Abstract
We investigated the effects of conditions often encountered during handling, transit, and storage of blood specimens on the quantity of detectable human immunodeficiency virus (HIV) RNA in plasma. HIV RNA copy numbers were measured with a commercially available assay (the Amplicor HIV-1 Monitor test kit). Variables examined were the time to processing of blood and plasma, the holding temperature of blood and plasma prior to processing, the effect of freezing and thawing of plasma, and the use of different anticoagulants. The relationship between the HIV RNA copy number and the HIV isolation rate by peripheral blood mononuclear cell (PBMC) coculture was also examined. We found that RNA copy numbers were maintained to within 0.5 log10 (approximately threefold) in blood and plasma samples held at room temperature or 4°C for up to 3 days and remained stable despite (limited) freezing and thawing of the plasma. HIV RNA copy numbers were also maintained after long-term storage of plasma at −70°C. The ability to isolate HIV from PBMCs was directly proportional to the HIV RNA copy number.
The virion-associated human immunodeficiency virus (HIV) RNA level has been shown to directly reflect the level of viral replication in vivo (3, 18) and to be a prognostic marker of clinical disease (4, 14, 15, 20). HIV RNA levels can also be utilized to predict clinical outcome early in infection (9), to indicate when antiretroviral therapy should be initiated, and to monitor response to the treatment (2, 5, 23). Thus, accurate and reliable quantitation of HIV RNA is an essential aspect of the management of patients infected with the virus.
While the magnitude of the HIV RNA level in an individual patient is likely to be predictive of his or her clinical outcome (15), levels of HIV RNA vary between individuals, irrespective of CD4 count and disease status (6, 21). However, by monitoring changes in HIV RNA levels in sequential specimens, response to therapy can be assessed for an individual patient (5, 6, 10, 13, 26). It is therefore important that measurements of HIV RNA load in sequential specimens be true reflections of the levels at particular times and not of differences resulting from deterioration of HIV RNA associated with suboptimal handling, transport, and/or storage of specimens.
Laboratories performing HIV RNA load determinations are often physically separate from blood collection clinics, thus necessitating transport of specimens. This invariably results in delays and, sometimes, different holding temperatures prior to specimen processing. Furthermore, blood may be collected in tubes containing a variety of anticoagulants or, in some cases, no anticoagulant. Once in the laboratory, other factors encountered that may affect the reproducibility of HIV RNA quantitation include inter- and intraassay variability, operator performance, the long-term storage of frozen plasma, and the freezing and thawing of plasma. Using a commercially available assay (Amplicor HIV-1 Monitor test kit; Roche Diagnostic Systems, Inc., Branchburg, N.J.), we examined several of these parameters under controlled laboratory conditions to assess whether they influence the accuracy of HIV RNA quantitation.
Previous studies have found the intraassay variability for a given sample assessed by the above assay to be less than or equal to 0.2 log10 (6, 13). Biological variability within an individual receiving a stable antiretroviral drug regimen has been estimated to be 0.3 log10 (26). Therefore, when interpreting our results we considered only changes greater than the sum of these two factors (0.5 log10; approximately threefold) to be significant (22).
MATERIALS AND METHODS
Specimens.
Blood specimens were collected from HIV-seropositive individuals attending outpatient clinics at several hospitals and private clinics in Melbourne, Australia. The individuals were predominantly male (97%) and between the ages of 18 and 30 years (97%). These specimens arrived at the testing laboratory within 6 h after being drawn, the time limit for separation of plasma recommended by the manufacturer. Except where otherwise stated, 8 to 10 ml of whole blood was collected in tubes containing 1.3% (wt/vol) acid citrate dextrose (ACD) (Greiner, Labortechnik) and transported to the laboratory by courier. When used, EDTA tubes were obtained from Greiner, Labortechnik. Plasma was collected by centrifugation at 400 × g for 10 min at room temperature (RT; 21°C) and stored at −70°C for between 5 and 10 days prior to being tested.
Quantitation of RNA viral load in plasma.
The Amplicor HIV-1 Monitor assay was used according to the manufacturer’s instructions in all tests requiring HIV RNA quantitation. Briefly, target RNA was prepared by guanidine thiocyanate lysis of HIV virions in plasma, followed by isopropanol precipitation. The enzyme rTth DNA polymerase was used to reverse transcribe the RNA into cDNA and to amplify DNA by PCR. The biotinylated products of amplification were diluted in fivefold steps and hybridized to target-specific oligonucleotide probes immobilized in the wells of a microtiter plate. This procedure was followed by enzyme immunoassay-based colorimetric detection of the bound products. Quantitation was accomplished by the inclusion of a known concentration of synthetic RNA (containing the same primer binding sites as the target HIV RNA but with a unique probe sequence), which was reverse transcribed and coamplified with the target RNA.
HIV isolation.
