Abstract
Coenzyme Q5 (COQ5), a C-methyltransferase, modifies coenzyme Q10 (COQ10) during biosynthesis and interacts with polyA-tail regulating zinc-finger protein ZC3H14 in neural development. Here, we present a fifth patient (a third family) worldwide with neurodevelopmental and physiological symptoms including COQ10 deficiency. Our patient harbors one novel c.681+1G>A and one recurrent p.Gly118Ser variant within COQ5. The patient’s mRNA profile reveals multiple COQ5 splice-variants. Subsequently, we comprehensively described patient’s clinical features as compared to phenotype and symptoms of other known congenital coenzyme Q5-linked cases. A core spectrum of COQ5-associated symptoms includes reduced COQ10 levels, intellectual disability, encephalopathy, cerebellar ataxia, cerebellar atrophy speech regression/dysarthria, short stature, and developmental delays. Our patient additionally displays dysmorphia, microcephaly, and regressive social faculties. These results formally establish causal association of biallelic COQ5 mutation with pathology, outline a core COQ5-linked phenotype, and identify mRNA mis-splicing as the molecular mechanism underlying all COQ5 variant-linked pathology to date.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13353-023-00773-9.
Keywords: COQ5, COQ10, Molecular mechanism, Expansion of the phenotype
Introduction
COQ5 is expressed in all human tissues (Nguyen et al. 2014) and affects fundamental developmental and cellular biology. As a protein, COQ5 interacts with poly-A regulating zinc-finger protein ZC3H14 affecting transcript stability, polypeptide levels in central nervous system (CNS) development (Pak et al. 2011; Najmabadi et al. 2011) and its methyl transferase activity provides essential modifications during biosynthesis of COQ10, a molecule crucial to mitochondrial and cellular eukaryotic metabolism (Nguyen et al. 2014; Yen et al. 2016; Hargreaves 2021).
Studies in yeast identified a minimum of 11 gene products (COQ1–11), including COQ5 which directly supports COQ10 biosynthesis. COQ1–9 form a multiprotein complex, the COQ synthome, found on the inner mitochondrial membrane (Nguyen et al. 2014; Yen et al. 2016; Hargreaves 2021). The COQ10 benzoquinone ring and 10-unit polyisoprenoid tail moieties synthesized in different subcellular locations are position together in the mitochondrion, where COQ5, 7, and 9 further modify COQ10. In addition, different COQ transcripts and polypeptides interact, interregulating stabilities, protein levels, and activities (Nguyen et al. 2014; Hargreaves 2021). These interactions impact both individual physiological COQ coenzyme functions not directly related to COQ10 biosynthesis (Hargreaves 2021), such as COQ5 interactions with polyA-tail regulating protein ZC3H14 during neural development (Pak et al. 2011), as well as COQ5 stability, influenced by COQ8, affecting COQ10 biosynthesis and COQ10-dependent metabolism (Nguyen et al. 2014; Yen et al. 2016; Hargreaves 2021).
Patients with identified biallelic mutation in any of the supporting COQ-enzymes (COQ1–2, 4–9) present with a range of developmental and physiological pathologies with a preponderance of neurodevelopmental manifestations and COQ10 tissue deficiency (Yen et al. 2016; Hargreaves 2021; Malicdan et al. 2018; Online Mendelian inheritance in man n.d.). These are defined as primary COQ10 deficiencies. Indirect factors causing a COQ10 deficiency such as aging, disease or certain drugs, resulting in malabsorption, or disrupted bio-distribution, for example, are secondary. Nonetheless, some symptomatic patients with biallelic mutations have normal COQ10 levels in tissues tested, indicating that their condition is not a straightforward related to COQ10 deficiency; others with established COQ10 deficits do not respond to oral COQ10 supplementation, suggesting involvement of additional factors.
