Abstract
The nuclear factor kappa B (NF-κB) family of transcription factors orchestrates signal-induced gene expression in diverse cell types. Cellular responses to NF-κB activation are regulated at the level of cell and signal specificity, as well as differential use of family members (subunit specificity). Here we used time-dependent multi-omics to investigate the selective functions of Rel and RelA, two closely related NF-κB proteins, in primary B lymphocytes activated via the B cell receptor. Despite large numbers of shared binding sites genome wide, Rel and RelA directed kinetically distinct cascades of gene expression in activated B cells. Single-cell RNA sequencing revealed marked heterogeneity of Rel- and RelA-specific responses, and sequential binding of these factors was not a major mechanism of protracted transcription. Moreover, nuclear co-expression of Rel and RelA led to functional antagonism between the factors. By rigorously identifying the target genes of each NF-κB subunit, these studies provide insights into exclusive functions of Rel and RelA in immunity and cancer.
Subject terms: Gene regulation in immune cells, Chromatin
Sen et al. provide in-depth temporal multi-omic analyses with single-cell RNA-sequencing (scRNA-seq) profiles of nuclear factor kappa B (NF-κB)-regulated gene expression in B cells upon B cell receptor (BCR) activation. Their findings reveal distinct kinetic patterns of gene expression mediated by RelA and Rel and functional antagonism between the closely related NF-κB subunits.
Main
The nuclear factor kappa B (NF-κB) family of transcription factors mediate responses to cell stimulation1. NF-κB responses have been most extensively studied in response to proinflammatory signals such as tumor necrosis factor alpha (TNFα), interleukin 1 beta (IL1β) and bacterial lipopolysaccharides (LPS)2,3. These agents induce NF-κB by the post-translational (classical) pathway via degradation of inhibitor of NF-κB (IκB) proteins. RelA, Rel and Nfkb1 family members are major mediators of classical NF-κB activation. Other stimuli, such as lymphotoxin β, B cell activating factor (BAFF) and CD40 activate the nonclassical pathway by elevating levels of NF-κB-inducing kinase (NIK). Thereafter, NIK-mediated phosphorylation and processing of Nfkb2 permit nuclear translocation of RelB-containing heterodimers for gene expression. Given the multitude of signals that activate it, a fundamental question in NF-κB biology is the basis by which the specificity of cellular responses is assured.
The specificity question can be parsed in three ways. First, how is cellular specificity achieved? This pertains to circumstances where the same signal induces NF-κB in different cell types, evoking different responses. Second, how is stimulus specificity achieved? This applies when distinct NF-κB stimuli induce NF-κB in the same cell type yet evoke distinct responses. Third, how is subunit specificity achieved? This is most pertinent in the comparison of Rel and RelA, both of which are activated by the classical pathway and coexpressed in most hematopoietic cells. The importance of subunit specificity is evident from the distinct phenotypes of Rel- or Rela-gene knockouts in mice4–8.
NF-κB-dependent gene expression has been thoroughly explored in macrophages. Using stringent criteria, a study discussed in ref. 9 identified a subset of primary response genes that were dependent on RelA for expression in response to lipid A. A smaller subset of these bound RelA at their promoters and were proposed to be direct targets of RelA. Sites of RelA recruitment in LPS-activated macrophages also bind PU.1, a B- and macrophage-specific transcription factor, and are marked by DNase 1 hypersensitive sites before LPS treatment10. These observations have led to the working model that cell-specific responses are directed by cell-specific accessibility of NF-κB to selected sites in the genome. However, this reasonable model has not been systematically tested by comparing gene expression and genome-wide binding in different cell types. The basis for stimulus specificity of NF-κB-dependent responses has also been explored in the context of inflammatory signaling in macrophages. Accumulating evidence indicates that differences in kinetic patterns of NF-κB induction by specific stimuli contribute to the pattern of inducible gene expression11,12.
By contrast, the basis for subunit specificity is least explored. According to a study discussed in ref. 13, careful analysis of the affinity of different NF-κB family members to related κB motifs suggests that subunit specificity could, in part, be achieved via distinct κB motifs in RelA- or Rel-specific target genes. Due to inadequate definition of RelA or Rel target genes, this model has also not been experimentally validated. It even remains unclear whether RelA and Rel bind to disparate κB sites in cells, as suggested by in vitro studies.
RelA and Rel have distinct functions in B cell biology, although neither protein is essential for B cell development. Rel deficiency alone, or in combination with Nfkb1, severely reduces germinal center (GC) formation during immune responses14. Thus, RelA cannot compensate for Rel in GC responses. Additionally, ectopic expression of transgenic Rel has been shown to increase GC responses15. NF-κB proteins, Rel in particular, have also been implicated in B cell lymphomas16–18. Despite involvement in critical physiological phenomena, NF-κB subunit specificity that distinguishes essential roles of Rel in GC responses or cancer is not known.
In this study, we combined time-dependent multi-omics to gain insights into the basis for subunit specificity of NF-κB family members in splenic B lymphocytes activated via the B cell receptor (BCR). RelA and Rel bound at early and late activation time points, respectively, and subsets of genomic-binding sites were selective for each factor. Bulk and single-cell RNA-sequencing (scRNA-seq) studies revealed marked heterogeneity of RelA- or Rel-specific gene expression and temporally distinct sets of target genes for each factor. The list of target genes identified included many that had not been previously linked to NF-κB. Additionally, we found that marginal zone (MZ) B cells were refractory to early NF-κB signaling, and nuclear co-expression of RelA and Rel resulted in mutual antagonism between the two factors. Our studies define kinetically distinct cascades of gene expression regulated by specific NF-κB subunits in BCR-activated B cells and thereby provide insights into exclusive functions of RelA and Rel in immunity and cancer.
Results
NF-κB target genes are poorly defined, especially in primary untransformed cells. Additionally, the extent to which RelA or Rel compensate for each other to regulate gene expression has not been previously characterized. To address these questions, we examined the responses of mouse splenic B cells activated via the BCR. We considered genes to be direct targets of NF-κB if the following criteria were observed: (1) they bound NF-κB in response to stimulation, (2) their expression changed in response to cell stimulation and (3) their inducible activity was altered by the absence of NF-κB (ref. 19). We applied these criteria to each NF-κB subunit to identify RelA- or Rel-specific target genes. A substantial subset of genes that satisfied only the latter two criteria was considered ‘indirect’ targets of NF-κB.
Genome-wide recruitment of RelA and Rel
BCR crosslinking initiates two phases of NF-κB activation in mouse B splenic B cells20. The first transient phase occurs by translocation of both RelA and Rel from cytoplasmic pools. Nuclear levels of RelA peak around 1 h and are restored back to the cytoplasm by 4 h. The second phase is dominated by newly translated Rel, with nuclear levels increasing between 6 h and 24 h. To account for the biphasic response, we examined NF-κB binding, chromatin accessibility, histone modifications and gene expression at 1, 4 and 18 h after continuous BCR crosslinking.
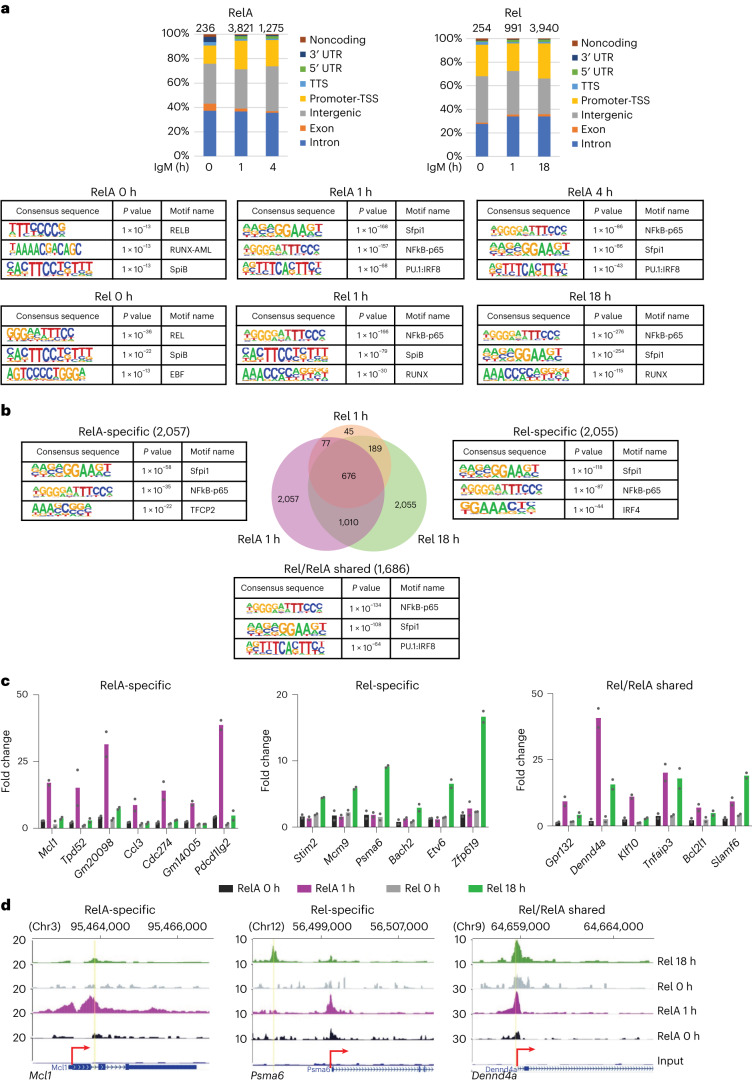
We carried out chromatin immunoprecipitation followed by sequencing (ChIP–seq) with anti-RelA and anti-Rel antibodies in naïve B cells treated with anti-IgM (Fab′2). At optimal anti-IgM crosslinking conditions, we determined that approximately half the cells induced RelA nuclear translocation (Supplementary Fig. 1a,b). Specificity of each antibody for ChIP was substantiated by quantitative PCR at previously characterized NF-κB target genes (Supplementary Fig. 1c). After peak calling using CisGenome (Supplementary Fig. 1d), we focused on peaks that were present in two biological replicate experiments (Supplementary Fig. 1e). RelA was recruited to 3,821 sites, which were mostly located in intronic and intergenic regions, after 1 h BCR crosslinking of wild-type (WT; C57BL/6) splenic B cells (Fig. 1a, left). RelA-bound sites dropped sharply at 4 h, closely following the pattern of bulk nuclear RelA. Hypergeometric Optimization of Motif EnRichment10 (HOMER, findMotifsGenome.pl program) analysis (Fig. 1a, bottom) identified motifs for Sfpi1, NF-κB-RelA and PU.1/IRF composite motifs as the top three sequences associated with regions of inducible RelA binding (see also Supplementary Fig. 1f). By contrast, maximum Rel binding (3,940 sites) occurred after 18 h of activation (Fig. 1a, right), also to intronic and intergenic regions that were enriched for NF-κB, Sfpi1 and RUNX binding motifs (Fig. 1a, bottom, and see also Supplementary Fig. 1g). HOMER analyses were substantiated using TFmotifview21 (Supplementary Fig. 1h). Inducible Rel-binding sites at 1 h largely overlapped with RelA binding at 1 h (Supplementary Fig. 1i). We conclude that the first and second phases of NF-κB-induced signaling lead to genome-wide recruitment of RelA and Rel, respectively. More restricted recruitment of Rel at early time points occurred to sites that also bound RelA.
Fig. 1. Identification of RelA- and Rel-binding sites in BCR-activated B cells.
a–d, Splenic B cells from C57BL/6 mice were activated with anti-IgM (F(ab)′2) for 0, 1, 4 and 18 h. ChIP was carried out using anti-RelA and anti-Rel antibodies. Coprecipitated genomic DNA was used to generate libraries for sequencing. Data from two independent ChIP–seq experiments were processed as described in Methods. Results shown are for RelA- and Rel-binding sites that were common to both replicates with threshold peak scores as described in Methods. a, Genomic locations of RelA (left) and Rel (right) binding sites across multiple genomic regions, as annotated by HOMER. Total number of peaks at each activation time point are noted above the bars. Top substantial transcription factor binding motifs enriched within RelA peaks (top panels) and Rel peaks (lower panels) at different time points were identified using HOMER (see also Supplementary Fig. 1f,g). b, RelA and Rel ChIP–seq libraries were merged to identify shared and unique binding sites for each NF-κB subunit at each different time points (Venn diagram). Sequence motifs within RelA-specific, Rel-specific and RelA/Rel shared peaks are shown alongside (see also Supplementary Fig. 1k; a and b, Default statistical setting used to obtain P value by HOMER). c, Rel or RelA binding to select target genes was verified by ChIP–PCR. Data shown are the average of two additional ChIP experiments with cells activated for the indicated times; fold change = 2(CT(input)−CT(target))/2(CT(input)−CT(neg)). d, Representative browser tracks based on mm9 annotation of ChIP–seq profiles of genes that bind only RelA (Mcl1), only Rel (Psma6) or both Rel and RelA (Dennd4a). The y axis represents normalized reads per 10 million aligned reads (see also Supplementary Fig. 1l).