HIV was isolated from patient peripheral blood mononuclear cells (PBMC) by coculture with phytohemagglutinin-stimulated donor PBMC (17). Patient PBMC were separated from 8 to 10 ml of ACD-treated blood by density gradient centrifugation. Patient and donor PBMC were cocultured in a one-to-one ratio in a medium containing RPMI 1640 (ICN Biomedicals, Inc., Costa Mesa, Calif.), 10% heat-inactivated fetal calf serum (Cytosystems, Castle Hill, Australia), and 10% recombinant human interleukin-2 (Boehringer Mannheim Australia Pty Ltd., Castle Hill, Australia). Cocultures were maintained at 37°C in 5% CO2 for up to 28 days. During this time, fresh medium was added twice every 7 days, and fresh donor PBMC were added once every 7 days. A detectable increase in p24 antigen levels in supernatant fluid, measured with a commercially available enzyme immunoassay kit (Vironstika HIV-1 Antigen Microelisa system; Organon Teknika Corp., Durham, N.C.), was considered to indicate active virus replication.
Statistical analysis.
A paired t test analysis was used to determine the statistical significance of relationships between log10 RNA copy number and the effects of time, holding conditions, anticoagulants, and freezing-thawing (the null hypothesis being that the difference between the first and last measurements was equal to zero). Statistical analysis of the relationship between the RNA copy number in plasma and the virus isolation rate was undertaken with a chi-square test for linear trend in proportions.
RESULTS
Effect of time and temperature on the quantitation of virion-associated HIV RNA in ACD-treated blood and plasma.
The following experiments were undertaken to assess the stability of HIV RNA in ACD-treated blood and plasma over a 72-h period. Blood specimens (n = 20) were centrifuged, and an aliquot (220 μl) of each plasma sample was stored at −70°C (considered to be time zero from arrival in the laboratory). The remainder of the blood was resuspended by gentle inversion of the tube, and the contents were left at RT (n = 10) or 4°C (n = 10). Aliquots of plasma were recovered as described above from each of these tubes after 24, 48, and 72 h and stored at −70°C. All aliquots were then removed from storage, and the HIV RNA copy number in each was determined by a single operator. A change of up to 0.5 log10 in the copy number was observed in blood specimens held for up to 72 h at either RT or 4°C (Table 1). For 15 of 20 specimens evaluated the change at any time point was less than 0.4 log10 from the copy numbers obtained at time zero. Neither the extent nor the direction of the fluctuation (i.e., increase or decrease in the copy number) was consistent or reflective of the delay in processing. Statistical analysis by the paired t test showed that over the 72-h evaluation period there was no significant change in log10 copy number for any holding condition (P = 0.06 to 0.70).
TABLE 1.
Effect of time and temperature on HIV RNA levels in whole blood and plasmaa
| Specimen no. and type | Holding conditions | HIV RNA copies (103)/ml of plasma (log10) at h:
|
P valuec | |||
|---|---|---|---|---|---|---|
| 0 | 24 | 48 | 72 | |||
| Whole blood | ||||||
| 1 | RT | 0.4 (2.6) | 0.5 (2.7) | 0.4 (2.6) | 1.0 (3.0) | 0.70 |
| 2 | RT | 1.8 (3.3) | 4.5 (3.7) | 1.5 (3.2) | 1.9 (3.3) | |
| 3 | RT | 2.7 (3.4) | 1.7 (3.2) | 1.8 (3.3) | 1.9 (3.3) | |
| 4 | RT | 10.8 (4.0) | 8.0 (3.9) | 7.5 (3.9) | 7.4 (3.9) | |
| 5 | RT | 15.2 (4.2) | 9.5 (4.0) | 7.5 (3.9) | 4.6 (3.7) | |
| 6 | RT | 49.8 (4.7) | 38.3 (4.6) | 59.2 (4.8) | 50.9 (4.7) | |
| 7 | RT | 59.2 (4.8) | 76.3 (4.9) | 69.3 (4.8) | 82.5 (4.9) | |
| 8 | RT | 87.3 (4.9) | 110.5 (5.0) | 86.5 (4.9) | 89.0 (5.0) | |
| 9 | RT | 97.5 (5.0) | 116.0 (5.1) | 59.1 (4.8) | 70.4 (4.9) | |
| 10 | RT | 130.6 (5.1) | 87.7 (4.9) | 101.0 (5.0) | 94.1 (5.0) | |
| 11 | 4°C | <0.4 (<2.6)b | N.D.d | N.D. | <0.4 (<2.6) | 0.06 |
| 12 | 4°C | 0.4 (2.6) | 0.4 (2.6) | 0.7 (2.8) | 0.4 (2.6) | |
| 13 | 4°C | 6.0 (3.8) | N.D. | N.D. | 2.2 (3.3) | |
| 14 | 4°C | 6.2 (3.8) | N.D. | N.D. | 3.2 (3.5) | |