Correlation of symptomology with a molecular basis of COQ10 biosynthesis/deficiency is clinically and therapeutically highly relevant. For example, neurodevelopmental defects associated with COQ10 deficiency due to a biallelic COQ10 disruption are mostly effectively treated with appropriate oral COQ10 supplementation, particularly if identified early in childhood. COQ5 mutations (Malicdan et al. 2018), which destabilizes the COQ5 polypeptide, leading to disrupted COQ10 biosynthesis, cause COQ10 deficiency that respond well to supplementation, within weeks. Accurate and rapid differential molecular diagnoses are crucial for treatment of clinically overlapping syndromes and enable for proper prognosis, planning, therapeutic measures, and quality of life, for both patient and family.
Here, we identified a novel COQ5 pathogenic variant and investigated the molecular mechanism underlying its pathology. We also characterized a specific constellation of clinical traits linked to mutations at the COQ5 locus, identified so far.
Materials and methods
RNA extraction, cDNA synthesis, PCR, cloning, and exome sequencing were done according to manufacturer’s instructions. For more details, please see supplementary data.
Results
Patient history
We present a Polish female patient current age 10 years, born at term by natural delivery at the 39th week of the mother’s first pregnancy. Parents were healthy, non-consanguineous, Caucasian in origin. At the time of birth, the mother was 29 years old; the father was 33 years old.
Family history was negative for individuals with neurodevelopmental disorders. Patient birth body parameters were the following: body weight: 3380 g (50 pc); body length: 54 cm (<95 pc); occipital frontal circumference: 31 cm (<3 pc). Apgar scores were 10 points at 1 and 5 min of life.
Presented girl displayed normal, uneventful postnatal adaptation and development from birth to 5 months of life.
At 22 weeks, directly after third dose-vaccination with DTaP (Infanrix-Hexa), motor and cognitive regression, deterioration of eye contact, and the lack of a smile were noticed. Consequently, further vaccinations were suspended. At the age of 18 months, further developmental regression became apparent. She was intensively rehabilitated, but no progress was observed. In clinical evaluation, she could not stand up or walk independently and she could pronounce a few simple syllables and try to combine them into words. Detailed clinical observations from 1.5 to 10 years of age are described in supplementary data.
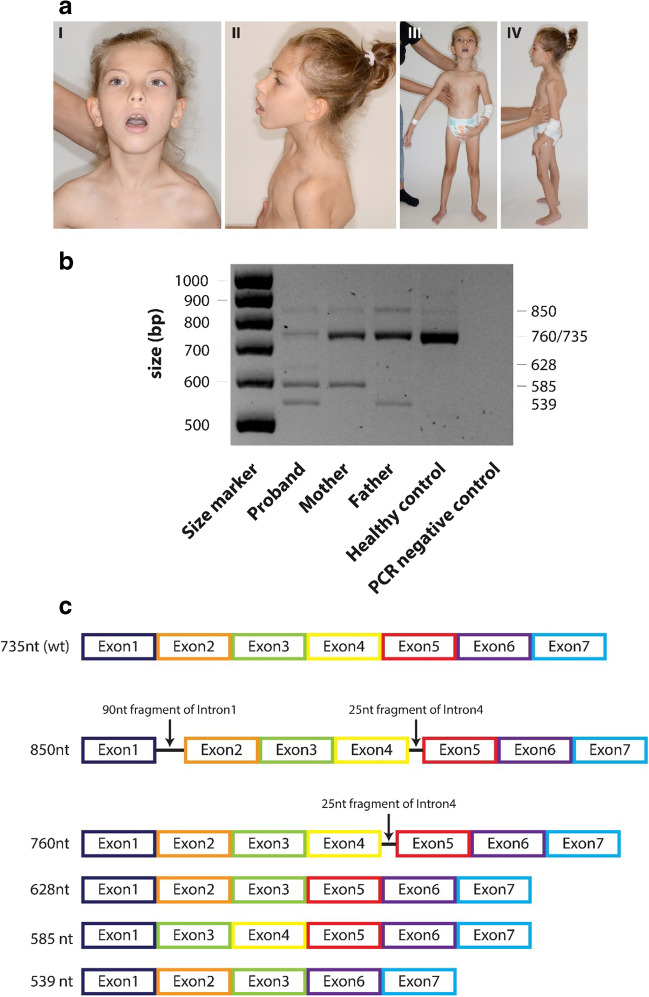
At the age of 10 years, she had not acquired the ability to walk; cognitive development remains at a low level. Summary of neurological examinations showed global developmental delay, tetraparesis with hypotonia and symmetrical tendon reflexes, limited eye contact, postural defects including a “rounded” back, protruding shoulder blades, and funnel-shaped chest (Fig. 1aIII–IV). Phenotypic evaluation of the face revealed slight ptosis, a tendency to open the mouth and tilt the head back, without other specific features of dysmorphism (Fig. 1aI).
