Abstract
APOE4, encoding apolipoprotein E4 (apoE4), is the greatest genetic risk factor for Alzheimer’s disease (AD), compared to the common APOE3. While the mechanism(s) underlying APOE4-induced AD risk remains unclear, increasing the lipidation of apoE4 is an important therapeutic target as apoE4-lipoproteins are poorly lipidated compared to apoE3-lipoproteins. ACAT (acyl-CoA: cholesterol-acyltransferase) catalyzes the formation of intracellular cholesteryl-ester droplets, reducing the intracellular free cholesterol (FC) pool. Thus, inhibiting ACAT increases the FC pool and facilitates lipid secretion to extracellular apoE-containing lipoproteins. Previous studies using commercial ACAT inhibitors, including avasimibe (AVAS), as well as ACAT-knock out (KO) mice, exhibit reduced AD-like pathology and amyloid precursor protein (APP) processing in familial AD (FAD)-transgenic (Tg) mice. However, the effects of AVAS with human apoE4 remain unknown. In vitro, AVAS induced apoE efflux at concentrations of AVAS measured in the brains of treated mice. AVAS treatment of male E4FAD-Tg mice (5xFAD+/-APOE4+/+) at 6–8 months had no effect on plasma cholesterol levels or distribution, the original mechanism for AVAS treatment of CVD. In the CNS, AVAS reduced intracellular lipid droplets, indirectly demonstrating target engagement. Surrogate efficacy was demonstrated by an increase in Morris water maze measures of memory and postsynaptic protein levels. Amyloid-beta peptide (Aβ) solubility/deposition and neuroinflammation were reduced, critical components of APOE4-modulated pathology. However, there was no increase in apoE4 levels or apoE4 lipidation, while amyloidogenic and non-amyloidogenic processing of APP were significantly reduced. This suggests that the AVAS-induced reduction in Aβ via reduced APP processing was sufficient to reduce AD pathology, as apoE4-lipoproteins remained poorly lipidated.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13311-023-01375-3.
Keywords: Alzheimer’s disease, APOE4, ACAT-1, Oligomeric Aβ, Neuroinflammation, APP processing
Introduction
Lipid dysregulation is involved in the pathogenesis and progression of neurodegenerative diseases including Alzheimer’s disease (AD) [1–3]. Accumulation of neutral lipids, particularly cholesteryl esters (CE) and triacylglycerol (TAG) stored as lipids droplets (LD), is found in aged and AD brains [4–8]. In addition, brains of AD patients [9] and familial AD-transgenic (FAD-Tg) mice [4, 5] have elevated cholesterol levels. Thus, repurposing drugs targeting lipid pathways may provide effective AD therapeutics.
The greatest genetic risk factor for sporadic AD is inheritance of the ε4 allele of the human (h)-APOE gene compared to the common ε3 and the neuroprotective but rare ε2 allele [10]. Apolipoprotein E (apoE), the protein encoded by the APOE gene, is the only lipoprotein-competent apolipoprotein expressed in the brain, where it is secreted primarily by astrocytes [11, 12]. As the protein component of CNS-lipoproteins, apoE-containing lipoproteins enable lipid transport within the interstitial space of the brain, available for uptake by both neurons and glia as apoE is a ligand for receptor-mediated endocytosis by the low-density lipoprotein receptor (LDLR) family. Although expressed at comparable levels, apoE4 levels in brain are lower than apoE3 levels in both humans and Tg mouse models [13–15], suggesting that apoE4 is unstable and undergoes accelerated degradation, likely due to the observed reduced lipidation state of apoE4-lipoproteins compared to apoE3-lipoproteins [16–23].
One of the pathological hallmarks of Alzheimer’s disease (AD) is amyloid plaques composed primarily of amyloid-beta (Aβ) peptide, although Aβ also exists in soluble forms, specifically soluble oligomeric Aβ (oAβ) considered a proximal neurotoxin [24–31]. Importantly, APOE4 is associated with accelerated Aβ accumulation, both as deposited amyloid and soluble Aβ [32–34]. Aβ peptides are produced primarily by neurons via the sequential cleavage of amyloid precursor protein (APP) by β- and γ-secretases (amyloidogenic pathway) at lipid rafts in the plasma membrane (PM) [35–37]. Outside of lipid rafts, APP is processed at the PM by α-secretase and -secretases producing sAPP-α (non-amyloidogenic pathway) [38–40]. The APP processing pathway is highly regulated by the mobilization of cholesterol between the lipid rafts and PM [41, 42]. Recent data suggest that the amount of cholesterol in neuronal lipid rafts is regulated by astrocytic apoE-lipoproteins promoting or limiting APP processing by β- and γ-secretases, and hence the amount of Aβ production [40]. ApoE is also known to interact with both amyloid plaques and soluble Aβ [43], an association that may be key in Aβ clearance [44], and studies have demonstrated that apoE4 does not clear plaques or soluble Aβ as well as apoE3 [45, 46]. Thus, increasing the lipidation of apoE4 represents a promising therapeutic target for modulating Aβ levels in the CNS.
To correct the structure of apoE4, and thus its function, several therapeutics targeted at enzymatic activities known to alter lipoprotein biogenesis and remodeling in the periphery have been explored for repurposing as CNS therapeutics [47]. Over a decade of evidence demonstrates that the cell-surface lipid transporter ATP binding cassette subfamily A member 1 (ABCA1) modulates AD pathogenesis, with ABCA1 antagonism increasing, and agonism decreasing, pathology [48–51]. We previously demonstrated that RXR agonists induce an increase in ABCA1 expression with an increase in apoE4 lipidation and a decrease in both total Aβ42 and soluble Aβ in EFAD (5xFAD+/-/APOE4+/+)-Tg mice [13]. However, severe hepatomegaly is associated with RXR agonism due to an induction of hepatic lipogenic pathways by sterol-regulatory element-binding protein 1C (SREBP1C) [52, 53].
Inhibition of ACAT (acyl-CoA: cholesterol-acyltransferase or sterol O-acyltransferase) is another potential pathway for increasing apoE lipidation and modifying APP processing. ACAT exists as two isoforms, ACAT1 is distributed ubiquitously, and ACAT2 is expressed primarily by enterocytes and hepatocytes [54, 55]. The endoplasmic reticulum (ER)-resident ACAT catalyzes intracellular cholesterol esterification, critical in this context as it reduces the ER-free cholesterol (ER-FC) pool by the formation of CE stored as LD [56]. By repurposing a cardiovascular drug that inhibits ACAT, specifically avasimibe (AVAS), LD formation is inhibited, allowing FC build-up, thus promoting ABCA1-mediated cholesterol efflux to nascent apoE4-lipoproteins [57–63]. ACAT activity and LD levels increase with age [64–66], AD, and APOE4 [67, 68], suggesting low levels of FC that will limit apoE lipidation and its downstream effects. Importantly, previous research demonstrates that ACAT inhibition reduces Aβ-induced neurotoxicity in vitro and AD pathology in FAD-Tg mouse models, including significant reductions in plaque burden and gliosis (reviewed in [69, 70]). In vitro, blocking ACAT promotes formation of the autophagosome/lysosome that contributes to microglia-associated Aβ clearance ([71]; reviewed in [72]). In vivo, attenuating ACAT activity increases the levels of 24-hydroxycholesterol and decreases the content of APP and Aβ in the brain of FAD-Tg mice [73, 74]. Using non-isoform specific ACAT inhibitors like AVAS, APP processing is reduced in brains of FAD-Tg mice ([75, 76]; reviewed in [77]). A key factor influencing the effect of ACAT inhibition is its location in the ER [78], next to key players: (1) in microglia, inhibition of ACAT causes an increase in ER-FC, promoting formation of autophagosomes/lysosomes [71]; (2) in neurons, an increase in ER-FC promotes cholesterol turnover by activating ER-resident cytochrome P450 family 46 subfamily A member 1 (CYP46A1) [73] and 3) stimulates APP anchoring in the ER, thus reducing neuronal APP processing at the PM [41].
The observations reported have been confined to FAD-Tg models expressing endogenous mouse apoE (m-apoE), not addressing the role of h-apoE4 in these processes. Mouse and human apoE differ at ~ 100/300 amino acids, and CNS m-apoE lipoproteins remain uncharacterized as to size and level of lipidation. Additionally, no effects have been reported for m-apoE lipidation state with the inhibition of ACAT. Importantly, m-apoE and h-APP combine in FAD-Tg mice to produce severe Aβ pathology, pathology that is reduced by knocking-out (KO) the m-APOE, further reduced by knocking-in (KI) h-APOE4, and again reduced with h-APOE3-KI (review [79–87]).