Approximately half (42%) of Rel-binding sites at 18 h overlapped RelA binding at 1 h (Fig. 1b); several thousand other sites were selective for either RelA or Rel. These assignments were validated for a subset of genes via independent ChIP followed by quantitative PCR assays (Fig. 1c). Genomic locations of RelA- and Rel-specific binding sites were similar (Supplementary Fig. 1j). However, Rel-bound sites were enriched for motifs of IRF proteins (Supplementary Fig. 1k). Representative genome browser tracks are shown in Fig. 1d and Supplementary Fig. 1l. We conclude that RelA or Rel recognizes similar sequences in vivo and many sites sequentially bind RelA (early) and Rel (late).
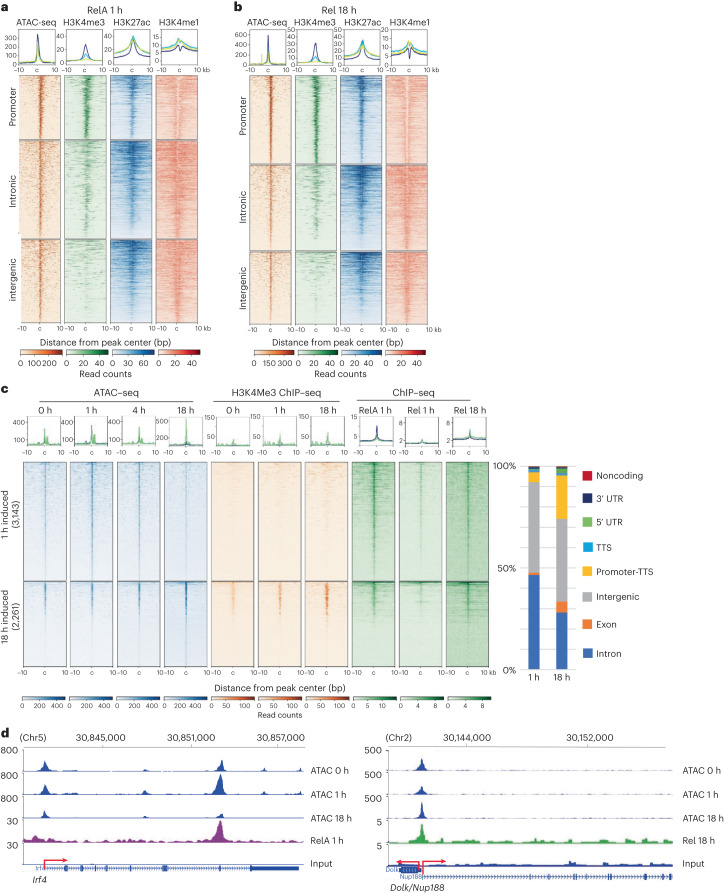
To examine epigenetic features of NF-κB responses, we carried out assay for transposase-accessible chromatin with sequencing (ATAC-seq) and histone ChIP–seq over the same time course (Supplementary Fig. 2a,b). Sites of both early RelA and late Rel recruitment were marked by accessible chromatin and histone modifications associated with active promoters (H3K4me3) or enhancers (H3K27ac) before cell activation (Fig. 2a,b). From time-dependent ATAC-seq, we identified several thousand sites at which pre-existing chromatin accessibility was transiently increased (Fig. 2c, top). Most of these sites bound RelA at 1 h and were not marked by H3K4me3, indicating that they fell in introns and intergenic regions. Transient ATAC sensitivity at these sites was consistent with changes being induced by transiently activated transcription factors such as RelA. An example is the Irf4 gene (Fig. 2d, left). Increased chromatin accessibility with 18 h activation correlated with Rel binding to a subset of H3K4me3+ promoter sites (Fig. 2c,d, bottom and right, respectively). Inducible and constitutive ATAC sites were enriched for distinct transcription factor motifs (Supplementary Fig. 2c,d). We conclude that RelA or Rel binding to enhancers and promoters, respectively, can induce chromatin accessibility changes in response to BCR activation.
Fig. 2. Epigenomic characteristics of inducible RelA- and Rel-binding regions in activated B cells.
a–d, Chromatin accessibility and histone modifications were assessed by ATAC-seq and ChIP–seq, respectively, in splenic B cells activated with anti-IgM for 0, 1, 4 and 18 h. a,b, Pre-existing chromatin state in unactivated B cells at sites of inducible RelA binding at 1 h (left) and Rel binding at 18 h (right). Heatmaps were centered on summits of RelA or Rel peaks, ordered based on the strength of ATAC signal and segregated by genomic locations as indicated to the left of each panel. Boxed plots above each heatmap show the average profile across all sites at each genomic location (blue = promoters (defined as −1 kb to +0.1 kb relative to the transcription start site), yellow = intergenic regions and green = introns). c, Chromatin accessibility changes induced by activation were determined using DiffBind analysis of ATAC-seq data (fold change ≥ 1.5, FDR ≤ 0.05). Transiently induced ATAC peaks (at 1 h) are shown on the top half (3,143 total peaks) and those induced at 18 h are shown at the bottom (2,261 total peaks), ordered according to the strength of the ATAC signal. H3K4me3 and RelA or Rel ChIP–seq profiles of the corresponding sites at the indicated time points are shown in adjacent panels. Boxed plots above each heatmap show the average ChIP–seq or ATAC-seq profile for each category of inducible ATC peaks (blue = 1 h induced ATAC peaks and green = 18 h induced ATAC peaks). Induced ATAC peaks at 1 h and 18 h were annotated to genomic regions shown on the right. d, Representative browser tracks of ATAC-seq peaks induced at 1 h (Irf4) or 18 h (Dolk/Nup188) and associated RelA or Rel binding at the indicated times. Transcriptional direction and transcription initiation sites are shown by red arrows. mm9 annotations are shown above the tracks.
Transcriptional responses to BCR stimulation
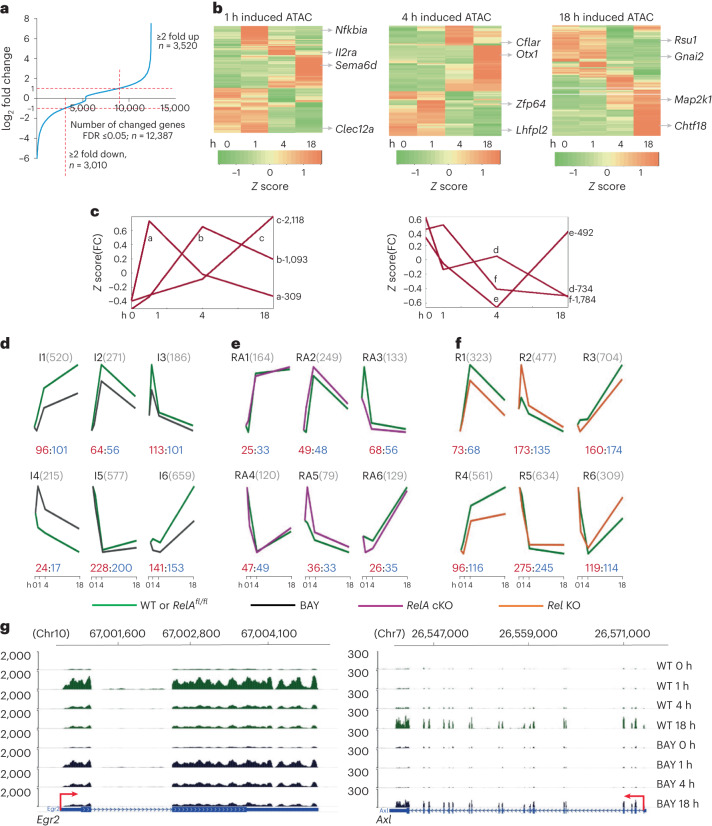
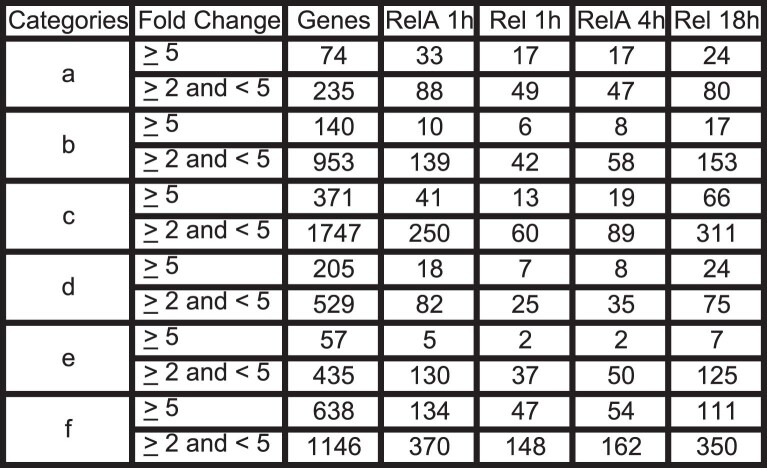
To decipher relationships between chromatin changes, RelA/Rel binding and gene expression, we carried out RNA-seq analyses of splenic B cells activated with anti-IgM (Supplementary Fig. 3a). We identified approximately 3,000 genes by EBSeq22 that were either upregulated or downregulated more than twofold over the time course (FDR ≤ 0.05; Fig. 3a). The timing of gene expression changes mostly coincided with or followed changes in chromatin accessibility (Fig. 3b). Upregulated genes (≥2-fold change) were further classified by k-means clustering into three categories based on the peak expression at 1, 4 or 18 h (Fig. 3c). After annotating NF-κB binding to genes using HOMER (annotatePeak.pl algorithm), we observed inducible RelA binding to 39% of genes in category ‘a’ (rapidly and transiently induced) and 13% of genes in categories ‘b’ and ‘c’ (Extended Data Table 1). RelA binding decreased after 4 h of anti-IgM treatment, whereas numbers of Rel-bound genes increased at 18 h, concordant with the overall patterns of RelA and Rel ChIP–seq (Extended Data Table 1). For parity, downregulated genes (≤2-fold change) were also distributed into three categories that broadly coincided with rapid, intermediate and transient downregulation (Fig. 3c, labeled d, e and f). Both RelA and Rel bound to several hundred genes that were downregulated by anti-IgM treatment (Extended Data Table 1).
Fig. 3. NF-κB-dependent transcriptional responses in activated B cells.
a–g, Differential gene expression was analyzed using EBSeq (FDR ≤ 0.05) from two biological replicate experiments. a, Fold changes in gene expression at either 1, 4 or 18 h after activation compared to unactivated cells. Horizontal red dotted lines correspond to fold change threshold of ≥2. Numbers of genes upregulated or downregulated by ≥2× are indicated. b, Heatmaps represent expression levels of genes that were (1) differentially expressed by twofold at any time point with anti-IgM treatment and (2) associated with induced ATAC-seq peaks at either 1, 4 or 18 h time points as indicated above the heatmaps. Representative genes are shown on the right. c, Inducible transcripts were partitioned into three upregulated (patterns a–c) and three downregulated (patterns d–f) kinetic patterns by k-means clustering. Each graph represents the centroid profile of the average of z score of fold change (FC) compared with untreated cells at 1, 4 and 18 h. Total number of genes in each pattern are indicated on the right. d–f, RNA-seq analyses in activated B cells treated with IKK2 inhibitor BAY, or B cells that lack RelA or Rel. Differentially expressed genes were identified by DESeq2 (FDR ≤ 0.05). Genes that were induced at least twofold in control cells in each group were further separated into six kinetic patterns by k-means clustering. Graphs represent the centroid profile of the average row z score of fold change for genes in each cluster color coded as described below. Names of clusters and numbers of genes in each cluster are shown on top. The numbers of genes to which RelA (1 h) and Rel (18 h) were bound are indicated in red and blue font, respectively, below the patterns. Activation times are noted on the x axis. d, Kinetic patterns of BAY-responsive transcripts in activated C57BL/6 WT B cells in the absence (green) or presence of BAY (black). e, Kinetic patterns of differentially expressed transcripts in activated B cells from control RelAfl/fl (green) mice and RelA cKO mice (purple) activated with anti-IgM for times indicated on the x axis. f, Kinetic patterns of differentially expressed transcripts in activated B cells from control C57BL/6 mice (green) and Rel KO mice (orange) activated with anti-IgM for the indicated times. g, Representative browser tracks (mm9 annotation) from RNA-seq analyses of early (Egr2) and late (Axl) NF-κB target genes in WT and BAY-treated cells. Anti-IgM treatment times are shown on the right; transcriptional direction and transcription initiation sites are shown by red arrows.
Extended Data Table 1.
