 | 5.40 | ||||||||||||
| Cholesterol and derivatives | 8.27 | 8.43 | 2.92 | ||||||||||||
| Non-metal oxoanionic compounds | 4.07 | 4.04 | 0.41 | ||||||||||||
| Organic sulfuric acids and derivatives | 23.64 | 25.3 | 27.37 | ||||||||||||
| Organic sulfonic acids and derivatives | 7.08 | 10.9 | 6.10 | ||||||||||||
| Organic carbonic acids and derivatives | 7.67 | 9 | 4.39 | ||||||||||||
| Organic phosphoric acids and derivatives | 8.74 | 8.28 | NA | ||||||||||||
| Benzothiazepines | 22.99 | 22.49 | 2.09 | ||||||||||||
| Bilirubins | 10.37 | 13.28 | NA | ||||||||||||
| Dihydrofurans | 10.09 | 6.84 | 2.42 | ||||||||||||
| Alkyl halides | 7.17 | 5.97 | 2.77 | ||||||||||||
| Sulfinic acids and derivatives | 9.57 | 21.87 | NA | ||||||||||||
| Azoles | 17.19 | 12.88 | 7.62 | ||||||||||||
| Azolidines | 14.11 | 17 | NA | ||||||||||||
| Cinnamic acids and derivatives | 48.32 | 49.73 | NA | ||||||||||||
| Peptidomimetics | 22.76 | 21.34 | NA | ||||||||||||
| Piperidines | 53.36 | 63.22 | NA | ||||||||||||
| Pyrrolidines | 10.55 | 13.2 | NA | ||||||||||||
| Coumarins and derivatives | 30.57 | 17.89 | NA | ||||||||||||
Notes: “High” precision is shown in green (≤10%), “moderate” in yellow (10% < x < 20%), and “low” in red (≥20%).
Table 3.
Reporting Accuracy (%) compared with NIST Metabolites in Frozen Human Plasma (SRM 1950).
| Accuracy (%) | Biocrates | HMT | Nightingale | ||||
|---|---|---|---|---|---|---|---|
| Analyte | NIST Value (uM) | Reported Value (uM) | Percent Difference | Reported Value (uM) | Percent Difference | Reported Value (uM) | Percent Difference |
| Fatty Acids | |||||||
| C18:2 n-6 (Z,Z)-9,12-Octadecadienoic Acid (Linoleic Acid) | 2838 | 2960 | 4.30% | ||||
| C22:6 n-3. (Z,Z,Z,Z,Z,Z)-4,7,10,13,16,19-Docosahexaenoic Acid (DHA) | 118 | 136 | 15.25% | ||||
| Amino Acids | |||||||
| Alanine | 300 | 331 | 10.17% | 211 | −29.61% | 312.246 | 4.08% |
| Glycine | 245 | 288 | 17.72% | 250 | 1.97% | 240.87 | −1.69% |
| Histidine | 72.6 | 80 | 10.08% | 59 | −18.25% | 70.0707 | −3.48% |
| Isoleucine | 55.5 | 66 | 18.92% | 46 | −17.02% | 44.604 | −19.63% |
| Leucine | 100.4 | 114 | 13.05% | 102 | 1.25% | 87.7893 | −12.56% |
| Lysine | 140 | 151 | 7.73% | 129 | −7.60% | ||
| Methionine | 22.3 | 22 | −0.94% | 14 | −38.50% | ||
| Proline | 177 | 199 | 12.30% | 138 | −22.19% | ||
| Serine | 95.9 | 98 | 2.14% | 69 | −27.57% | ||
| Threonine | 119.5 | 127 | 6.10% | 92 | −23.08% | ||
| Tyrosine | 57.3 | 61 | 6.48% | 49 | −13.95% | 61.8318 | 7.91% |
| Valine | 182.2 | 174 | −4.40% | 152 | −16.30% | 185.06 | 1.57% |
| Arginine | 81.4 | 95 | 16.45% | ||||
| Cysteine | 44.3 | 46 | 4.50% | ||||
| Cystine | 7.8 | 8.0 | 2.76% | ||||
| Phenylalanine | 51 | 57 | 12.54% | 47 | −8.27% | 53.0223 | 3.97% |
| Clinical Markers | |||||||
| Creatinine | 60 | 65 | 7.69% | 43 | −28.14% | 58.1642 | −3.06% |
| Glucose | 4560 | 4679.41 | 2.62% | ||||
| Homocysteine | 8.5 | 8.5 | 0.58% | ||||
| Cortisol | 0.23 | 0.19 | −17.92% | ||||
| Cholesterol | 3917 | 3620 | −7.58% | ||||
Notes: “High” accuracy is shown in green (≤10%), “moderate” in yellow (10% < x < 20%), and “low” in red (≥20%). Accuracy assessed only for vendors that reported quantitative units and not relative units; accuracy estimated using Shipment 1 data. All percent accuracy values are versus NIST COA values, such that a negative value is below the NIST-provided reference value.
