Abstract
To facilitate the investigation of hepatitis B virus (HBV) sequence variation, we recently established a method for functional analysis of PCR-amplified full-length HBV genomes. This study aimed at estimating the number of mutations introduced during amplification of genomes from samples from patients with low levels of viremia and their influence on replication and antigen expression. Wild-type HBV DNA template molecules in concentrations like those present in samples from patients with very low levels of viremia were amplified, sequenced (30 kb total), and functionally tested. We found that Taq polymerase and a Taq-Pwo polymerase mixture introduced an average of 5.7 and 3.1 mutations per genome, respectively, corresponding to polymerase error rates of 12.1 × 10−5 and 6.0 × 10−5. One of 8 genomes (12%) amplified with Taq polymerase, but 7 of 17 genomes amplified with Taq-Pwo polymerases (41%), remained replication competent. All replication-competent genomes expressed HBs and HBe antigens and had an average of only 0.9 mutations per genome. In contrast, replication-defective genomes had an average of 5.4 mutations, which frequently also disturbed viral antigen expression. From these data we conclude that many of the replication-competent HBV genomes from a clinical specimen will retain their replication and antigen expression phenotypes even after extensive amplification with Taq-Pwo polymerases. Because replication competence is highly sensitive to random mutations, it is the best marker for the identification of HBV genomes with few or no PCR-introduced mutations.
Hepatitis B virus (HBV) sequence variability is increasingly recognized as a factor which modulates the course and outcome of HBV infection. Variants with disturbed hepatitis B e antigen (HBeAg) synthesis (24, 30), large deletions in the nucleocapsid gene (12, 20), or novel hepatocyte nuclear factor 1 binding sites in the core promoter (14, 25) were found to be associated with severe liver disease. Furthermore, mutations in the major antigenic determinant of the hepatitis B surface antigen (HBsAg) are probably responsible for the failure of immunization (4, 21), and mutations in the virus polymerase protein render HBV resistant to therapy with nucleoside analogs (1, 19, 38).
To facilitate the analysis of natural HBV isolates, we recently established a method for efficient PCR amplification of full-length HBV genomes which allows the isolation of a large number of genomes even from clinical samples from patients with low levels of viremia (13). Only a single cloning step is required before the phenotype of an amplified genome can be tested by transfection into hepatoma cell lines. Alternatively, the cloning step can be omitted and the phenotype of the whole HBV genome population can be studied by directly transfecting the amplified genomes. Application of this method to structural analysis of HBV in clinical samples has already led to the identification of novel and highly diverse types of genomes with deletions of up to 2 kb in serum and liver samples (15, 31).
To extend knowledge of the molecular features of natural virus isolates, other research groups have also established full-length genome PCRs for human immunodeficiency virus type 1 (6, 8, 28, 29), human T lymphotropic virus type 1 (34), human papillomavirus (33), hepatitis A virus (35, 36), hepatitis C virus (35), tick-borne encephalitis virus (11), simian virus 40 (18), and simian foamy virus (17) by taking advantage of long-range PCR conditions described recently (2). A major concern noted in these studies as well as in our previous study is the introduction of artificial sequence errors during PCR (7) which may alter the phenotypes of the amplified genomes or even render them defective. In two studies this problem was addressed by sequencing amplified genomes of human papillomavirus (33) and human immunodeficiency virus type 1 (28), which revealed an error frequency of about one error per 700 bases. The functional consequences of PCR-introduced mutations have so far been analyzed only in a recent study with PCR-amplified simian virus 40 genomes which found no functional deficiencies in the amplified clones (18). However, the consequences of random mutations in HBV may be quite different.
Therefore, prior to application of our PCR method to clinical samples we aimed in this study to evaluate the frequency and possible functional relevance of PCR-introduced mutations and to elucidate the optimal amplification system. Because problems with artificial mutations are particularly likely to arise if virus genomes are present in small amounts and require a high number of cycles, the test conditions were chosen appropriately. A few wild-type molecules in concentrations like those present in samples from patients with low levels of viremia were extensively amplified in different PCR systems, and the replication competence as well as the antigen expression of the amplified genomes was tested. Furthermore, a large number of amplified and cloned genomes were sequenced, the error frequencies were correlated with the functional phenotypes, and the error rates of the polymerases used in the PCR were determined.
MATERIALS AND METHODS
Amplification, cloning, and sequencing of complete HBV genomes.
Plasmid pSM2, containing a dimeric wild-type HBV genome (10), was linearized, and defined amounts of it were amplified in 50-μl PCR assay mixtures containing 50 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 200 μM deoxynucleoside triphosphate (dNTP), 0.01% gelatin, 5 U of Taq polymerase (Boehringer Mannheim, Mannheim, Germany), and 0.3 μM (each) primers P1 (5′-CCGGAAAGCTTGAGCTCTTCTTTTTCACCTCTGCCTAATCA) and P2 (5′-CCGGAAAGCTTGAGCTCTTCAAAAAGTTGCATGGTGCTGG) (HBV positions 1821 to 1841 and 1823 to 1806, respectively; SstI and SapI sites are underlined) or by using the Expand high-fidelity PCR assay (Boehringer Mannheim) containing 1.5 mM MgCl2, 200 μM dNTP, 2.6 U of Taq and Pwo polymerase mixture, and 0.3 μM (each) primers P1 and P2 (13). PCRs with Pfu polymerase (Stratagene) and Pwo polymerase (Boehringer Mannheim) were performed in the commercial buffers supplemented with 200 μM (each) dNTP and 0.3 μM (each) primers P1 and P2. The PCR was run for 40 cycles at 94°C for 40 s, 60°C for 1.5 min, and 68°C for 3 min, with an increment of 2 min after every 10 cycles, in a Robocycler (Stratagene). For cloning, the amplified HBV genomes were digested with SstI within the heterologous primer sequences, gel purified, and inserted into vector pUC19. The HBV DNA insert was sequenced with vector- and HBV-specific primers (2357 to 2380, GGCAGGTCCCCTAGAAGAAGAACT; 477 to 455, GGACAAACGGGCAACATACCTTG; 1121 to 1100, AGAAAGGCCTTGTAAGTTGGCG; 676 to 699, TTTACTAGTGCCATTTGTTCAGTG; 66 to 90, GCTCCAGTTCAGGAACAGTAAACCC; and 2432 to 2408, ATTGAGATCTTCTGCGACGCGGCGA), using the SequiTherm sequencing kit (Epicentre Technologies, Madison, Wis.) and an automated sequencer (LI-COR, Lincoln, Nebr.).