This study was designed to determine the effect of ACAT inhibition by AVAS on h-apoE4 lipidation and AD pathology in male E4FAD-Tg mice [79, 88]. Our hypothesis is that in vivo, AVAS, via inhibition of ACAT, will increase the local ER-FC pool, promoting ABCA1-mediated efflux of cholesterol to nascent apoE4-lipoproteins, in addition to reducing APP processing. This correction of apoE4 structure via lipidation will restore its function in the CNS and reduce measures of AD pathology.
AVAS treatment significantly increased MWM measures of memory and postsynaptic protein levels, indicating surrogate efficacy, reduced intracellular LDs, demonstrating indirect target engagement, and reduced pathological changes in Aβ solubility/deposition, and neuroinflammation, all critical components of APOE4-modulated AD pathology. However, there was no increase in apoE4 levels or lipidation, while APP processing was significantly reduced. This suggests that the AVAS-induced reduction in processing of APP to Aβ was sufficient to establish surrogate efficacy and reduce AD pathology, as apoE4-lipoproteins remained poorly lipidated.
Methods
All reagents used in the methods are listed in Table S1.
Cell Culture
Primary glial cultures (~ 95% astrocytes, 5% microglia) were isolated from cerebral cortex (CX) of 2-day(d) old E4FAD-non carrier (5xFAD−/−/APOE4+/+) pups (previously described [89, 90]). Briefly, after harvest, cells reached confluency at 10–12 d in vitro (DIV) and were trypsinized and plated into two 175 cm2 tissue culture flasks. After 20 DIV, secondary mixed glia cultures were seeded in 96-well plates for 24 h (h) with 10% fetal bovine serum. The cultures were changed to serum-free media for 24 h prior to treatment for 6 h and 24 h with increasing concentrations of AVAS. Cell viability and secreted apoE levels were measured as described below.
MTT for Cell Viability
Briefly, after treatment with 0.05–10.0 μg/ml AVAS for 6 h and 24 h, the media was removed, and the cells incubated with 5% MTT and processed as per the manufacturer’s instructions. The absorbance values are expressed as % of vehicle control (VC)-treated cells (% control).
ApoE ELISA
After treatment with 0.05–10 μg/ml AVAS for 6 h and 24 h, the media was removed and apoE levels measured with an in-house apoE ELISA that utilizes α-apoE as capture antibody and α-apoE biotin-gt as the detection antibody (previously described [91]). A recombinant human apoE3 standard was prepared and diluted to produce the standard curves. Samples were diluted to read within the linear range of the standard curve.
PK Analysis
3-month (M) old male C57BL/6 (BL6) mice (Charles River, Wilmington, MA) were treated by gavage with AVAS (30 mg/kg) for 3 h, 6 h, and 24 h, followed by sacrifice and analysis of AVAS levels in blood and brain. Briefly, stock solutions of AVAS that were dissolved in DMSO were prepared daily. The drug was formulated in 0.5% w/v sodium carboxymethyl cellulose, 9% Tween 80 in water with 1% final DMSO concentration (vehicle solution). Mice were sacrificed 3 h, 6 h, and 24 h after treatment. Spot checks to determine whether male E4FAD mice differed from male BL6 were made at 3 h with 100 mg/kg AVAS treatment by gavage, and the level of the drug was measured in both blood and brain. Mice were sacrificed using CO2 asphyxiation, blood was immediately drawn from the inferior vena cava, and mice were intracardially perfused with ice-cold PBS, decapitated, and brains were removed, separated into 2 hemispheres (hemis) and flash frozen. Blood samples were immediately transferred into heparin-coated tubes, centrifuged for 6 min (m) at 3220 g at 4 °C. Plasma supernatants were flash-frozen and immediately stored at − 80 °C until analysis. Brain and plasma concentrations of AVAS were quantified by LC–MS/MS analysis on a SCIEX 5500 system equipped with an Agilent 1200 HPLC, using warfarin as an internal standard [52].
Animals, Study Design, and Treatment
All experiments follow the Institutional Animal Care and Use Committee protocols of the University of Illinois at Chicago. As previously described, the EFAD carriers are 5xFAD+/−/APOE+/+ and the non-carrier is 5xFAD−/−/APOE+/+ [13]. Male E4FAD mice (n = 20) were randomized into 2 treatment groups (n = 10): vehicle control (VC) and AVAS (AVAS). AVAS (30 mg/kg weight) was suspended at 4.8 mg/ml in vehicle solution and administered via daily gavage for 60 d from 6 to 8 M; VC-treated mice were treated with vehicle solution. Mice were weighed daily prior to gavage treatment to determine drug dose.
Behavior and Mouse Sacrifice
In the week prior to sacrifice, mouse behavior was evaluated using the open field test (OFT) and an adapted Morris water maze (MWM) protocol (previously described [92, 93]). Behavior was tracked in real time by an overhead camera and videos analyzed using ANY-maze video tracking software (Stoelting Co., Wood Dale, IL, USA). Briefly, for OFT, the mice were placed in the center of a white box (l38.5 w × 30 h × 30 cm) and allowed to move around freely for 10 m. The total distance traveled and the time spent in the periphery and center were measured. For MWM, acquisition trials (learning) consisted of recording the latency to reach a visible platform in 4 trials/day (1 min each) for 5 consecutive days. On day 6, in the absence of the platform, reference memory was assessed with a single probe trial test recording the latency to reach the target location or target quadrant. A 24 h interval between the last training trial and the probe trial allows reference memory to be tested, independent of the short-term memory of the last training session [94]. After the probe trial, mice were sacrificed and brains were removed and dissected at the midline to produce two hemi-brains, one each for immunohistochemical and biochemicalanalysis (previously described [13]).
Immunostaining
Serial sagittal sections (35 μm thick and separated by 280 μm) from E4FAD mouse brains were used for staining/immunostaining measures. For lipid staining, the sections were washed in TBS, mounted on glass microscope slides, and dried for 1 h. In the dark, 1X LipidSpot-610, “a fluorogenic neutral lipid stain that rapidly accumulates in lipid droplets where it becomes brightly fluorescent” (Biotium product sheet), was applied directly to the slides and incubated for 1 h (previously described [95]). The stained sections were imaged at 63X with a Zeiss Fluorescent Microscope, one field for subiculum (SB) and three fields for CX: visual CX, somatosensory CX, and frontal CX. Images were analyzed by counting the number of LD per nuclei (10–14 nuclei per field) and for the CX, averaged per mouse. For amyloid deposition, the sections were washed in TBS and stained with Thio-S. For Aβ deposition, astrogliosis, and microgliosis, sections were immunostained using MOAB-2, GFAP, and Iba1 antibodies, respectively (previously described [13, 96]). Whole sections were imaged at 10X magnification with a Zeiss Fluorescence microscope and analyzed for area covered by Thio-S, Aβ, GFAP, and Iba-1 in CX using ImageJ software.
Plasma Lipoprotein Profiles
Blood was collected in heparin-coated tubes and centrifuged (3500 rpm, 15 m) to separate plasma. The total-, HDL-, and LDL-cholesterol and apoA1 and apoB levels in the plasma were measured using Beckman Coulter AU480 chemistry analyzer [97, 98]. ApoE was measured by in-house ELISA as previously described.
Sequential Protein Extraction Fractions
For biochemical analysis, hemi-brains were dissected into CX and hippocampus (HP). Cortices were processed using a three-step-sequential protein fractionation method, resulting in soluble (Tris-buffered saline: TBS), non-ionic detergent (TBS + 1%Triton X-100: TBSX), and insoluble (neutralized formic acid: FA) (previously described [99]). Total protein in the TBS and insoluble extracts was quantified using the Bradford assay, and total protein in the TBSX extracts was quantified with the BCA Protein Assay (previously described [99]). Specific fractions were used for subsequent biochemical analyses.
Western Blots
For Western blot analysis, 15 μg of protein from TBS-X fractions was separated on 4–12% Bis–Tris NuPAGE precast gels and transferred to 0.2 μm PVDF membranes. Membranes were incubated in 5% non-fat dry milk in TBS + Tween-20 (0.0625%) for 1 h and incubated overnight with primary antibodies against PSD95, drebrin, β-actin, β-tubulin, ABCA1, APP, and C-terminal fragments (CTF) at 4 °C. After 3 washes with TBS + Tween-20, membranes were incubated with HRP-conjugated secondary antibodies for 45 min, washed with TBS + Tween-20, developed with Pierce chemiluminescence reagents, and visualized with an Odyssey FC Imaging System (previously described [92, 99]).
Native Gels
For native gels, 15 μg of protein from TBS fractions was separated on 4–20% Tris–glycine gels following the manufacturer’s instructions and transferred to 0.2 μm PVDF at 30 V for 16 h. After transfer, blots were treated with Ponceau S Staining Solution (0.1% (w/v) Ponceau S in 5% (v/v) acetic acid) to visualize the molecular mass markers. Membranes were incubated in 5% non-fat dry milk in TBS for 1 h and incubated overnight with primary goat anti-apoE antibody in 1% non-fat dry milk overnight at 4 °C, followed by HRP-conjugated secondary antibodies for 45 min in 1% non-fat dry milk, developed with Pierce chemiluminescence reagents, and visualized with an Odyssey FC Imaging System (previously described [22]).