Inducible number of genes and RelA or Rel binding across differentially expressed genes in each k-means category. Each category was further segregated into two groups with >5 or >2 and <5-fold change. Numbers correspond to RelA or Rel ChIP–seq peaks at the indicated times that were annotated to genes in each group by HOMER
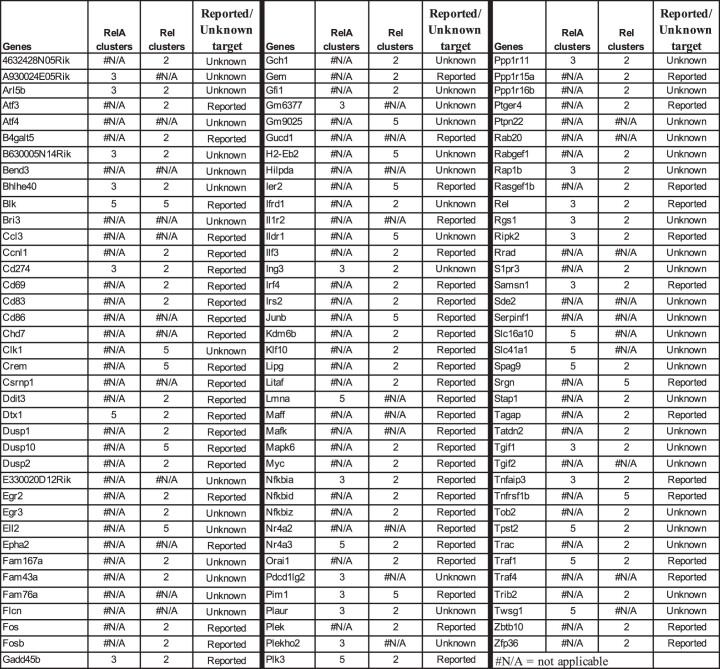
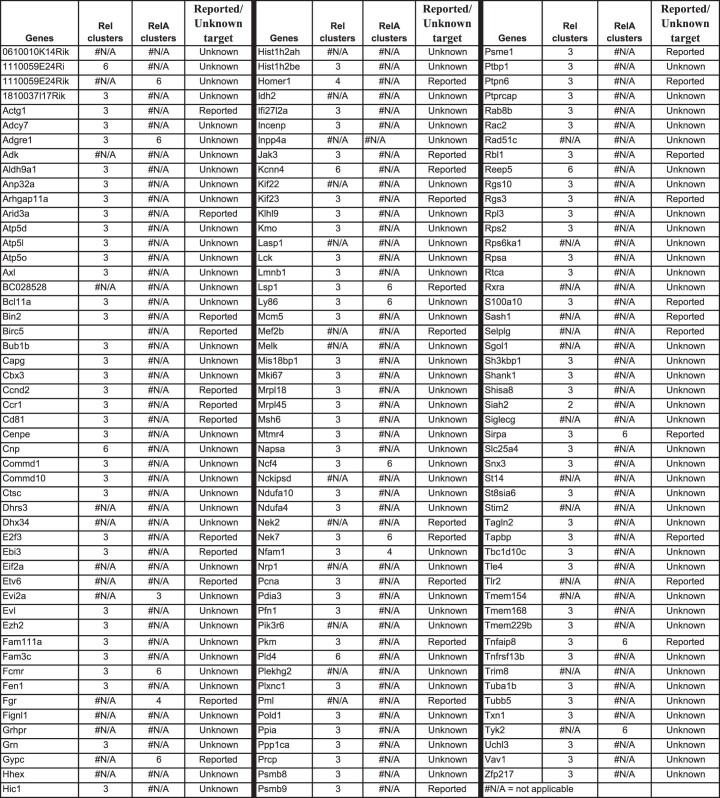
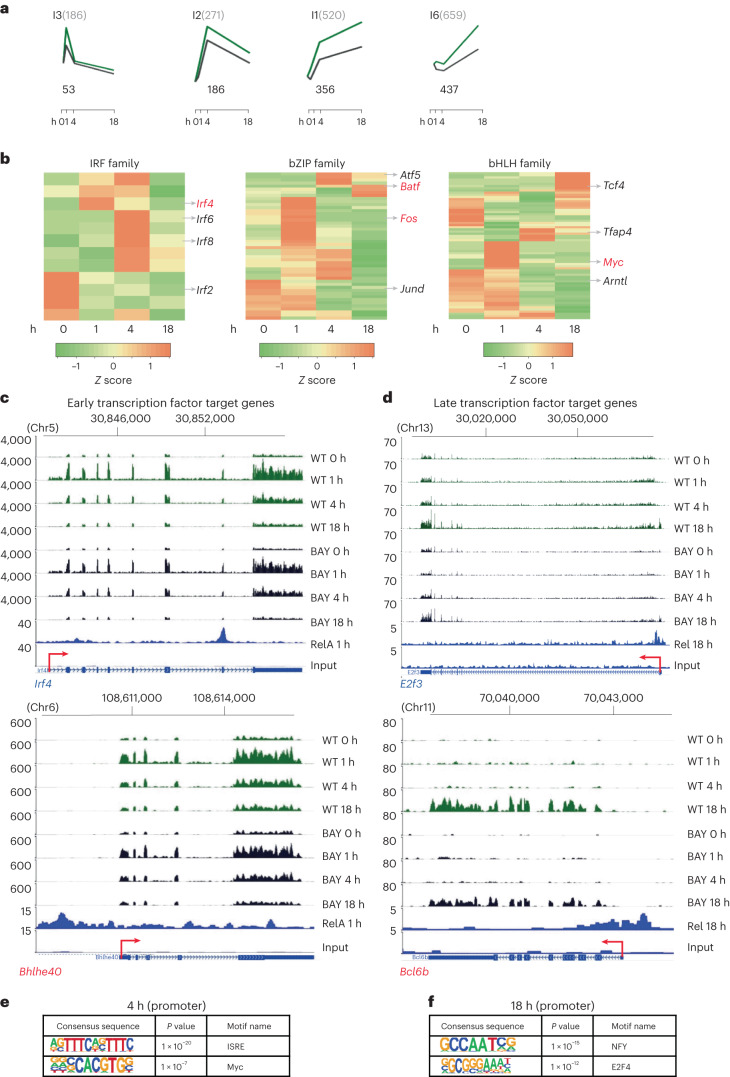
To identify NF-κB target genes, we carried out RNA-seq analyses with WT splenic B cells treated with anti-IgM in the presence of BAY 11-7082 (abbreviated as BAY hereafter), an inhibitor of IKK2 (ref. 23). By blocking classical NF-κB activation, the inhibitor suppressed BCR-induced RelA and Rel at early time points (Supplementary Fig. 3b, top). We found that BAY treatment also abrogated late Rel activation (Supplementary Fig. 3b, bottom), thereby permitting its use throughout the experimental time course. Differential gene expression analysis by DESeq2 followed by k-means clustering identified six kinetic patterns of gene expression (Fig. 3d, clusters I1–I6). A total of 186 transiently induced RNAs were suppressed by the inhibitor (Fig. 3d, cluster I3); 113 of these genes bound RelA at 1 h (red numbers), implicating them as direct NF-κB targets (Extended Data Table 2). A total of 101 of these genes also bound Rel at 18 h (blue numbers), although RNA levels did not change. A total of 271 genes with intermediate activation profiles were suppressed by the inhibitor (Fig. 3d, cluster I2), of which 64 bound RelA at 1 h. A total of 1,179 late-induced genes were suppressed by the inhibitor (Fig. 3d, clusters I1 and I6), of which 254 bound Rel at 18 h. Although many of these genes also bound RelA at 1 h, maximal RNA expression coincided with late Rel binding. For a small subset of target genes, we confirmed that RNA induction was accompanied by increased protein expression (Supplementary Fig. 3c). Taken together, these experiments identify early and late NF-κB target genes in BCR-activated B cells (Fig. 3g and Extended Data Tables 3–5).
Extended Data Table 2.
Direct NF-κB targets from Fig. 3d, cluster I3
Extended Data Table 3.
Direct NF-κB targets from Fig. 3d, cluster I1
Extended Data Table 5.
Direct NF-κB targets from Fig. 3d, cluster I2
In each cluster of differentially expressed genes, we found many genes that did not inducibly bind either RelA or Rel (Fig. 3d, difference between gray and red/blue numbers). Our interpretation is that such genes were activated by transcription factors induced by NF-κB and reflect transcriptional cascades initiated by NF-κB activation (see subsection ‘NF-κB-induced cascades’). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses revealed distinct biological functions of early and late NF-κB activation (Supplementary Fig. 3d). The early transient response (cluster I3) captured pathways such as ‘NF-κB signaling’ and various cancer-related pathways as the most significant. By contrast, long-term responses (clusters I1 and I6) were enriched for aspects of cell cycle regulation, metabolism and neurodegenerative diseases (Supplementary Fig. 3d). Finally, RelA and Rel also bound to many genes that were activated by BCR crosslinking but unaffected by the inhibitor (Supplementary Fig. 3e) or unaffected by activation (Supplementary Fig. 3f). Such genes may be activated by NF-κB in response to other stimuli.
Subunit selective NF-κB target genes
To distinguish between the effects of RelA and Rel, we carried out time-dependent RNA-seq in RelA-(RelAfl/flxCd19-cre mice; Supplementary Fig. 4a) or Rel- (Rel−/−) deficient B cells following BCR crosslinking. Previous studies have shown that germline or conditional Rel knockouts have similar B cell subsets and functional responses24. Differential gene expression analyses were carried out with DESeq2 and displayed after k-means clustering (Fig. 3e,f). The most prominent effects of RelA deficiency were observed at 1 h (Fig. 3e, clusters RA3 and RA5), whereas those of Rel deficiency were observed at 4 h and 18 h (Fig. 3f, clusters R1, R3 and R4). Absence of Rel also increased the inducible activation of many genes at 1 h (Fig. 3f, cluster R2). We conclude that RelA and Rel regulate temporally distinct phases of inducible gene expression in B cells.
Rapidly activated genes
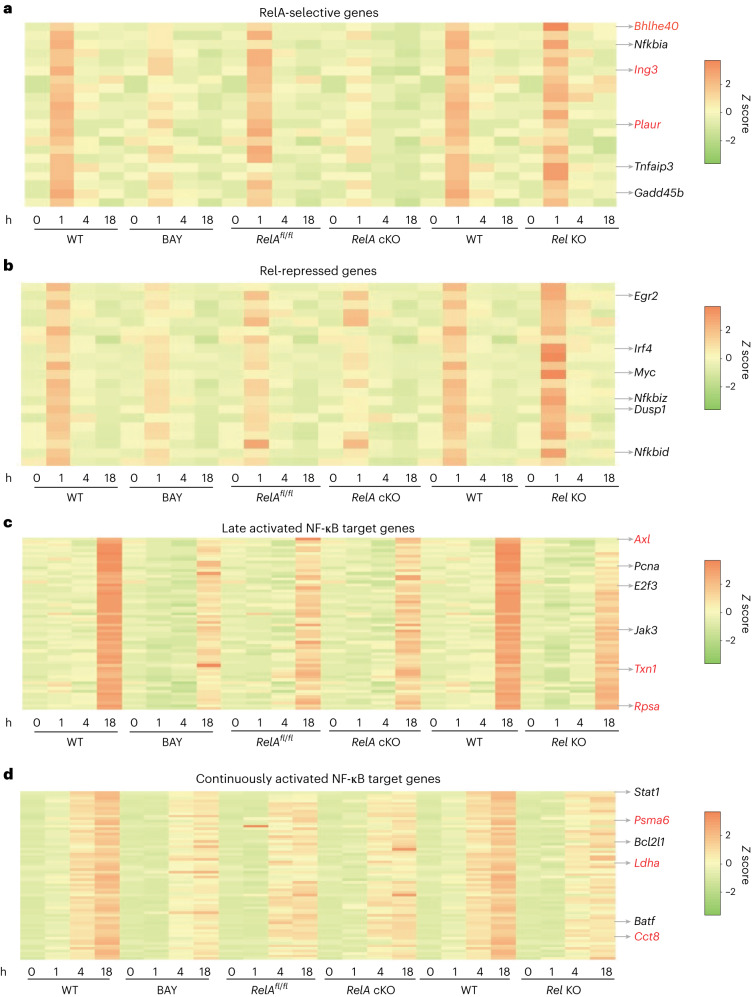
Twenty-one of 113 RelA-binding genes in cluster I3 were shared with cluster RA3 (Fig. 4a and Extended Data Table 2). These genes were classified as being RelA-selective. Some of the best documented NF-κB target genes, such as Tnfaip3, Gadd45b and Nfkbia, were in this subset. This list also contained many genes, such as Bhlhe40, Arl5b and Plaur, that had not been previously associated with RelA. Many RelA-selective genes were upregulated in the absence of Rel (cluster R2), indicating that nuclear co-expression of both factors attenuated RelA-dependent early gene activation (Extended Data Table 2).
Fig. 4. Subunit selective expression patterns of NF-κB target genes.
a–d, RNA-seq z score heatmaps of selected genes affected by BAY 11-7082 treatment (columns 1–8), RelA deficiency (columns 9–16) or Rel deficiency (columns 17–24). Each set of eight columns shows kinetic expression patterns in control (WT or RelAfl/fl) and test cells (BAY, RelA cKO and Rel KO) after 0, 1, 4 and 18 h of activation. Representative genes are noted on the right of each panel. Newly identified NF-κB target genes are noted in red font. a, Expression patterns of 20 RelA-selective genes defined as those whose induction was reduced by the inhibitor and RelA deficiency (overlap between clusters I3 and RA3 in Fig. 3d,e). Expression patterns of the same genes in Rel-deficient B cells are shown in the last eight columns (most of these genes fell in cluster R2 in Fig. 3f). b, Additional genes whose expression was increased in Rel-deficient compared to WT B cells (cluster R2 in Fig. 3f). c, Expression patterns of NF-κB target genes that were maximally induced between 4 h and 18 h of activation (cluster I6 in Fig. 3d). d, Those that were discernibly upregulated at 4 h but reached maximal expression 18 h (cluster I1 in Fig. 3d).
