Abstract
Cervical cancer is the most prevalent tumor in developing countries and the second most frequent cancer among females worldwide. Specific human papillomaviruses (HPVs) and, most notably, HPV types 16 and 18 are recognized as being causally associated with this malignancy. Antibodies against early HPV proteins E6 and E7 have been found more often in patients with tumors than in controls. Existing peptide enzyme-linked immunosorbent assays (ELISAs) for the detection of anti-E6 and anti-E7 antibodies in human sera have low levels of sensitivity and specificity and thus are not suitable for use as diagnostic tools. Based on highly purified recombinant native proteins, we developed four sandwich ELISAs for the detection of antibodies against HPV type 16 and 18 E6 and E7 proteins. We demonstrate their sensitivities and high degrees of specificity for cervical cancer. Among a total of 501 serum specimens from unselected patients with invasive cervical cancer, 52.9% reacted positively in at least one of the four assays. In contrast, among 244 serum specimens from control subjects without cervical cancer, only 2 reactive serum specimens (0.8%) were found. For 19 of 19 antibody-positive patients, the HPV type indicated by seroreactivity was identical to the HPV DNA type found in the tumor, which also indicates a high degree of specificity for antibody detection with respect to HPV type. In a direct comparison of 72 serum specimens from patients with cervical cancer, 56% of the specimens reacted in at least one of the four protein ELISAs, whereas 40% reacted in at least one of seven peptide ELISAs covering the four antigens. These assays could be of value for the detection of invasive cervical cancer in settings in which cytology-based early tumor screening is not available, for the clinical management of patients diagnosed with cervical cancer, and for the immunological monitoring of E6 and E7 vaccination trials.
Certain types of human papillomaviruses (HPVs), mainly HPV types 16 and 18, have been recognized as major etiological factors for the development of cervical cancer (4, 10). Parts of the viral genomes are specifically expressed in tumor tissues. The active molecules are the early HPV proteins E6 and E7, which have oncogenic properties in human keratinocytes (1). They interact with different cellular proteins which are known to be involved in the control of the cell cycle and of DNA repair, most notably, the tumor suppressor proteins p53 and Rb (12, 25). Antibodies against the E6 and E7 proteins of HPV types 16 and 18 have been found to be strongly associated with cervical cancer (9, 17, 24) but the value of E6- and E7-specific serology for the diagnosis of this disease is still questionable. Peptide enzyme-linked immunosorbent assays (ELISAs) that use small, linear epitopes of the proteins for antibody detection have low levels of sensitivity and specificity for the detection of disease (17, 24). Radioimmunoprecipitation assays (RIPAs) with whole native proteins can also detect antibodies against conformational epitopes and have increased sensitivity and disease specificity (17, 24). However, the handling procedures for RIPAs are complex and require sophisticated laboratory methods and therefore are not suited for routine testing of large numbers of samples. In the present study sandwich ELISAs with full-length, native recombinant E6 and E7 proteins of HPV types 16 and 18 have been developed. With a large series of sera from unselected cervical cancer patients and control subjects, the sensitivity of the assay for invasive cervical cancer was 53%, with a specificity of disease detection of greater than 99% (2 positive subjects among 244 control subjects).
MATERIALS AND METHODS
Sandwich protein ELISAs.
The affinity-purified mouse monoclonal tag antibody was coated overnight at 4°C to the solid phase of a 96-well Polysorb plastic plate (Nunc, Roskilde, Denmark) (500 ng/100 μl/well in 0.05 M carbonate buffer [pH 9.6]). After blocking the plate with phosphate-buffered saline (PBS) containing 0.2% (wt/vol) casein and 0.05% (vol/vol) Tween 20 (1 h at 37°C) purified and refolded E6-tag or E7-tag fusion proteins were bound to the capture antibody via their tag peptide (1 h at room temperature). The tag peptide consists of the carboxy-terminal undecapeptide (amino acid sequence, KPPTPPPEPET) of the simian virus 40 large T antigen. E7-tag proteins were diluted 1:100 (200 ng/100 μl/well) in blocking buffer, E6-tag proteins were used undiluted (300 ng/100 μl/well) in refolding buffer. In each well 100 μl of human serum diluted 1:50 in blocking buffer was incubated for 1 h at room temperature. Bound human antibodies were detected by donkey anti-human immunoglobulin G polyclonal antibody conjugated to horseradish peroxidase (diluted 1:10,000 in blocking buffer and incubation for 1 h at room temperature; Dianova, Hamburg, Germany) by using tetramethylbenzidine as the substrate. All washing steps to remove excess reagents were done with PBS (pH 7.2) containing 0.05% (vol/vol) Tween 20. After 8 min the enzyme reaction was stopped with sulfuric acid and the absorbance at 450 nm was determined.