Aβ and IL-1β ELISAs
ELISAs for apoE (previously described) and Aβ42 levels were measured in TBS, TBSX, and FA extraction fractions, and oAβ (in-house, previously described [34]) and IL-1β in the TBS extract. oAβ was prepared and diluted to produce a standard curve (previously described [91, 100]). IL-1β was measured following the manufacturer’s instructions (previously described [96]). All samples were diluted to read within the sensitivity of the ELISAs.
Statistical Analysis
All data are expressed as mean ± SEM. Data were analyzed by Student’s t-test using GraphPad Prism version 8 (for Mac, GraphPad Software, La Jolla, CA). With significance defined as p < 0.05, † is the treatment effect from comparison of the VC- to AVAS-treatment.
Results
AVAS Induces a Dose-Dependent Increase in Secreted apoE Levels in Primary Glial Cultures
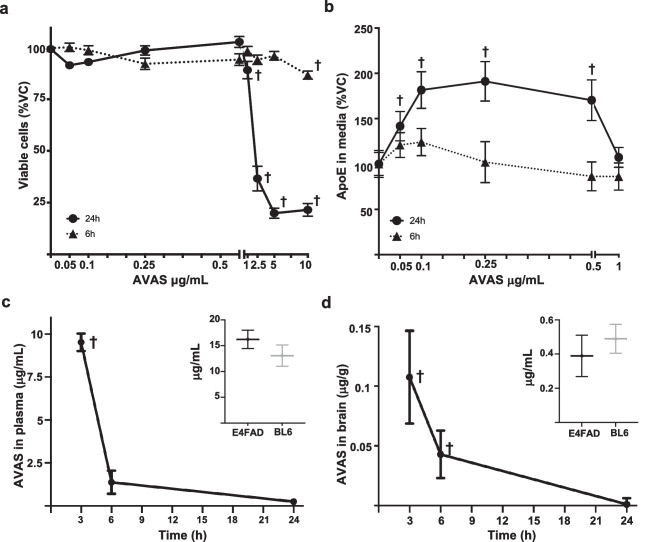
To determine the toxicity of AVAS in vitro, primary glial cells were treated for 6 h and 24 h with 0.05–10.0 μg/ml AVAS (Fig. 1a). For both the 6 h and 24 h incubations, an AVAS dose from 0.05–1.0 μg/ml was not toxic. From 2.5–10.0 μg/ml, there was a small but significant reduction in viability for the 6 h treatment while viability for 24 h treatment dropped to ~ 25%. Primary glial cells treated for 24 h with non-cytotoxic concentrations of AVAS (0.05–1.0 μg/ml) induced a 50% (0.05 μg/ml) and 2-fold (0.1 and 0.5 μg/ml) increase in apoE levels in the media, with no treatment-effect at 6 h (Fig. 1b). These results demonstrate that ACAT inhibition by AVAS can promote an increase in extracellular apoE levels in glial cells and/or promote apoE stability in the media of glial cells.
Fig. 1.
In vitro screening and pharmacokinetics of AVAS. In vitro: Mixed glial cultures from EFAD-non-carrier (5xFAD−/−/APOE+/+) mice treated with vehicle control (VC) or AVAS (0.05–10 μg/mL) for 6 h or 24 h. a Cell viability after AVAS treatment measured by MTT. b ApoE levels in the media after AVAS treatment measured by apoE ELISA. In vivo: c Plasma concentration of AVAS (30 mg/kg) by gavage in male BL6 mice after 3 h, 6 h, and 24 h treatment. Insert c Plasma concentration of AVAS (100 mg/kg) by gavage in male E4FAD (black) or male BL6 (gray) mice after 3 h treatment. d Brain concentration of AVAS (30 mg/kg) by gavage in male BL6 mice after 3 h, 6 h, and 24 h treatment. Insert d Brain concentrations of AVAS (100 mg/kg) by gavage in male E4FAD (black) or male BL6 (gray) mice after 3 h treatment. AVAS was quantified by LC–MS/MS analysis. Data are expressed as mean ± SEM (n = 4–8), analyzed by Student’s t test: p < 0.05, ✝ = vs. VC
AVAS Reaches Effective Concentrations in Mouse Brain
AVAS has previously been administered to FAD-Tg mice by subcutaneous pellet [76]. To establish an effective oral dose for in vivo experiments, pharmacokinetics (PK) in plasma and brain were correlated with the AVAS-induced increase in apoE in glial cell cultures, an in vitro pharmacodynamic (PD) measure significant at 100 nM AVAS (Fig. 1b). Administration of AVAS by gavage (30 mg/kg p.o.) yielded plasma concentrations of 9.0 μg/ml at 3 h and 1.25 μg/ml at 6 h post-administration (Fig. 1c). Although the brain/blood bioavailability ratio was low (≈0.01), measured brain concentrations were above the desired PD threshold at 3 h and 6 h (Fig. 1d). To our knowledge, this is the first measurement of AVAS concentrations in the brain after oral drug delivery, showing that total brain concentrations reach 100 ng/g at 3 h after administration. To control for potential difference between the wildtype mice (male C57BL/6) used for PK and the male E4FAD mice to be used in the study, 100 mg/kg AVAS was administrated, and brain and plasma concentrations measured, with no significant differences at 3 h in plasma (inset Fig. 1c) or brain (inset Fig. 1d).
AVAS Treatment Has Minimal Peripheral Effects on Plasma Lipid Profiles and Mouse Body Weights
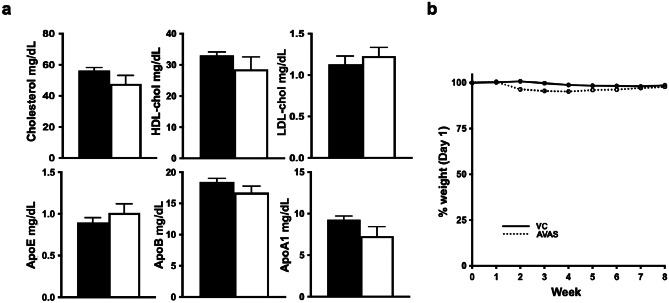
AVAS is a drug repurposed from the cardiovascular field. It was initially designed to promote reverse cholesterol transport, reducing atherosclerotic plaques via a reduction in blood cholesterol levels [101]. AVAS treatment is expected to increase the levels of HDL while reducing LDL, thus reducing total plasma cholesterol [102]. AVAS treatment did not modify the cholesterol-specific plasma lipid profile, including total-, HDL-, and LDL-cholesterol levels (Fig. 2a, top). As well, there was no change in peripheral levels of apoE, apoB, or apoA1 (Fig. 2a, bottom). In the AVAS-treated mice compared to VC, body weights decreased significantly but to a maximum of 5% from week 2–6, with no significant difference in weeks 7–8 (Fig. 2b). These data suggest that the AVAS-specific effects observed in this study are confined to the CNS, independent of potential peripheral effects.
Fig. 2.