Preparation of PCR products for transfection.
HBV genomes were amplified for 35 cycles from HBV DNA from the serum of a patient or from plasmid-integrated wild-type HBV DNA with the Expand high-fidelity PCR assay. The 3.2-kb PCR products were gel purified with the QIAquick gel extraction kit (Qiagen, Hilden, Germany). Subsequently, 0.5 μl (1%) of each product was reamplified for 20 cycles in five 50-μl assay mixtures with the same primers and conditions described above. The amplification products were pooled, extracted with phenol-chloroform (1:1) and chloroform, and washed five times with water in Centricon-100 concentrators (Amicon, Beverly, Mass.) to remove residual dNTPs and primers. The DNA was then quantified spectrophotometrically (a ratio of the optical density at 260 nm to that at 280 nm of 1.7 to 1.8) and by fluorescence intensity measurement of the ethidium bromide-stained 3.2-kb DNA fragments separated on agarose gels. For standardization, defined amounts of 3.2-kb HBV DNA fragments were used.
Transfection of HBV DNA by calcium phosphate precipitation.
Linear HBV monomers with SapI sticky ends were released by cleavage with 1 U of SapI/μg of DNA for 12 h from plasmids containing P1- and P2-amplified HBV genomes and from plasmid pHBV-SapI (13). The latter (designated WT) contains the HBV wild-type genome (10) flanked by the primers P1 and P2. For direct transfection of amplified 3.2-kb HBV genomes, the heterologous primer sequences were cleaved with 1.5 U of SapI (New England Biolabs) per μg of PCR product for 12 h. HuH7 cells were plated at a density of 1.3 × 106 per 50-mm-diameter petri dish (Falcon) 1 day before transfection. The medium was changed 4 h before transfection. Without further purification the SapI-digested DNA (5 μg/dish in 0.1 volume of the final transfection mix) was mixed with CaCl2, added dropwise to 2× HEPES-buffered saline (pH 7.05), and transfected into confluent HuH7 cells. The medium was changed again 16 h after transfection, and 500 U of alpha interferon (IFN-α)/ml of medium was added where appropriate. The cells were harvested 3 to 4 days after transfection. Transfection efficiency was measured by cotransfection of 1 μg of reporter plasmid expressing secreted alkaline phosphatase and determination of secreted alkaline phosphatase enzymatic activity in the cell culture supernatant (5). HBsAg and HBeAg in the cell culture supernatant were assayed with commercially available kits (Abbott).
Purification of HBV DNA from intracellular core particles and Southern blot analysis.
Cells were washed and lysed in 1 ml of lysis buffer (50 mM Tris-HCl [pH 7.4], 1 mM EDTA, 1% Nonidet P-40) per dish (14). The lysate was incubated on ice for 15 min, and nuclei were pelleted by centrifugation. The supernatant was treated with 10 mM MgCl2–100 μg of DNase I/ml for 30 min at 37°C, adjusted to 25 mM EDTA, and digested with 1% sodium dodecyl sulfate–0.5 mg of proteinase K/ml for 2 h at 37°C. Nucleic acids were phenol-chloroform extracted and ethanol precipitated after the addition of 20 μg of tRNA. DNAs were separated on a 1.5% agarose gel, blotted onto Hybond N membranes (Amersham), and hybridized with a 32P-labelled full-length HBV probe. Autoradiography was analyzed with FUJIX BAS 2000 (Fuji, Tokyo, Japan), and signals were quantified with TINA software (Raytest, Straubenhardt, Germany).
Calculation of polymerase error rates and distribution of mutations.
The calculations were based on the following simplified PCR model. During one PCR cycle an individual single-stranded HBV genome is duplicated, and Pgen is the likelihood that the polymerase introduced an error into the newly polymerized strand. One of the two strands present at the end of this reaction (template or polymerized strand) is chosen by chance (the probability of choosing the polymerized strand with an error introduced in this cycle is Pgen/2) and is used as the template in the next duplication cycle. After repeating this procedure n times (n = number of duplication cycles) the likelihood that a genome has accumulated k mutations (0 ≤ k ≤ n) is described by the binomial distribution b(k;n;Pgen/2). The mean number of mutations that accumulate per genome (μgen) after n cycles is therefore given by the equation μgen = nPgen/2. Division of this equation by the nucleotide number of the HBV genome reveals the relation μpos = nPpos/2, where μpos is the mean number of mutations that accumulate per nucleotide position and Ppos is the error rate of the polymerase (errors per polymerized position) (7). μpos corresponds to the experimentally determined mean error frequency f (mutations per bases sequenced), and n was calculated by using the equation 2n = Nf/Ni, where Ni is the initial template number and Nf is the final copy number at the end of the PCR (both estimated from the mass and the molecular weight of the template and the PCR product). For calculation of Ppos the equations were combined as Ppos = 0.6 × f/log10(Nf/Ni).
RESULTS
Influence of the type of DNA polymerase on PCR sensitivity.