To circumvent possible developmental abnormalities in B cells from RelAfl/flxCd19-cre mice, we used Tat-Cre25 to delete RelA in mature B cells (Supplementary Fig. 4b). As controls, WT (C57BL/6) B cells were treated identically. We activated control and RelA-depleted cells with anti-IgM and evaluated the effects on RelA-selective gene induction. We found that activation profiles after RelA deletion in mature B cells closely resembled that of B cells from RelAfl/flxCd19-cre mice (Supplementary Fig. 4c).
Eighty-one of 113 NF-κB target genes in cluster I3 were not differentially regulated in RelA-deficient B cells (Extended Data Table 2), suggesting that they were activated by either RelA or Rel. Yet, many of these genes fell in cluster R2 indicating that RelA was a better activator compared to Rel (Fig. 4b). Our observations demonstrate differential activation potential of RelA or Rel on endogenous genes. This subset included genes such as Dusp1, Nfkbid, Nfkbiz, Myc and Irf4. A few genes in cluster I3 fell in RA5, indicating that they were positively regulated by Rel and suppressed by RelA at 1 h. We conclude that the simultaneous nuclear presence of both RelA and Rel sets up a state of mutual antagonism at early activation time points.
Intermediate and late time points
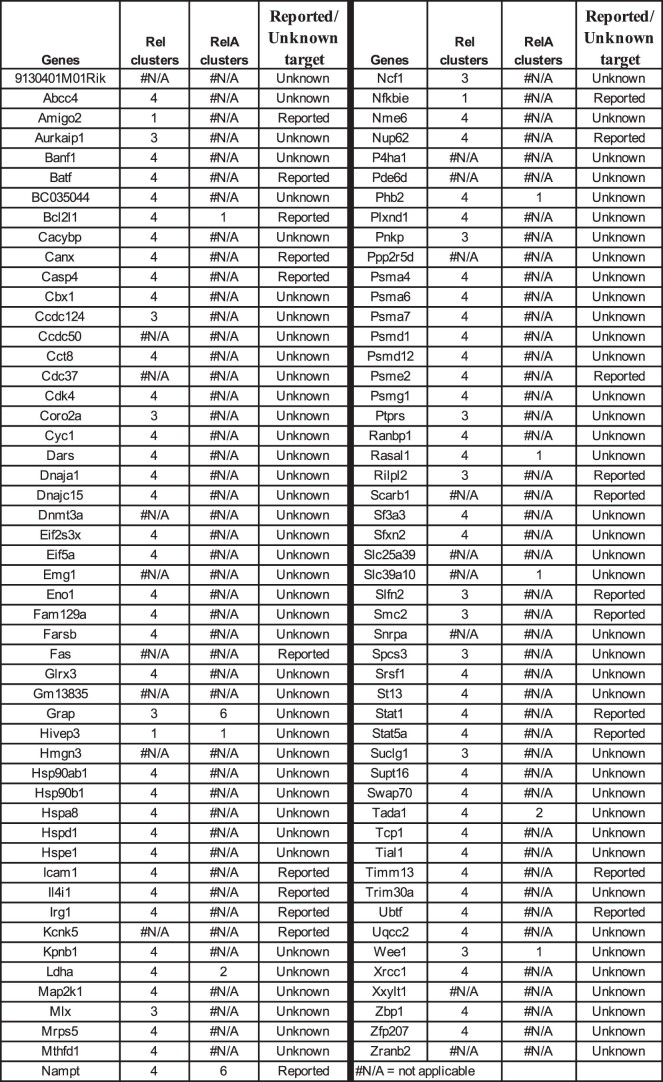
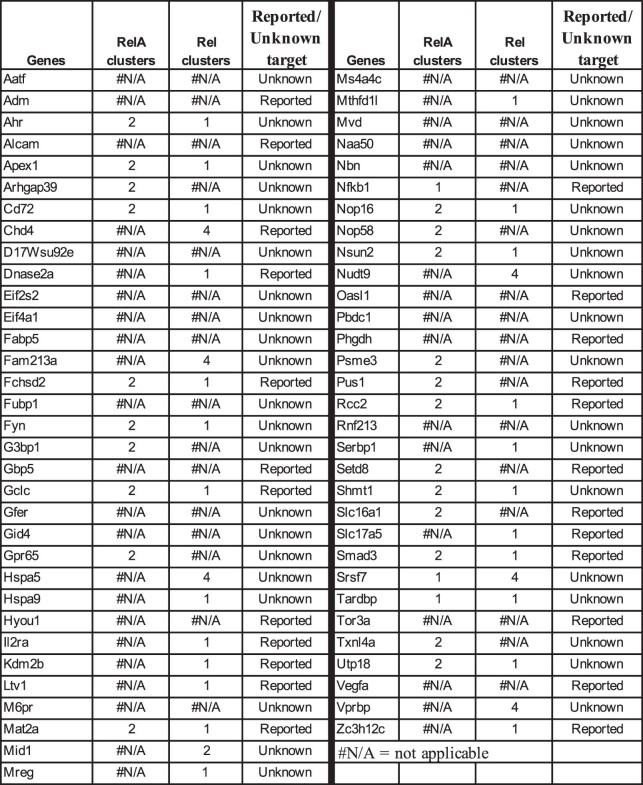
Most RelA/Rel-binding genes with maximal expression at 18 h (clusters I1 and I6) were unaffected by RelA deficiency and located in Rel-responsive clusters R3 and R4 (Figs. 3f and 4c,d and Extended Data Tables 3 and 4). We propose that these genes are direct targets of Rel. These included well-accepted Rel target genes (such as Bcl2l1), putative NF-κB target genes (such as Icam1) and many other genes that were not previously associated with Rel/NF-κB (such as Axl, Ldha and Timm13). Rel binding to a subset of these genes (Supplementary Fig. 4d) was further confirmed by ChIP–qPCR experiments (Supplementary Fig. 4e). Approximately one-third of NF-κB-responsive genes that were maximally expressed at 4 h (cluster I2) fell in cluster R1, marking them as Rel targets (Extended Data Table 5). We conclude that Rel-dependent gene expression occurs as early as 4 h after stimulation and dominates the NF-κB transcriptional landscape at longer time points. Most Rel target genes identified here have not been previously associated with NF-κB (Extended Data Tables 3–5).
Extended Data Table 4.
Direct NF-κB targets from Fig. 3d, cluster I6
To distinguish between NF-κB target gene expression in follicular and MZ B cells, we activated each cell type purified by fluorescence-activated cell sorting (FACS) (Supplementary Fig. 5a) with anti-IgM and carried out time-dependent RNA-seq. We found twofold fewer rapid, transiently induced genes in MZ cells (Supplementary Fig. 5b,c), and the NF-κB signaling pathway was missing in MZ cells at early time points (Supplementary Fig. 5d). Attenuated early NF-κB activation in MZ cells was substantiated by quantitative RT–PCR of select early induced NF-κB target genes (Supplementary Fig. 5e). These observations indicate that our present studies of NF-κB-dependent gene activation largely reflect responses of follicular B cells.
Singe-cell analysis of NF-κB-dependent gene expression
To further dissect NF-κB responses, we carried out scRNA-seq in activated B cells from WT, conditional RelA knockout (RelAfl/flxCd19-cre, RelA cKO) and Rel knockout (Rel−/−) mice. To focus on short- and long-term targets of RelA and Rel, respectively, we activated RelA cKO cells for 0, 1 and 4 h and Rel−/− B cells for 0, 1 and 18 h. Uniform Manifold Approximation and Projection (UMAP) visualization showed relatively minor differences between genotypes, presumably reflecting alterations in only small subsets of genes due to the absence of specific NF-κB family members (Supplementary Fig. 6a). We focused on the behavior of RelA and Rel target genes identified by bulk RNA-seq analyses described above.
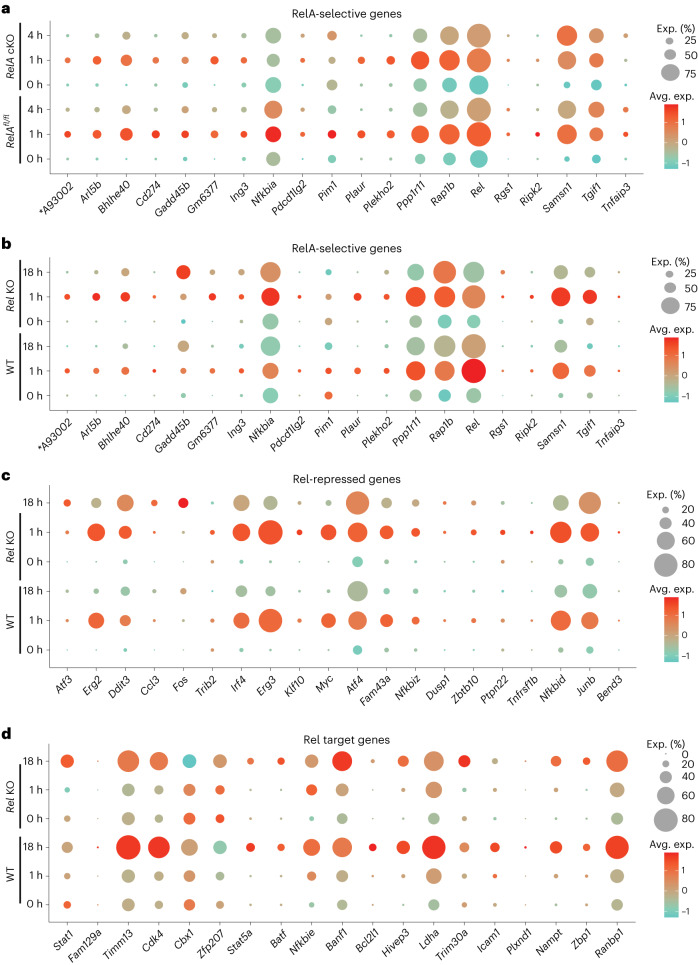
We observed markedly heterogenous responses of 20 RelA-selective genes (Fig. 5a). Some of the best-known NF-κB targets, such as Tnfaip3, Gadd45b and Pim1, were expressed in small proportions (5–25%) of activated cells. By contrast, Rap1b and Rel were expressed in relatively large proportions (70–90%) of cells. Single-cell analyses further corroborated reduced NF-κB activation in MZ B cells. Strongly RelA-dependent genes, such as Nfkbia and Pim1, were poorly induced in a cluster tentatively identified as MZ (based on the expression of Cd1d1, Cd9, Tm6sf1, Dtx1, S1pr3 and Cr2) regardless of the RelA genotype (Supplementary Fig. 6b). However, other early NF-κB target genes that are less RelA-dependent, such as Samsn1 and Bhlhe40, were inducible in this subset of cells. Additional studies will be required to rigorously define the contribution of BCR-induced NF-κB signaling in MZ B cells. We quantified the effects of RelA deficiency by measuring the average scaled expression levels in control and RelA-deficient cells (Supplementary Fig. 6c). We found that kinetic patterns and RelA selectivity of genes identified by bulk RNA-seq were also evident at single-cell resolution.
Fig. 5. scRNA-seq analysis of NF-κB target genes in activated B cells.
a–d, scRNA-seq was carried out with anti-IgM-activated B cells from RelAfl/fl (control) and RelAfl/flxCd19-cre (RelA cKO) for 0, 1 and 4 h or from C57BL/6 (WT) and Rel−/− B cells treated with anti-IgM for 0, 1 and 18 h. In total, 10,000 B cells were assayed from each condition. Data analysis was carried out by Seurat as described in Methods. Dotplot visualizations of differential gene expression patterns of select NF-κB target genes are shown. Gene expression changes are plotted by both scaled average expression (avg. exp., illustrated by dot color) and the percentage of cells found to express that gene (exp (%), depicted by the size of the dot). Cellular origin of libraries is indicated on the y axis. a, Single-cell analysis of RelA-selective genes from Fig. 4a (x axis). Expression levels and proportions of gene-expressing cells in control and RelA cKO B cells are depicted at 0, 1 and 4 h. b, Expression patterns of RelA-selective genes from Fig. 4a are depicted in WT and Rel-deficient B cells activated for 0, 1 and 18 h. c, Single-cell expression patterns of additional Rel-repressed genes (from cluster R2 in Fig. 3f) in WT and Rel-deficient B cells activated for 0, 1 and 18 h. d, Single-cell analysis of Rel target genes (from clusters R3 and R4 in Fig. 3f) in WT and Rel-deficient B cells activated for 0, 1 and 18 h. Additional quantitation for all patterns is shown in Supplementary Fig. 6c–f. An asterisk indicates that A93002 = A930024E05Rik.