Peripheral effects of AVAS: No change in plasma lipoprotein profile or mouse body weights with AVAS treatment. a Plasma levels of total-, HDL-, and LDL-cholesterol measured by enzymatic assay (top) and apoE, apoB, and apoA1 measured by turbidimetry (bottom). Data are expressed as mean ± SEM (n = 10), analyzed by Student’s t test, p < 0.05, ✝ = vs. VC. b. Mouse weight expressed as % weight on day 1 of treatment. Body weight expressed as mean ± SEM (n = 10), analyzed by 2-way ANOVA
MWM Memory Performance Improved, and Postsynaptic Protein Levels Increased with AVAS Treatment
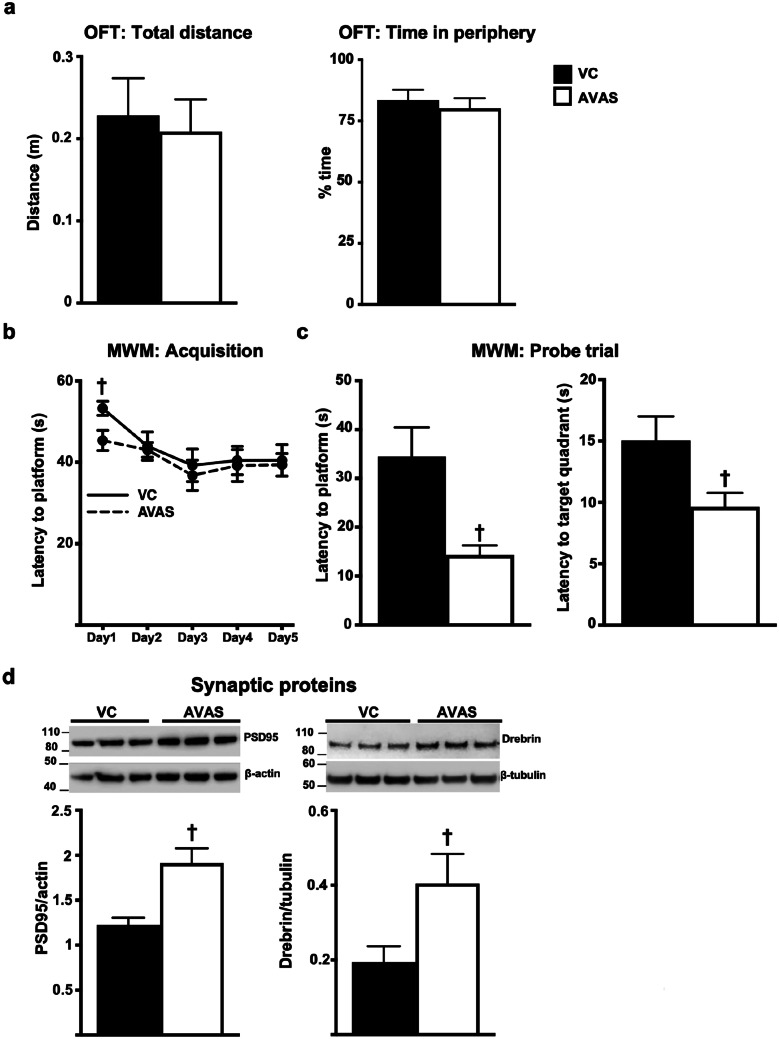
For an AD therapeutic, efficacy is defined as enhanced or retained cognition. We operationally defined surrogate efficacy via MWM performance + synaptic protein levels. Based on results from OFT, AVAS did not affect the general locomotion of the mice as the total distance traveled and the time spent in the periphery by AVAS-treated mice did not significantly differ from the VC mice (Fig. 3a). With the MWM, all the mice learned to reach the platform with no significant differences between the VC- and AVAS-treated mice during the acquisition trials (Fig. 3b). However, in the probe trials (memory), AVAS induced a significant ~ 50% decrease in the latency to target platform and to target quadrant compared to VC group (Fig. 3c). Representative Western blot of synaptic proteins, our second defined component of efficacy, are quantified and normalized to β-actin and β-tubulin (Fig. 3d). Levels of the post-synaptic proteins PSD95 and drebrin are significantly increased 50% and 2-fold, respectively, with AVAS treatment compared to VC. Thus, the AVAS-induced improvement in MWM memory is consistent with the increase in post-synaptic proteins, indications that AVAS exhibits surrogate efficacy.
Fig. 3.
MWM measures of memory improve, and postsynaptic protein levels increase with AVAS treatment. a Open field (OFT) measure of total distance covered and % time in the periphery. b MWM acquisition/learning latency by training day c probe/memory trial latencies to target platform (left) and to target quadrant (right). d Western blot (top) for synaptic protein levels in the CX normalized to β-actin for quantification of PSD95 (left) and drebrin (right). Data are expressed as mean ± SEM (n = 10), analyzed by Student’s t test, p < 0.05, ✝= vs. VC
Intracellular Lipid Droplets Are Reduced by AVAS Treatment
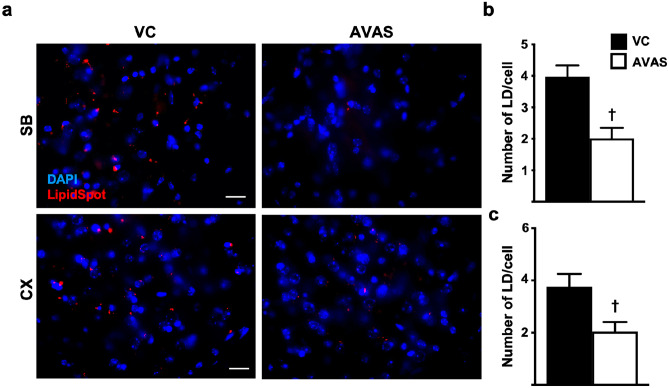
As ACAT catalyzes intracellular cholesterol esterification, facilitating the formation of intracellular LD [103], inhibition of AVAS is predicted to reduce intracellular LD. Thus, to determine target engagement for AVAS, the number of LD/cell was measured in the subiculum (SB) and CX. Figure 4a shows representative 63X sagittal images from SB (left) and CX (right) from VC (top) and AVAS (bottom) of treated mice stained with LipidSpot (LD) and DAPI (DNA). Compared to VC, AVAS induced a significant ~ 50% decrease in LD/cell in both the SB (Fig. 4b) and CX (Fig. 4c), an indication that AVAS exhibits direct target engagement.
Fig. 4.
Intracellular lipid droplets are reduced by AVAS treatment. a Representative images of staining for lipid droplets (LD) (LipidSpot) and nuclei (DAPI) in subiculum (SB, top) and cortex (CX, bottom) for VC (left) and AVAS (right) treatment. Scale bars: 50 mm. b Quantification of LD per nuclei in SB (b) and CX (c). Data are expressed as mean ± SEM (n = 3), analyzed by Student’s t test: p < 0.05, ✝ = vs. VC
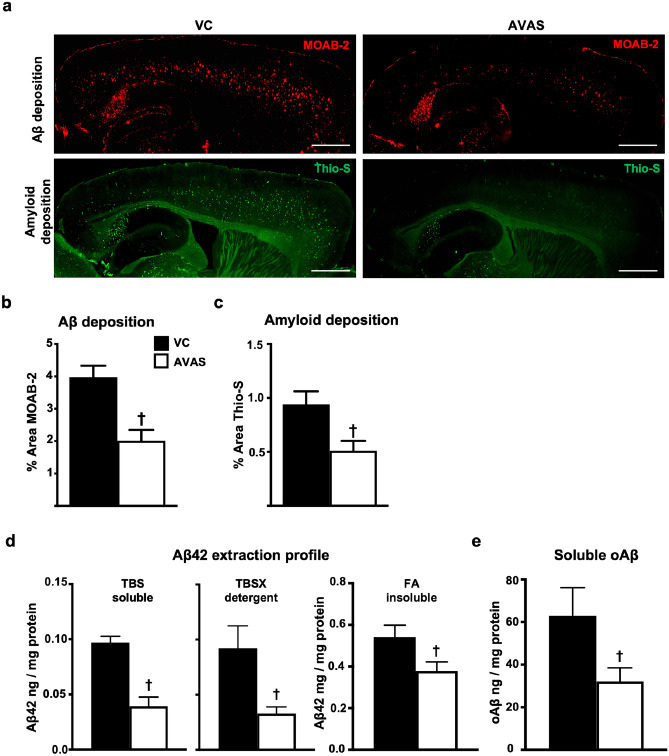
Deposited, Soluble, and Insoluble Aβ Are Reduced by AVAS Treatment
AVAS treatment reduced both Aβ and amyloid deposition. Figure 5a shows representative 10X sagittal images from VC (left) and AVAS (right) treated mice immunostained with the anti-Aβ antibody MOAB-2 (top) to detect Aβ deposition and stained with Thio-S (bottom) to detect amyloid. Both Aβ (Fig. 5b) and amyloid deposition (Fig. 5c) are significantly reduced by ~ 50% in the CX of AVAS vs. VC-treated mice.
Fig. 5.
Deposited, soluble, and insoluble Aβ are reduced by AVAS treatment. a Representative images of immunohistochemistry for Aβ deposition (MOAB-2) (top) and amyloid staining (Thio-S) (bottom) for VC (left) and AVAS (right) treatment. Scale bars: 1000 μm. b Quantification of % area of Aβ deposition in the CX. c Quantification of % area of amyloid deposition in the CX. d Aβ42 extraction profile from the CX measured by Aβ42 ELISA. e Soluble oligomeric Aβ (oAβ) from the soluble extraction fraction measured by oAβ ELISA. Data are expressed as mean ± SEM (n = 6–10), analyzed by Student’s t test, p < 0.05, ✝ = vs. VC
By Aβ42 ELISA, Aβ42 in the soluble TBS, detergent TBSX, and insoluble FA extraction fractions are significantly reduced in AVAS vs. VC (Fig. 5d). The decrease in the insoluble FA extraction fraction, primarily Aβ42 from the insoluble amyloid plaques, is consistent with the decrease in Aβ deposition (Fig. 5a). Importantly, AVAS reduced soluble oAβ levels by 50% compared to VC (Fig. 5e), consistent with the 50% decrease in soluble Aβ42 in the TBS extraction fraction (Fig. 5d).