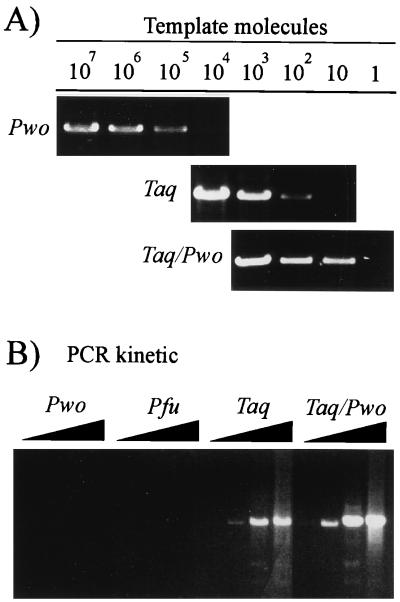
To determine which PCR system can be used for amplification of HBV genomes in sera from patients with low levels of viremia, defined copy numbers of plasmid-integrated HBV genomes were amplified with Pwo polymerase, which possesses proofreading activity (9), Taq polymerase, which lacks proofreading activity (37), and a Taq-Pwo polymerase mixture. The Pwo polymerase did not amplify less than 105 template molecules to a level detectable in an ethidium bromide-stained gel (Fig. 1A). In contrast, reactions with Taq polymerase and Taq-Pwo polymerases efficiently amplified as few as 102 and 10 molecules, respectively (Fig. 1A). To confirm these enzyme type-dependent differences in sensitivity, a PCR kinetic was performed with approximately 105 to 106 HBV virion DNA molecules as a template. The same set of enzymes as described above and the proofreading activity-possessing Pfu polymerase were used. Pwo and Pfu polymerases did not amplify the HBV virion DNA to levels detectable in an ethidium bromide-stained gel even after 35 cycles, whereas Taq and Taq-Pwo polymerases produced a signal after 25 cycles (Fig. 1B). These data indicate that amplification of HBV genomes from sera from patients with low levels of viremia requires Taq polymerase either alone or in combination with a thermostable polymerase with proofreading activity.
FIG. 1.
Sensitivity of the HBV full-length genome PCR with different thermostable polymerases. (A) Defined amounts of cloned HBV DNA (plasmid pSM2) were amplified for 40 cycles. The 3.2-kb PCR products were resolved in an agarose gel and stained with ethidium bromide. (B) HBV DNA from virus particles in the supernatant of HBV DNA-transfected hepatoma cells was amplified (105 to 106 template molecules). After 20, 25, 30, and 35 cycles an aliquot was removed from the PCR mixture, separated in an agarose gel, and stained with ethidium bromide. All assays produced a signal with 1 ng of HBV DNA as a positive control.
Influence of PCR-introduced mutations on the phenotypes of the amplified HBV genomes.
The impact of PCR-introduced mutations was studied by extensive amplification of wild-type HBV genomes with both PCR systems which efficiently amplify HBV genomes and by functional testing of the amplified genomes as follows. From a cloned dimeric wild-type HBV genome, 900 and 90 molecules were amplified with Taq polymerase and Taq-Pwo polymerase mixture, respectively. An aliquot of the PCR products was cloned, and 8 Taq polymerase-amplified and 17 Taq-Pwo polymerase-amplified HBV genomes were randomly selected for functional analysis. After transfection of these genomes into HuH7 hepatoma cells, core particle-associated replicative HBV DNA was analyzed by Southern blotting and the secretion of HBsAg and HBeAg into the cell culture supernatant was determined by enzyme immunoassays.
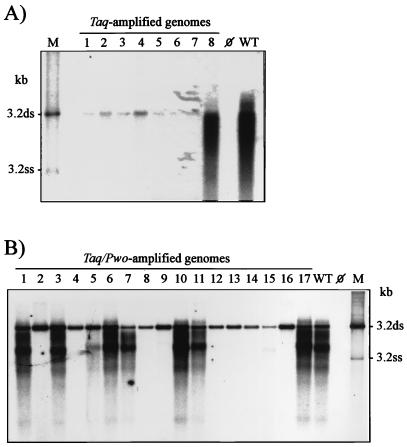
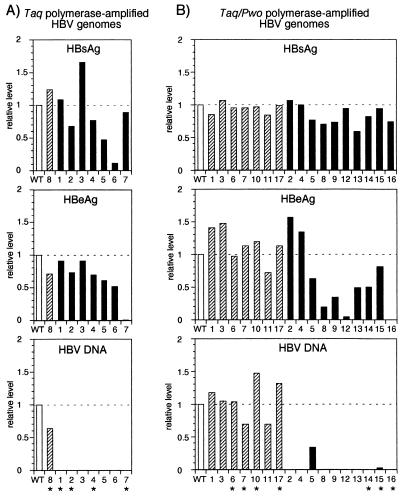
Only 1 of the 8 Taq polymerase-amplified genomes (12%) was replication competent (Fig. 2A and 3A), whereas 7 of the 17 Taq-Pwo polymerase-amplified HBV genomes (41%) produced replicative HBV DNA in amounts comparable to that of the nonamplified wild-type genome (Fig. 2B and 3B). In contrast to the replication level, expression and secretion of HBs- Ag and HBeAg were much less frequently affected. Most of the Taq polymerase-amplified genomes and all of the Taq-Pwo polymerase-amplified genomes produced HBsAg at about the wild-type level (Fig. 3). Similarly, 87% of the Taq polymerase-amplified genomes and 65% of the Taq-Pwo polymerase-amplified genomes efficiently produced HBeAg (Fig. 3). All genomes which showed the authentic replication phenotype also expressed HBsAg and HBeAg at preamplification levels, whereas genomes with defects in replication frequently also had defects in antigen production (Table 1).
FIG. 2.
Southern blot analysis of intracellular replicative HBV DNA intermediates produced by amplified genomes after transfection into HuH7 cells. (A) Analysis of cloned genomes amplified from 900 template molecules of wild-type HBV DNA (10) (plasmid pSM2) with Taq polymerase. (B) Analysis of cloned genomes amplified from 90 template molecules of wild-type HBV DNA with Taq-Pwo polymerase mixture. WT, nonamplified wild-type genome (10) (plasmid pHBV-SapI); ⊘, mock transfection; M, marker lane with 3.2-kb double-stranded (ds) and single-stranded (ss) HBV DNA. Note that the bands at the 3.2-kb position seen in all lanes of the amplified genomes represent input rather than progeny HBV DNA.
FIG. 3.
Levels of HBsAg, HBeAg, and intracellular replicative HBV DNA produced by HBV genomes amplified with Taq polymerase (A) or Taq-Pwo polymerase mixture (B). HBsAg and HBeAg were measured in the cell culture supernatant by enzyme immunoassays (Abbott), and the HBV DNA signals shown in Fig. 2 were quantified with TINA software (Raytest). The designation of the clones is the same as in Fig. 2. The values are given relative to that of the nonamplified wild-type genome (WT; open bar). Genomes which replicate at about wild-type genome level (range, WT/1.5 to WT × 1.5) are grouped (shaded bars), and those which were partially or completely sequenced are marked with an asterisk. Solid bars represent defective genomes.