| 15 | 4°C | 12.2 (4.1) | N.D. | N.D. | 11.7 (4.1) | |
| 16 | 4°C | 18.2 (4.3) | N.D. | N.D. | 22.2 (4.4) | |
| 17 | 4°C | 44.9 (4.7) | N.D. | N.D. | 25.8 (4.4) | |
| 18 | 4°C | 49.2 (4.7) | N.D. | N.D. | 47.2 (4.7) | |
| 19 | 4°C | 49.8 (4.7) | 39.6 (4.6) | 43.9 (4.6) | 23.7 (4.4) | |
| 20 | 4°C | 167.9 (5.2) | N.D. | N.D. | 48.5 (4.7) | |
| Plasma | ||||||
| 21 | RT | 0.4 (2.6) | 0.4 (2.6) | <0.4 (<2.6) | 0.4 (2.6) | 0.59 |
| 22 | RT | 5.1 (3.7) | N.D. | N.D. | 6.2 (3.8) | |
| 23 | RT | 8.9 (4.0) | N.D. | N.D. | 8.7 (3.9) | |
| 24 | RT | 15.6 (4.2) | N.D. | N.D. | 16.4 (4.2) | |
| 25 | RT | 16.5 (4.2) | N.D. | N.D. | 10.6 (4.0) | |
| 26 | RT | 38.9 (4.6) | N.D. | N.D. | 34.3 (4.5) | |
| 27 | RT | 46.5 (4.7) | 19.3 (4.3) | 29.8 (4.5) | 40.8 (4.6) | |
| 28 | RT | 122.7 (5.1) | 108.9 (5.0) | 122.9 (5.1) | 176.2 (5.2) | |
| 29 | RT | 124.4 (5.1) | N.D. | N.D. | 161.3 (5.2) | |
| 30 | 4°C | <0.4 (<2.6) | N.D. | N.D. | <0.4 (<2.6) | 0.60 |
| 31 | 4°C | <0.4 (<2.6) | N.D. | N.D. | <0.4 (<2.6) | |
| 32 | 4°C | 0.4 (2.6) | 0.4 (2.6) | 0.3 (2.5) | <0.4 (<2.6) | |
| 33 | 4°C | 2.3 (3.4) | N.D. | N.D. | 2.7 (3.4) | |
| 34 | 4°C | 5.6 (4.8) | N.D. | N.D. | 7.2 (4.9) | |
| 35 | 4°C | 18.4 (4.3) | N.D. | N.D. | 14.2 (4.2) | |
| 36 | 4°C | 20.1 (4.3) | N.D. | N.D. | 21.1 (4.3) | |
| 37 | 4°C | 36.8 (4.6) | N.D. | N.D. | 42.9 (4.6) | |
| 38 | 4°C | 46.5 (4.7) | 36.8 (4.6) | 33.2 (4.5) | 24.5 (4.4) | |
| 39 | 4°C | 122.7 (5.1) | 152.3 (5.2) | N.D. | 247.2 (5.4) | |
Whole blood was held at either RT or 4°C. After centrifugation at 400 × g for 10 min, plasma was held at RT or 4°C.
The level of HIV RNA was below the level of detection of the assay (400 copies per ml). Where a value of 0.4 is quoted, the copy number obtained was between 400 and 440 per ml.
The P values are calculated based on the difference between time zero and 72-h data for each specimen type and holding condition.
N.D., not done.
In an experiment similar to that carried out on whole-blood samples, plasma samples obtained following centrifugation of ACD-treated blood were aliquoted and stored at −70°C for up to 10 days prior to being tested, after being held for 0, 24, 48, or 72 h at RT (n = 9) or 4°C (n = 10) (Table 1). Similar to that observed with whole blood, the maximum difference in HIV RNA copy numbers was 0.4 log10 (Table 1, sample 27). This was not statistically significant by paired t test analysis (P = 0.59 to 0.60).
The stability of HIV RNA in blood during the first 6 h after the blood was drawn was also assessed. Blood specimens (n = 8) were processed at the site of collection, at the time of drawing, and 2 and 6 h after being drawn. Because −70°C storage was not available at the site of collection, plasma samples were immediately frozen in dry ice prior to storage at −70°C on return to the laboratory. They were subsequently tested by a single operator in a single test. For the eight specimens evaluated, there was no more than a 0.4-log10 fluctuation in copy number over the 6-h period (Table 2). This was not statistically significant by paired t test analysis (P = 0.50).
TABLE 2.
Quantitation of HIV RNA copy number in whole blood during the first 6 h postcollection
| Specimen no. | HIV RNA copies (103)/ml of plasma (log10) at ha
|
||
|---|---|---|---|
| 0 | 2 | 6 | |
| 1 | <0.4 (<2.6) | <0.4 (<2.6) | <0.4 (<2.6) |
| 2 | 0.5 (2.7) | 0.4 (2.6) | 0.8 (2.9) |
| 3 | 1.1 (3.0) | 0.9 (3.0) | 0.7 (2.9) |
| 4 | 1.4 (3.2) | 1.3 (3.1) | 0.9 (3.0) |
| 5 | 3.7 (3.6) | 8.0 (3.9) | 4.7 (3.7) |
| 6 | 5.0 (3.7) | 7.3 (3.9) | 3.5 (3.5) |
| 7 | 13.3 (4.1) | 15.2 (4.2) | 14.1 (4.2) |
| 8 | 129.6 (5.1) | 92.5 (5.0) | 135.4 (5.1) |
P = 0.50; calculated based on the difference between the time zero and 6-h values.
Effect of anticoagulants on HIV RNA copy number.
The manufacturer recommends that the Amplicor HIV-1 Monitor assay be performed on the plasma component of blood treated with either EDTA or ACD. We assessed the effect on HIV RNA copy number of these two anticoagulants, as well as assaying the copy number in anticoagulant-free blood (serum).