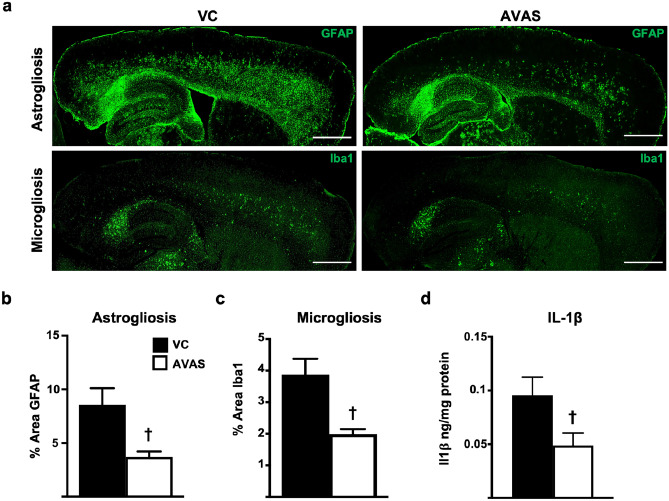
Neuroinflammation Is Reduced by AVAS Treatment
To investigate the effect of ACAT inhibition on neuroinflammation, astrogliosis and microgliosis were analyzed in male E4FAD mice after AVAS treatment. Figure 6a shows representative 10X sagittal images of from VC (left) and AVAS (right) treated mice immunostained with GFAP (top) to detect astrogliosis and Iba1 (bottom) to detect microgliosis. Both astrogliosis (Fig. 6b) and microgliosis (Fig. 6c) are significantly reduced by 50% in the CX in AVAS vs. VC treated mice. In addition, an ELISA for IL-1β, a key proinflammatory cytokine secreted by both astrocytes and microglia [104], revealed a 50% decrease in the CX with AVAS vs. VC-treated mice (Fig. 6d). Thus, AVAS exhibited both target engagement and efficacy, as well as inhibition of Aβ deposition and solubility, and neuroinflammation.
Fig. 6.
Neuroinflammation is reduced by AVAS treatment. a Representative images of immunohistochemistry for astrogliosis (GFAP) (top) and microgliosis (Iba1) (bottom) for VC (left) and AVAS (right) treatment. Scale bars: 1000 μm. b Quantification of % area of astrogliosis in the CX. c Quantification of % area of microgliosis in the CX. d by IL-1β ELISA. Data are expressed as mean ± SEM (n = 6–10), analyzed by Student’s t test, p < 0.05, ✝ = vs. VC
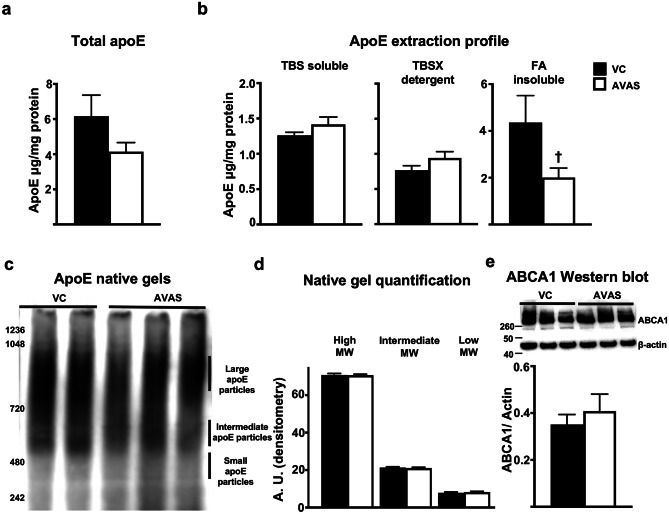
ApoE4 Levels, Lipidated apoE, and ABCA1 Levels Are Not Affected by AVAS Treatment
AVAS treatment does not have a significant effect on total apoE4 levels (Fig. 7a). Importantly, there is no difference in the levels of apoE4 extracted in the TBSX fraction from AVAS vs. VC-treated mice (Fig. 7b). The protocol for this sequential protein extraction method was originally optimized to extract lipoprotein-associated apoE to the TBSX fraction by using 1%Triton X-100, a non-ionic detergent that is less stringent compared to SDS or Tween 20, allowing the intact lipoproteins to extract in this fraction [99]. Thus, the lack of an AVAS-induced increase in apoE4 levels in the TBSX extraction fraction is evidence that apoE4 lipidation did not increase. To further interrogate whether AVAS effects the lipidation of apoE4-lipoproteins, apoE native gels were run (Fig. 7c) and no changes in the high, intermediate, or low molecular weight particles were observed with AVAS vs. VC-treated mice (Fig. 7d). In the insoluble-FA fraction, there is a 50% reduction in apoE4 with AVAS treatment, consistent with the AVAS-induced reduction in Aβ deposition (Fig. 5a–c) and insoluble-FA Aβ42 (Fig. 5d), as apoE is known to seed, or co-deposit, with Aβ in plaques. As measured by Western blot and normalized to β-actin (Fig. 7e), ABCA1 levels did not change with AVAS treatment, evidence that cholesterol transport to the apoE4-lipoproteins was not increased [52]. These data are evidence that our hypothesis for the mechanism by which AVAS effects AD pathology likely does not involve lipidation of apoE4-lipoproteins.
Fig. 7.
ApoE4 levels, lipidated apoE4, and ABCA1 levels are not affected by AVAS treatment. a Total apoE and b apoE extraction profile from the CX measured by apoE ELISA. c Native gel for apoE. d Quantification of apoE particles by size from native gel. e Western blot (top) for ABCA1 levels in the CX, normalized to β-actin for quantification (bottom). Data are expressed as mean ± SEM (n = 6–10), analyzed by Student’s t test, p < 0.05, † = vs. VC
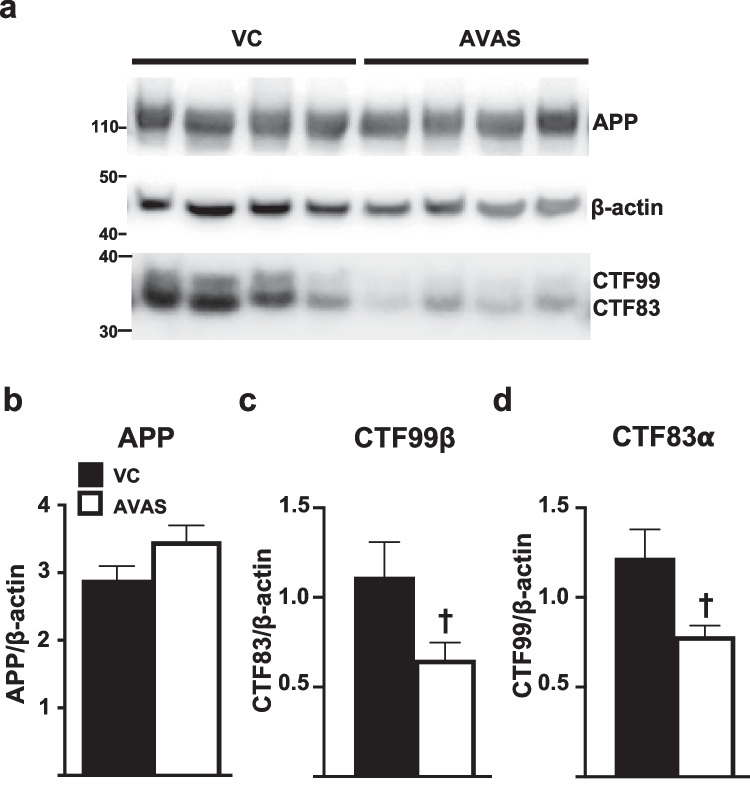
APP Processing Is Reduced by AVAS Treatment
We next tested the hypothesis that AVAS-induced inhibition of AD pathology is mediated by a decrease in APP processing by β-amyloid cleavage enzyme (β-secretase), reducing Aβ levels. To evaluate if this pathway is modulated by AVAS in E4FAD mice, TBSX brain fractions were analyzed by Western blot for full length APP and APP-CTF (Fig. 8a). APP, CTFα, and CTFβ normalized to β-actin for quantification. APP levels do not change with AVAS treatment (Fig. 8b). AVAS induced a 50% decrease in both APP-CTF83 (Fig. 8c, α-secretase/non-amyloidogenic cleavage) and APP-CTF99 (Fig. 8d, β-secretase/amyloidogenic cleavage) compared to VC-treated mice. Thus, AVAS treatment reduces APP processing, reducing the production of Aβ, consistent with previous studies using FAD-Tg mouse models expressing m-apoE [76, 105].
Fig. 8.