TABLE 1.
Correlation of the replication competence of the amplified HBV genomes with HBsAg and HBeAg levels and the number of PCR-introduced mutations
| Replication competence | No. of genomes (%) expressing:
|
PCR-introduced mutations
|
|||
|---|---|---|---|---|---|
| HBsAg | HBeAg | HBsAg and HBeAg | Errors/kb sequenced (%) | Errors/genome (avg) | |
| Authentic (n = 8) | 8 (100) | 8 (100) | 8 (100) | 4/13.7 (0.029) | 0.9 |
| Defective (n = 17) | 15 (88) | 10 (59) | 8 (47) | 28/16.6 (0.169) | 5.4 |
Since many genomes in clinical samples may be naturally defective in replication, we wondered whether a fraction of replication-competent genomes similar to that in the PCR test system is obtained with HBV DNA from patients. Therefore, 102 to 104 HBV genomes (as estimated by PCR) from sera of eight patients with fulminant hepatitis (32) were amplified with Taq-Pwo polymerases, cloned, and analyzed by transfection. Of 35 genomes tested, 24 (68%) were replication competent. This fraction is even larger than that obtained after the amplification of cloned wild-type genomes, indicating that the predominant virus population in the sera of these patients was replication competent. Taken together, after extensive amplification, as required for sera from patients with very low levels of viremia, the fraction of HBV genomes that exhibit the original phenotype of replication and antigen expression is about 40% if the PCR is performed with Taq-Pwo polymerase mixture. Taq polymerase alone produces a much larger fraction of defective genomes. The replication competence of a genome correlates with intact HBsAg and HBeAg expression.
Number and type of mutations introduced during PCR.
To elucidate which and how many mutations accumulate during PCR and whether the number differs between genomes which showed the authentic phenotype and those which were rendered defective during PCR, a total of 30 kb of the amplified genomes was sequenced (Table 2). Taking the data of all analyzed genomes together, those with the authentic phenotype had an average of 0.9 mutations whereas the defective ones had an average of 5.4 mutations (Table 1). Nearly all of the mutations (27 of 32) were transitions, usually T→C or A→G (Table 2), which are specific for Taq polymerase (37). They frequently led to amino acid changes in at least one HBV reading frame and created a translational stop codon in the polymerase (P) and nucleocapsid (C) genes of genomes Taq-2 and Taq/Pwo-16, respectively. The latter mutations explain the replication defects of these genomes as well as the lack of HBeAg expression of genome Taq/Pwo-16 (Fig. 3B). Because the defective genomes Taq-1, Taq/Pwo-14, and Taq/Pwo-15 lack amino acid changes in the nucleocapsid protein, those in the polymerase protein are most likely responsible for the replication defect (Table 2). Taken together, the data indicate a correlation between the number of PCR-introduced mutations and the phenotypes of the amplified genomes. In defective genomes six times more mutations accumulated than in those which retained the authentic phenotype. The results also show that the random introduction of only a few mutations into a HBV genome can render it replication defective.
TABLE 2.
Mutations introduced into amplified HBV genomes during PCR
| Genome (replication competence)a | Bases sequenced (kb) | No. of errors | % Errors | Nucleotide change
|
Amino acid change in HBV gene:
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Position | Type | C | S | X | P | ||||
| Taq amplified | |||||||||
| 1 (−) | 3.0 | 6 | 0.20 | 43 | A→G | R→G | |||
| 226 | A→G | N→D | |||||||
| 641 | T→C | W→R | M→T | ||||||
| 709 | G→A | A→T | |||||||
| 2549 | T→C | ||||||||
| 2965 | A→G | T→A | H→R | ||||||
| 2 (−) | 3.0 | 4 | 0.13 | 786 | C→G | P→R | |||
| 1059 | T→C | ||||||||
| 2941 | T→A | W→R | L→STOP | ||||||
| 3133 | C→A | P→T | P→H | ||||||
| 4 (−) | 0.5 | 3 | 0.60 | 1369 | T→C | F→L | |||
| 1717 | G→A | C→Y | |||||||
| 1768 | T→C | F→S | |||||||
| 7 (−) | 0.6 | 1 | 0.17 | 1383 | A→T | R→W | |||
| 8 (+) | 0.9 | 0 | 0.00 | ||||||
| Taq-Pwo amplified | |||||||||
| 14 (−) | 3.2 | 6 | 0.19 | 308 | A→G | N→D | K→R | ||
| 887 | T→A | V→D | |||||||
| 1560 | T→C | F→L | |||||||
| 1564 | C→T | S→L | H→Y | ||||||
| 1626 | G→A | A→T | |||||||
| 2389 | T→C | L→P | |||||||
| 15 (−) | 3.2 | 5 | 0.16 | 119 | T→C | S→P | V→A | ||
| 1044 | T→C | ||||||||
| 1181 | T→C | V→A | |||||||
| 1474 | T→C | L→P | |||||||
| 1750 | A→G | E→G | |||||||
| 16 (−) | 3.2 | 3 | 0.09 | 810 | T→C | F→S | |||
| 1843 | C→T | ||||||||
| 2206 | G→A | W→STOP | |||||||
| 6 (+) | 3.2 | 0 | 0.00 | ||||||
| 7 (+) | 3.2 | 3 | 0.09 | 1343 | T→C | V→A | |||
| 1976 | T→C | S→P | |||||||
| 2137 | A→G | ||||||||
| 10 (+) | 3.2 | 1 | 0.03 | 1071 | T→C | ||||
| 17 (+) | 3.2 | 0 | 0.00 | ||||||
−, defective; +, authentic.
Determination of the polymerase error rate.