Background reactions of the individual human serum specimens, e.g., reactivity with the capture antibody, were determined in control wells without the antigen. Control wells were coated with tag antibody and blocked, and instead of the antigen solution only the respective buffer (blocking buffer for E7 and E6 refolding buffer for E6) was used. For each serum specimen and assay the specific reactivity (net optical density [OD]) was calculated as the absorbance in the antigen-containing well minus the absorbance in the respective control well.
The cutoff value to define antibody-positive sera was calculated separately for each protein as the arithmetic mean of the net OD values of all control sera plus three standard deviations, excluding the outliers, as described previously (16). To reduce the probability of false-positive reactions, borderline sera with net OD values of <0.150 and >0.025 were retested in duplicate and were classified according to the mean net OD of the duplicates by using the same cutoff value. Of 2,980 initial reactions (745 serum specimens from Tanzania and Mexico were tested in the four assays), 430 (14.4%) were classified as borderline.
Capture antibody.
Mouse monoclonal antibody which recognizes the tag peptide was purified from a cell culture supernatant of the hybridoma cell line KT3 (14) by affinity chromatography. Anti-tag antibody was bound to tag-Sepharose and was eluted with 0.1 M glycine (pH 2.7). The eluate was adjusted to pH 7.2, dialyzed against PBS (pH 7.2), and stored at −20°C.
Recombinant E6-tag and E7-tag protein expression, purification, and refolding.
Recombinant E6-tag and E7-tag fusion proteins were expressed in Schizosaccharomyces pombe (mutant leu1-32). Cells were transformed by heat shock with the S. pombe shuttle vector pREP-L20, as modified by Tommasino et al. (23), containing the E6 or E7 DNA sequences of the original HPV type 16 (HPV-16) isolate (10) or of the HPV-18 isolate from the cervical cancer cell line SW 756 (11), upstream of the tag-coding sequence. The following HPV sequences were used in the constructs: nucleotides (N) 83 to 556 for HPV-16 E6, N 562 to 855 for HPV-16 E7 (sequence numbering is from a previous report [19]), N 105 to 578 for HPV-18 E6, and N 590 to 904 for HPV-18 E7 (7). Proteins were purified from cell extracts under denaturing conditions. E7-tag proteins were purified and refolded as previously described in detail (5). Briefly, the proteins were extracted under denaturing conditions in 8 M urea, purified by hydroxyapatite and anion-exchange chromatography, and refolded by dialysis. The chromatographic purification and refolding of E6-tag proteins are described in detail elsewhere (15). The identities of the purified proteins were verified by Western blotting and immunostaining with the anti-tag antibody and rabbit polyclonal anti-E6 and anti-E7 hyperimmune sera generated by immunization with ovalbumin-peptide conjugates or bacterial fusion proteins. E6-tag proteins were refolded overnight directly before use in ELISA; refolded E7-tag proteins were stored at −70°C.
E6-tag and E7-tag functionality assays.
The E6 function to facilitate ubiquitination of p53 was assayed as described previously (18), and data are presented elsewhere (15). The E7 function to bind to the Rb protein was shown for the purified and refolded E7 tag proteins by precipitation with glutathione S-transferase (GST)–Rb fusion protein bound to glutathione-Sepharose (data are presented elsewhere [5]). Precipitated E7-tag was identified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting of blots immunostained with monoclonal anti-tag antibody. The specificity of binding was shown by control precipitations without E7-tag, without GST-Rb, or with GST alone.
Human sera.