Figure 4.
Platform-specific, log-transformed, average metabolite levels in control samples for vendors reporting absolute units; each point represents mean ± SEM for 11 control samples in total: 9 control samples from 6 individuals (with 3 technical replicates), and 2 NIST pooled reference plasma samples. Each panel depicts the range of covered metabolites, across all assays, for an exemplar metabolite class: (A) amino acids, (B) fatty acids, (C) lysophosphatidylcholines (LPC), (D) hydroxy acids, (E) ceramides, and (F) triglycerides. Depicted data are from the second sample shipment. NIST = concentrations reported in the National Institute of Standards and Technology (NIST) SRM 1950 Certificate of Analysis (COA, revised June 2020).
Text Correction
There was an error in the original publication. NIST standards were used for part of the study to compare the accuracy of the different measurements. In the measurement of fatty acids and cholesterol, the NIST standards reflect the total concentration of fatty acids and cholesterol. The vendors in the published manuscript were reporting free fatty acids and free cholesterol, and therefore the comparison represented in Table 3 is not valid. This discrepancy was brought to our attention after the paper was published. The fatty acid and cholesterol values and percent accuracy for Biocrates and Lipotype were removed, and mention of these measurements was removed from the results section.
Updated links and data location information have been added to the appropriate sections.
A correction has been made to
1. Section 2.4, First Paragraph:
The accuracy of metabolite measurements, in comparison to NIST SRM 1950 pooled reference plasma reference values in the NIST certificate of analysis (COA), is provided in Table 3. Assessments of accuracy were constrained by (i) the fraction of classes represented in the NIST COA, (ii) vendor-specific coverage of metabolites, and (iii) the use of relative units which excluded Metabolon. Accuracy was evaluated for a set of amino acids listed in the NIST COA, which showed roughly similar high or moderate accuracy across plat-forms. Normalization methods informed by platform-specific normative levels could inform efforts to compare or merge datasets across metabolomics approaches. The majority of metabolites across all platforms were detected with excellent linearity across the dilution curve (i.e., coefficient of determination values near 1, suggesting that abundance is not a core obstacle in current metabolomics technologies; depicted in Supplementary Figure S1).
2. Supplementary Materials:
The following are available online at https://www.mdpi.com/article/10.3390/metabo11090609/s1, Figure S1: Linearity across the dilution curve of NIST pooled reference plasma (SRM 1950), Table S1: Metabolites and lipids implicated in posttraumatic stress disorder (PTSD) in previously published metabolomics studies, Table S2: List of samples sent to each metabolomics vendor in two identical shipments, Table S3: Metabolomics bake-off sample information.
3. Data Availability Statement:
Data and the Metabolomics Platform Exploration Tool will be made available in the BRAIN Commons, a cloud-based platform for computational discovery designed for the brain health community at https://www.braincommons.org/publications/doi-10-3390-metabo-11090609/ accessed on 28 September 2022.
The authors state that the scientific conclusions are unaffected. This correction was approved by the Academic Editor. The original publication has also been updated.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
Reference
- 1.Chaby L.E., Lasseter H.C., Contrepois K., Salek R.M., Turck C.W., Thompson A., Vaughan T., Haas M., Jeromin A. Cross-Platform Evaluation of Commercially Targeted and Untargeted Metabolomics Approaches to Optimize the Investigation of Psychiatric Disease. Metabolites. 2021;11:609. doi: 10.3390/metabo11090609. [DOI] [PMC free article] [PubMed] [Google Scholar]