APP processing is reduced by AVAS treatment. a Western blot of APP processing to APP-CTFs. APP, CTFα, and CTFβ normalized to β-actin for quantification; b APP, c CTFα, and d CTFβ. Data are expressed as mean ± SEM (n = 4), analyzed by Student’s t test, p < 0.05, ✝ = vs. VC
Discussion
In pre-clinical studies, regulation of brain cholesterol levels through different strategies has successfully reduced Aβ pathology. Examples of these strategies include (1) reducing de novo synthesis of cholesterol by inhibiting the rate limiting enzyme HMG-CoA reductase [106], (2) increasing cholesterol turnover via the activation of the rate limiting enzyme CYP46A1 [107–109], and (3) enhancing cholesterol efflux via increasing levels and activity of ABCA1 [19, 46, 53, 110]. Interestingly, interventions that promote these processes reduce Aβ pathology, including increasing ER-FC via inhibiting CE formation by ACAT1 inhibition [74, 75, 103, 111].
For an AD therapeutic, efficacy is defined as enhanced or retained cognition. In this study, surrogate efficacy was defined by MWM performance in acquisition (learning) and probe (memory) trials and levels of post synaptic proteins. With MWM, previous studies demonstrate that ACAT inhibition improves MWM learning [75]. While we did not see an overall improvement during MWM acquisition trials with AVAS treatment, the latency to platform on day 1 was lower in AVAS- vs. VC-treated mice, suggesting that the AVAS treatment enhanced learning on that day only; the latency to platform was not significantly different between VC- and AVAS-treated mice for days 2–5 (Fig. 3b), an effect observed with other AVAS-treated FAD-Tg models [75]. However, with the probe trial for memory (platform removed), the latency to the platform and target quadrant was significantly less for AVAS-treated compared to VC-treated mice (Fig. 3c), a phenomenon previously reported in the literature for FAD-Tg mouse models [112–117]. An improvement in memory is consistent with the effect of memory measured by CFC in APP/ACAT−/− mice [73]. Importantly, we demonstrate AVAS-induced increases in the levels of postsynaptic proteins PSD95 and drebrin (Fig. 3d).
In the current study, although AVAS treatment did not modify the cholesterol-specific plasma lipoprotein profile (Fig. 2a), brain-specific readouts did change. AVAS induced a significant reduction in LD, indirectly demonstrating target engagement (Fig. 4). LD are composed of neutral lipids, primarily CE and TAG, thus our data are consistent with previous findings that inhibition of ACAT reduces brain CE levels [75, 76]. These data suggest that the AVAS-specific effects observed in this study are confined to the CNS, independent of potential peripheral effects.
To evaluate if the observed surrogate efficacy and indirect target engagement are related to a reduction in AD pathology, we assessed Aβ pathology and neuroinflammation after AVAS treatment in E4FAD mice. AD pathology is reduced by ACAT inhibition, consistent with previous studies. Specifically, we show a reduction in soluble oAβ, soluble and insoluble Aβ42 levels, Aβ deposition, amyloid deposition, and neuroinflammation (Figs. 5 and 6) [75, 76]. Remarkably, this occurs in a FAD-Tg-mouse expressing h-apoE4, not m-apoE, and without modifying the lipidation state of apoE-particles (Fig. 7).
AVAS-induced inhibition of peripheral cholesterol esterification was originally a therapeutic target for atherosclerosis as it facilitated cholesterol efflux to apoA1-HDL in plasma, thus reducing the development of foam cells from macrophage uptake of oxidized LDL. This decrease in endothelial cell disruption and atherosclerotic plaque development was dependent on apoA1 as the plasma cholesterol acceptor [57–63]. However, little is known about the effect of ACAT inhibition of cholesterol efflux in the brain where apoE will be the primary cholesterol acceptor. To investigate this effect in vitro and in vivo, it is important to consider the difference between h-apoE and m-apoE. While humans express three primary isoforms that vary at only two of the 299 amino acids, mice express one form of apoE, the same as apoE4 at residues Arg112/Arg158, although the homology between m- and h-apoE is only 70% [118]. M-apoE and h-apoE differ in both structure [119] and functions ranging from lipid binding and transport, to effects on AD pathology [22, 84, 85, 119, 120].
Driven by the hypothesis that ACAT inhibition by AVAS will increase ABCA1-mediated FC efflux to apoE-particles in the brain, we examined the effect of ACAT inhibition in vitro on APOE4 glia cultures. This study demonstrated an AVAS-induced increase in apoE4 levels in the media compared to VC (Fig. 1b), likely by an increase in apoE4 stability from increased lipidation of the apoE4-particles. Conversely, AVAS treatment in E4FAD mice does not appear to modify brain apoE levels or lipidation in either the plasma (Fig. 2) or the brain (Fig. 7). This may be the result of additional regulatory pathways acting in vivo and not in vitro as shown previously with ACAT inhibition exhibiting a cell-specific response, either inhibition or induction of cholesterol efflux and ABCA1 levels [57–61, 103, 121–124]. In vitro, ACAT inhibition increases ABCA1 levels and cholesterol efflux of cholesterol-loaded macrophages, while decreasing ABCA1 levels in adipocytes [125] and decreasing ABCA1 levels, lipid catabolism, and cholesterol efflux in APOE4 microglia [126–128], thus decreasing the capacity to lipidate apoE4-particles [129]. Potentially important for the EFAD model, APOE4 astrocytes chronically exposed to fatty acids (a possible mimic for aged astrocytes rich in LD) produce TAG-rich apoE-particles rather than cholesterol-rich particles [130]. It is possible that the drastic reduction in LD after AVAS treatment (Fig. 4) is the result of LD-CE loss, in the absence of sufficient levels of LD-TAG for formation of apoE4-TAG particles.
The primary hypothesis for the AVAS-induced reduction in AD pathology is a decrease in APP processing. Kovacs et al. described this mechanism, centered on a novel pathway in which APP is palmitoylated at the ER for mobilization to the PM lipid rafts to be processed by γ and β secretases [41]. While the specific mechanism remains unclear, ACAT inhibition significantly reduces palmitoylated APP in lipid rafts, thus reducing APP processing [75–77, 105, 131]. Although we did not measure APP palmitoylation or APP localization to lipid rafts, we did see a reduction in both amyloidogenic and non-amyloidogenic processing of APP, with no change in total APP (Fig. 8) [75, 76]. Thus, the reduction of AD pathology in EFAD mice after AVAS treatment is predominantly due to a reduction in APP processing/Aβ secretion.
Another mechanism for the AVAS-induced reduction in Aβ pathology is microglia-driven phagocytosis. Chang et al. propose that, in addition to reducing Aβ production, cholesterol mobilization by ACAT inhibition in microglia can promote autophagy and lysosomal biogenesis, leading to an increased clearance of oAβ [69–71, 77]. However, Aβ degradation by autophagosome formation has been reported for the ACAT potent inhibitor K604 and in ACAT-KO [71], suggesting that autophagocytic activity is present predominantly with the total inhibition of ACAT activity. Further, microglia from AD patients and APOE4 carriers exhibit impaired autophagy [132–134]; thus, the cholesterol mobilization by ACAT-inhibition may not be enough to overcome an already impaired process.
This study confirms the central role of lipid homeostasis in modifying AD pathology. Overall, our results suggest that the AVAS-induced redistribution of cholesterol within in the cell is sufficient to modify LDs and lipid raft formation/distribution, resulting in increased synaptic function and reduced APP processing. However, these AVAS-induced changes are not sufficient to increase lipid efflux and modify apoE4-particles. Further, evidence suggests that the LD are rich in CE and poor in TAG, thus not supporting the formation of apoE4-TAG particles [130]. Although AVAS failed phase III clinical trials for cardiovascular efficacy [101], it has an acceptable safety profile [135, 136] and is currently being proposed as an anti-cancer agent [137]. The efficacy and potential for disease modification observed in this study with human apoE4 supports further testing of the ACAT-inhibitor AVAS for treatment of AD. In the future, it will be important to evaluate AVAS efficacy in the context of the universal biological variables (UBV) of AD risk, specifically age, APOE, and sex, thus understanding the effect of AVAS in the presence of apoE3 vs. apoE4, using both male and female and young vs. old EFAD mice. For instance, we cannot eliminate the possibility that AVAS treatment may increase apoE3 lipidation and further decrease Aβ levels via a clearance pathway within an APOE3 context [138].
Supplementary Information
Below is the link to the electronic supplementary material.
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
Author Contribution
MJL designed the current study. YW and GRJT performed PK analysis and analyzed the data. JMY maintained the EFAD mouse colony and executed the behavioral tests. AV, DB, and EA treated the mice. AV and DB supervised the biochemical and immunostaining experiments, respectively, including data analysis and figure design. NF isolated and maintained the cultures for the in vitro experiments. CP performed the Western blots. AA analyzed the behavioral data. AV and MJL wrote the manuscript. GRJT critically reviewed the manuscript, giving valuable feedback. AbbVie contributed to the study design, research, and interpretation of data, reviewing, and approving the manuscript. All authors read and approved the final manuscript.
