Abstract
The accuracy of pooling urine samples for the detection of genital Chlamydia trachomatis infection by ligase chain reaction (LCR) was examined. A model was also developed to determine the number of samples to be pooled for optimal cost savings at various population prevalences. Estimated costs included technician time, laboratory consumables, and assay costs of testing pooled samples and retesting individual specimens from presumptive positive pools. Estimation of population prevalence based on the pooled LCR results was also applied. After individual urine specimens were processed, 568 specimens were pooled by 4 into 142 pools and another 520 specimens were pooled by 10 into 52 pools. For comparison, all 1,088 urine specimens were tested individually. The sample-to-cut-off ratio was lowered from 1.0 to 0.2 for pooled samples, after a pilot study which tested 148 samples pooled by 4 was conducted. The pooling algorithm was 100% (48 of 48) sensitive when samples were pooled by 4 and 98.4% (61 of 62) sensitive when samples were pooled by 10. Although 2.0% (2 of 99) of the negative pools of 4 and 7.1% (1 of 14) of the negative pools of 10 tested presumptive positive, all samples in these presumptive-positive pools were negative when retested individually, making the pooling algorithm 100% specific. In a population with 8% genital C. trachomatis prevalence, pooling by four would reduce costs by 39%. The model demonstrated that with a lower prevalence of 2%, pooling eight samples would reduce costs by 59%. Pooling urine samples for detection of C. trachomatis by LCR is sensitive, specific, and cost saving compared to testing individual samples.
There are 89.9 million cases of genital Chlamydia trachomatis infection every year worldwide (13), 4.5 million of which occur in the United States (4). Although many C. trachomatis infections are asymptomatic (16), the sequelae from infection, including pelvic inflammatory disease (PID), and infertility, represent a large burden for populations worldwide. Furthermore, inflammatory sexually transmitted diseases, such as those caused by C. trachomatis, increase the risk of both human immunodeficiency virus (HIV) transmission and infection (7, 11). Together, the high percentage of asymptomatic infections, the sequelae of infections, and the increased association with HIV transmission underscore the importance of screening as a necessary intervention to reduce the burden of diseases caused by C. trachomatis.
Detection of genital C. trachomatis infection by ligase chain reaction (LCR) with first-void urine is a noninvasive, highly sensitive, and highly specific procedure (2, 8). Although the cost of LCR is higher than that of other tests such as direct fluorescent antibody, antigen detection by enzyme immunoassay, and nucleic acid probe tests, LCR is more sensitive and more specific (15, 17). Culture has been considered to be the “gold standard” in the past but costs more and is less sensitive than either LCR or PCR (3, 5, 10, 12, 14).
Pooling serum samples for HIV testing was found to be accurate and has been used to reduce the cost of enzyme-linked immunosorbent assays for detection of antibody to HIV (1, 6). Pooling for HIV testing has been used to develop both population estimates and, in a multiple-step procedure, to determine which individual sample is positive. Pooling has also been applied to the PCR detection of C. trachomatis in endocervical and urethral scrapes (9), but in that study the sample size was small. The investigators acknowledged the need for subsequent studies to rule out the possibility of reduced sensitivity by diluting out individual specimens in the pool.
The screening of women at risk for C. trachomatis has been recommended by the Institute of Medicine as a cost-effective program which would prevent the high cost of untreated infections which lead to PID (4). As a screening and treatment intervention reduces the prevalence of C. trachomatis infection over time, the cost per specimen tested with the pooling protocol algorithm would be further decreased. The reduction in price occurs for two reasons: (i) as prevalence decreases, pooling a greater number of samples increases cost savings and (ii) the samples from fewer pools would test presumptive positive such that fewer samples would be retested individually. Therefore, the cost for finding one case does not increase dramatically as prevalence decreases, as is the case when samples are tested individually.
In this study we examined the accuracy and cost-saving ability of pooling urine specimens for the detection of genital C. trachomatis infections by LCR. A cost analysis of the pooling protocol was conducted to determine the number of specimens it would be necessary to pool in order to provide the highest cost savings, taking into account the prevalence of infection in the population screened.