Sera were collected from patients with clinically diagnosed invasive cervical cancer at the Hospital de Oncologica in Mexico City, Mexico, in 1992 and at the Tanzania Tumor Center, Ocean Road Hospital in Dar es Salaam, Tanzania, from 1988 to 1991. Sera were collected from age-matched control subjects at the same time that sera were obtained from the patients. In Tanzania sera were obtained from female patients with nongynecological tumors or from gynecological inpatients without malignant tumors who visited the same institution. In Mexico sera were collected from healthy women in the same area. After collection, the sera were stored at −20°C until transport (at ambient temperature) to the laboratory. The sera were then heat inactivated (56°C, 30 min) and stored in aliquots at −20°C until use. Sera from these collections have been used in previous studies (2, 17, 22). In this study the mean age of the Mexican patients was 52.7 years (range, 29 to 81 years), and the mean age of the Mexican controls was 49.3 years (range, 26 to 69 years); for the Tanzanian patients the mean age was 47.9 years (range, 29 to 75 years), and for the Tanzanian controls the mean age was 43.6 years (range, 30 to 69 years). To test for native protein conformation after refolding, sera known from RIPAs to contain antibodies against conformational HPV-16 E6 (n = 7) and E7 (n = 3) epitopes were used. For validation of the sandwich protein ELISAs, 72 serum samples from patients with cervical carcinoma for which peptide ELISA data were available and 25 serum samples from patients with cervical carcinoma who had HPV-16 or -18 DNA-positive tumors were used. To determine the sensitivities and specificities of the four protein ELISAs for cervical cancer, 129 serum samples from Mexican cervical cancer patients, 372 serum samples from Tanzanian cervical cancer patients, 59 serum samples from Mexican controls and 185 serum samples from Tanzanian controls were used.
RESULTS
Principle, design, and optimization of the sandwich protein ELISAs.
We used complete native recombinant proteins purified from the fission yeast S. pombe to establish a new generation of HPV ELISAs. The proteins carry at their carboxy termini an undecapeptide (tag) derived from the simian virus 40 large T antigen. This tag mediates attachment to ELISA plates coated with affinity-purified tag-specific monoclonal antibodies. The capture method was chosen to prevent partial denaturation of the small E6 and E7 proteins. It has been described that for other proteins denaturation can occur as a consequence of direct binding to plastic surfaces and can decrease sensitivity and specificity (8). We compared antibody reactivities after direct binding of unfused HPV-16 E7 protein and indirect binding of HPV-16 E7-tag fusion protein to the plate. With 73 serum samples from patients with cervical carcinoma and 60 control serum samples we found an increased sensitivity (38 versus 27%) and specificity (100 versus 97%) of the sandwich ELISA.
During establishment of optimized conditions for the assays, all parameters were evaluated by ELISA, with different sets of appropriate sera used as internal standards. The antibody purification protocol and the appropriate blocking reagent yielding the lowest background reactivity with a set of 11 human serum samples were determined first. The amount of the capture antibody necessary for saturating the protein binding capacity of the plate was evaluated with HPV-16 E7-tag as the antigen and four serum samples positive by the peptide ELISA for the HPV-16 E7 (16). To identify the optimal renaturation conditions for E6 and E7 proteins, equal amounts of each of the purified HPV-16 antigens were subjected to different protocols and were subsequently tested in the sandwich ELISA. For HPV-16 E6, six serum samples were used, and for HPV-16 E7, three serum samples were used; all serum samples were assumed to react with conformational epitopes only due to their positive reactions by RIPAs (17) but negative reactions by the peptide ELISA (16) and Western blotting. The optimal renaturation protocols found with the HPV-16 proteins were also used with the respective HPV-18 proteins. The amounts saturating the capture antibody in the plates were determined by titration of all four antigens by using two reactive serum samples for each antigen.
Recombinant E6-tag and E7-tag protein expression, purification, and refolding.
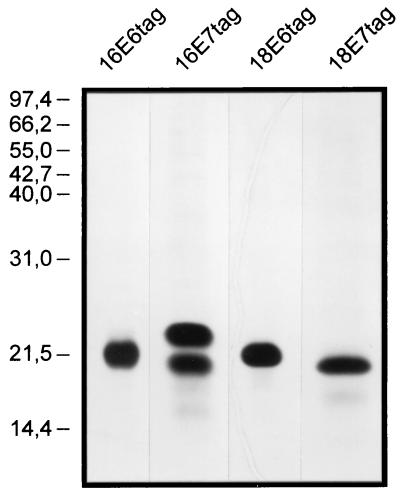
HPV E7 proteins are phosphorylated in human tissues (20, 21). To ensure correct protein folding and epitope presentation, S. pombe was chosen as the expression system because of its reported ability to phosphorylate recombinant HPV-16 E7 protein (23). The E6 and E7 proteins of HPV types 16 and 18 were expressed with the tag peptide fused to their carboxy termini. Under denaturing conditions, the recombinant proteins were purified in two chromatographic steps to greater than 99% purity (Fig. 1) and were refolded in physiological buffers. E7 fusion proteins of both HPV types were purified and refolded by following an established general protocol (5). For the E6 fusion proteins, a different, generally applicable purification and refolding protocol was established (15). Like cervical cancer cells, S. pombe coexpresses minor amounts of an N-terminal truncated variant HPV-16 E7 that starts from a second, internal ATG codon. A slightly smaller HPV-16 E6 protein that probably also represents an N-terminal truncated variant is coexpressed. These variant proteins were copurified (Fig. 1).