To probe the basis for increased expression of many NF-κB target genes in Rel-deficient B cells, we first examined the status of RelA-selective genes in scRNA-seq from WT and Rel-deficient B cells. Many of these genes, such as Arl5b, Nfkbia and Samsn1, were upregulated on a per-cell basis in Rel−/− B cells (Fig. 5b and Supplementary Fig. 6d). Increased expression in Rel−/− B cells was also evident for genes that were not RelA-selective, including Nfkbid, Trib2 and Ptpn22 (Fig. 5c and Supplementary Fig. 6e). In addition to higher expression per cell, many of these genes were also expressed in a greater proportion of Rel−/− B cells (for example, Bhlhe40, Samsn1, Atf4 and Irf4; Fig. 5b,c). We conclude that nuclear co-expression of RelA and Rel attenuates RelA-dependent transcription. Reduced expression of Rel in Rel−/− B cells may reflect positive autoregulation of the Rel gene promoter by Rel26 or reduced stability of the noncoding transcript.
Rel target genes also showed substantial heterogeneity at the single-cell level (Fig. 5d and Supplementary Fig. 6f). Their responsiveness to Rel was evident in both reduced expression per cell and reduced proportions of cells expressing these genes at the 18 h time point (for example, Timm13, Ldha and Bcl2l1). A minority of genes showed the opposite trend in scRNA-seq from that predicted by bulk RNA-seq (for example, Banf1). Mechanisms underlying this pattern remain to be explored. Overall, single-cell studies strongly corroborated target gene predictions and kinetic patterns observed in bulk RNA-seq analyses and revealed wide variations in NF-κB responses in BCR-activated B cells.
NF-κB-initiated cascades
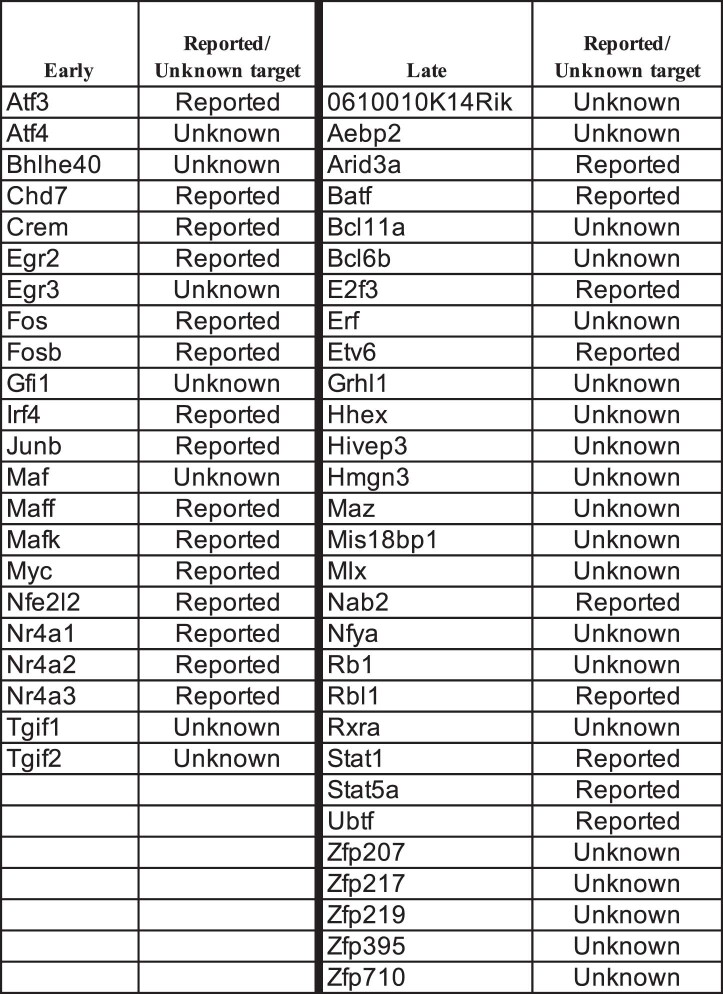
Many genes that were activated in B cells and inhibited by BAY did not bind RelA or Rel in ChIP–seq assays (Fig. 6a and Supplementary Fig. 7). We reasoned that these genes were targets of transcription factors activated by NF-κB and referred to as indirect targets of NF-κB (ref. 19). Of the many transcription factor genes induced in response to BCR signaling (Fig. 6b), a subset met our criteria for being direct NF-κB targets (Extended Data Table 6). Early activated transcription factor genes included previously known NF-κB targets such as Fos/Fosb, Myc and Irf4 (http://www.bu.edu/NF-kB/gene-resources/target-genes/), as well as new genes such as Tgif1/2, Bhlhe40 and Atf4 (Fig. 6c). Late-induced NF-κB-responsive transcription factor genes were targets of Rel, and most had not been previously associated with NF-κB (Fig. 6d and Extended Data Table6).
Fig. 6. NF-κB-activated transcriptional cascades.
a–f, Genes whose expression was altered by blocking both RelA and Rel induction with BAY 11-7082 but did not bind either RelA or Rel were inferred to be targets of NF-κB-induced transcription factors. a, Such genes were identified in four of the six k-means clusters in BAY-treated cells (Fig. 3d). Graphs show average row z score of fold change for genes in each cluster in activated WT B cells in the absence (green) or presence of BAY (black; from Fig. 3d). Activation times are noted on the x axis. Numbers on the top of each plotted kinetic pattern denote total number of genes in that cluster; numbers below each plot denote the numbers of genes that did not score for RelA or Rel binding at any activation time by ChIP–seq analyses. b, Heatmap profiles of three transcription factor gene families induced in response to B cell activation. Representative NF-κB target genes are indicated to the right of each panel (red font). See Extended Data Table 6 for a complete list of NF-κB target genes that encode transcription factors. c,d, Browser tracks of early (c) and late (d) transcription factor genes that were affected by BAY treatment. The top eight lines show RNA-seq tracks at different time points of anti-IgM treatment in the presence (BAY) or absence (WT) of BAY. The bottom two lines show RelA 1 h or Rel 18 h ChIP–seq and associated input tracks. e,f, DNA-binding motifs enriched in promoters (−400 to +100 bp relative to the transcription start site) of indirect NF-κB targets at 4 h (e) and 18 h (f). Motifs were identified using the known motifs setting in the HOMER algorithm. Significance was calculated using default statistical setting provided by HOMER.
Extended Data Table 6.
Direct transcription factor targets of NF-κB
To test the idea that indirect NF-κB target genes were activated by NF-κB-induced transcription factors, we searched for DNA-binding motifs in promoters (−400 bp to +100 bp relative to transcription start site) of genes that were upregulated at 4 h (clusters I2 + I1) and 18 h (clusters I1 + I6). Interferon-stimulated regulatory elements and Myc-binding sites were enriched in promoters of genes induced at 4 h (Fig. 6e), suggesting that a subset of indirect NF-κB target genes were direct targets of Irf4 and Myc. Promoters of indirect target genes that reached the highest levels of expression at 18 h were enriched for binding sites NF-Y and E2F4 (Fig. 6f). Rel targets included the gene encoding the closely related factor E2f3 (Fig. 6d). The basis for the emergence of the NF-Y motif among late activated indirect target genes remains unclear. We hypothesize that Rel-regulated transcription factors, such as Arid3a, Bcl11a, Rxra, Bcl6b, Hhex and several zinc finger proteins, whose motifs were not identified in this analysis, activate gene expression from promoter distal sequences. We conclude that early and late NF-κB induce an array of transcription factor genes that activate cascades of NF-κB-regulated gene expression in activated B cells. Both direct and indirect NF-κB target genes were implicated in oncogenic processes in the CancerSea database (Supplementary Fig. 8a,b).
Discussion
Our studies reveal that two closely related NF-κB family members drive kinetically distinct patterns of gene expression in activated B cells. Rigorous identification of many previously unknown targets of each factor revealed transcriptional cascades initiated by NF-κB activation that impact B cell function and cancer. We found that both RelA and Rel contributed to early transient gene expression. Simultaneous nuclear presence of RelA and Rel resulted in attenuated RelA-dependent gene expression that was manifested both at the expression level per cell and in proportions of cells that activated specific target genes. By contrast, late NF-κB target genes were largely Rel-dependent. We also noted that NF-κB transcriptional responses were markedly heterogeneous across individual cells. Indeed, some of the best-known target genes, such as Tnfaip3 and Gadd45b among early genes and Bcl2l1 among late genes, were induced in less than 20% of cells. Implications of our findings for B cell biology and cancer follow.
Genome-wide RelA and Rel binding was highest at times of maximal nuclear expression of each factor, at 1 h and 18 h postactivation, respectively. Residual RelA binding at 4 h occurred largely at sites that were also RelA-bound at 1 h. Most Rel binding at 1 h overlapped with RelA, setting up competition for binding at a subset of sites. Bulk and scRNA-seq analyses showed that nuclear co-expression of Rel and RelA down modulated RelA-dependent gene expression. Rel has been previously proposed to repress select inflammatory genes in TNFα-activated fibroblasts by recruiting HDAC1 (ref. 27). We have not investigated whether this mechanism applies in BCR-activated B cells. However, we favor the idea that repressive functions of Rel result from competition for binding sites, with Rel being a poorer transcriptional activator compared to RelA. About 40% of sites that bound RelA at 1 h also bound Rel at 18 h. In principle, early RelA binding followed by late Rel binding could confer persistent NF-κB activity as has been noted for sequential RelA and RelB recruitment28. This proved not to be the case.
It is interesting to note that approximately 2,000 sites were exclusively occupied by either RelA (at 1 h) or Rel (at 18 h; note that the use of ‘exclusive’ here is constrained by thresholds used to define ChIP–seq peaks during analysis). This discrimination was not based on sequences underlying RelA- or Rel-specific peaks because both sets were enriched for Sfpi1 and NF-κB motifs. Nor was it governed by chromatin accessibility because both regions were premarked with active chromatin marks in resting B cells. One possibility is that levels of post-translationally induced Rel at 1 h may be lower than de novo translated Rel present at 18 h, leading to binding site selectivity by mass action. Alternatively, site selectivity may be determined by subunit composition. Pre-existing RelA or Rel that translocate early have been proposed to largely consist of p50-containing heterodimers29, whereas higher levels of Rel at later time points may have substantial proportions of Rel homodimers.
Among 113 transiently induced NF-κB target genes that bound RelA at 1 h, only 21 were adversely affected in RelA-deficient B cells. We infer that Rel does not effectively substitute for the absence of RelA at these genes and termed them as being RelA-selective. The relatively few RelA-selective genes identified in this study may be a special feature of activated B cells that express robust levels of Rel. In cells that express lower levels of Rel, RelA-containing NF-κB may be the dominant factor activating these genes. Most of the 113 early NF-κB target genes also bound Rel at 18 h, yet RNA levels peaked at 1 h. Thus, sequential binding of RelA and Rel did not confer protracted transcriptional activity of these genes.
We identified an additional 254 NF-κB target genes that reached maximal expression at late time points (clusters I1 and I6). These genes were negatively affected by the absence of Rel but not RelA, marking them as Rel targets. Approximately 60% of late target genes (cluster I6) bound RelA at 1 h, yet maximal RNA levels occurred coincident with Rel binding at 18 h. Despite RelA binding at 1 h, expression of these genes was unaltered by RelA deficiency, indicating that early RelA binding was not required for their maximal expression at later times. These observations demonstrate that RelA or Rel binding per se is not an accurate measure of transcriptional activity induced by these factors.
We hypothesize that the late transcriptional activity of Rel target genes is determined by the nature of neighboring sequences. We found that RelA-bound regions associated with the 113 early and transiently induced NF-κB target genes (cluster I3) were enriched for NF-κB and Sfpi1 binding motifs, whereas Rel peaks associated with 153 late activated NF-κB-responsive genes (cluster I6) were enriched for NF-κB and IRF binding motifs. Because several Irf genes were induced in response to BCR stimulation, this pattern is consistent with the notion that late Rel works with factors that are upregulated at earlier stages. Interestingly, some Irf genes are themselves early NF-κB targets, suggesting that early NF-κB target genes may cooperate with late-induced Rel to activate late gene expression.
Indirect NF-κB targets were affected by combined blockade of RelA and Rel but did not bind either protein. Our working hypothesis is that the majority of these genes are regulated by NF-κB-induced transcriptional regulators. Indeed, many genes encoding transcription factors were among the few hundred direct NF-κB targets we identified. Some are well-established, such as Myc and Irf4. Many more were newly identified targets, including Tgif1 and Tgif2 that are implicated in different kinds of cancers30–34, stress-responsive transcription factors (Atf3 and Atf4) that are implicated in crosstalk with NF-κB in tumors35–38 and Bhlhe40, a transcription factor implicated in immune function39.
The group of new Rel-regulated transcription factors was also biologically meaningful. Two new direct targets included Hhex and Bcl6b, both of which have been shown to be involved in optimal GC responses40–42. Although GC responses are severely reduced in germline Rel knockout mice14,24, Rel target genes that manifest their critical role in GCs are not known. Our evidence that Rel directly activates two key GC factors provides the first insight into pathways by which Rel impacts GC responses. Another Rel target identified here, Bcl11a, also links Rel to the GC. Bcl11a has been shown to be highly expressed in GCs18,43, but its functional role has not been evaluated. It is not our intention to claim that 18 h activated B cells are equivalent to GC B cells. We surmise that connections between Rel and Hhex/Bcl6b/Bcl11a established here may be strengthened in the GC microenvironment to optimize Rel-dependent responses.