Blood from nine HIV-seropositive individuals was collected in tubes containing ACD (1.5 ml) or EDTA (spray coated) and in plain tubes. Plasma or serum samples recovered from this blood were stored for between 5 and 10 days at −70°C prior to being tested. Specimens obtained from each individual were evaluated in a single assay.
No more than a 0.3-log10 difference in HIV RNA copy numbers was observed in plasma samples treated with either of the anticoagulants (Table 3). The log10 difference in HIV RNA copy numbers remained unchanged when the results were adjusted to allow for a dilution factor of 15% associated with the volume of anticoagulant in the ACD tube (data not shown). HIV RNA copy numbers were, in general, lower in serum specimens, although in eight of nine specimens there was no more than a 0.4-log10 difference between the copy numbers measured in serum and in blood treated with either anticoagulant (Table 3). In one of nine serum specimens (specimen 3) there was a decrease in the HIV RNA copy number of between 0.5 and 0.8 log10 compared to that obtained for the plasma samples. Statistical analysis revealed that, although there was no statistically significant difference between log10 copy numbers in samples collected in ACD tubes and those of samples collected in EDTA tubes or between those of samples in ACD tubes and those of samples in plain tubes (P = 0.08 and 0.40, respectively), there was a statistically significant difference between copy numbers in EDTA tubes and those in plain tubes (P < 0.001 by paired t test analysis).
TABLE 3.
Effect of anticoagulants on HIV RNA levels
| Specimen no. | HIV RNA copies (103)/ml of plasma in:
|
||
|---|---|---|---|
| ACD tubesa | EDTA tubesb | Plain tubes | |
| 1 | <0.4 (<2.6) | <0.4 (<2.6) | <0.4 (<2.6) |
| 2 | 1.0 (3.0) | 2.1 (3.3) | 0.8 (2.9) |
| 3 | 3.3 (3.5) | 6.1 (3.8) | 1.0 (3.0) |
| 4 | 3.8 (3.6) | 3.8 (3.6) | 2.0 (3.3) |
| 5 | 4.6 (3.7) | 7.4 (3.9) | 5.5 (3.7) |
| 6 | 8.4 (3.9) | 13.6 (4.1) | 8.4 (3.9) |
| 7 | 25.8 (4.4) | 21.1 (4.3) | 11.8 (4.1) |
| 8 | 28.7 (4.5) | 37.4 (4.6) | 29.8 (4.5) |
| 9 | 262.8 (5.4) | 267.1 (5.4) | 266.0 (5.4) |
P = 0.08 comparing data from ACD and EDTA tubes; P = 0.40 comparing data from ACD and plain tubes.
P < 0.001 comparing data from EDTA and plain tubes.
Effect of freezing and thawing of plasma on the stability of HIV RNA copy number.
The stability of HIV RNA in plasma subjected to multiple cycles of freezing and thawing at −70°C was assessed. Aliquots of eight samples of plasma stored for 7 days at −70°C were frozen and thawed up to three times and then analyzed in the same assay by a single operator. Three cycles of freezing and thawing did not result in more than a 0.2 log10 change in HIV RNA copy numbers in any of the plasma samples tested (Table 4), and this was not a statistically significant change (P = 0.12 by paired t test analysis).
TABLE 4.
Effect of freezing and thawing on stability of HIV RNA in plasma
| Specimen no. | HIV RNA copies (103)/ml of plasma (log10) aftera:
|
||
|---|---|---|---|
| 1 FT cycle | 2 FT cycles | 3 FT cycles | |
| 1 | 0.4 (2.6) | 0.4 (2.6) | 0.7 (2.8) |
| 2 | 0.7 (2.8) | 0.7 (2.8) | 0.78 (2.9) |
| 3 | 17.7 (4.2) | 18.5 (4.3) | 25.6 (4.4) |
| 4 | 40.8 (4.6) | 27.6 (4.4) | 28.4 (4.5) |
| 5 | 119.4 (5.1) | 162.4 (5.2) | 95.0 (5.0) |
| 6 | 228.5 (5.4) | 144.6 (5.2) | 167.2 (5.2) |
| 7 | 225.6 (5.4) | 194.7 (5.3) | 301.3 (5.5) |
| 8 | 305.2 (5.5) | 368.7 (5.6) | 268.9 (5.4) |
P = 0.12 comparing data after one and three cycles of freezing and thawing (FT).
Effect of long-term storage of plasma at −70°C on HIV RNA copy number.