MATERIALS AND METHODS
Sample size and parameters.
As part of an ongoing study to determine chlamydia prevalence in asymptomatic U.S. Army females with a mean age (± standard deviation [SD]) of 22 (±4 years), urine samples were tested by LCR to ascertain genital C. trachomatis infection. A sample of 568 processed urine specimens was pooled by 4 into 142 pools, and 520 specimens were pooled by 10 into 52 pools. Pools were formed by order of consecutive laboratory accession number. All 1,088 pooled urine samples were also tested individually. For all discrepant individual and pool results, both the individual samples and the pools were retested to confirm results.
Urine specimen, collection, preparation and assay setup.
Specimen, collection, preparation, and assay setups were performed according to the manufacturer’s instructions for the urine-based chlamydia LCR assay (Abbott Laboratories, Abbott Park, Ill.).
Specimens were refrigerated immediately after collection and shipped overnight delivery with wet packs to maintain refrigerator temperature. Specimens were either processed immediately on arrival at the laboratory or refrigerated and processed within 2 days. The total time before processing never exceeded 4 days as per the LCR package insert. Processed refrigerated specimens were amplified the day after processing. Processed urine can be refrigerated or frozen for up to 60 days before testing. We refrigerated our processed specimens for up to 7 days in case retesting was needed.
One milliliter of urine was centrifuged at ≥9,000 × g for 15 min (±2 min) at room temperature. The supernatant was removed, and the pellet was resuspended into 1.0 ml of LCR urine specimen resuspension buffer and vortexed. Preparations were then boiled at 97°C (±2°C) for 15 min (±1 min) to extract the DNA and stored at 2 to 8°C for up to 7 days until tested. Processed urine specimens were subsequently tested individually and tested pooled.
When specimens were tested individually, a volume of 100 μl of processed urine specimen was placed into its own LCR chlamydia amplification vial (unit dose). For each pool of four, 25 μl of each of the four processed specimens was placed into a single unit dose. For each pool of 10, 10 μl of each of the ten processed specimens was placed into a single unit dose. The total specimen volume was then 100 μl for each unit dose. Two negative controls, two positive calibrators, and a positive processing control were included in every amplification run in accordance with the manufacturer’s instructions.
DNA amplification and detection.
Unit dose tubes containing DNA preparations were amplified under the following conditions: 40 cycles of denaturation at 93°C for 1 s, annealing at 59°C for 1 s, extension at 62°C for 1 min, 10 s, and soaking at 25°C in an LCR thermocycler (Abbott Laboratories). Amplified DNA was detected in an LCR-automated machine which performed a particle-based enzyme immunoassay with a fluorescent signal. For individually tested samples, a sample-to-cutoff ratio (S/CO) of ≥1.0 was considered positive, and borderline negative samples (0.80 to 0.99 S/CO) were retested, according to manufacturer’s instructions.
Pilot study.
Because the volume for each individual urine specimen is decreased in the pooled assay, a pilot study was conducted to determine an appropriate S/CO for the pooled assays. The desired S/CO would detect all positive pools while not detecting most, if not all, negative pools. The pilot study consisted of 148 processed urine samples from the ongoing study of female U.S. Army recruits. The technician, blinded to the individual test results, pooled and tested these 148 samples by four. By lowering the S/CO from 1.0 to 0.2, all of the positive pools were detected (100%) (25 of 25) and only 2.7% (1 of 37) of the negative pools tested presumptive positive. Since all pools which test positive are retested, specificity with the pooling algorithm is 100%, i.e., no different than with testing processed specimens individually.
Cost analysis.