NF-κB has been shown to be necessary for a subset of human B cell lymphomas44,45; translocations of REL and NFKB2 have been noted in others16,46. Target genes that mediate NF-κB’s oncogenic potential are not well characterized. Both early and late target genes identified here provide insights into this question. Early target genes were linked to hypoxia, inflammation and quiescence in the CancerSea database, suggesting a role for NF-κB in initiating oncogenesis. By contrast, late target genes were associated with cell cycle, DNA damage and repair and invasion, suggesting continued impact of NF-κB during cancer progression. Several genes in our list of Rel targets, such as Arid3a, Bcl11a and Bcl6b, could serve as downstream effectors of ectopic Rel activation during lymphomagenesis47. Additionally, the Rel target gene Maz has been implicated in cancers48,49 and as a multiple sclerosis susceptibility gene50. Most early induced NF-κB target genes associated with cancers (such as Tgif1,2 and Atf3,4) were also Rel-responsive, indicating that early and late Rel target genes may drive oncogenesis. Because many of these target genes encode transcription factors, NF-κB-initiated transcriptional cascades may further amplify its oncogenic functions.
Methods
Mice, B cell preparation, cell culture and antibodies
Primary B cells were purified from the spleens of 6- to 12-week-old WT (C57BL/6), RelAfl/fl (ref. 51), RelA cKO (RelAfl/flxCd19-cre; generated by crossing RelAfl/fl with Cd19-cre (Jackson Laboratory, 006785)) and Rel KO (Rel−/−) (ref. 17). For each experiment, five to six age-matched mice (mixed male/female) were used. Additional randomization was not required for these studies because the data were obtained ex vivo from pooled B cells obtained from multiple mice. Data collection and analysis were not performed blind to the conditions of the experiments. No animals or data points were excluded from the analyses for any reason.
All studies were conducted under the guidance of the Institutional Animal Welfare Assurance Number—NIH Intramural Research Program, D16-00602; NIA AAALAC unit, 000401. Mice were housed in an intramural NIH vivarium with full AAALAC International accreditation. The circadian light cycle is 12 h light and 12 h dark that are on automatic timers with electronic monitoring monitored by the Siemens building control system. The temperature is set at 72 °F (range, 69–75 °F) with a humidity range of 30–70%. The mice are fed an ad libitum diet from Envigo (Envigo 2018SX Global 18% protein extruded rodent diet). The Baltimore municipal water is filtered by reverse osmosis and then chlorinated at 2–3 ppm. All housing standards are as per the Guide for the Care and Use of Laboratory Animals.
Single-cell suspensions from the spleen were prepared in RoboSep buffer (Stemcell Technologies, 20104) and enriched for B cells by negative selection using EasySep Mouse B cell Isolation Kit (Stemcell Technologies, 19854). B cells were cultured at 2 × 106 cells per ml in RPMI 1640 medium supplemented with 10% heat-inactivated FBS (HyClone, SH30088.03), penicillin–streptomycin–glutamine (Invitrogen, 10378016) and 55 nM 2-mercaptoethanol (Sigma-Aldrich, M6250). Follicular and MZ B cells were FACS sorted, as previously described52, using BD FACS-Aria Fusion. Follicular B cells were sorted as B220+ (BioLegend, 103240), AA4.1− (BioLegend, 136510), CD21lo (BioLegend, 123410) and CD23hi (BioLegend, 101606). MZ B cells were sorted as B220+, AA4.1− and CD21hiCD23lo. TAT-Cre treatment was as follows: splenic B cells (10 × 106 ml−1) from RelAfl/fl and C57BL6 (WT, as a control) were treated with 100 μg of TAT-Cre (Excellgen, EG-1001) for 45 min at 37 °C, in humidified CO2 incubator and then washed. Using a dead cell removal kit (Miltenyi Biotec, 130-090-101), live cells were isolated after TAT-Cre treatment and recultured for 72 h in RPMI 10% FBS supplemented with 50 ng ml−1 of BAFF (R&D Systems, 2149-BF). After 72 h, cultures were washed using warm RPMI, and cells were cultured at 2 × 106 cells per ml until the next steps. Cells were activated with 10 μg ml−1 goat anti-mouse IgM Fab′2 (Jackson Laboratory, 115-006-075) for 1, 4 and 18 h. After activation, dead cells were removed with dead cell removal magnetic microbeads (Miltenyi Biotec, 130-090-101). To inhibit IKK signaling, cells were pretreated with 1 µM BAY 11-7082 (Calbiochem, B5556) for 1 h before anti-mouse IgM stimulation. The following antibodies were used for ChIP, ChIP–seq or western blot: anti-RelA (Santa Cruz Biotechnology, sc-372), anti-Rel (Santa Cruz Biotechnology, sc-71), anti-hnRNP A1 (Santa Cruz Biotechnology, sc-32301), anti-H3K27ac (Active Motif, 39133), anti-H3K4me1 (Abcam, ab8895) and anti-H3K4me3 (Abcam, ab8580). Horseradish peroxidase-coupled goat anti-mouse IgG (Santa Cruz Biotechnology, sc-2005) and goat anti-rabbit IgG (Santa Cruz Biotechnology, sc-2004) were used for immunoblotting.
Immunoblotting
Proteins were separated by electrophoresis through 10% SDS–PAGE and electrophoretically transferred to nitrocellulose membrane. After blocking with 5% dried milk in Tris–HCl (pH 7.4)-buffered saline/0.05% Tween (TBST) for 1 h, membranes were incubated with primary antibodies overnight at 4 °C, washed in TBST and incubated for 1 h with horseradish peroxidase-coupled secondary antibodies. Proteins were detected by using the enhanced chemiluminescence systems (Pierce Biotechnology, 32106) and imaged using Syngene Imaging System.
Flow cytometry and image cytometry
Isolated B cells (0.5 × 106 cells) from spleens were fixed with 2% paraformaldehyde (Electron Microscopy, 15713) for 10 min in the dark at room temperature (23 °C). Fixed cells were washed with PBS and resuspended in ice-cold Perm Buffer III (BD Biosciences, 558050) and incubated overnight at −20 °C. Cells were washed with PBS and resuspended in BD perm/wash buffer (BD Biosciences, 554723). Cells were either stained with rabbit-anti mouse RelA (Cell Signaling Technology, 8242S; 1:1,500) or isotype (Cell Signaling Technology, 3900; 1:1,000) antibody for 1 h at 4 °C. Cells were washed with perm/wash buffer and restained with anti-rabbit Fab′2-AF647 antibody (Cell Signaling Technology, 4414; 1:1,000) for 30 min. Similarly activated cells were stained with the following antibodies: Myc-AF-647 (Cell Signaling Technology, 13871; 1:50), Egr2-APC (eBiosciences, 17-6691-82; 1:50), Rel-PE (eBiosciences, 12-6111-80; 1:200), Ikba-PE (Cell Signaling Technology, 7523; 1:100), Dec1-AF-594 (Novus Biologicals, 1800AF594; 1:100), Ezh2-PE (BD Biosciences, 562478; 1:50), CD72-BV421 (BD Biosciences, 740058; 1:200) and HSP90-β (Invitrogen, PA3-012; 1:1,500). HSP90-β-stained cells were further stained with anti-rabbit Fab′2-PE antibody (Cell Signaling Technology, 79408; 1:1,000) for 30 min. Finally, cells were washed and acquired on BD Aria Fusion or BD Symphony flow cytometer, and data were analyzed with FlowJo v10.1.
For image cytometry, cells were stained as above for RelA after anti-IgM (F(ab)′2) or PMA (Sigma-Aldrich, P1585) and Ionomycin (Sigma-Aldrich, I0634) treatment and later resuspended in perm/wash buffer solution containing DAPI (500 ng ml−1; Thermo Fisher Scientific, 62248) and RNase A (1 μg ml−1; Invitrogen, 912091021). Stained cells were acquired on Amnis ImageStream X Mk II Imaging Flow Cytometer (Luminex), and data were analyzed using IDEAS 6.0 software (Luminex).
ChIP and ChIP–seq library construction
All ChIP experiments (n = 2) were performed as previously described19. Briefly, for H3K27ac and H3K4me3 ChIP, cells were washed twice with PBS, fixed at room temperature (23 °C) with 1% formaldehyde in PBS for 10 min. For RelA and Rel ChIP, cells were crosslinked with 1.5 mM EGS (Pierce Biotechnology, 21565) for 30 min followed by 1% formaldehyde (Sigma-Aldrich, F8775) at room temperature (23 °C) for 15 min in PBS. Reactions were quenched by adding glycine to a final concentration of 0.125 M, and cells were washed twice with cold PBS. Cells were resuspended in cell lysis buffer (50 mM HEPES–KOH (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate and 0.1% SDS), rotated for 15 min at 4 °C and then centrifuged at 1,200g for 10 min at 4 °C. The nuclear pellet was resuspended in nuclear lysis buffer (50 mM HEPES–KOH (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate and 1% SDS) and rotated for 15 min at 4 °C. After centrifugation, the pellet was lysed in buffer containing 50 mM HEPES–KOH (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS and protease inhibitors. The crosslinked chromatin was fragmented by sonication (Branson sonicator) to a shear size of 200–2,000 bp. ChIP was performed with 2.5 μg anti-RelA or anti-Rel antibody, 3 μg anti-H3K27ac or anti-H3K4me3 antibody prebound to 50 μl Protein A (Invitrogen, 10001D) or G Dynabeads (Invitrogen, 10003D). Sonicated chromatin was added to antibody-bound beads and incubated at 4 °C overnight. Beads were collected by centrifugation, washed and incubated at 65 °C for 4 h in elution buffer (50 mM Tris–HCl (pH 7.5), 10 mM EDTA and 1% SDS) to reverse crosslinking. ChIP DNA was purified by phenol–chloroform extraction followed by ethanol precipitation. ChIP–qPCR was performed using Sybr Green Fast (Applied Biosystems, 4385610). The y axis for Fig. 1c was calculated by the following formula: fold change = 2(CT(input)−CT(target))/2(CT(input)−CT(neg control)). The y axis for Supplementary Fig. 4e represents ChIP signals at target amplicons compared to input control. Primer sequences are provided in Supplementary Table 1.
For sequencing, adapters were ligated to the precipitated DNA fragments or the input DNA to construct a sequencing library according to the manufacturer’s protocol for the NuGEN Ovation Ultralow DNA-seq library preparation kit (0344-32, Tecan). A 75 bp single-end read sequencing was performed on Illumina NextSeq 500 instrument. Two biological replicate ChIP–seq experiments were carried out for each antibody.
ATAC-seq library construction
ATAC-seq library construction (n = 2) was performed as previously described with minor modifications53. In total, 100,000 B cells were lysed with 100 µl of lysis buffer (10 mM Tris–HCl (pH 7.4), 10 mM NaCl, 3 mM MgCl2, 0.1% NP-40 and 0.1% Tween-20). After centrifuging at 500x g for 10 min at 4 °C, pelleted nuclei were resuspended with 50 µl of transposition mix (FC-121-1030, Illumina; 1× Tagment DNA buffer, Tn5 Transposase, nuclease-free H2O) and incubated for 30 min at 37 °C in a thermomixer. Transposed DNA was purified using MinElute columns (Qiagen, 28004) and subsequently amplified with Nextera sequencing primers and NEB high fidelity 2× PCR master mix for 12 cycles (New England Biolabs, M0541). PCR-amplified DNA was purified using MinElute columns and sequenced using the Illumina NextSeq sequencer with paired-end reads of 150 bases.
RNA isolation for quantitative RT–PCR and RNA-seq of bulk B cells and Fo/MZ cells
From resting or activated B cells, follicular and MZ B cells, total RNA (n = 2) was isolated using the RNeasy Plus Mini Kit (Qiagen, 74134). For total B cells, cDNA was synthesized with the SuperScript First-Strand Synthesis System (Invitrogen, 11904018), while for follicular and MZ B cells, cDNA was synthesized with SuperScript IV VILO Master Mix (Invitrogen, 11756050). PCR was performed in duplicate using the ABI ViiA 7 Real-Time PCR System using QuantStudio 1.6.1 (Thermo Fisher Scientific, 4453545) with iTaq Universal SYBR Green Supermix (BioRad, 1725150). The primers are listed in Supplementary Table 2.
For RNA-seq, total RNA from B cells was used for barcoded library preparation using Illumina TruSeq total RNA preparation kit (Illumina, 20040525). Samples were sequenced at the Johns Hopkins Transcriptomics and Deep Sequencing Core using the Illumina HiSeq 2000 with 75 bp single-end reads.
For follicular and MZ B cells, indexed libraries were generated from 10 ng total RNA from two biological replicates using the SMARTer Stranded Total RNA-seq Kit v2—Pico Input Mammalian (Takara Bio, 634412) per manufacturer’s protocol. The libraries were sequenced on an Illumina NovaSeq 6000 using 100 bp paired-end reads. BCL files were demultiplexed and converted to FASTQ files using Illumina’s bcl2fastq program (v2.20.0.422).