Retrospective analysis of plasma stored over long periods of time at −70°C is often undertaken for clinical or research purposes. We compared the levels of HIV RNA in 10 plasma samples stored at −70°C for 12 months. Because of the time between assays, we were unable to control for operator or test lot. Over the first 5 months of storage, there was a decrease in HIV RNA copy numbers of greater than 0.5 log10 in 2 of these 10 specimens (specimens 8 and 10) and between 0.3 and 0.5 log10 in another 3 specimens (specimens 2, 5, and 7) (Table 5). One specimen with an initial level of 500 RNA copies/ml dropped below the level of detection of the assay (less than 400 copies/ml) after 5 months of storage. No change in log10 copy numbers was observed in the remaining four specimens over the 5-month storage period. Overall, the difference in log10 copy numbers over the 5-month storage period was statistically significant (P = 0.002). Following 12 months of storage of the same specimens, no specimen contained an RNA copy number that varied by more than 0.4 log10 from that of the same specimen originally tested at time zero, and overall there was no statistically significant difference between log10 copy numbers obtained at time zero and after 12 months of storage (P = 0.80). However, one specimen (specimen 10) varied by 0.7 log10 from the result obtained after 5 months of storage, and similar to the storage data for time zero to 5 months, there was a statistically significant difference between log10 copy numbers in samples stored between 5 and 12 months at −70°C (P = 0.001).
TABLE 5.
Effect of long-term storage at −70°C on HIV RNA in plasma
| Specimen no. | HIV RNA copies (103)/ml of plasma (log10) at moa:
|
||
|---|---|---|---|
| 0 | 5 | 12 | |
| 1 | 0.5 (2.7) | <0.4 (<2.6) | 0.8 (2.9) |
| 2 | 4.2 (3.6) | 2.0 (3.3) | 3.8 (3.6) |
| 3 | 11.5 (4.1) | 9.1 (4.0) | 21.8 (4.3) |
| 4 | 17.6 (4.2) | 10.4 (4.0) | 6.3 (4.2) |
| 5 | 41.5 (4.6) | 15.9 (4.2) | 28.9 (4.5) |
| 6 | 50.8 (4.7) | 41.8 (4.6) | 50.9 (4.7) |
| 7 | 71.9 (4.9) | 22.8 (4.4) | 30.8 (4.5) |
| 8 | 180.6 (5.3) | 46.0 (4.7) | 81.7 (4.9) |
| 9 | 335.5 (5.5) | 355.5 (5.5) | 581.6 (5.8) |
| 10 | 643.0 (5.8) | 74.9 (4.9) | 365.8 (5.6) |
P = 0.002 comparing values for time zero and 5 months of storage at −70°C; P = 0.001 comparing values for 5 and 12 months of storage at −70°C; P = 0.80 comparing values for time zero and 12 months of storage at −70°C.
Relationship between HIV RNA copy number in plasma and virus isolation rate.
During clinical trials it is often necessary to isolate HIV to allow drug susceptibility testing and/or genomic sequence analysis. We compared the rate of recovery of HIV from PBMC as measured by the detection of p24 antigen in coculture supernatant fluids with the HIV RNA copy number in the plasma derived from the same blood specimen.
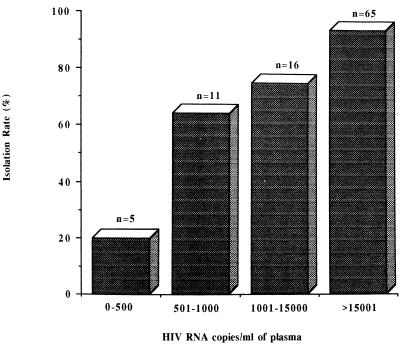
Ninety-seven specimens from which we attempted to isolate HIV were stratified according to their HIV RNA copy numbers. Figure 1 shows a correlation between our ability to isolate HIV from PBMC and the amount of HIV RNA in the plasma of the patient. The rate of isolation from 65 specimens with HIV RNA levels greater than 15,000 copies/ml was 94%. Of specimens with 1,000 to 15,000 copies/ml (n = 16), 500 to 1,000 copies/ml (n = 11), and less than 500 copies/ml (n = 5), the isolation rates were 75, 64, and 20%, respectively. According to a chi-square test for linear proportion, this was a highly significant trend (P < 0.001).
FIG. 1.
Relationship between HIV RNA copy number and virus isolation rate in PBMC cocultures. HIV isolation and quantitation of HIV RNA levels in plasma were attempted with 97 specimens. Results are expressed as percentages of isolation-positive cultures versus the HIV RNA copy number per milliliter of plasma.
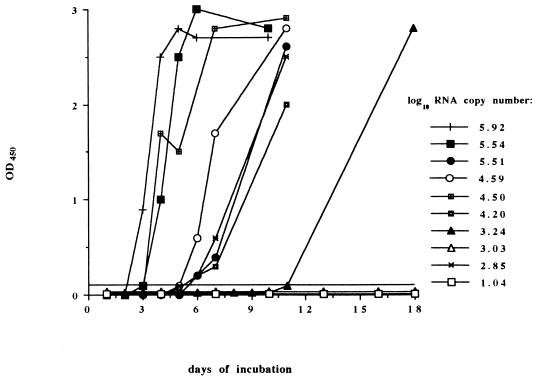
Our results also showed that specimens with high plasma HIV RNA copy numbers became culture positive earlier than those with lower RNA levels (Fig. 2). Two specimens with HIV RNA levels of 346,200 and 831,000 RNA copies/ml were culture positive (as indicated by detectable levels of p24 antigen) by day 3 postinfection. Four specimens with HIV RNA levels between 15,000 and 330,000 RNA copies/ml were culture positive within 4 to 6 days postinfection. HIV was not isolated from two of four specimens with less than 2,000 RNA copies/ml, and the remaining two specimens took longer than 7 days to become culture positive. The trend toward a higher rate of isolation from PBMC in blood samples where plasma copy numbers were high was also highly significant (P < 0.001).