A model was developed to determine the pool size that yielded the highest cost savings. The binomial distribution was used to estimate the number of pools that are likely to be positive given a selected pool size and population disease prevalence. Next, the optimal pooling number for a range of disease prevalences was calculated. For a dichotomous outcome (i.e., positive or negative test result for a genital C. trachomatis infection), independence was assumed (i.e., the order of the samples received was random with regard to the distribution of the positive or negative samples in the population). The expected percentage of positive pooled assays was determined using the following equation: s = [(l − r/n)c] × 100%, where s is the expected number of positive pools, r is the number of positive samples tested, n is the total number of samples tested, r/n is the prevalence of disease, and c is the number of specimens pooled. This equation accounted for the probability that from 1 to c samples in the pool were positive.
A baseline total cost of $12.76 per individual sample which included $0.36 for laboratory consumables, $3.56 for technician cost, and $8.84 for the LCR assay was used. Laboratory consumables include gloves and supplies used for handling samples. Technician cost was calculated assuming an average of 10 runs per week (i.e., 380 samples), an annual salary of $30,000 with an additional 28% of salary in benefits, and a 69% laboratory or university overhead (i.e., $30,000 × 1.28 × 1.69 = $64,896). The cost of the five controls used for each 19 specimens tested was calculated into the LCR assay cost, in which the base cost per unit dose was $7. A sensitivity analysis was also done first, with the base cost per unit dose ranging from $5 to $15, technician cost set at $3.56, and cost for laboratory consumables set at $0.36 per specimen tested. In the second sensitivity test, the annual salary of the technician ranged from $20,000 with 20% overhead to $40,000 with 69% overhead, unit dose cost was set at $7.00, and cost for laboratory consumables set at $0.36 per specimen tested. Low technician and low assay costs as well as high technician and high assay costs were also calculated.
Estimation of population prevalence with pooled data.
Pooling can also be used to reduce the cost of estimating population prevalence. Based on calculations from a previous study, the estimated population prevalence and 95% confidence interval (CI) were back calculated from the pooled data (6). Separate estimates were made for samples pooled by 4 and by 10. Calculations were based on the following equations: (i) Estimated prevalence: p = 1 − [1 − (s/n)]1/c (ii) (SD): SD = {[(s/n) × (1 − s/n)(2/c) − 1]/(n × c2)}0.5 (iii) 95% CI: p ± 1.96 (SD) where s is the total number of presumptive-positive pools, n is the total number of pools, and c is the number of specimens in each pool.
RESULTS
Sensitivity and specificity of the pooled assays.
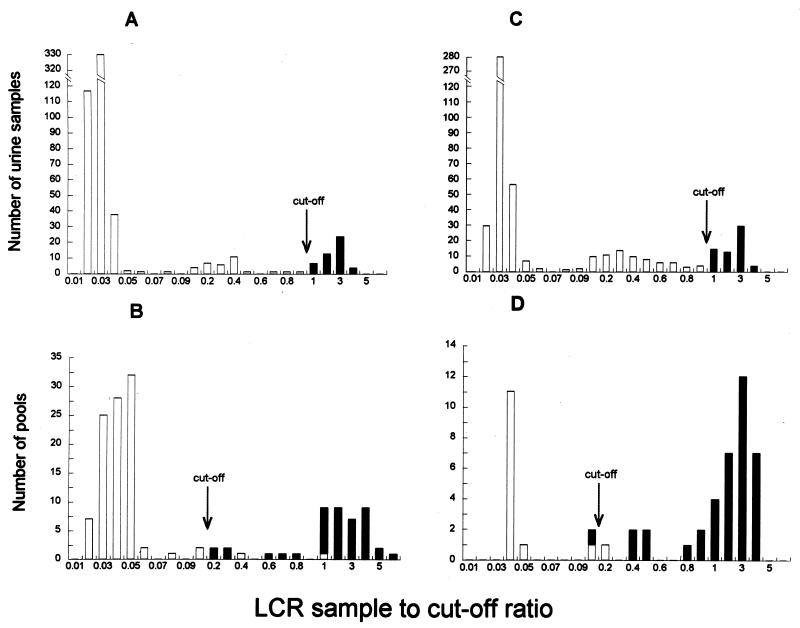
A comparison of the distribution of S/COs for the individual and pooled samples indicated that lowering the S/CO from 1.0 to 0.20 for determining positive pools resulted in high sensitivity with a low proportion of specimens from negative pools that need to be retested individually (Fig. 1). There were two weakly positive individual specimens (i.e., an S/CO of ≥1 but <2.0) in the pilot study (pooled by 4), six weak positives in the study pooled by 4, and four low positives in the study pooled by 10. These weak positives were the only positive specimens in the pool. These pools all tested between 0.2 and 1.0 and sometimes higher.