RNA-seq analysis
RSEM54 (v1.3.2) was used to align RNA-seq reads to the mouse genome and quantify transcript abundance. EBSeq22 (v1.24.0) was used to produce normalized reads for all samples and to identify differential RNA expression in WT, follicular and MZ samples across the entire time course. Comparisons between WT and Rel KO or RelA cKO or BAY 11-7082-treated samples were carried out in DESeq2 (ref. 55). Differentially expressed genes were identified using DESeq2 (v1.22.1) following the model: ddsTC <- DESeqDataSetFromMatrix(countData = countData, colData = colData, design = ~Time+Treat+Time:Treat; ddsTC <- DESeq(ddsTC, test=“LRT”, reduced = ~ Treat+Time). Two filtering steps were followed to define differentially expressed genes in cells with modified NF-κB expression. First, all differentially changed genes were selected based on an FDR ≤ 0.05 after DESeq2 analysis; second, only those genes were selected that were more than twofold upregulated or downregulated in wild samples after anti-IgM treatment. K-means analysis of RNA expression data was carried out in MATLAB using fold change compared with 0 h, with correlation as the distance metric, repeat clustering set to five and other parameters set to default. RNA-seq analyses were performed with activated B cells treated with IKK2 inhibitor BAY or B cells that lack RelA or Rel. Genes that were induced at least twofold in control cells in each group were further separated into six kinetic patterns by k-means clustering. Names of clusters and numbers of genes in each cluster are shown in Fig. 3d–f. A number of genes to which RelA (1 h) and Rel (18 h) binding were annotated are indicated in red and blue font, respectively, below the patterns. Activation times are noted on the x axis. For visualization, bigwig files were generated. RelAfl/fl and RelA cKO samples were aligned to mouse genome assembly mm10, all other bigwig files were aligned to mm9. All samples were normalized to 1 million aligned reads for visualization. CancerSEA56 analysis in Supplementary Fig. 8a,b was analyzed in the link http://biocc.hrbmu.edu.cn/CancerSEA/ with default parameters set. Enriched KEGG pathways from the genes were identified using the web server tool ShinyGO 0.77 (http://bioinformatics.sdstate.edu/go/)57.
Analysis of ChIP–seq data
Bowtie1 (v1.0) software was used to map RelA and Rel quality-filtered reads from demultiplexed FASTQ files to mouse genome assembly mm9 (Ensembl v67) with default options. CisGenome software (http://www.biostat.jhsph.edu/~hji/cisgenome/) was used for peak calling with a window size of 200 bases and a threshold of four reads (two from each strand), FDR ≤ 0.1 and fold change ≥ 2. Peaks separated by less than 200 bases were merged into one peak. For RelA ChIP–seq peak calls, we selected the peaks that were more than tenfold changed compared to input, and for Rel peak calls, we selected the peaks that are more than fivefold changed compared to input in the Rel ChIP–seq. For histone ChIP–seq, we used Bowtie2 (v2.9.2) (ref. 58) with default parameters, aligned on the mouse genome assembly mm9. We used HOMER annotatePeaks.pl to annotate peaks with default parameters (promoter regions were defined from −1 kb to +100 bp). We then combined the two annotated gene lists together. Motif finding was carried out with HOMER using the de novo setting for underpeaks area and known motif setting for promoter motif analysis. TFmotifView program (http://bardet.u-strasbg.fr/tfmotifview/) was used to verify the RelA 1 h and Rel 18 h peaks (Supplementary Fig. 1h). The programs ‘findMotifsGenome.pl’ and ‘findMotifs.pl’ were used to identify transcription factor binding motifs within peaks or promoter regions (−400 to +100 bp from TSSs). All samples were normalized to 10 million reads for visualization.
Analysis of ATAC-seq data
Quality control of sequenced reads was performed by FastQC (v0.11.8) (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and adapter filtration was performed by Trimmomatic (v0.33) (ref. 59), with the Nextera paired-end adapter annotation provided in the software. Paired ends were aligned to mm9 using Bowtie2 (v2.9.2) (ref. 58) with parameter -X 2000, which specifies that paired reads with insertion up to 2,000 bp were allowed to align. Reads with mapping quality score < 10 were filtered out using Samtools (v1.9), and PCR duplicates were removed using Picard tools (v1.127; http://broadinstitute.github.io/picard/). Model based analysis of ChIP-Seq (MACS) 2 (v2.1.1) (ref. 60) was used for removing duplicate reads and calling peaks with parameter options –shift 100 –extsize 200. Differentially accessible peaks after anti-IgM treatment at 1 h, 4 h or 18 h were analyzed by DiffBind (v3.8.4; http://bioconductor.org/packages/release/bioc/vignettes/DiffBind/inst/doc/DiffBind.pdf), with FDR ≤ 0.05 and fold change ≥ 1.5 to define induced peaks at different time point compared with 0 h. HOMER findMotifsGenome.pl was used to investigate the motif enrichment of induced peaks compared with 0 h. For genome tracks, bigwig files were created from bam files using BEDTools61 and UCSC Genome Browser Utilities62. Genome tracks were explored using the WashU Epigenome Browser63.
Heatmap visualizations
Heatmaps displaying normalized read densities of ATAC-seq and ChIP–seq samples were generated with the computeMatrix and plotHeatmap modules of the deepTools package v3.4.1 (ref. 64).
Droplet-based single-cell sequencing
Using a Chromium Single Cell 3ʹ GEM Kit v3 (10x Genomics, 1000094), Chromium Single Cell 3ʹ Gel Bead Kit v3 (10x Genomics, 1000093) and the Chromium Chip B Single Cell Kit (10x Genomics, 1000153), the anti-IgM Fab′2 activated splenic B cell suspensions in RPMI plus 2.0% FBS, at 1,000 cells per µl, were loaded onto a chromium single-cell controller (10x Genomics) to generate single-cell gel beads-in-emulsion (GEMs) according to the manufacturer’s protocol. Briefly, approximately 10,000 cells per sample (n = 2) were added to chip B to create GEMs. Cells were lysed, and the bead captured poly(A) RNA was barcoded during reverse transcription in Thermo Fisher Scientific Veriti 96-well thermal cycler at 53 °C for 45 min, followed by 85 °C for 5 min. cDNA was generated and amplified. Quality control and quantification of the cDNA were conducted using Agilent’s High Sensitivity DNA Kit (5067-4626) in the 2100 Bioanalyzer. scRNA-seq libraries were constructed using a Chromium Single Cell 3ʹ Library Kit v3 (10x Genomics, 1000095) and indexed with Chromium i7 Multiplex Kit (10x Genomics, 220103). The libraries were sequenced using an Illumina HiSeq2500 sequencer with a paired-end, single-indexing strategy consisting of 28 cycles for read 1 and 91 cycles for read 2. Multiple sequencing runs were performed to achieve greater sequencing depth for each barcode.
scRNA-seq data processing
Demultiplexing of raw base call files into FASTQ files was completed with 10x Genomics Cell Ranger (v.3.0.2) mkfastq coupled with mouse reference version mm10. Cell Ranger count combine was used to aggregate multiple sequencing run reads to achieve increased sequencing depth. The results from Cell Ranger were processed in R v3.6.3 with Seurat v3.2.3 (ref. 65) using default parameters, unless otherwise specified. Quality control filtering was applied to each sample to eliminate downstream analysis of empty droplets, low-quality cells and potential doublets. Filtering excluded genes detected in less than ten cells and cells containing more than 10% mitochondrial genes. For the RelAfl/fl and RelA cKO datasets, the filtering also excluded cells expressing less than 400 or more than 5,000 transcripts and below 800 or above 18,000 counts. While, for 0-, 1- and 4-h datasets, cells express less than 300 or more than 4,000 transcripts and below 800 or above 15,000 counts. For the WT and Rel−/− 18-h dataset, 300 or 6,000 transcripts and 800 or 40,000 counts, respectively, were filtered out. Subsequently, each sample was normalized using the NormalizeData function. The top 2,000 highly variable genes, selected with the FindVariableFeatures function, were used for integrated analysis. Following scaling, principal component analysis was performed, and then 20 dimensions were used for the FindNeighbors and RunUMAP functions to generate UMAP, setting a resolution of 0.3 to group cells into clusters with distinct gene expression profiles.
Statistical analysis
Statistical tests were performed on qRT–PCR, ChIP–qPCR and flow cytometry data using GraphPad Prism 9.2.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41590-023-01561-7.
Supplementary information
Supplementary Figs. 1–8 with associated legends, full blot image for Supplementary Fig. 3b and full blot image depicting anti-Rel antibody (sc-71) validation.
ChIP–PCR primers used in Fig. 1c and Supplementary Figs. 1c and 4d.
qRT–PCR primers used in Supplementary Figs. 4c and 5e.
Supporting data for Supplementary Fig. 1b. Nuclear Translocation_Similarity index between RelA and DAPI.
Supporting data for Supplementary Fig. 3c. Median fluorescence intensity (MFI) for NF-κB targets.
Supporting data for Supplementary Fig. 4c. Gene expression analysis of RelA-selective genes in ex vivo RelA deleted B cells (by qRT–PCR CT values provided).
Supporting data for Supplementary Fig. 4e. ChIP–PCR analysis of Rel target genes at 18 h after anti-IgM treatment (CT values provided).
Supporting data for Supplementary Fig. 5e. Gene expression analysis of NF-κB target genes in follicular and MZ B cells (by qRT–PCR, CT values provided).
Source data
Statistical source data.
Acknowledgements
We thank all the members of Sen Lab in Gene Regulation Section (GRS) for valuable discussions. We thank M. Meffert (Johns Hopkins School of Medicine), C. Bassing (University of Pennsylvania) and W. Khan (University of Miami) for critical comments on the manuscript. We also thank E. Lehrmann for processing and tracking our GEO submittal. H.J. and W.Z. are partially supported by NIH grant R01HG009518. The remaining authors are supported by the Intramural Research Programs of the National Institute on Aging (NIA) and the National Institute of Allergy and Infectious Diseases (to M.Z. and P.L.).
Extended data
Author contributions
M.Z. and R.S. designed research; M.K. generated mice; M.Z. performed ATAC-seq, ChIP–seq, ChIP–PCR, bulk RNA-seq, western blots and qRT–PCR experiments; C.A.S. performed bulk and scRNA-seq; J.J. carried out RNA analyses; A.S. performed FACS sorting and Amnis analysis; P.C. performed ChIP–PCR and RT–PCR; J.F. sequenced libraries; S.N. initiated studies with Tat-Cre. M.Z., S.D., C.A.S., Y.Z., K.M.-M., K.G.B., W.Z., P.L. and H.J. analyzed data, and M.Z., C.A.S., A.S. and R.S. prepared the manuscript.
Peer review
Peer review information
Nature Immunology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary handling editor: L. A. Dempsey in collaboration with the Nature Immunology team. Peer reviewer reports are available.
Data availability
The datasets generated and analyzed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Superseries accession number GSE197035 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE197035). Source data for Fig. 1c are provided. ChIP–seq data are also available at https://wangftp.wustl.edu/~dli/nih-mingming/biowulf/ and may be visualized at WashU Epigenome Browser: https://epigenomegateway.wustl.edu/. TFmotifView results are available here: http://bardet.u-strasbg.fr/tfmotifview/?results=734fKxPAnCh8w1 (RelA 1 h); http://bardet.u-strasbg.fr/tfmotifview/?results=H1bHoIdGkHrKPy (Rel 18 h). Supplementary Data 1–5 and Source data are provided with this paper.
Code availability
No custom code was used in these studies.
Competing interests
The authors declare no conflicts of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
is available for this paper at 10.1038/s41590-023-01561-7.
Supplementary information
The online version contains supplementary material available at 10.1038/s41590-023-01561-7.