FIG. 2.
Relationship between virion-associated HIV RNA copy number and time to HIV isolation positive in PBMC coculture. The level of p24 antigen in the culture supernatant was monitored for 10 specimens for which HIV RNA levels in plasma had been quantitated. HIV RNA copy numbers for each specimen are expressed as log10 RNA copies per milliliter of plasma. OD450, optical density at 450 nm.
DISCUSSION
One of the features of HIV infection has been the variable nature of the disease between individuals. For example, the time between seroconversion and the development of AIDS (16, 19, 24), the immune response at the time of seroconversion and its effect on progression to AIDS (27), and the response to antiviral therapy and subsequent development of drug resistance (11, 12) vary from patient to patient, making management problematic. Viral load measurements at the time of seroconversion have been shown to predict clinical outcome (4, 9, 14, 15, 20) and to reflect the response to antiretroviral therapy, including the development of resistance (2, 5, 6, 23). Hence, accurate viral load measurements are important in the management of infected patients, particularly when sequential specimens are involved. Several commercial assays are available for this purpose (for example, Amplicor HIV-1 Monitor [Roche Diagnostic Systems], Quantiplex HIV-RNA assay [Chiron Corp.], and Q-NASBA [Organon Teknika]) and provide comparable results (21). While intra- and interassay variability contributes an approximately 0.2-log10 variation between test results (26), delays in the transport and processing of specimens by the laboratory may also contribute to variable results. Our examination of a number of variables has allowed us to make some preliminary decisions regarding how long HIV RNA levels in blood and/or plasma remain stable and, therefore, suitable for HIV RNA quantitation. Examination of a greater number of specimens should enable conclusions to be made that could ultimately constitute more formal guidelines.
Our results showed that the HIV RNA copy numbers in blood and plasma maintained at either 4°C or RT for between 6 and 72 h postcollection was relatively stable, not changing by more than 0.5 log10 (threefold) (Table 1). However, we were concerned that the HIV RNA copy number might decline at an accelerated rate during the first 6 h after blood was drawn (the maximum time recommended by the manufacturer for processing of blood specimens) than at subsequent times. Testing demonstrated only a moderate fluctuation in copy number (<0.4 log10, or less than twofold) during the first 6 h (Table 2). Therefore, laboratories receiving specimens subjected to these conditions should be able to report results with relative confidence. Our results are in agreement with those obtained by the manufacturer, showing that the storage of plasma at 2 to 8°C or at RT for up to 7 days had little or no effect on HIV RNA copy number (27a).
Our experiments showed that both EDTA and ACD were acceptable anticoagulants for use with the Amplicor HIV Monitor assay. A maximum difference of 0.3 log10 RNA copies/ml in plasma derived with these anticoagulants was demonstrated, but this was not statistically significant. This result is similar to that reported by Holodniy and colleagues (7), who found a maximum difference in plasma RNA levels of 0.17 log10 when anticoagulants such as EDTA, ACD, and sodium citrate were evaluated by the bDNA method.
The manufacturer and others have found consistently lower HIV RNA levels in serum than in plasma, although there is a strong correlation between the two (7, 21a). Copy numbers in serum approximately half those found in plasma (21a) may be a result of the entrapment of virions within the fibrin network of the clot (1). However, in the majority of serum specimens we tested (eight of nine), the reduction in RNA copy number in serum compared to that in plasma derived from ACD tubes was less than 0.5 log10 and any differences were not statistically significant. Of note, however, was the significant difference between copy numbers in blood collected in ACD versus plain tubes.
Retrospective analysis of biological markers often requires the testing of specimens that have been stored frozen for long periods and sometimes subjected to multiple rounds of freezing and thawing. Our findings support the results of others, which suggest that there is no consistent effect on the HIV RNA copy number following up to three cycles of freezing and thawing at −70°C (27). In addition, long-term storage of plasma for up to 12 months at −70°C did not result in statistically significant changes in the HIV RNA copy number. We have no obvious explanation for the somewhat variable copy numbers obtained after 5 months of storage, which resulted in statistically significant differences between copy numbers obtained between time zero and 5 months and those obtained between 5 and 12 months. Our inability to control for test lot and operator variability during these periods may have contributed to this result. Overall, our 12-month stability data supports that of Winters and colleagues (26), who reported stable RNA levels in plasma stored for up to 12 months at −70°C. However, the protocol used by that group for plasma preparation included an additional centrifugation step to remove platelets, any remaining cells, and cellular debris, a procedure which was likely to have minimized the presence of RNases. Other groups have added guanidinium or other inhibitors to plasma prior to storage to prevent similar deterioration of RNA through the action of RNases (5, 28).