FIG. 1.
Detection of genital C. trachomatis infection by LCR. Graphs A and C show the distribution of individual urine samples from two groups of 568 and 520 women taken from the study population. Graph B shows the distribution of the samples in A pooled by 4 (n = 142), and graph D shows the distribution of the samples in C pooled by 10 (n = 52). The S/CO for individual samples, which was 1.0, was lowered to 0.2 for pooled samples, as indicated.
The pooling algorithm was 100% (48 of 48) sensitive when pooling by 4 and 98.4% (61 of 62) sensitive when pooling by 10 (Table 1). Although 2.0% (2 of 99) of the negative pools of four and 7.1% (1 of 14) of the negative pools of 10 tested presumptive positive, all of the samples in these pools would be retested individually, according to the pooling algorithm. Retesting the individual samples in the presumptive-positive pools resulted in no false-positive specimens (100% specificity).
TABLE 1.
Accuracy of pooling urine samples for the detection of genital C. trachomatis infection in asymptomatic women by LCRa
| Parameter | Result for pool size with indicated no. of samples
|
|
|---|---|---|
| 4 | 10 | |
| Total no. of urine samples | 568 | 520 |
| No. of positive specimens (%) | 48 (8.6) | 62 (11.9) |
| No. of pools | 142 | 52 |
| No. of presumptive-positive pools | 45 | 39 |
| Estimated population prevalence calculated from pooled data (95% CI) | 9.1 (6.5, 11.6) | 12.9 (8.8, 17.0) |
| No. of samples retested individually (%) | 180 (31.7) | 390 (75.0) |
| No. of positive pools (%) | 43 (30.3) | 38 (73.1) |
| Sensitivity of the pools (%) | 43/43 (100) | 37/38 (97.4) |
| Pooling algorithm sensitivity (%) | 48/48 (100) | 61/62 (98.4) |
| Specificity of the pools (%) | 97/99 (98.0) | 13/14 (92.9) |
| Pooling algorithm specificity (%) | 520/520 (100) | 458/458 (100) |
| Total no. of assays performed | 322 | 442 |
| No. of assays saved | 246 | 78 |
For the pooling algorithm, samples were first tested pooled and then presumptive-positive pools were retested individually.
Cost analysis.
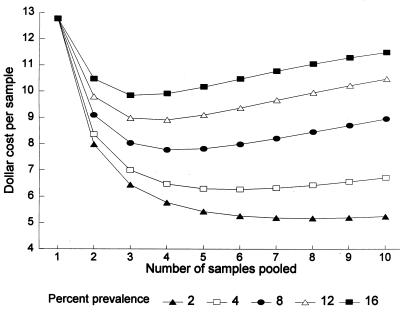
For a population with 8% genital C. trachomatis prevalence, which is close to the 8.5% prevalence found in our study population of female U.S. Army recruits, pooling by four provided the highest cost savings. The reduction of total assay costs per specimen, which included technician time, decreased from $12.76 to $7.78, i.e., by 39%. The model demonstrated that with a 2% prevalence, pooling eight samples would reduce the cost per sample by 59%. A population prevalence graph was constructed from the model to determine the number of pooled samples that would achieve the highest cost savings (Fig. 2).
FIG. 2.
Cost-saving ability of pooling processed urine specimens before the performance of the LCR test for the detection of genital C. trachomatis infections. The graph shows the cost per sample when the pooling algorithm was used, depending on the number of specimens pooled and taking into account various prevalences of infection in the population screened. A baseline total cost of $12.76 per individual sample which included laboratory consumables, technician time, and LCR unit dose costs was used.