References
- 1.Hayden, M. S. & Ghosh, S. Regulation of NF-κB by TNF family cytokines. Semin. Immunol.26, 253–266 (2014). 10.1016/j.smim.2014.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dorrington, M. G. & Fraser, I. D. C. NF-κB signaling in macrophages: dynamics, crosstalk, and signal integration. Front Immunol.10, 705 (2019). 10.3389/fimmu.2019.00705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smale, S. T. & Natoli, G. Transcriptional control of inflammatory responses. Cold Spring Harb. Perspect. Biol.6, a016261 (2014). 10.1101/cshperspect.a016261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beg, A. A., Sha, W. C., Bronson, R. T., Ghosh, S. & Baltimore, D. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-κB. Nature376, 167–170 (1995). 10.1038/376167a0 [DOI] [PubMed] [Google Scholar]
- 5.Doi, T. S., Takahashi, T., Taguchi, O., Azuma, T. & Obata, Y. NFκB RelA-deficient lymphocytes: normal development of T cells and B cells, impaired production of IgA and IgG1 and reduced proliferative responses. J. Exp. Med.185, 953–961 (1997). 10.1084/jem.185.5.953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kontgen, F. et al. Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity, and interleukin-2 expression. Genes Dev.9, 1965–1977 (1995). 10.1101/gad.9.16.1965 [DOI] [PubMed] [Google Scholar]
- 7.Boffa, D. J. et al. Selective loss of c-Rel compromises dendritic cell activation of T lymphocytes. Cell Immunol.222, 105–115 (2003). 10.1016/S0008-8749(03)00114-X [DOI] [PubMed] [Google Scholar]
- 8.Tumang, J. R. et al. c-Rel is essential for B lymphocyte survival and cell cycle progression. Eur. J. Immunol.28, 4299–4312 (1998). [DOI] [PubMed] [Google Scholar]
- 9.Tong, A. J. et al. A stringent systems approach uncovers gene-specific mechanisms regulating inflammation. Cell165, 165–179 (2016). 10.1016/j.cell.2016.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell38, 576–589 (2010). 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng, Q. J. et al. NF-κB dynamics determine the stimulus specificity of epigenomic reprogramming in macrophages. Science372, 1349–1353 (2021). 10.1126/science.abc0269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adelaja, A. et al. Six distinct NFκB signaling codons convey discrete information to distinguish stimuli and enable appropriate macrophage responses. Immunity54, 916–930 e917 (2021). 10.1016/j.immuni.2021.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siggers, T. et al. Principles of dimer-specific gene regulation revealed by a comprehensive characterization of NF-κB family DNA binding. Nat. Immunol.13, 95–102 (2011). 10.1038/ni.2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zarnegar, B. et al. Unique CD40-mediated biological program in B cell activation requires both type 1 and type 2 NF-κB activation pathways. Proc. Natl Acad. Sci. USA101, 8108–8113 (2004). 10.1073/pnas.0402629101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kober-Hasslacher, M. et al. c-Rel gain in B cells drives germinal center reactions and autoantibody production. J. Clin. Invest.130, 3270–3286 (2020). 10.1172/JCI124382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang, C. C., Zhang, J., Lombardi, L., Neri, A. & Dalla-Favera, R. Rearranged NFKB-2 genes in lymphoid neoplasms code for constitutively active nuclear transactivators. Mol. Cell. Biol.15, 5180–5187 (1995). 10.1128/MCB.15.9.5180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng, S., Hsia, C. Y., Leone, G. & Liou, H. C. Cyclin E and Bcl-xL cooperatively induce cell cycle progression in c-Rel−/− B cells. Oncogene22, 8472–8486 (2003). 10.1038/sj.onc.1206917 [DOI] [PubMed] [Google Scholar]
- 18.Kober-Hasslacher, M. & Schmidt-Supprian, M. The unsolved puzzle of c-Rel in B cell lymphoma. Cancers (Basel)11, 941 (2019). 10.3390/cancers11070941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao, M. et al. Transcriptional outcomes and kinetic patterning of gene expression in response to NF-κB activation. PLoS Biol.16, e2006347 (2018). 10.1371/journal.pbio.2006347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Damdinsuren, B. et al. Single round of antigen receptor signaling programs naive B cells to receive T cell help. Immunity32, 355–366 (2010). 10.1016/j.immuni.2010.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leporcq, C. et al. TFmotifView: a webserver for the visualization of transcription factor motifs in genomic regions. Nucleic Acids Res.48, W208–W217 (2020). 10.1093/nar/gkaa252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leng, N. et al. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics29, 1035–1043 (2013). 10.1093/bioinformatics/btt087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamthong, P. J. & Wu, M. Inhibitor of nuclear factor-κB induction by cAMP antagonizes interleukin-1-induced human macrophage-colony-stimulating-factor expression. Biochem. J.356, 525–530 (2001). 10.1042/bj3560525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heise, N. et al. Germinal center B cell maintenance and differentiation are controlled by distinct NF-κB transcription factor subunits. J. Exp. Med.211, 2103–2118 (2014). 10.1084/jem.20132613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peitz, M., Pfannkuche, K., Rajewsky, K. & Edenhofer, F. Ability of the hydrophobic FGF and basic TAT peptides to promote cellular uptake of recombinant Cre recombinase: a tool for efficient genetic engineering of mammalian genomes. Proc. Natl Acad. Sci. USA99, 4489–4494 (2002). 10.1073/pnas.032068699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grumont, R. J., Richardson, I. B., Gaff, C. & Gerondakis, S. rel/NF-κB nuclear complexes that bind kB sites in the murine c-rel promoter are required for constitutive c-rel transcription in B-cells. Cell Growth Differ.4, 731–743 (1993). [PubMed] [Google Scholar]
- 27.de Jesus, T. J. & Ramakrishnan, P. NF-κB c-Rel dictates the inflammatory threshold by acting as a transcriptional repressor. iScience23, 100876 (2020). 10.1016/j.isci.2020.100876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saccani, S., Pantano, S. & Natoli, G. Modulation of NF-κB activity by exchange of dimers. Mol. Cell11, 1563–1574 (2003). 10.1016/S1097-2765(03)00227-2 [DOI] [PubMed] [Google Scholar]
- 29.Liou, H. C., Sha, W. C., Scott, M. L. & Baltimore, D. Sequential induction of NF-κB/Rel family proteins during B-cell terminal differentiation. Mol. Cell. Biol.14, 5349–5359 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang, T. et al. Aberrant super-enhancer landscape reveals core transcriptional regulatory circuitry in lung adenocarcinoma. Oncogenesis9, 92 (2020). 10.1038/s41389-020-00277-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weng, C. C. et al. Loss of the transcriptional repressor TGIF1 results in enhanced Kras-driven development of pancreatic cancer. Mol. Cancer18, 96 (2019). 10.1186/s12943-019-1023-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shah, A., Melhuish, T. A., Fox, T. E., Frierson, H. F. Jr. & Wotton, D. TGIF transcription factors repress acetyl CoA metabolic gene expression and promote intestinal tumor growth. Genes Dev.33, 388–402 (2019). 10.1101/gad.320127.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang, J. et al. TGIF2 promotes cervical cancer metastasis by negatively regulating FCMR. Eur. Rev. Med Pharm. Sci.24, 5953–5962 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Du, R. et al. TGIF2 promotes the progression of lung adenocarcinoma by bridging EGFR/RAS/ERK signaling to cancer cell stemness. Signal Transduct. Target Ther.4, 60 (2019). 10.1038/s41392-019-0098-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ku, H. C. & Cheng, C. F. Master regulator activating transcription factor 3 (ATF3) in metabolic homeostasis and cancer. Front Endocrinol. (Lausanne)11, 556 (2020). 10.3389/fendo.2020.00556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson, M. R., Xu, D. & Williams, B. R. ATF3 transcription factor and its emerging roles in immunity and cancer. J. Mol. Med (Berl.)87, 1053–1060 (2009). 10.1007/s00109-009-0520-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lebeaupin, C., Yong, J. & Kaufman, R. J. The impact of the ER unfolded protein response on cancer initiation and progression: therapeutic implications. Adv. Exp. Med Biol.1243, 113–131 (2020). 10.1007/978-3-030-40204-4_8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmitz, M. L., Shaban, M. S., Albert, B. V., Gokcen, A. & Kracht, M. The crosstalk of endoplasmic reticulum (ER) stress pathways with NF-κB: complex mechanisms relevant for cancer, inflammation and infection. Biomedicines6, 58 (2018). 10.3390/biomedicines6020058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emming, S. et al. A molecular network regulating the proinflammatory phenotype of human memory T lymphocytes. Nat. Immunol.21, 388–399 (2020). 10.1038/s41590-020-0622-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mlynarczyk, C., Fontan, L. & Melnick, A. Germinal center-derived lymphomas: the darkest side of humoral immunity. Immunol. Rev.288, 214–239 (2019). 10.1111/imr.12755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laidlaw, B. J., Duan, L., Xu, Y., Vazquez, S. E. & Cyster, J. G. The transcription factor Hhex cooperates with the corepressor Tle3 to promote memory B cell development. Nat. Immunol.21, 1082–1093 (2020). 10.1038/s41590-020-0713-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takeda, R. et al. HHEX promotes myeloid transformation in cooperation with mutant ASXL1. Blood136, 1670–1684 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Faumont, N. et al. c-Rel is the pivotal NF-κB subunit in germinal center diffuse large B-cell lymphoma: a LYSA study. Front Oncol.11, 638897 (2021). 10.3389/fonc.2021.638897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lim, K. H., Yang, Y. & Staudt, L. M. Pathogenetic importance and therapeutic implications of NF-κB in lymphoid malignancies. Immunol. Rev.246, 359–378 (2012). 10.1111/j.1600-065X.2012.01105.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Compagno, M. et al. Mutations of multiple genes cause deregulation of NF-κB in diffuse large B-cell lymphoma. Nature459, 717–721 (2009). 10.1038/nature07968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neri, A. et al. B cell lymphoma-associated chromosomal translocation involves candidate oncogene lyt-10, homologous to NF-κB p50. Cell67, 1075–1087 (1991). 10.1016/0092-8674(91)90285-7 [DOI] [PubMed] [Google Scholar]
- 47.Houldsworth, J. et al. Relationship between REL amplification, REL function, and clinical and biologic features in diffuse large B-cell lymphomas. Blood103, 1862–1868 (2004). 10.1182/blood-2003-04-1359 [DOI] [PubMed] [Google Scholar]
- 48.Triner, D. et al. Myc-associated zinc finger protein regulates the proinflammatory response in colitis and colon cancer via STAT3 signaling. Mol. Cell. Biol.38, e00386-18 (2018). 10.1128/MCB.00386-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bataller, L., Wade, D. F., Graus, F., Rosenfeld, M. R. & Dalmau, J. The MAZ protein is an autoantigen of Hodgkin’s disease and paraneoplastic cerebellar dysfunction. Ann. Neurol.53, 123–127 (2003). 10.1002/ana.10434 [DOI] [PubMed] [Google Scholar]
- 50.Andlauer, T. F. et al. Novel multiple sclerosis susceptibility loci implicated in epigenetic regulation. Sci. Adv.2, e1501678 (2016). 10.1126/sciadv.1501678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steinbrecher, K. A., Harmel-Laws, E., Sitcheran, R. & Baldwin, A. S. Loss of epithelial RelA results in deregulated intestinal proliferative/apoptotic homeostasis and susceptibility to inflammation. J. Immunol.180, 2588–2599 (2008). 10.4049/jimmunol.180.4.2588 [DOI] [PubMed] [Google Scholar]
- 52.Singh, A. et al. Transcription factor TFII-I fine tunes innate properties of B lymphocytes. Front Immunol.14, 1067459 (2023). 10.3389/fimmu.2023.1067459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods10, 1213–1218 (2013). 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics12, 323 (2011). 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15, 550 (2014). 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yuan, H. et al. CancerSEA: a cancer single-cell state atlas. Nucleic Acids Res.47, D900–D908 (2019). 10.1093/nar/gky939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ge, S. X., Jung, D. & Yao, R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics36, 2628–2629 (2020). 10.1093/bioinformatics/btz931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods9, 357–359 (2012). 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics30, 2114–2120 (2014). 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang, Y. et al. Model-based analysis of ChIP-seq (MACS). Genome Biol.9, R137 (2008). 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics26, 841–842 (2010). 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kent, W. J., Zweig, A. S., Barber, G., Hinrichs, A. S. & Karolchik, D. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics26, 2204–2207 (2010). 10.1093/bioinformatics/btq351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou, X. et al. The human epigenome browser at Washington University. Nat. Methods8, 989–990 (2011). 10.1038/nmeth.1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ramirez, F., Dundar, F., Diehl, S., Gruning, B. A. & Manke, T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res.42, W187–W191 (2014). 10.1093/nar/gku365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stuart, T. et al. Comprehensive integration of single-cell data. Cell177, 1888–1902 (2019). 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figs. 1–8 with associated legends, full blot image for Supplementary Fig. 3b and full blot image depicting anti-Rel antibody (sc-71) validation.
ChIP–PCR primers used in Fig. 1c and Supplementary Figs. 1c and 4d.
qRT–PCR primers used in Supplementary Figs. 4c and 5e.
Supporting data for Supplementary Fig. 1b. Nuclear Translocation_Similarity index between RelA and DAPI.
Supporting data for Supplementary Fig. 3c. Median fluorescence intensity (MFI) for NF-κB targets.
Supporting data for Supplementary Fig. 4c. Gene expression analysis of RelA-selective genes in ex vivo RelA deleted B cells (by qRT–PCR CT values provided).
Supporting data for Supplementary Fig. 4e. ChIP–PCR analysis of Rel target genes at 18 h after anti-IgM treatment (CT values provided).
Supporting data for Supplementary Fig. 5e. Gene expression analysis of NF-κB target genes in follicular and MZ B cells (by qRT–PCR, CT values provided).
Statistical source data.
Data Availability Statement
The datasets generated and analyzed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Superseries accession number GSE197035 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE197035). Source data for Fig. 1c are provided. ChIP–seq data are also available at https://wangftp.wustl.edu/~dli/nih-mingming/biowulf/ and may be visualized at WashU Epigenome Browser: https://epigenomegateway.wustl.edu/. TFmotifView results are available here: http://bardet.u-strasbg.fr/tfmotifview/?results=734fKxPAnCh8w1 (RelA 1 h); http://bardet.u-strasbg.fr/tfmotifview/?results=H1bHoIdGkHrKPy (Rel 18 h). Supplementary Data 1–5 and Source data are provided with this paper.
No custom code was used in these studies.