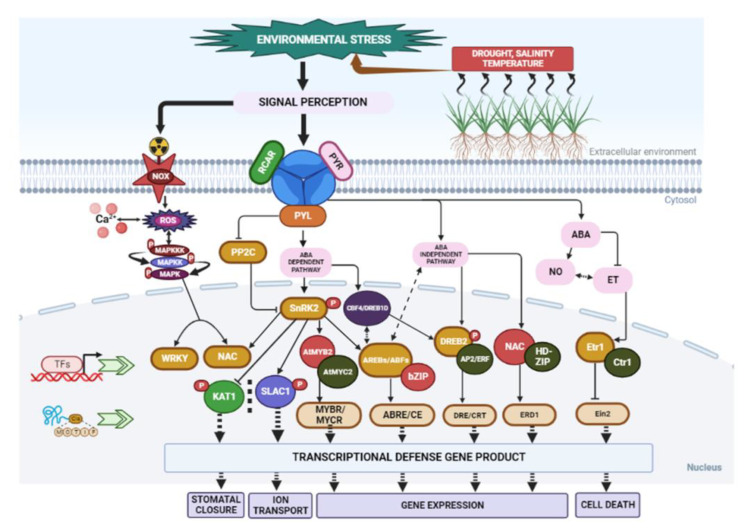
Figure 3.
Schematic overview of signal perception, transduction, and transcriptional regulatory networks under environmental stresses, including drought, heat, and other stresses. In plants with and without ABA, abiotic stress activates the ABA signalling system. In the absence of ABA, PP2C phosphatases (negative regulators) interact with SnRK2 kinases (positive regulators) and inhibit their activation. Signal transduction is blocked due to the inactivation of SnRK2s. The presence of ABA permits receptors to bind ABA and interact with PP2Cs, allowing SnRK2s to be released from binding. Autophosphorylation activates the SnRK2s, allowing for transduction to take place. SnRK2s that have been activated include phosphorylate downstream substrate proteins, such as transcription factors. Abbreviations: Abscisic acid (ABA); dehydration-response element-binding protein/C-repeat binding factor (DREB/CBF); ethylene response 1 (ETR1); Arabidopsis NAC domain-containing protein (ANAC); mitogen-activated protein kinase (MAPK); sucrose nonfermenting-1 related kinase(SnRK); transcription factors (TFs); reactive oxygen species (ROS); protein phosphatase 2C (PP2C); ABA receptors (PYR/PYL/RCAR); ethylene response factors (ERFs); nitric oxide (NO)-ethylene (ET) interplay; ABA-responsive element-binding proteins/ABA-responsive element -binding factor (AREB/ABF); Etr, ethylene-resistant; Ctr, constitutive triple response; Ein, ethylene-insensitive; basic-domain leucine zipper (bZIP); dehydration-responsive elements/C-repeat (DRE/CRT); ABA-responsive elements (ABREs); potassium channel protein in Arabidopsis thaliana 1 (KAT1); slow anion channel 1 (SLAC1).