FIG. 1.
Purified recombinant E6-tag and E7-tag fusion proteins of HPV-16 (16E6tag and 16E7tag) and HPV-18 (18E6tag and 18E7tag) from S. pombe in silver-stained sodium dodecyl sulfate-polyacrylamide gels. The positions of size markers (in kilodaltons) are shown on the left.
After refolding by the final protocols, the native conformations of the recombinant proteins were evaluated by three criteria: solubility, interaction with specific cellular proteins, and reaction with antibodies against conformational epitopes. First, no loss of reactivity in the respective ELISA was observed after spinning the proteins at 100,000 × g for 1 h (data not shown). Second, both refolded E7-tag proteins were specifically precipitated by Rb (5), and both refolded E6-tag proteins catalyzed the ubiquitination of p53 (15). Third, for the HPV-16 proteins, sera positive by RIPA but negative by peptide ELISA and Western blotting reacted with the respective refolded protein in the protein ELISA. Six E6 antibody-positive serum samples showed net OD values of between 0.2 and 0.55, and three E7 antibody-positive serum samples showed net OD values of between 0.2 and 0.5.
Specificity and sensitivity for cervical cancer.
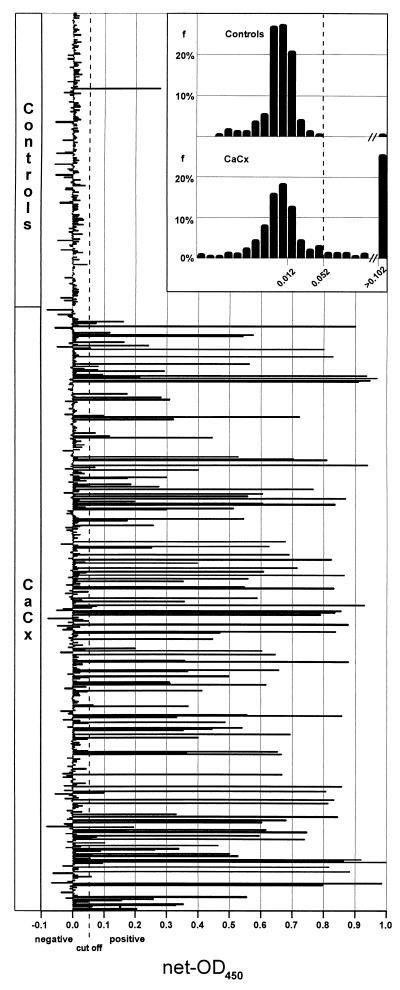
The prevalence of antibodies to the four proteins was determined with a group of 501 serum samples from Tanzanian (n = 372) and Mexican (n = 129) patients clinically diagnosed with invasive cervical cancer (the serum samples were untyped with respect to HPV DNA type) and a group of 244 serum samples from age-matched Tanzanian (n = 185) and Mexican (n = 59) control patients attending the same hospital for a variety of other diseases. To classify the sera as antibody positive or negative, a cutoff value was calculated from the reactions of the control sera (Fig. 2). All four protein ELISAs showed an extremely high specificity for cervical cancer (>99%) (Table 1). Of the 244 control serum samples, only 2 reacted positively (one assay each). Overall, 53% of the serum samples from patients with cervical carcinomas reacted in at least one of the assays, with 35% being HPV-16 E6 and/or E7 antibody positive and with 20% recognizing at least one of the two HPV-18 proteins. In the case of HPV-16, the prevalence of E6 reactions was nearly twice as high as that of E7 reactions, whereas for HPV-18, the frequency of E7 reactions surpassed that for E6 reactions. There were no gross differences in reactivities with respect to the geographic origins of the sera. For the four assays in combination, a sensitivity for cervical carcinoma detection of 53% and a specificity of greater than 99% were calculated. The association of HPV seroreactivity with cervical cancer is highly significant (odds ratio, 135; 95% confidence interval, 36 to 1,137; P < 0.001).