The fidelity of polymerases is usually determined in assays which are based on one polymerization reaction or a few PCR cycles (7). However, the fidelity of Taq polymerase may be different if the reaction runs for a large number of cycles. This may be due, for example, to a change in the reaction conditions on which the fidelity of Taq polymerase strongly depends (7). Therefore, we calculated the error rate of Taq polymerase and Taq-Pwo polymerase mixture in the 40-cycle full-length PCR. Based on the number of initial template molecules, the number of final reaction products, and the number of mutations found per number of bases sequenced, the error rate of Taq polymerase was determined to be 12.1 × 10−5 misincorporations per polymerized nucleotide and that of the Taq-Pwo polymerase mixture was determined to be 6.0 × 10−5 (Table 3). The calculated error rate of Taq polymerase agrees well with several published values ranging from 8.9 × 10−5 to 13.4 × 10−5 (2, 3, 37), which indicates no decrease in fidelity during full-length PCR. The combination of Pwo polymerase with Taq polymerase improved the fidelity of polymerization twofold, which is close to the threefold increase previously determined with a lacI-based assay (9).
TABLE 3.
Error rates of Taq polymerase and Taq-Pwo polymerase mixture in the HBV full-length PCR
| Enzyme | Nia | Nfb | Total sequenced (kb) | Errors/genome (avg) | Error rate (Ppos) |
|---|---|---|---|---|---|
| Taq | 900 | 6 × 1011 | 7.9 | 5.7 | 12.1 × 10−5 |
| Taq-Pwo | 90 | 6 × 1011 | 22.4 | 3.1 | 6.0 × 10−5 |
Ni, initial template number.
Nf, final copy number (approximately 2 μg of PCR product).
Functional analysis of amplified HBV genomes without cloning: optimization and influence of PCR on the HBV phenotype.
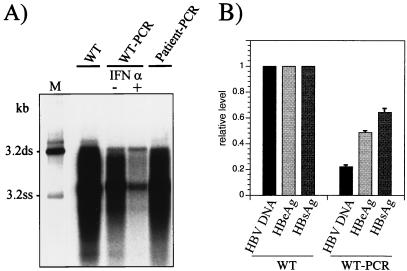
The HBV full-length PCR also allows functional analysis of the amplified genomes by transfection without prior cloning (13). This technique had not been tested with HBV DNA from samples from patients with low levels of viremia because a single PCR did not yield sufficient DNA for transfection. However, when the 3.2-kb PCR products were gel purified with the Qiagen kit and reamplified with the Taq-Pwo polymerase mixture, between 5 and 7 μg of DNA per 50-μl PCR assay mixture was obtained. The 3.2-kb HBV DNA represented about 50% of the total DNA: the rest was nonspecific background. HBV genome populations amplified by two rounds of PCR from a small number of molecules of cloned wild-type HBV DNA as well as from HBV DNA from the serum of a patient worked well in transfection experiments. They expressed HBsAg as well as HBeAg (not shown) and produced replicative HBV DNA intermediates (Fig. 4A, lanes WT-PCR and Patient-PCR). The potential of this system for the analysis of the sensitivity of clinical HBV isolates to antiviral drugs was evaluated by testing the IFN-α responsiveness of the amplified HBV genomes. Replication of an amplified wild-type genome population was inhibited by IFN-α (Fig. 4A, compare lanes IFN-α + and −), as is known for nonamplified wild-type HBV in stable transfection systems (16). Thus, a two-step PCR method was established for amplification of HBV genomes from material from patients with low levels of viremia which yields sufficient product for transfection without prior cloning. The amplified genome populations replicate, express viral antigens, and are susceptible to IFN-α when transfected into cells.
FIG. 4.
(A) Southern blot analysis of intracellular replicative HBV DNA intermediates produced after transfection of cloned genomes (WT) or PCR products (WT-PCR and Patient-PCR). WT, nonamplified cloned wild-type genome pHBV-SapI; WT-PCR, HBV genomes produced during 60 cycles with 0.3 pg of wild-type genomes integrated in plasmid pSM2 (105 template molecules); Patient-PCR, HBV genomes synthesized during 60 cycles with approximately 104 to 105 HBV virion DNA template molecules isolated from the serum of a patient; M, length marker as described in the legend to Fig. 2; ds, double stranded; ss, single stranded. (B) Comparison of the levels of secreted HBsAg and HBeAg as well as that of intracellular replicative HBV DNA produced by WT and WT-PCR. The WT levels were defined as 1.0. The means and standard deviations of two experiments are shown.
As shown above, individual amplified DNA molecules are rather frequently rendered completely defective by PCR-introduced mutations. Such drastic effects are not necessarily expected when the whole population of amplified molecules is functionally analyzed. However, amplification may quantitatively change phenotypic features of the amplified DNA population according to the number of mutations (and thus, of defective genomes) that accumulate during PCR. To estimate this effect, a nonamplified, cloned wild-type genome as well as extensively amplified wild-type genomes was transfected and its replication and antigen expression levels were compared. In general, the PCR products produced lower levels of replicative HBV DNA, HBsAg, and HBeAg than the nonamplified genome (Fig. 4A and B, compare lanes WT-PCR and WT). Interestingly, the levels of replicative HBV DNA were reduced two to three times more than the antigen levels, which is probably a consequence of PCR-introduced mutations.
DISCUSSION
The efficiency of HBV full-length genome PCR as well as the frequency and functional relevance of PCR-introduced mutations in different polymerase assays was evaluated in this study. The Taq-Pwo polymerase mixture was found to be the most sensitive, and it introduced fewer mutations when amplifying low copy numbers. With this assay, about 40% of the replication-competent HBV genomes from a clinical specimen from a patient with a low level of viremia can be expected to retain their replication and antigen expression phenotypes after amplification. Because replication competence was found to be highly sensitive to random mutations, it is a useful marker to identify HBV genomes with few or no PCR-introduced mutations.
Previous studies demonstrated an increase in PCR sensitivity and fidelity if a polymerase that exhibits proofreading activity is combined with an enzyme lacking this activity (2, 9). Here, we confirmed this by testing Taq and Pwo polymerases alone and combined in the HBV full-length PCR. However, additional enzyme combinations are currently available, and further experiments are needed to test whether they perform similarly to or even better than the Taq-Pwo polymerase system.