Fig. 1.
a Photographic documentation of the patient at age 10 years old. b COQ5 cDNA gel electrophoresis. c Graphical representation of the splicing products of the COQ5 gene in a patient, her parents, and healthy control. For more details, please see supplementary data
Exploratory assays
Magnetic resonance imaging (MRI) of the brain at the age of 8 years revealed clear cortico-subcortical atrophy of the cerebellum–enlargement of the 4th ventricle and widening of cerebellar sulci (Supp.Fig.1a-d, arrows) as compared to the normal appearance in a healthy individual (Supp.Fig.1e-h, arrows). The cerebellar sulci were wider in our patient at age 8 years than at 18 months, as well as in comparison to an age-matched healthy control. This indicates progressive atrophy of cerebellar tissue. Atrophy involved both vermis and cerebellar hemispheres. The supratentorial structures are normal. Echocardiogram, abdominal ultrasonography, and ophthalmologic investigation were also uneventful.
Low COQ10 level tests showing 0.6 mg/l (normal range >0.67 mg/l) and corroborated twice at age 9 after 2 months of irregular QuinoMit 32.5 mg/day nasal spray administration, level of COQ10 in leukocytes remained low (Table 1 p.55–56). For the last 1.5 years, she has been treated with QuinoMit Q10 Fluid at a dose of 26 mg every second day, with good tolerance. We are observing a gradual improvement in the patient’s motor condition—muscle strength is better, the patient sits up on her own from a lying position, stands on her own, and the ptosis has decreased. However, she has not acquired the ability to walk; cognitive development remains at a low level.
Table 1.
Clinical features for COQ5 patient 5 compared to reported symptomology for 4 previous COQ5 patients
| # | Features | Najmabadi et al. (2011) | Malicdan et al. (2018) | Present report | ||
|---|---|---|---|---|---|---|
| Patient | Patient 1 | Patient 2 (III.4) | Patient 3 (III.3) | Patient 4 (III.6) | Patient 5 | |
| Mutation | c.352G>A p.(Gly118Ser)hom | c.575-1761_*1489dup p.(?) | c.575-1761_*1489dup p.(?) | c.575-1761_*1489dup p.(?) | c.352G>A/681+1G>A p.Gly118Ser/? | |
| Gender | nd | Female | Female | Female | Female | |
| Current age | nd | nd | nd | nd | 10 years | |
| Parental consanguinity | Yes | No | No | No | No | |
| Ethnicity | nd | Iraqi-Jewish | Iraqi-Jewish | Iraqi-Jewish | Polish | |
| 1 | Encephalomyopathy | nd | + | + | + | − |
| 2 | Cerebellar ataxia (non-progressive) | nd | + | + (mild) | + | + |
| 3 | Encephalopathy | nd | + | + | + | + |
| 4 | Generalized tonic clonic seizures | nd | + (17 years old several) | + (22 years old single) | + | − |
| 5 | Developmental delay | nd | + | + | + | + |
| 6 | Short stature | nd | + | nd | + | + (−3.19 SDS 9 years old) |
| 7 | Delay in motor milestones development | nd | + (moderate) | nd | + (mild) | + |
| 8 | Delay in cognitive milestones | nd | + (moderate) | nd | nd | + |
| 9 | Dysarthria | nd | + | + (mild) | + (mild) | na (lack of speech) |