Funding
This study was partially sponsored by AbbVie, Inc. Additionally, the LaDu lab was funded by National Institute of Aging grants (R01 AG058068, R01 AG057008), institutional funds from the College of Medicine at the University of Illinois, Chicago, and philanthropic support from Louis and Christine Friedrich.
Data Availability
The data that support the findings of this study are available from the corresponding author, GRJT, upon reasonable request.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Gregory R. J. Thatcher, Email: grjthatcher@arizona.edu
Mary Jo LaDu, Email: mladu@uic.edu.
References
- 1.Ralhan I, Chang CL, Lippincott-Schwartz J, Ioannou MS. Lipid droplets in the nervous system. J Cell Biol. 2021;220(7). [DOI] [PMC free article] [PubMed]
- 2.Picard C, Julien C, Frappier J, Miron J, Theroux L, Dea D, et al. Alterations in cholesterol metabolism-related genes in sporadic Alzheimer's disease. Neurobiol Aging. 2018;66(180):e1–e9. doi: 10.1016/j.neurobiolaging.2018.01.018. [DOI] [PubMed] [Google Scholar]
- 3.Yin F. Lipid metabolism and Alzheimer's disease: clinical evidence, mechanistic link and therapeutic promise. FEBS J. 2022. [DOI] [PMC free article] [PubMed]
- 4.Chan RB, Oliveira TG, Cortes EP, Honig LS, Duff KE, Small SA, et al. Comparative lipidomic analysis of mouse and human brain with Alzheimer disease. J Biol Chem. 2012;287(4):2678–2688. doi: 10.1074/jbc.M111.274142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tajima Y, Ishikawa M, Maekawa K, Murayama M, Senoo Y, Nishimaki-Mogami T, et al. Lipidomic analysis of brain tissues and plasma in a mouse model expressing mutated human amyloid precursor protein/tau for Alzheimer's disease. Lipids Health Dis. 2013;12:68. doi: 10.1186/1476-511X-12-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alzheimer A, Stelzmann RA, Schnitzlein HN, Murtagh FR. An English translation of Alzheimer's 1907 paper, "Uber eine eigenartige Erkankung der Hirnrinde". Clin Anat. 1995;8(6):429–431. doi: 10.1002/ca.980080612. [DOI] [PubMed] [Google Scholar]
- 7.van der Kant R, Langness VF, Herrera CM, Williams DA, Fong LK, Leestemaker Y, et al. Cholesterol metabolism is a druggable axis that independently regulates tau and amyloid-beta in iPSC-derived Alzheimer's disease neurons. Cell Stem Cell. 2019;24(3):363–375. doi: 10.1016/j.stem.2018.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marschallinger J, Iram T, Zardeneta M, Lee SE, Lehallier B, Haney MS, et al. Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nat Neurosci. 2020;23(2):194–208. doi: 10.1038/s41593-019-0566-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feringa FM, van der Kant R. Cholesterol and Alzheimer's disease; from risk genes to pathological effects. Front Aging Neurosci. 2021;13:690372. doi: 10.3389/fnagi.2021.690372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alzheimer’s Association. 2023 Alzheimer's disease facts and figures. Alzheimers Dement. 2023;19(4). 10.1002/alz.13016 [DOI] [PubMed]
- 11.Elliott DA, Weickert CS, Garner B. Apolipoproteins in the brain: implications for neurological and psychiatric disorders. Clin Lipidol. 2010;51(4):555–573. doi: 10.2217/clp.10.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LaDu MJ, Gilligan SM, Lukens JR, Cabana VG, Reardon CA, Van Eldik LJ, et al. Nascent astrocyte particles differ from lipoproteins in CSF. J Neurochem. 1998;70(5):2070–2081. doi: 10.1046/j.1471-4159.1998.70052070.x. [DOI] [PubMed] [Google Scholar]
- 13.Youmans KL, Tai LM, Nwabuisi-Heath E, Jungbauer L, Kanekiyo T, Gan M, et al. APOE4-specific changes in abeta accumulation in a new transgenic mouse model of Alzheimer disease. J Biol Chem. 2012;287(50):41774–41786. doi: 10.1074/jbc.M112.407957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beffert U, Cohn JS, Petit-Turcotte C, Tremblay M, Aumont N, Ramassamy C, et al. Apolipoprotein E and beta-amyloid levels in the hippocampus and frontal cortex of Alzheimer's disease subjects are disease-related and apolipoprotein E genotype dependent. Brain Res. 1999;843(1–2):87–94. doi: 10.1016/S0006-8993(99)01894-6. [DOI] [PubMed] [Google Scholar]
- 15.Sullivan PM, Han B, Liu F, Mace BE, Ervin JF, Wu S, et al. Reduced levels of human apoE4 protein in an animal model of cognitive impairment. Neurobiol Aging. 2011;32(5):791–801. doi: 10.1016/j.neurobiolaging.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 16.Minagawa H, Gong JS, Jung CG, Watanabe A, Lund-Katz S, Phillips MC, et al. Mechanism underlying apolipoprotein E (ApoE) isoform-dependent lipid efflux from neural cells in culture. J Neurosci Res. 2009;87(11):2498–2508. doi: 10.1002/jnr.22073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gong JS, Morita SY, Kobayashi M, Handa T, Fujita SC, Yanagisawa K, et al. Novel action of apolipoprotein E (ApoE): ApoE isoform specifically inhibits lipid-particle-mediated cholesterol release from neurons. Mol Neurodegener. 2007;2:9. doi: 10.1186/1750-1326-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu J, Liu CC, Chen XF, Zhang YW, Xu H, Bu G. Opposing effects of viral mediated brain expression of apolipoprotein E2 (apoE2) and apoE4 on apoE lipidation and abeta metabolism in apoE4-targeted replacement mice. Mol Neurodegener. 2015;10(1):6. doi: 10.1186/s13024-015-0001-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boehm-Cagan A, Bar R, Liraz O, Bielicki JK, Johansson JO, Michaelson DM. ABCA1 Agonist reverses the ApoE4-driven cognitive and brain pathologies. J Alzheimers Dis. 2016;54(3):1219–1233. doi: 10.3233/JAD-160467. [DOI] [PubMed] [Google Scholar]
- 20.Bar R, Boehm-Cagan A, Luz I, Kleper-Wall Y, Michaelson DM. The effects of apolipoprotein E genotype, alpha-synuclein deficiency, and sex on brain synaptic and Alzheimer's disease-related pathology. Alzheimers Dement (Amst) 2018;10:1–11. doi: 10.1016/j.dadm.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boehm-Cagan A, Michaelson DM. Reversal of apoE4-driven brain pathology and behavioral deficits by bexarotene. J Neurosci. 2014;34(21):7293–7301. doi: 10.1523/JNEUROSCI.5198-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fagan AM, Holtzman DM, Munson G, Mathur T, Schneider D, Chang LK, et al. Unique lipoproteins secreted by primary astrocytes from wild type, apoE (-/-), and human apoE transgenic mice. J Biol Chem. 1999;274(42):30001–30007. doi: 10.1074/jbc.274.42.30001. [DOI] [PubMed] [Google Scholar]
- 23.Heinsinger NM, Gachechiladze MA, Rebeck GW. Apolipoprotein E genotype affects size of ApoE complexes in cerebrospinal fluid. J Neuropathol Exp Neurol. 2016. [DOI] [PMC free article] [PubMed]
- 24.Klein WL, Stine WB, Jr, Teplow DB. Small assemblies of unmodified amyloid beta-protein are the proximate neurotoxin in Alzheimer's disease. Neurobiol Aging. 2004;25(5):569–580. doi: 10.1016/j.neurobiolaging.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 25.Larson ME, Lesne SE. Soluble abeta oligomer production and toxicity. J Neurochem. 2012;120(Suppl 1):125–139. doi: 10.1111/j.1471-4159.2011.07478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tu S, Okamoto S, Lipton SA, Xu H. Oligomeric abeta-induced synaptic dysfunction in Alzheimer's disease. Mol Neurodegener. 2014;9:48. doi: 10.1186/1750-1326-9-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klyubin I, Cullen WK, Hu NW, Rowan MJ. Alzheimer's disease abeta assemblies mediating rapid disruption of synaptic plasticity and memory. Mol Brain. 2012;5:25. doi: 10.1186/1756-6606-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferreira ST, Lourenco MV, Oliveira MM, De Felice FG. Soluble amyloid-beta oligomers as synaptotoxins leading to cognitive impairment in Alzheimer's disease. Front Cell Neurosci. 2015;9:191. doi: 10.3389/fncel.2015.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cline EN, Bicca MA, Viola KL, Klein WL. The amyloid-beta oligomer hypothesis: beginning of the third decade. J Alzheimers Dis. 2018;64(s1):S567–S610. doi: 10.3233/JAD-179941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roychaudhuri R, Huynh TV, Whitaker TR, Hodara E, Condron MM, Teplow DB. A critical role of Ser26 hydrogen bonding in abeta42 assembly and toxicity. Biochemistry. 2017;56(48):6321–6324. doi: 10.1021/acs.biochem.7b00772. [DOI] [PubMed] [Google Scholar]