A sensitivity analysis for the cost savings model was conducted with ranges of both technician and LCR unit dose costs. For specimens tested individually, raising the base cost of the LCR unit dose from $7 to $15 resulted in an increase of the total cost per specimen tested from $12.76 to $22.87, whereas lowering the cost of the LCR unit dose to $5 reduced the total cost per specimen tested to $10.24. Similarly, raising the annual salary of the technician from $30,000 to $40,000 and assuming 28% benefits and 69% overhead increased the total cost per specimen tested from $12.76 to $13.95, whereas lowering the technician’s annual salary to $20,000 and the overhead to 20% reduced the total cost per specimen tested from $12.76 to $10.89. The cost per specimen tested with the low unit dose and low technician costs was $11.43, while the high unit dose and technician costs yielded a cost of $24.06 per specimen tested.
For a population prevalence of 8% and pooling by four, ranging the unit dose cost from $5 to $15 would result in a total cost of $6.43 and $13.17, respectively, per specimen tested. Ranging technician cost from low to high would result in a total cost increase of $6.36 and $8.68, respectively, per specimen tested. The overall savings of the pooling algorithm over individual testing ranged from 37 to 42% when low to high unit dose cost was considered. Similarly, the overall savings ranged from 42 to 38% when a low-to-high technician cost was considered. The total cost per specimen with the pooling by four algorithm with both the low unit dose and low technician cost was $5.01 per specimen tested, and that with the high unit dose and high technician cost was $14.07 per specimen tested. In all of these scenarios, pooling provided a cost savings compared with individual testing.
Estimation of population prevalence with pooled data.
The observed prevalence for the individual samples in the 142 pools of 4 was 8.5% (48 of 568), and that for the 52 pools of 10 was 11.9% (62 of 520) (Table 1). The estimated population prevalence, back calculated from the number of positive pools, for the 142 pools of 4 was 9.1 (95% CI: 6.5 and 11.6), and for the 52 pools of 10 it was 12.9 (95% CI: 8.8 and 17.0). Each 95% CI included the observed prevalence of the subsample, 10.1% (110 of 1,088). Additionally, each 95% CI included values within 8 to 9%, the overall prevalence measured in a much larger sample (>10,000) of this population.
DISCUSSION
In this study we evaluated pooling of processed urine specimens for LCR detection of C. trachomatis for both accuracy and cost-saving ability. The high sensitivity and specificity of LCR was not affected by pooling up to 10 samples when the S/CO was adjusted from 1.0 to 0.2. Although a small percentage of negative pools tested presumptive positive, no specificity was lost with the pooling algorithm, since all specimens in pools which test presumptive positive are retested individually with the manufacturer’s specified S/CO for the individual test. Since retesting negative pools does increase costs, the specificity of pools must be high.
The cost analysis model showed that depending on the prevalence of C. trachomatis, the number of specimens that should be pooled for optimal cost savings varies. As prevalence decreases, the pooling protocol for screening could save more than 59% of the cost per specimen compared to that for testing individual samples only. Also, early studies have shown that C. trachomatis screening and treatment programs are cost effective; the Centers for Disease Control and Prevention has estimated that for every dollar spent on prevention, $12 is saved in treating sequelae (4). The use of the pooling algorithm for testing samples obtained during screening could further increase savings in health care costs.
Since C. trachomatis prevalence levels have ranged from 4 to 20% in various populations in the United States, pooling three to four samples is likely to provide the highest cost savings. Furthermore, the cost saved does not significantly change the sensitivity or specificity of the assay. In the event that screening is not conducted, pooling can be used to determine population prevalences over time in order to measure the benefits of disease interventions such as mass treatment or behavioral interventions. The population prevalence back calculation, described previously (6), gave an accurate estimate of the observed population prevalence in this study.
Use of the pooling algorithm would benefit investigators and program planners in two ways: (i) money saved from the use of the pooling algorithm could be applied to other areas of disease prevention and/or (ii) the amount of money allocated to screening would allow more specimens to be tested for the same total cost. Pooling samples for the detection of genital C. trachomatis infection in urine samples is cost saving and simple to perform and could be applicable in screening programs in the United States and in population-based research worldwide.