FIG. 2.
HPV-16 E6 antibody reactivities in sera from Tanzanian and Mexican cervical cancer (CaCx) patients (n = 501) and controls (n = 244) measured by sandwich protein ELISA. Each bar represents the net OD value for an individual serum specimen. The cutoff value of 0.052 for sera with positive reactions is indicated by an interrupted vertical line. (Inset) Frequency distribution of net OD values. Each bar represents the frequency of sera (f; percentage of all sera from the group) with net OD values, in ranges of 0.01. Sera with net OD values greater than 0.102 are summarized in one bar.
TABLE 1.
Sensitivity and cervical cancer specificity of the E6 and E7 sandwich protein ELISAs
| Serum group | % Positive sera
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| HPV-16 ELISA
|
HPV-18 ELISA
|
Any ELISA | |||||||
| E6 | E7 | E6 and E7 | E6 and/or E7 | E6 | E7 | E6 and E7 | E6 and/or E7 | ||
| Mexico | |||||||||
| Patients with CaCxa (n = 129) | 25.6 | 18.6 | 11.6 | 32.6 | 7.8 | 15.5 | 4.7 | 18.6 | 50.4 |
| Controls (n = 59) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Tanzania | |||||||||
| Patients with CaCx (n = 372) | 30.4 | 16.9 | 11.8 | 35.5 | 13.2 | 14.2 | 7.5 | 19.9 | 53.8 |
| Controls (n = 185) | 0.5 | 0 | 0 | 0.5 | 0 | 0.5 | 0 | 0.5 | 1.1 |
| Total | |||||||||
| Patients with CaCx (n = 501) | 29.1 | 17.4 | 11.8 | 34.7 | 11.8 | 14.6 | 6.8 | 19.6 | 52.9 |
| Controls (n = 244) | 0.4 | 0 | 0 | 0.4 | 0 | 0.4 | 0 | 0.4 | 0.8 |
CaCx, cervical carcinoma.
HPV type specificity.
Only about one-third of the serum samples reactive with one protein also reacted with the second protein of the same HPV type, indicating, as has been seen previously (17), an independent antibody response to E6 and E7 in the majority of the patients. There was a very high HPV type specificity in the protein ELISAs. Of the total of 265 positive serum samples only 7 (2.6%) reacted with proteins from both HPV types. For three samples high reactivity with one protein and low reactivity with the corresponding protein of the other HPV type was present, consistent with antibody cross-reactivity. For the other four samples the reactions with proteins from both HPV types were rather strong, which could be the result of early-region expression of both HPV types in the cervical carcinoma or an additional HPV-related lesion.
There was a strong correlation between the HPV type determined by DNA analysis of the tumor and the HPV type reactivity of the corresponding serum sample (Table 2). Of 19 serum samples from patients with HPV-16 DNA-positive tumors, none reacted in the HPV-18 ELISAs but 15 of them reacted in at least one of the HPV-16 ELISAs. Similarly, none of six serum samples from patients with HPV-18 DNA-positive tumors reacted in the HPV-16 ELISAs, whereas four of them reacted in at least one of the HPV-18 ELISAs. In summary, for 19 of 19 antibody-positive samples, the HPV type indicated by seroreactivity was identical to the HPV DNA type in the tumor.
TABLE 2.
HPV DNA type and seroreactivity
| DNA type in tumor | No. of positive serum specimens
|
|||||
|---|---|---|---|---|---|---|
| HPV-16 ELISA
|
HPV-18 ELISA
|
|||||
| E6 | E7 | E6 and/or E7 | E6 | E7 | E6 and/or E7 | |
| HPV-16 DNA (n = 19) | 10 | 10 | 15 | 0 | 0 | 0 |
| HPV-18 DNA (n = 6) | 0 | 0 | 0 | 4 | 2 | 4 |
Comparison with peptide ELISA.