| 10 | Mild-moderate cognitive disability | nd | + | below average intelligence | nd | regression after 6 months |
| Behavioral problems | ||||||
| 11 | Impulsivity | nd | + | nd | nd | + |
| 12 | Attention deficiency | nd | + | nd | nd | + |
| 13 | Oppositional defiant disorder | nd | + | nd | nd | − |
| 14 | Myoclonic jerks | nd | + (12 years old) | + (20 years old) | nd | − |
| 15 | Epilepsy | nd | + | nd | nd | − |
| 16 | Microcephaly | nd | − | − | − | + (−3.56 SDS 9 years old) |
| 17 | Nystagmus | nd | + | + (horizontal) | + (horizontal) | − |
| 18 | Slow saccades | nd | + | nd | nd | − |
| 19 | Saccadic movements | nd | + | nd | + | − |
| 20 | Apraxic gaze | nd | + | nd | nd | − |
| 21 | Fundoscopy | nd | normal | nd | nd | − |
| 22 | Dysarthric cerebellar speech | nd | + | nd | nd | na (lack of speech) |
| 23 | Dysmetria in finger-to-nose test | nd | + | nd | − | − |
| 24 | Postural and intention tremor | nd | + | nd | − | − |
| 25 | Ataxic gait | nd | + | + (mild) | nd | na (lack of gait) |
| 26 | Dysmetria and oculomotor apraxia | nd | + (mild) | nd | − | |
| 27 | Abnormal gait pattern | nd | + | + | abnormal tandem walking identified at 14 years old | na (lack of gait) |
| 28 | Lower limb spasticity | nd | + (mild) | nd | + | + |
| 29 | Cerebellar atrophy | nd | + (non-progressive) | + (mild) | nd | + |
| 30 | EEG | nd | infrequent generalized polyspike-wave discharges | infrequent generalized polyspike-wave discharges | nd | normal |
| 31 | Nerve conduction study | nd | normal | nd | nd | nd |
| 32 | Cardiac echocardiogram | nd | normal | nd | nd | normal |
| Morphology and biochemistry | ||||||
| 33 | Complete blood count | nd | normal | nd | nd | normal |
| 34 | Electrolyte levels | nd | normal | nd | nd | normal |
| 35 | Creatine phosphokinase | nd | normal | nd | nd | normal |
| 36 | Liver and renal function tests | nd | normal | nd | nd | normal |
| 37 | Carnitine and acyl-carnitine | nd | normal | nd | nd | normal |
| 38 | Copper | nd | normal | nd | nd | normal |
| 39 | Ceruloplasmin | nd | normal | nd | nd | nd |
| 40 | Thyroid function tests | nd | normal | nd | nd | normal |
| 41 | Lactate | nd | normal | nd | nd | normal |
| 42 | Pyruvate | nd | normal | nd | nd | nd |
| 43 | Ammonia | nd | normal | nd | nd | nd |
| 44 | Blood amino acid profile | nd | normal | nd | nd | normal |
| 45 | Very long chain fatty acids | nd | normal | nd | nd | nd |
| 46 | Phytanic acid | nd | normal | nd | nd | nd |
| 47 | Homocysteine | nd | normal | nd | nd | normal |
| 48 | Isoelectric focusing of transferrin | nd | normal | nd | nd | nd |
| 49 | Alpha-fetoprotein | nd | normal | nd | nd | nd |
| 50 | Quantitative immunoglobulin levels | nd | normal | nd | nd | nd |
| 51 | Vitamin E levels | nd | normal | nd | nd | nd |
| 52 | Urine for protein | nd | normal | nd | nd | normal |
| 53 | Urine for organic acids | nd | normal | nd | nd | normal |
| 54 | Dysmorphia | nd | − | nd | nd | + |
| 55 | COQ10 enzyme tests values in leukocytes before COQ treatment | nd | 65 pmol/μg* | 78 pmol/μg* | 72 pmol/μg* | 0.6 mg/l** |