We were able to demonstrate a direct correlation between the HIV isolation rate from PBMC by using coculture techniques and the HIV RNA copy number in plasma derived from the same blood. A previous study has demonstrated a correlation between the copy number and the overall isolation rate from plasma (but not infectious titer in plasma) (25). Significant correlation between plasma RNA levels and cell dilution culture has also been shown for patients receiving didanosine therapy (8). Our results therefore provide further evidence that viral load measurements in plasma are reflections of the amount of infectious virus present in vivo, a conclusion strengthened by our observation that HIV replication in vitro tends to be detected earlier in cocultures of PBMC derived from specimens with high HIV RNA copy numbers.
In conclusion, we have assessed the stability of HIV RNA in whole blood and plasma subjected to conditions often encountered in specimen handling, transport, and storage. We found that the RNA copy number did not change significantly when blood or plasma was maintained at RT or 4°C for up to 72 h, when plasma was subjected to three cycles of freezing and thawing, when blood was collected in different (or no) anticoagulants, or after long-term (12 months) storage at −70°C. We also found a close association between the HIV viral load and the ease of isolation in culture.
ACKNOWLEDGMENTS
We thank the Melbourne Red Cross Blood Bank for the provision of blood packs, Dianne Young (Roche Laboratories) for technical advice, and Penny Tresize and Nick Crofts for statistical analysis. We also thank the personnel of the Melbourne Sexual Health Centre for allowing the on-site specimen processing necessary as part of this study.
REFERENCES
- 1.Coombs R W, Henrard D R, Mehaffey W F, Gibson J, Eggert E, Quinn T C, Phillips J. Cell-free plasma human immunodeficiency virus type 1 titer assessed by culture and immunocapture-reverse transcription-polymerase chain reaction. J Clin Microbiol. 1993;31:1980–1986. doi: 10.1128/jcm.31.8.1980-1986.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harrigan, R. 1995. Measuring viral load in the clinical setting. J. Acquired Immune Defic. Syndr. 10(Suppl. 1):S34–S40. [PubMed]
- 3.Ho D D, Neumann A U, Perelson A S, Chen W, Leonard J M, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 4.Ho D D. Viral counts count in HIV infection. Science. 1996;272:1124–1125. doi: 10.1126/science.272.5265.1124. [DOI] [PubMed] [Google Scholar]
- 5.Holodniy M, Katzenstein D A, Israelski D M, Merigan T C. Reduction in plasma human immunodeficiency virus ribonucleic acid after dideoxynucleoside therapy as determined by the polymerase chain reaction. J Clin Invest. 1991;88:1755–1759. doi: 10.1172/JCI115494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holodniy M, Mole L, Winters M, Merigan T C. Diurnal and short-term stability of HIV virus load as measured by gene amplification. J Acquired Immune Defic Syndr. 1994;7:363–368. [PubMed] [Google Scholar]
- 7.Holodniy M, Mole L, Yen-Lieberman B, Margolis D, Starkey C, Carroll R, Spahlinger T, Todd J, Jackson J B. Comparative stabilities of quantitative human immunodeficiency virus RNA in plasma from samples collected in VACUTAINER CPT, VACUTAINER PPT, and standard VACUTAINER tubes. J Clin Microbiol. 1995;33:1562–1566. doi: 10.1128/jcm.33.6.1562-1566.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katzenstein D A, Winters M, Bubp J, Israelski D, Winger E, Merigan T C. Quantitation of human immunodeficiency virus by culture and polymerase chain reaction in response to didanosine after long-term therapy with zidovudine. J Infect Dis. 1994;169:416–419. doi: 10.1093/infdis/169.2.416. [DOI] [PubMed] [Google Scholar]
- 9.Katzenstein T L, Pedersen C, Nielson C, Lundgren J D, Jakobsen P H, Gerstoft J. Longitudinal serum HIV RNA quantification: correlation to viral phenotype at seroconversion and clinical outcome. AIDS. 1996;10:167–173. doi: 10.1097/00002030-199602000-00006. [DOI] [PubMed] [Google Scholar]
- 10.Kojima, E., T. Shirasaka, B. Anderson, S. Chokekijchai, S. Sei, R. Yarchoan, and H. Mitsuya. 1993. Monitoring the activity of antiviral therapy for HIV infection using a polymerase chain reaction method coupled with reverse transcription. AIDS 7(Suppl. 2):S101–S105. [DOI] [PubMed]
- 11.Land S, McGavin C, Lucas R, Birch C. Incidence of zidovudine-resistant human immunodeficiency virus isolated from patients before, during and after therapy. J Infect Dis. 1992;166:1139–1142. doi: 10.1093/infdis/166.5.1139. [DOI] [PubMed] [Google Scholar]
- 12.Larder B, Darby G, Richman D D. HIV and reduced sensitivity to zidovudine (AZT) isolated during prolonged therapy. Science. 1989;243:1731–1734. doi: 10.1126/science.2467383. [DOI] [PubMed] [Google Scholar]