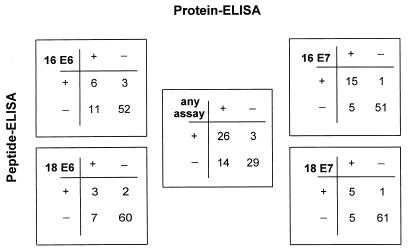
The sensitivities of the four protein ELISAs were further validated with a selected set of 72 serum samples from Mexican patients with cervical cancer for which ELISA data with peptides were available for comparison (13, 16). A similar picture was found for all four HPV antigens. A substantial number of serum samples reacted only in the protein ELISA, and some peptide ELISA-positive sera did not react in the corresponding protein ELISA (Fig. 3). The most striking difference was seen for HPV-16 E6, in which 11 of 17 protein ELISA-positive serum samples were detected only by this assay and 3 of 9 peptide ELISA-positive serum samples did not react in the protein ELISA. The low level of agreement of the two types of tests for E6 antibodies is demonstrated by Cohen’s (6) exact kappa value, which was only 0.36 (95% confidence interval, 0.10 to 0.61) for HPV-16 E6 and 0.34 (95% confidence interval, 0.01 to 0.66) for HPV-18 E6. A better agreement was found for the E7 assays, with kappa values of 0.78 (95% confidence interval, 0.61 to 0.95) for HPV-16 E7 and 0.58 (95% confidence interval, 0.28 to 0.88) for HPV-18 E7. The peptide ELISAs have also been reported to give positive reactions with a substantial fraction of serum samples from controls without cervical cancer (9, 17). This was not found with the protein ELISAs described here. We therefore assume that these peptide ELISA reactions are false positive and consider that almost no anti-E6 or anti-E7 antibodies are present in the healthy population. Such false-positive reactions are also expected to be detected by the previous tests in the groups with cervical cancer and could account in our comparison for those serum samples from patients with cervical cancer that were peptide ELISA positive but protein ELISA negative. The additional numbers of serum specimens found to be positive by the protein ELISA reflect the higher sensitivity of this assay type.
FIG. 3.
Comparison of sandwich protein and peptide ELISAs for detection of anti-HPV-16 and anti-HPV-18 E6 and E7 antibodies in sera from cervical cancer patients (n = 72). The numbers of serum samples positive or negative in the individual assays are shown in the four outer squares. In the central square (any assay) sera positive for at least one of the four antigens are grouped as positive.
DISCUSSION
The high specificity of all four sandwich protein ELISAs for cervical cancer compared to those of the peptide ELISAs and ELISAs in which the protein is coupled directly to the plate is facilitated by the background control used to correct for reactions not specific for HPV proteins. The conditions in wells containing the E6-tag and E7-tag fusion proteins bound to the anti-tag antibody and in the control wells containing only the anti-tag antibody are very similar. For negative sera this results in the very low difference in absorption (net OD) observed for the group of control sera. During development of the assays we have seen that the high biochemical purities of the antigens and the anti-tag antibody and the capture principle both contribute significantly to the low level of reactivity of control sera. This allowed a stringent definition of low cutoff values even for sera of African origin, which are known to have high background reactivities in serological assays. Thus, even weakly reacting sera from patients with cervical cancer can be reliably distinguished from negative sera. Optimized renaturation protocols for the antigens were seen to drastically enhance the reactivities of positive sera. Thus, low cutoff values and a high proportion of antigen in the native conformation contribute to the high sensitivity.
However, despite this increased sensitivity, the four assays still detected disease in only 53% of the group of unselected patients with invasive cervical cancer. In Tanzania, HPV-16 DNA has been found in 38 to 45% of biopsy specimens from patients with cervical cancer, and HPV-18 DNA has been found in 25 to 32% of biopsy specimens from patients with cervical cancer (3, 22). In Mexico, 43% of tumors have been found to contain HPV-16 DNA (17). It appears reasonable to assume that these type distributions are also valid for the large groups of patients with cervical cancer analyzed here. From the observed low rate of antibody cross-reactions between HPV-16 and HPV-18 early proteins and from the agreement of the HPV type specificity of serological and DNA data, we expect antibodies against E6 and E7 proteins from yet other HPV types to react only rarely in our assays. On the basis of these assumptions we estimate a serological HPV type-specific detection rate of 76 to 95% for HPV-16 DNA-positive patients with cervical cancer and 57 to 80% for HPV-18 DNA-positive Tanzanian patients with tumors.
The possibility that the sensitivities of our assays could be increased by technical improvements cannot be excluded. On the other hand, unreactive sera might reflect immunological nonreactivity to these HPV proteins, as has been discussed previously (24). In view of the clear HPV type specificity of our assays, higher overall detection rates might also be obtained by the development of additional protein ELISAs for E6 and E7 proteins of the less prevalent carcinoma-associated HPV types such as HPV-45. Despite the sequence heterogeneity among the E6 and E7 proteins of different HPV types, the general purification and renaturation protocols developed for E6 and E7 should be applicable without major variation. We are analyzing whether equally sensitive multiplex assays that detect antibodies against E6 or E7 proteins from different HPV types simultaneously can be developed.