Two observations have important implications for the functional analysis of genomes amplified from clinical samples. First, nearly all amplified wild-type genomes either replicated at the authentic preamplification level or were completely defective, and second, the replication competence of an amplified genome correlated with intact HBsAg and HBeAg expression and a low number of artificial mutations. Therefore, it can be expected that the majority of genomes which remain replication competent after amplification show the authentic replication and antigen expression phenotypes and contain no or very few PCR-introduced mutations. Replication-competent genomes in which mutations have accumulated during amplification will most likely be defective and can easily be excluded from a detailed analysis when one screens first for replication competence.
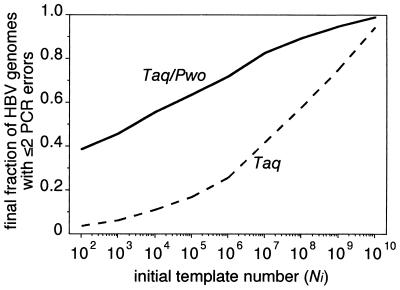
The identification of replication-defective genomes in sera is not easy. An indication whether a genome is naturally or artificially defective may be provided by determining the type and prevalence of the mutation(s) by sequencing. All artificial mutations found in the amplified genomes in our study were nucleotide changes, and the same one was never found in more than one genome. Therefore, a mutation(s) shared by a substantial virus subpopulation or deletions and insertions is not likely to be PCR derived. Furthermore, the data provided by this study allow a rough estimate of the fraction of naturally defective genomes in clinical specimens. If far more genomes are replication defective than would be expected from artificial mutations after amplification (Fig. 5 shows an estimation of this fraction according to the template number), this indicates a large fraction of naturally defective genomes in the analyzed sample. A first analysis revealed that this is obviously not the case for HBV populations in sera of patients with fulminant hepatitis, as the fraction of genomes which were replication competent after amplification corresponded to the expected level.
FIG. 5.
Calculated fraction of genomes with ≤2 artificial mutations within the final population of amplified genomes when initial template molecule amounts (Ni), as indicated on the x axis, were amplified to 2 μg of PCR product (approximately Nf = 6 × 1011 final molecules) with Taq or Taq-Pwo polymerases. The fraction of genomes with ≤2 artificial mutations may roughly correspond to the fraction of genomes which remain replication competent after PCR, since most replication-competent genomes will contain either no, one, or two mutations after PCR, according to our experiments (an average of one mutation per genome). Consistent with this assumption, the calculated fractions of genomes with ≤2 mutations (5 and 40% after PCR with Taq polymerase [900 templates] and Taq-Pwo polymerases [90 templates], respectively) are in good agreement with the fractions of replication-competent genomes determined experimentally (12 and 41%). Based on a simplified PCR model the fractions were calculated by using the distribution function of the binomial distribution B(2;n;Pgen/2) (see Materials and Methods). n was calculated with Ni and Nf, and Pgen = Ppos × 3,200. The error rate, Ppos, was experimentally determined for Taq polymerase and the Taq-Pwo polymerase mixture (Table 3).
The need for analysis of individual genomes can be circumvented if the amplified genomes are directly transfected without prior cloning, as was previously shown for genomes amplified from high-titer sera (13). As an extension of these previous results we present evidence that this method works even after extensive amplification of a small number of template molecules. However, for correct interpretation of data obtained by the described method one has to take into account that amplification selectively reduces the level of replication compared to that of antigen expression. This is probably the result of PCR-introduced mutations, which affect replication more frequently than antigen expression, as is evident from the analysis of the cloned genomes. This effect need not be considered if data are compared that were obtained with the same HBV DNA preparation analyzed under different conditions, as exemplified here by testing HBV replication with and without the addition of IFN-α to the transfected cells. Thus, the PCR product transfection assay may prove particularly useful for monitoring the possible resistance of clinical HBV isolates to antiviral drugs before and during therapy (1, 19, 38).
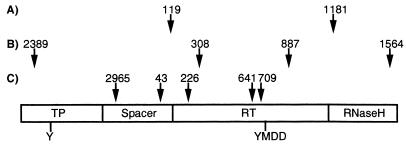
From a biological point of view, it is surprising that the replication-competent genomes contained only an average of one artificial mutation. This suggests that the random introduction of very few mutations can severely interfere with HBV replication. This would not be expected, considering the high sequence heterogeneity of HBV, with several genotypes which differ by more than 10% on the nucleotide level (23). Replication can be affected by mutations which inactivate RNA or DNA elements involved in transcription, pregenome encapsidation, and replication or by mutations which alter the function of the nucleocapsid or HBV polymerase proteins. In two of the defective genomes one of the latter proteins is predictably truncated due to premature stop codons, which explains the replication defect. In most other defective genomes inactivation of the HBV polymerase function is most likely responsible for the replication defect, for two reasons. First, the coding sequence of the HBV polymerase covers about two-thirds of the HBV genome, which leads to a high probability that a random mutation will change the amino acid sequence of the HBV polymerase. Consistent with this speculation, most of the amino acid changes in the amplified genomes were found in the polymerase. Second, the HBV polymerase is a multifunctional protein with a function in assembly—encapsidation of RNA—and three enzymatic functions: DNA synthesis priming and DNA polymerase and RNase H activity (22, 26). All these functions are probably highly susceptible to changes in the protein structure. This speculation is supported by our observation that in three amplified genomes the defect in replication can be attributed to only a few amino acid changes in the polymerase (Fig. 6). This interpretation is consistent with a study showing that most of a variety of single amino acid changes rendered the HBV polymerase inactive (27). In conclusion, our data point to a rather limited sequence flexibility of the HBV genome, probably because of the highly mutation-sensitive polymerase protein encoded by a large part of the genome.
FIG. 6.
Schematic map of the positions of amino acid changes in the polymerase protein of replication-defective genomes Taq/Pwo-15 (A), Taq/Pwo-14 (B), and Taq-1 (C). The nucleocapsid protein, as well as known sequence elements essential for replication, is not affected in these genomes (Table 2). The functional domains (TP, terminal protein for DNA synthesis priming; RT, reverse transcriptase) (26), as well as functionally important amino acid residues involved in HBV DNA synthesis (Y, priming tyrosine; YMDD, RT catalytic site), are indicated.