- 13.Lin H J, Myers L E, Yen-Lieberman B, Hollinger F B, Henrard D, Hooper C J, Kokka R, Kwok S, Rasheed S, Vahey M, Winters M A, McQuay L J, Nara P L, Reichelderfer P, Coombs R W, Jackson J B. Multicenter evaluation of quantification methods for plasma human immunodeficiency virus type 1 RNA. J Infect Dis. 1994;170:553–562. doi: 10.1093/infdis/170.3.553. [DOI] [PubMed] [Google Scholar]
- 14.Mellors J W, Kingsley L A, Rinaldo C R, Jr, Todd J A, Hoo B S, Kokka R P, Gupta P. Quantitation of HIV-1 RNA in plasma predicts outcome after seroconversion. Ann Intern Med. 1995;122:573–579. doi: 10.7326/0003-4819-122-8-199504150-00003. [DOI] [PubMed] [Google Scholar]
- 15.Mellors J W, Rinaldo C R, Jr, Gupta P, White R M, Todd J A, Kingsley L A. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 16.Munoz A, Wang M-C, Bass S, Taylor J M G, Kingsley L A, Chmiel J S, Polk F the Multicenter AIDS Cohort Study Group. Acquired immunodeficiency syndrome (AIDS)-free time after human immunodeficiency virus type-1 (HIV-1) seroconversion in homosexual men. Am J Epidemiol. 1989;130:530–539. doi: 10.1093/oxfordjournals.aje.a115367. [DOI] [PubMed] [Google Scholar]
- 17.Neate E V, Pringle R C, Jowett J B M, Healey D S, Gust I D. Isolation of HIV from Australian patients with AIDS, AIDS related conditions and healthy positive individuals. Aust N Z J Med. 1987;17:461–466. doi: 10.1111/j.1445-5994.1987.tb00096.x. [DOI] [PubMed] [Google Scholar]
- 18.Perelson A S, Neumann A U, Markowitz M, Leonard J M, Ho D D. HIV-1 dynamics in vivo: virion clearance rate, infected cell life span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 19.Phair J, Jacobson L, Detels R, Rinaldo C, Saah A, Schrager L, Monoz A. Acquired immune deficiency syndrome occurring within 5 years of infection with human immunodeficiency virus type-1: the multicenter cohort study. J Acquired Immune Defic Syndr. 1992;5:490–496. [PubMed] [Google Scholar]
- 20.Piatak M, Saag M S, Yang L C, Clark S J, Kappes J C, Luk K-C, Hahn B H, Shaw G M, Lifson J D. High levels of HIV-1 in plasma during all stages of infection determined by competitive PCR. Science. 1993;259:1749–1754. doi: 10.1126/science.8096089. [DOI] [PubMed] [Google Scholar]
- 21.Revets H, Marissens D, De Wit S, Lacor P, Clumeck N, Lauwers S, Zissis G. Comparative evaluation of NASBA HIV-1 RNA QT, AMPLICOR-HIV Monitor, and QUANTIPLEX HIV RNA assay, three methods for quantification of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1996;34:1058–1064. doi: 10.1128/jcm.34.5.1058-1064.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21a.Roche Diagnostic Systems, Inc. Unpublished data.
- 22.Saag M S, Holodniy M, Kuritzkes D R, O’Brien W A, Coombs R, Poscher M E, Jacobsen D M, Shaw G M, Richman D D, Volberding P A. HIV viral load markers in clinical practice. Nat Med. 1996;2:625–629. doi: 10.1038/nm0696-625. [DOI] [PubMed] [Google Scholar]
- 23.Semple M, Loveday C, Weller I, Tedder R. Direct measurement of viraemia in patients infected with HIV-1 and its relationship to disease progression and zidovudine therapy. J Med Virol. 1991;35:38–45. doi: 10.1002/jmv.1890350109. [DOI] [PubMed] [Google Scholar]
- 24.Sheppard H W, Lang W, Ascher M S, Vittinghoff E, Winkelstein W. The characterization of non-progressors: long-term HIV-1 infection with stable CD4+ T-cell levels. AIDS. 1993;7:1159–1166. [PubMed] [Google Scholar]
- 25.Van Kerckhoven I, Fransen K, Peeters M, De Beenhouwer H, Piot P, Van Der Groen G. Quantification of human immunodeficiency virus in plasma by RNA PCR, viral culture, and p24 antigen detection. J Clin Microbiol. 1994;32:1669–1673. doi: 10.1128/jcm.32.7.1669-1673.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winters M A, Tan L B, Katzenstein D A, Merigan T C. Biological variation and quality control of plasma human immunodeficiency virus type 1 RNA quantitation by reverse transcriptase polymerase chain reaction. J Clin Microbiol. 1993;31:2960–2966. doi: 10.1128/jcm.31.11.2960-2966.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong M T, Dolan M J, Kozlow F, Doe R, Melcher G P, Burke D S, Boswell R N, Vahey M. Patterns of virus burden and T-cell phenotype are established early and are correlated with the rate of disease progression in human immunodeficiency virus type 1 infected persons. J Infect Dis. 1996;173:877–887. doi: 10.1093/infdis/173.4.877. [DOI] [PubMed] [Google Scholar]
- 27a.Young, D. (Roche Laboratories). Personal communication.
- 28.Zhu Y S, Gong Y, Cimino G D. Quantitative analysis of HIV-1 RNA in plasma preparations. J Virol Methods. 1995;52:287–299. doi: 10.1016/0166-0934(94)00149-b. [DOI] [PubMed] [Google Scholar]