We started analyzing a small number of serum samples from patients with premalignant cervical lesions. The frequencies of HPV-16 E6- and/or HPV-16 E7-positive reactions were far lower with these sera than with the sera from patients with invasive cervical carcinoma, but some were clearly positive. Studies are in progress to determine the predictive value of E6- and E7-specific antibodies for the progression of such lesions. The high degree of association of the anti-E6 and anti-E7 antibodies with disease and the absence of these antibodies in the population without cancer strongly suggest that E6 and E7 are not expressed in sufficient amounts and/or are not at the appropriate site to be accessible to the immune system during primary and latent infections. By using the newly developed assays, the time of seroconversion during the development of cervical neoplasia can now be determined.
HPV-specific sandwich protein ELISAs with high degrees of sensitivity and specificity might be of value for the detection of invasive cervical cancer in developing countries where the logistics for systematic screening by cytology are difficult to establish. In addition, they might contribute to the management and follow-up of patients diagnosed with cervical cancer and also immunological monitoring of E6- and E7-specific vaccination trials.
ACKNOWLEDGMENTS
We thank J. Chang-Claude for help with statistical analysis and R. Pipkorn for tag peptide synthesis, M. Tommasino for the S. pombe expression system, and W. Deppert for the gift of the KT3 cell line. The contributions of our clinical colleagues in establishing the serum collections in Tanzania and Mexico are gratefully acknowledged.
REFERENCES
- 1.Barbosa M S, Vass W C, Lowy D R, Schiller J T. In vitro biological activities of the E6 and E7 genes vary among human papillomaviruses of different oncogenic potential. J Virol. 1991;65:292–298. doi: 10.1128/jvi.65.1.292-298.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bleul C, Müller M, Frank R, Gausepohl H, Koldovsky U, Mgaya H N, Luande J, Pawlita M, ter Meulen J, Viscidi R, Gissmann L. HPV type 18 E6 and E7 antibodies in human sera: increased anti-E7 prevalence in cervical cancer patients. J Clin Microbiol. 1991;29:1579–1588. doi: 10.1128/jcm.29.8.1579-1588.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosch X F, Manos M M, Munoz N, Sherman M, Janson A M, Peto J, Schiffmann M H, Moreno V, Kurman R, Shah K V. Prevalence of human papillomavirus in cervical cancer; a worldwide perspective. J Natl Cancer Inst. 1995;87:796–802. doi: 10.1093/jnci/87.11.796. [DOI] [PubMed] [Google Scholar]
- 4.Boshart M, Gissmann L, Ikenberg H, Kleinheinz A, Scheurlen W, zur Hausen H. A new type of papillomavirus DNA, its presence in genital cancer biopsies and in cell lines derived from cervical cancer. EMBO J. 1984;3:1151–1157. doi: 10.1002/j.1460-2075.1984.tb01944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braspenning J, Manetti R, Zumbach K, Meschede W, Gissmann L, Tommasino M. A general purification protocol for E7 proteins from ‘high and low risk’ human papillomavirus types expressed in the yeast Schizosaccharomyces pombe. J Prot Exp Puri. 1997;10:192–201. doi: 10.1006/prep.1997.0731. [DOI] [PubMed] [Google Scholar]
- 6.Cohen J A. Coefficient of agreement for nominal scales. Ed Psychol Measure. 1960;20:37–46. [Google Scholar]
- 7.Cole S T, Danos O. Nucleotide sequence and comparative analysis of the human papillomavirus type 18 genome. J Mol Biol. 1987;193:599–608. doi: 10.1016/0022-2836(87)90343-3. [DOI] [PubMed] [Google Scholar]
- 8.Darst S A, Robertson C R, Berzofsky J A. Adsorption of the protein antigen myoglobin affects the binding of conformation-specific monoclonal antibodies. Biophys J. 1988;53:533–539. doi: 10.1016/S0006-3495(88)83133-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dillner J, Wiklund F, Lenner P, Eklund C, Fredriksson-Shanazarian V, Schiller J T, Hibma M, Hallmans G, Stendahl U. Antibodies against linear and conformational epitopes of human papillomavirus type 16 that independently associate with incident cervical cancer. Int J Cancer. 1995;60:377–382. doi: 10.1002/ijc.2910600318. [DOI] [PubMed] [Google Scholar]
- 10.Dürst M, Gissmann L, Ikenberg H, zur Hausen H. A new type of papillomavirus DNA from a cervical carcinoma and its prevalence in cancer biopsies from different geographic regions. Proc Natl Acad Sci USA. 1983;60:3812–3815. doi: 10.1073/pnas.80.12.3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dürst M, Croce C M, Gissmann L, Schwarz E, Huebner K. Papillomavirus sequences integrate near cellular oncogenes in some cervical carcinomas. Proc Natl Acad Sci USA. 1987;84:1070–1074. doi: 10.1073/pnas.84.4.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dyson N, Howley P M, Münger K, Harlow E. The human papillomavirus 16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 13.Gissmann, L., and C. Rittmüller. Unpublished data.