Pooling is a technique which could be immediately used for significant cost savings in high-volume laboratories such as state labs and referral labs. Laboratories which are currently using less sensitive and specific and less costly techniques could introduce both LCR and pooling into their laboratories.
Specific populations or laboratories that might benefit from pooling include any lab in which the combination of turnaround time and volume allows at a minimum a combination of 19 pools and retests per day. With 96 specimens at a population prevalence of about 4%, pooling by six would fill up one full run (38 test unit doses) per day. The run would include, on average, 16 pools of six and 22 retests.
Laboratory managers should consider two points before using pooling. First, processed specimens from presumptive-positive pools need to be amplified and detected individually. This additional step adds a minimum of 3 hours until individual test results for specimens in presumptive-positive pools are known. Second, laboratory managers should estimate the cost savings they expect to gain for their laboratories. This estimate is a combination of both technicians’ salaries and their benefits, institutional overhead, and the prevalence of chlamydia in the populations served by the laboratory. Pooling a greater number than is recommended for certain population prevalences can cost more money than testing specimens individually.
A potential limitation of the pooling algorithm is the possibility of technician error while processed samples are pooled in the LCR run. The use of tray maps simplifies this process. Samples should be organized by skipping a space after each pooled group in the specimen rack. Thus, pooling adds no significant complexity to setting up unit doses. Additional technician error can be avoided when samples from presumptive-positive pools (detected in the previous run) are retested individually before the routine testing of the new pooled groups. Therefore, each run has a combination of samples that are retested individually and new pooled samples from the next batch of specimens.
The study laboratory has met Clinical Laboratory Improvement Act requirements for the modification of a clinical laboratory procedure from a Food and Drug Administration-approved diagnostic kit. Investigators consider performance documentation of the required study adequate for including the pooling protocol in testing clinical specimens in the study laboratory. Each laboratory that wishes to introduce pooling must meet the requirements to modify a Food and Drug Administration-approved package insert. These requirements include meeting the regulations as set forth in the Federal Register (3a).
Use of pooling processed urine samples for LCR testing of C. trachomatis will decrease the cost of screening, providing more evidence that screening programs can and should be implemented. Further applications of pooling include pooling urine specimens for the LCR detection of Neisseria gonorrhoeae. The cost savings of pooling urine for both N. gonorrhoeae and C. trachomatis should also be considered.
ACKNOWLEDGMENTS
We acknowledge D. Perkins for her collaboration and T. and A. Kacena for their assistance.
REFERENCES
- 1.Behets F, Bertozzi S, Kasali M, Kashamuka M, Atikala L, Brown C, Ryder R W, Quinn T C. Successful use of pooled sera to determine HIV-1 seroprevalence in Zaire with development of cost-efficiency models. AIDS. 1990;4:737–741. doi: 10.1097/00002030-199008000-00004. [DOI] [PubMed] [Google Scholar]