ACKNOWLEDGMENTS
We thank S. Polywka and R. Laufs for measuring HBsAg and HBeAg and A. Röhl of the Mathematisches Seminar der Universität Hamburg for advice. We appreciate the critical reading of the manuscript by V. Radwitz-Will.
This work was supported by grants from the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie (Verbundvorhaben, project 01KI9560) and the Wilhelm-Sander-Stiftung. The Heinrich-Pette-Institut für Experimentelle Virologie und Immunologie is supported by the Freie und Hansestadt Hamburg and the Bundesministerium für Gesundheit.
REFERENCES
- 1.Aye T T, Bartholomeusz A, Shaw T, Bowden S, Breschkin A, McMillan J, Angus P, Locarnini S. Hepatitis B virus polymerase mutations during antiviral therapy in a patient following liver transplantation. J Hepatol. 1997;26:1148–1153. doi: 10.1016/s0168-8278(97)80125-0. [DOI] [PubMed] [Google Scholar]
- 2.Barnes W M. PCR amplification of up to 35-kb DNA with high fidelity and high yield from lambda bacteriophage templates. Proc Natl Acad Sci USA. 1994;91:2216–2220. doi: 10.1073/pnas.91.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cariello N F, Swenberg J A, Skopek T R. Fidelity of Thermococcus litoralis DNA polymerase (Vent) in PCR determined by denaturing gradient gel electrophoresis. Nucleic Acids Res. 1991;19:4193–4198. doi: 10.1093/nar/19.15.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carman W F, Zanetti A R, Karayiannis P, Waters J, Manzillo G, Tanzi E, Zuckerman A J, Thomas H C. Vaccine-induced escape mutant of hepatitis B virus. Lancet. 1990;336:325–329. doi: 10.1016/0140-6736(90)91874-a. [DOI] [PubMed] [Google Scholar]
- 5.Cullen B R, Malim M H. Secreted placental alkaline phosphatase as a eukaryotic reporter gene. Methods Enzymol. 1992;216:326–368. doi: 10.1016/0076-6879(92)16033-g. [DOI] [PubMed] [Google Scholar]
- 6.Dittmar M T, Simmons G, Donaldson Y, Simmonds P, Clapham P R, Schulz T F, Weiss R A. Biological characterization of human immunodeficiency virus type 1 clones derived from different organs of an AIDS patient by long-range PCR. J Virol. 1997;71:5140–5147. doi: 10.1128/jvi.71.7.5140-5147.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckert K A, Kunkel T A. The fidelity of DNA polymerases used in the polymerase chain reaction. In: McPherson M J, Quirke P, Taylor G R, editors. PCR: a practical approach. Oxford, England: Oxford University Press; 1991. pp. 225–244. [Google Scholar]
- 8.Fang G, Weiser B, Visosky A A, Townsend L, Burger H. Molecular cloning of full-length HIV-1 genomes directly from plasma viral RNA. J Acquired Immune Defic Syndr Hum Retrovirol. 1996;12:352–357. doi: 10.1097/00042560-199608010-00004. [DOI] [PubMed] [Google Scholar]
- 9.Frey B, Suppmann B. Demonstration of the Expand PCR system’s greater fidelity and higher yields with a lacI-based PCR fidelity assay. Biochem Inf. 1995;2:34–35. [Google Scholar]
- 10.Galibert F, Mandart E, Fitoussi F, Tiollais P, Charnay P. Nucleotide sequence of the hepatitis B genome (subtype ayw) cloned in E. coli. Nature (London) 1979;281:646–650. doi: 10.1038/281646a0. [DOI] [PubMed] [Google Scholar]
- 11.Gritsun T S, Gould E A. Infectious transcripts of tick-borne encephalitis virus, generated in days by RT-PCR. Virology. 1995;214:611–618. doi: 10.1006/viro.1995.0072. [DOI] [PubMed] [Google Scholar]
- 12.Günther S, Baginski S, Kissel H, Reinke P, Krüger D H, Will H, Meisel H. Accumulation and persistence of hepatitis B virus core gene deletion mutants in renal transplant patients are associated with end-stage liver disease. Hepatology. 1996;24:751–758. doi: 10.1002/hep.510240401. [DOI] [PubMed] [Google Scholar]
- 13.Günther S, Li B C, Miska S, Krüger D H, Meisel H, Will H. A novel method for efficient amplification of whole hepatitis B virus genomes permits rapid functional analysis and reveals deletion mutants in immunosuppressed patients. J Virol. 1995;69:5437–5444. doi: 10.1128/jvi.69.9.5437-5444.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Günther S, Piwon N, Iwanska A, Schilling R, Meisel H, Will H. Type, prevalence, and significance of core promoter/enhancer II mutations in hepatitis B viruses from immunosuppressed patients with severe liver disease. J Virol. 1996;70:8318–8331. doi: 10.1128/jvi.70.12.8318-8331.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Günther S, Sommer G, Iwanska A, Will H. Heterogeneity and common features of defective hepatitis B virus genomes derived from spliced pregenomic RNA. Virology. 1997;238:363–371. doi: 10.1006/viro.1997.8863. [DOI] [PubMed] [Google Scholar]
- 16.Hayashi Y, Koike K. Interferon inhibits hepatitis B virus replication in a stable expression system of transfected viral DNA. J Virol. 1989;63:2936–2940. doi: 10.1128/jvi.63.7.2936-2940.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herchenröder O, Turek R, Neumann-Haefelin D, Rethwilm A, Schneider J. Infectious proviral clones of chimpanzee foamy virus (SFVcpz) generated by long PCR reveal close functional relatedness to human foamy virus. Virology. 1995;214:685–689. doi: 10.1006/viro.1995.0086. [DOI] [PubMed] [Google Scholar]
- 18.Lednicky J A, Jafar S, Wong C, Butel J S. High-fidelity PCR amplification of infectious copies of the complete simian virus 40 genome from plasmids and virus-infected cell lysates. Gene. 1997;184:189–195. doi: 10.1016/s0378-1119(96)00594-x. [DOI] [PubMed] [Google Scholar]