| 56 | COQ10 enzyme tests values in leukocytes after COQ treatment | nd | after 3-month treatment: 293 pmol/μg* | after 3-month treatment: 332 pmol/μg* | after 3-month treatment: 279 pmol/μg* | after 2-month treatment: 0.7 mg/l** |
| 58 | Other biochemical tests abnormalities | nd | III.4 | III.3 | III.6 | nd |
nd no data, na not applicable
*Normal range 119.86 ± 24.23
**Normal range > 0.67 mg/l
Molecular results
The aCGH analysis did not show any pathogenic copy number variations. Exome sequencing (ES) revealed one novel variant c.681+1G>A and one recurrent pathogenic variant c.352G>A (p.Gly118Ser) in COQ5 (Najmabadi et al. 2011). Biallelic origin was confirmed by analysis of the patient’s parents using Sanger sequencing (Supp.Fig.2c). Examination of COQ5 mRNA (Supp.Fig.3) showed that its level is markedly diminished in our patient (Supp.Fig.3 band 735 bp compared to the mRNA band in mother, father, and healthy control). Mis-spliced RNA forms in the patient indicate specific maternal (Supp.Fig.3 band 585 bp) and paternal (Supp.Fig.3 band 539 bp) mis-splice contributions. Exons, ribonucleotide sequence, and amino acid sequence for each band depicting mis-splices are shown in Supp.Fig.3. The patient also harbors a mis-splice that did not contain exon 4 (Supp.Fig.3, band 628 nt) undetected in parents and healthy control.
COQ5-linked syndrome phenotype
The available genetic and clinical information reported for all known patients with COQ5 mutations compared with our patient 5 is compiled in Table 1. This provides a core phenotype (Table 1: 2, 3, 5, 6, 9, 10, 29, 55) linked to COQ5 disruption.
Discussion
In this study, we identified a novel splicing c.681+1G>A COQ5 variant. Up to date, this is a third pathogenic variant found at the COQ5 locus. The variant was detected in the patient harboring known pathogenic p.Gly118Ser COQ5 variant on the other allele. Biallelic origin of the variants and recessive mode of inheritance was confirmed by Sanger sequencing of parent’s DNA. The patient’s mRNA profile indicated that both inherited COQ5 variants contribute to characteristic mis-spliced mRNA forms, deleting either exon 2, or exons 4 and 5. Further, we comprehensively described clinical features of our patient as compared to phenotype and symptoms of other known congenital coenzyme Q5-linked cases. These allowed us indicating a specific constellation of clinical traits linked to mutations at the COQ5 locus.
Our patient has COQ10 deficiency and pronounced neurodevelopmental traits. Evidently, neither COQ5 variant supports normal COQ5 function or healthy COQ10 levels. If either did, our patient would show healthy neurodevelopment with normal COQ10 levels. Each of these two variants therefore, directly confirms association of the other with pathology. Which symptoms in our patient result from COQ10 deficiency, versus other COQ5-associated enzymatic, regulatory, or structurally associated dysfunctions, such as binding interactions with CZ3H14, or COQ8, or the COQ-synthome itself, are elusive.