- 14.McArthur H, Walter G. Monoclonal antibodies specific for the carboxy terminus of simian virus 40 large T antigen. J Virol. 1984;145:483–491. doi: 10.1128/jvi.52.2.483-491.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meschede, W., J. Braspenning, M. Scheffner, L. Gissmann, and M. Pawlita. Purification and refolding of recombinant human papillomavirus early protein E6 expressed in Schizosaccharomyces pombe. Submitted for publication.
- 16.Müller M, Viscidi R P, Sun Y, Guerrero E, Hill P M, Shah F, Bosch F X, Munoz N, Gissmann L, Shah K V. Antibodies to HPV 16 E6 and E7 proteins as markers for HPV 16 associated invasive cancer. Virology. 1992;187:508–514. doi: 10.1016/0042-6822(92)90453-v. [DOI] [PubMed] [Google Scholar]
- 17.Nindl I, Benitez-Bribiesca L, Berumen J, Farmanara N, Fisher S, Gross G, Lopez-Carillo L, Müller M, Tommasino M, Vazquez-Curiel A, Gissmann L. Antibodies against linear and conformational epitopes of the human papillomavirus (HPV) type 16 E6 and E7 oncoproteins in sera of cervical cancer patients. Arch Virol. 1994;137:341–353. doi: 10.1007/BF01309480. [DOI] [PubMed] [Google Scholar]
- 18.Scheffner M, Huibregtse J M, Vierstra R D, Howley P M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 19.Seedorf K, Krammer G, Dürst M, Suhai S, Rowekamp W G. Human papillomavirus type 16 DNA sequence. Virology. 1985;145:181–185. doi: 10.1016/0042-6822(85)90214-4. [DOI] [PubMed] [Google Scholar]
- 20.Selvey L A, Dunn L A, Tindle R W, Park D S, Frazer I H. Human papillomavirus (HPV) type 18 E7 protein is a short-lived, steroid-inducible phosphoprotein in HPV-transformed cell lines. J Gen Virol. 1994;75:1647–1653. doi: 10.1099/0022-1317-75-7-1647. [DOI] [PubMed] [Google Scholar]
- 21.Smotkin D, Wettstein F O. The major human papillomavirus protein in cervical cancers is a cytoplasmic phosphoprotein. J Virol. 1987;61:1686–1689. doi: 10.1128/jvi.61.5.1686-1689.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.ter Meulen J, Eberhardt H C, Luande J, Mgaya H N, Chang-Claude J, Mtiro H, Mhina M, Kashaija P, Ockert S, Yu X, Meinhardt G, Gissmann L, Pawlita M. Human papillomavirus (HPV) infection, HIV infection and cervical cancer in Tanzania, East Africa. Int J Cancer. 1992;51:515–521. doi: 10.1002/ijc.2910510403. [DOI] [PubMed] [Google Scholar]
- 23.Tommasino M, Contorni M, Scarlato V, Bungnoli M, Maundrell K. Synthesis, phosphorylation and nuclear localization of human papillomavirus E7 in Schizosaccharomyces pombe. Gene. 1990;93:265–270. doi: 10.1016/0378-1119(90)90234-i. [DOI] [PubMed] [Google Scholar]
- 24.Viscidi R P, Yeping S, Tsuzaki B, Bosch F X, Munoz N, Shah K. Serologic response in human papillomavirus-associated invasive cervical cancer. Int J Cancer. 1993;55:780–784. doi: 10.1002/ijc.2910550515. [DOI] [PubMed] [Google Scholar]
- 25.Werness B A, Levine A J, Howley P M. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]