- 2.Chernesky M A, Chong S, Jang D, Luinstra K, Sellors J, Mahony J B. Ability of commercial ligase chain reaction and PCR assays to diagnose Chlamydia trachomatis infections in men by testing first void urine. J Clin Microbiol. 1997;35:982–984. doi: 10.1128/jcm.35.4.982-984.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chernesky M A, Jang D, Lee H, Burczak J D, Hu H, Sellors J, Tomazic-Allen S J, Mahony J B. Diagnosis of Chlamydia trachomatis infections in men and women by testing first-void urine by ligase chain reaction. J Clin Microbiol. 1994;32:2682–2685. doi: 10.1128/jcm.32.11.2682-2685.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Federal Register. Establishment and verification of method performance specifications, standard no. 492.1213. Fed Regist. 1992;40:7164. [Google Scholar]
- 4.Institute of Medicine. The hidden epidemic—confronting sexually transmitted diseases. Washington, D.C: National Academy Press; 1997. pp. 28–68. [Google Scholar]
- 5.Jaschek G, Gaydos C, Welsh L, Quinn T C. Direct detection of Chlamydia trachomatis in urine specimens from symptomatic and asymptomatic men by using a rapid polymerase chain reaction assay. J Clin Microbiol. 1993;31:1209–1212. doi: 10.1128/jcm.31.5.1209-1212.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kline R L, Brothers T A, Brookmeyer R, Zeger S, Quinn T C. Evaluation of human immunodeficiency virus seroprevalence surveys using pooled sera. J Clin Microbiol. 1989;27:1449–1452. doi: 10.1128/jcm.27.7.1449-1452.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laga M, Manoka A, Kivuvu M, et al. Non-ulcerative sexually transmitted diseases as risk factors for HIV-1 transmission in women: results from a cohort study. AIDS. 1993;7:95–102. doi: 10.1097/00002030-199301000-00015. [DOI] [PubMed] [Google Scholar]
- 8.Lee H H, Chernesky M A, Schachter J, Burczak J D, Andrews W W, Muldoon S, Leckie G, Stamm W E. Diagnosis of Chlamydia trachomatis genitourinary infection in women by ligase chain reaction assay of urine. Lancet. 1995;345:213–216. doi: 10.1016/s0140-6736(95)90221-x. [DOI] [PubMed] [Google Scholar]
- 9.Lisby G, Scheibel J, Abrahamsson L O, Christensen E S, Paloheimo S. Detection of Chlamydia trachomatis in individual and pooled endocervical and urethral scrapes by a commercially available polymerase chain reaction. APMIS. 1994;102:797–800. [PubMed] [Google Scholar]
- 10.Palmer H M, Gilroy C B, Thomas B J, Hay P E, Gilchrist C, Taylor-Robinson D. Detection of Chlamydia trachomatis by the polymerase chain reaction in swabs and urine from men with non-gonococcal urethritis. J Clin Pathol. 1992;44:321–325. doi: 10.1136/jcp.44.4.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plummer F A, Simonsen J N, Cameron D W, et al. Cofactors in male-female sexual transmission of human immunodeficiency virus type 1. J Infect Dis. 1991;163:233–239. doi: 10.1093/infdis/163.2.233. [DOI] [PubMed] [Google Scholar]
- 12.Quinn T C. Update on Chlamydia trachomatis infections. Infect Med. 1994;11:201–211. [Google Scholar]
- 13.Quinn T C, Cates W. Epidemiology of sexually transmitted diseases in the 1990’s. In: Quinn T C, editor. Advances in host defense mechanisms. Vol. 8. New York, N.Y: Raven Press; 1992. pp. 1–37. [Google Scholar]
- 14.Quinn T C, Zenilman J, Rompalo A. Advances in diagnosis and treatment. In: Schrier R W, Abboud F M, Baxter I D, Fauci A S, editors. Sexually transmitted diseases. St. Louis, Mo: Mosby; 1994. pp. 149–196. [PubMed] [Google Scholar]
- 15.Schwebke J R, Clark A M, Pettinger M B, Nsubga P, Stamm W E. Use of a urine enzyme immunoassay as a diagnostic tool for Chlamydia trachomatis antigens in urine as an alternative to swabs and cultures. J Clin Microbiol. 1991;29:2446–2449. doi: 10.1128/jcm.29.11.2446-2449.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stamm W E, Holmes K K. Chlamydia trachomatis infections of the adult. In: Holmes K K, Mardh P A, Sparling P F, Wiesner P J, editors. Sexually transmitted diseases. New York, N.Y: McGraw-Hill Co.; 1990. pp. 181–193. [Google Scholar]
- 17.Wu C, Lee M, Yin S, Yang D, Cheng S. Comparison of polymerase chain reaction, monoclonal antibody-based enzyme immunoassay, and cell culture for detection of Chlamydia trachomatis in genital specimens. Sex Transm Dis. 1992;91:193–197. doi: 10.1097/00007435-199207000-00002. [DOI] [PubMed] [Google Scholar]