- 19.Ling R, Mutimer D, Ahmed N, Boxall E H, Elias E, Dusheiko G M, Harrison T J. Selection of mutations in the hepatitis B virus polymerase during therapy of transplant recipients with lamivudine. Hepatology. 1996;24:711–713. doi: 10.1002/hep.510240339. [DOI] [PubMed] [Google Scholar]
- 20.Marinos G, Torre F, Günther S, Thomas M G, Will H, Williams R, Naoumov N V. Hepatitis B virus variants with core gene deletions in the evolution of chronic hepatitis B infection. Gastroenterology. 1996;111:183–192. doi: 10.1053/gast.1996.v111.pm8698197. [DOI] [PubMed] [Google Scholar]
- 21.McMahon G, Ehrlich P H, Moustafa Z A, McCarthy L A, Dottavio D, Tolpin M D, Nadler P I, Ostberg L. Genetic alterations in the gene encoding the major HBsAg: DNA and immunological analysis of recurrent HBsAg derived from monoclonal antibody-treated liver transplant patients. Hepatology. 1992;15:757–766. doi: 10.1002/hep.1840150503. [DOI] [PubMed] [Google Scholar]
- 22.Nassal M, Schaller H. Hepatitis B virus replication—an update. J Viral Hepat. 1996;3:217–226. doi: 10.1111/j.1365-2893.1996.tb00047.x. [DOI] [PubMed] [Google Scholar]
- 23.Norder H, Courouce A M, Magnius L O. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology. 1994;198:489–503. doi: 10.1006/viro.1994.1060. [DOI] [PubMed] [Google Scholar]
- 24.Omata M, Ehata T, Yokosuka O, Hosoda K, Ohto M. Mutations in the precore region of hepatitis B virus DNA in patients with fulminant and severe hepatitis. N Engl J Med. 1991;324:1699–1704. doi: 10.1056/NEJM199106133242404. [DOI] [PubMed] [Google Scholar]
- 25.Pult I, Chouard T, Wieland S, Klemenz R, Yaniv M, Blum H E. A hepatitis B virus mutant with a new hepatocyte nuclear factor 1 binding site emerging in transplant-transmitted fulminant hepatitis B. Hepatology. 1997;25:1507–1515. doi: 10.1002/hep.510250633. [DOI] [PubMed] [Google Scholar]
- 26.Radziwill G, Tucker W, Schaller H. Mutational analysis of the hepatitis B virus P gene product: domain structure and RNase H activity. J Virol. 1990;64:613–620. doi: 10.1128/jvi.64.2.613-620.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roychoudhury S, Faruqi A F, Shih C. Pregenomic RNA encapsidation analysis of eleven missense and nonsense polymerase mutants of human hepatitis B virus. J Virol. 1991;65:3617–3624. doi: 10.1128/jvi.65.7.3617-3624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salminen M O, Koch C, Sanders-Buell E, Ehrenberg P K, Michael N L, Carr J K, Burke D S, McCutchan F E. Recovery of virtually full-length HIV-1 provirus of diverse subtypes from primary virus cultures using the polymerase chain reaction. Virology. 1995;213:80–86. doi: 10.1006/viro.1995.1548. [DOI] [PubMed] [Google Scholar]
- 29.Sanchez G, Xu X, Chermann J C, Hirsch I. Accumulation of defective viral genomes in peripheral blood mononuclear cells of human immunodeficiency virus type 1-infected individuals. J Virol. 1997;71:2233–2240. doi: 10.1128/jvi.71.3.2233-2240.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato S, Suzuki K, Akahane Y, Akamatsu K, Akiyama K, Yunomura K, Tsuda F, Tanaka T, Okamoto H, Miyakawa Y, Mayumi M. Hepatitis B virus strains with mutations in the core promoter in patients with fulminant hepatitis. Ann Intern Med. 1995;122:241–248. doi: 10.7326/0003-4819-122-4-199502150-00001. [DOI] [PubMed] [Google Scholar]
- 31.Sommer, G., S. Günther, M. Sterneck, S. Otto, and H. Will. A new class of defective hepatitis B virus genomes with an internal poly(dA) sequence. Virology, in press. [DOI] [PubMed]
- 32.Sterneck M, Günther S, Santantonio T, Fischer L, Broelsch C E, Greten H, Will H. Hepatitis B virus genomes of patients with fulminant hepatitis do not share a specific mutation. Hepatology. 1996;24:300–306. doi: 10.1002/hep.510240203. [DOI] [PubMed] [Google Scholar]
- 33.Stewart A C, Gravitt P E, Cheng S, Wheeler C M. Generation of entire human papillomavirus genomes by long PCR: frequency of errors produced during amplification. Genome Res. 1995;5:79–88. doi: 10.1101/gr.5.1.79. [DOI] [PubMed] [Google Scholar]
- 34.Tamiya S, Matsuoka M, Etoh K, Watanabe T, Kamihira S, Yamaguchi K, Takatsuki K. Two types of defective human T-lymphotropic virus type I provirus in adult T-cell leukemia. Blood. 1996;88:3065–3073. [PubMed] [Google Scholar]
- 35.Tellier R, Bukh J, Emerson S U, Miller R H, Purcell R H. Long PCR and its application to hepatitis viruses: amplification of hepatitis A, hepatitis B, and hepatitis C virus genomes. J Clin Microbiol. 1996;34:3085–3091. doi: 10.1128/jcm.34.12.3085-3091.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tellier R, Bukh J, Emerson S U, Purcell R H. Amplification of the full-length hepatitis A virus genome by long reverse transcription-PCR and transcription of infectious RNA directly from the amplicon. Proc Natl Acad Sci USA. 1996;93:4370–4373. doi: 10.1073/pnas.93.9.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tindall K R, Kunkel T A. Fidelity of DNA synthesis by the Thermus aquaticus DNA polymerase. Biochemistry. 1988;27:6008–6013. doi: 10.1021/bi00416a027. [DOI] [PubMed] [Google Scholar]
- 38.Tipples G A, Ma M M, Fischer K P, Bain V G, Kneteman N M, Tyrrell D. Mutation in HBV RNA-dependent DNA polymerase confers resistance to lamivudine in vivo. Hepatology. 1996;24:714–717. doi: 10.1002/hep.510240340. [DOI] [PubMed] [Google Scholar]