Compilation of the phenotypes for all five biallelic COQ5 patients (Table 1), exposes common traits presenting together in each patient and which as a set, are largely non-overlapping with other COQ protein-associated pathologies (Online Mendelian inheritance in man n.d.). This suggests a specific constellation of clinical traits linked to mutations at the COQ5 locus, so far, encompassing cerebellar ataxia, encephalopathy, developmental delay, short stature, dysarthria, ID, cerebellar atrophy, and COQ10 deficiency in leucocyte assay (Table 1: 2, 3, 5, 6, 9, 10, 29, 55, respectively). Although each trait may occur individually in many unrelated neurodevelopmental disorders, and also in patient pathology profiles with other COQ loci biallelic mutation, we propose that a majority (5/8 or more) of these traits occurring together represents a core phenotype linked to COQ5 disruption. While not diagnostically definitive, a clinical perspective pointing strongly to a specific COQ-locus, in this case COQ5, should accelerate the diagnostic procedure and enable crucial interventions earlier in patient development. Eventually, gene-trait associations should allow mapping of specific traits to protein domains, specific functions, and particular alleles, informing etiologic gene test panels. These could facilitate rapid differential clinical diagnoses for specific traits, syndromes, or compound conditions, early on in developmental stages, potentially even in utero. For example, rapid in utero distinction of primary versus secondary COQ10 deficiencies in biallelic patients, eliminating the “trial and error” factor currently inherent in both diagnosis and COQ10 supplementation therapy, could be life-changing.
In conclusion, our data clarifies the intricate molecular biology underlying COQ-locus clinical pathologies and COQ10-related metabolism, with concomitant progress in clinical handling.
Supplementary information
(DOCX 644 kb)
Acknowledgements
We are grateful to the patient and her parents for participating in this study.
Author contribution
Conceptualization: P.G.; data curation: M.D., A.P., M.J., E.O., M.B.-F., A.G.-C., E.B.-O., D.L.G., W.W., and P.G.; formal analysis: M.D., W.W., and P.G.; funding acquisition: P.G.; investigation: M.D., W.W., and P.G.; methodology: P.G.; project administration: P.G.; resources: W.W. and P.G.; software: M.D.; supervision: P.G.; validation: M.D. and P.G.; visualization: P.G.; writing—original draft preparation: P.G.; writing—review and editing: M.D., M.J., E.O., M.B.-F., A.G.-C. E.B.-O., D.L.G., A.M.R., and P.G. All authors have read and agreed to the published version of the manuscript.
Declarations
Ethical approval
This study complies with the latest Declaration of Helsinki and was approved by the Ethics Committee of the Institute of Mother and Child in Warsaw.
Consent to participate
Written informed consent was obtained from the parents.
Consent for publication
The authors affirm that human research participants provided informed consent for publication of the images in Fig. 1a.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Hargreaves IP. Coenzyme Q10 in mitochondrial and lysosomal disorders. J Clin Med. 2021;10(9):1970. doi: 10.3390/jcm10091970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malicdan MCV, et al. A novel inborn error of the coenzyme Q10 biosynthesis pathway: cerebellar ataxia and static encephalomyopathy due to COQ5 C-methyltransferase deficiency. Hum Mutat. 2018;39(1):69–79. doi: 10.1002/humu.23345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najmabadi H, et al. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature. 2011;478(7367):57–63. doi: 10.1038/nature10423. [DOI] [PubMed] [Google Scholar]
- Nguyen TP, et al. Molecular characterization of the human COQ5 C-methyltransferase in coenzyme Q10 biosynthesis. Biochim Biophys Acta. 2014;1841(11):1628–1638. doi: 10.1016/j.bbalip.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Online Mendelian inheritance in man (n.d.) OMIM®. Johns Hopkins University, Baltimore, MD. MIM Number: PS607426. World Wide Web URL: COQ10 in Online Mendelian Inheritance in Man. https://www.omim.org/entry/607426
- Pak, et al. Mutation of the conserved polyadenosine RNA binding protein, ZC3H14/dNab2, impairs neural function in Drosophila and humans. Proc Natl Acad Sci USA. 2011;108(30):12390–12395. doi: 10.1073/pnas.1107103108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen HC, et al. Disruption of the human COQ5-containing protein complex is associated with diminished coenzyme Q10 levels under two different conditions of mitochondrial energy deficiency. Biochim Biophys Acta. 2016;1860(9):1864–1876. doi: 10.1016/j.bbagen.2016.05.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 644 kb)