- 31.Roychaudhuri R, Zheng X, Lomakin A, Maiti P, Condron MM, Benedek GB, et al. Role of species-specific primary structure differences in abeta42 assembly and neurotoxicity. ACS Chem Neurosci. 2015;6(12):1941–1955. doi: 10.1021/acschemneuro.5b00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261(5123):921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 33.Cerf E, Gustot A, Goormaghtigh E, Ruysschaert JM, Raussens V. High ability of apolipoprotein E4 to stabilize amyloid-beta peptide oligomers, the pathological entities responsible for Alzheimer's disease. FASEB J. 2011;25(5):1585–1595. doi: 10.1096/fj.10-175976. [DOI] [PubMed] [Google Scholar]
- 34.Tai LM, Bilousova T, Jungbauer L, Roeske SK, Youmans KL, Yu C, et al. Levels of soluble apolipoprotein E/amyloid-beta (Abeta) complex are reduced and oligomeric abeta increased with APOE4 and Alzheimer disease in a transgenic mouse model and human samples. J Biol Chem. 2013;288(8):5914–5926. doi: 10.1074/jbc.M112.442103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hampel H, Vassar R, De Strooper B, Hardy J, Willem M, Singh N, et al. The beta-secretase BACE1 in Alzheimer's disease. Biol Psychiatry. 2021;89(8):745–756. doi: 10.1016/j.biopsych.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cordy JM, Hussain I, Dingwall C, Hooper NM, Turner AJ. Exclusively targeting beta-secretase to lipid rafts by GPI-anchor addition up-regulates beta-site processing of the amyloid precursor protein. Proc Natl Acad Sci U S A. 2003;100(20):11735–11740. doi: 10.1073/pnas.1635130100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wahrle S, Das P, Nyborg AC, McLendon C, Shoji M, Kawarabayashi T, et al. Cholesterol-dependent gamma-secretase activity in buoyant cholesterol-rich membrane microdomains. Neurobiol Dis. 2002;9(1):11–23. doi: 10.1006/nbdi.2001.0470. [DOI] [PubMed] [Google Scholar]
- 38.Hartmann T, Kuchenbecker J, Grimm MO. Alzheimer's disease: the lipid connection. J Neurochem. 2007;103(Suppl 1):159–170. doi: 10.1111/j.1471-4159.2007.04715.x. [DOI] [PubMed] [Google Scholar]
- 39.Ehehalt R, Keller P, Haass C, Thiele C, Simons K. Amyloidogenic processing of the Alzheimer beta-amyloid precursor protein depends on lipid rafts. J Cell Biol. 2003;160(1):113–123. doi: 10.1083/jcb.200207113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang H, Kulas JA, Wang C, Holtzman DM, Ferris HA, Hansen SB. Regulation of beta-amyloid production in neurons by astrocyte-derived cholesterol. Proc Natl Acad Sci U S A. 2021;118(33). [DOI] [PMC free article] [PubMed]
- 41.Bhattacharyya R, Black SE, Lotlikar MS, Fenn RH, Jorfi M, Kovacs DM, et al. Axonal generation of amyloid-beta from palmitoylated APP in mitochondria-associated endoplasmic reticulum membranes. Cell Rep. 2021;35(7):109134. doi: 10.1016/j.celrep.2021.109134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chauhan NB. Membrane dynamics, cholesterol homeostasis, and Alzheimer's disease. J Lipid Res. 2003;44(11):2019–2029. doi: 10.1194/jlr.R300010-JLR200. [DOI] [PubMed] [Google Scholar]
- 43.Huynh TV, Davis AA, Ulrich JD, Holtzman DM. Apolipoprotein E and Alzheimer's disease: the influence of apolipoprotein E on amyloid-beta and other amyloidogenic proteins. J Lipid Res. 2017;58(5):824–836. doi: 10.1194/jlr.R075481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sagare A, Deane R, Bell RD, Johnson B, Hamm K, Pendu R, et al. Clearance of amyloid-beta by circulating lipoprotein receptors. Nat Med. 2007;13(9):1029–1031. doi: 10.1038/nm1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tokuda T, Calero M, Matsubara E, Vidal R, Kumar A, Permanne B, et al. Lipidation of apolipoprotein E influences its isoform-specific interaction with Alzheimer's amyloid beta peptides. Biochem J. 2000;348(Pt 2):359–365. doi: 10.1042/bj3480359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tai LM, Koster KP, Luo J, Lee SH, Wang YT, Collins NC, et al. Amyloid-beta pathology and APOE genotype modulate retinoid X receptor agonist activity in vivo. J Biol Chem. 2014;289(44):30538–30555. doi: 10.1074/jbc.M114.600833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tai LM, Ghura S, Koster KP, Liakaite V, Maienschein-Cline M, Kanabar P, et al. APOE-modulated abeta-induced neuroinflammation in Alzheimer's disease: current landscape, novel data and future perspective. J Neurochem. 2015. [DOI] [PMC free article] [PubMed]
- 48.Wahrle SE, Jiang H, Parsadanian M, Hartman RE, Bales KR, Paul SM, et al. Deletion of Abca1 increases Aβ deposition in the PDAPP transgenic mouse model of Alzheimer disease. J Biol Chem. 2005;280(52):43236–43242. doi: 10.1074/jbc.M508780200. [DOI] [PubMed] [Google Scholar]
- 49.Wahrle SE, Jiang H, Parsadanian M, Kim J, Li A, Knoten A, et al. Overexpression of ABCA1 reduces amyloid deposition in the PDAPP mouse model of Alzheimer disease. J Clin Invest. 2008;118(2):671–682. doi: 10.1172/JCI33622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wahrle SE, Jiang H, Parsadanian M, Legleiter J, Han X, Fryer JD, et al. ABCA1 is required for normal central nervous system ApoE levels and for lipidation of astrocyte-secreted apoE. J Biol Chem. 2004;279(39):40987–40993. doi: 10.1074/jbc.M407963200. [DOI] [PubMed] [Google Scholar]
- 51.Riddell DR, Zhou H, Comery TA, Kouranova E, Lo CF, Warwick HK, et al. The LXR agonist TO901317 selectively lowers hippocampal abeta42 and improves memory in the Tg2576 mouse model of Alzheimer's disease. Mol Cell Neurosci. 2007;34(4):621–628. doi: 10.1016/j.mcn.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 52.Tai LM, Koster KP, Luo J, Lee SH, Wang Y-T, Collins NC, et al. Amyloid-β pathology and APOE genotype modulate retinoid X receptor agonist activity in vivo. J Biol Chem. 2014;289(44):30538–30555. doi: 10.1074/jbc.M114.600833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koster KP, Smith C, Valencia-Olvera AC, Thatcher GR, Tai LM, LaDu MJ. Rexinoids as therapeutics for Alzheimer's disease: role of APOE. Curr Top Med Chem. 2017;17(6):708–720. doi: 10.2174/1568026616666160617090227. [DOI] [PubMed] [Google Scholar]
- 54.Meiner V, Tam C, Gunn MD, Dong LM, Weisgraber KH, Novak S, et al. Tissue expression studies on the mouse acyl-CoA: cholesterol acyltransferase gene (Acact): findings supporting the existence of multiple cholesterol esterification enzymes in mice. J Lipid Res. 1997;38(9):1928–1933. doi: 10.1016/S0022-2275(20)37168-6. [DOI] [PubMed] [Google Scholar]
- 55.Cases S, Novak S, Zheng YW, Myers HM, Lear SR, Sande E, et al. ACAT-2, a second mammalian acyl-CoA:cholesterol acyltransferase. Its cloning, expression, and characterization. J Biol Chem. 1998;273(41):26755–26764. doi: 10.1074/jbc.273.41.26755. [DOI] [PubMed] [Google Scholar]
- 56.Chang TY, Li BL, Chang CC, Urano Y. Acyl-coenzyme A:cholesterol acyltransferases. Am J Physiol Endocrinol Metab. 2009;297(1):E1–9. doi: 10.1152/ajpendo.90926.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sugimoto K, Tsujita M, Wu CA, Suzuki K, Yokoyama S. An inhibitor of acylCoA: cholesterol acyltransferase increases expression of ATP-binding cassette transporter A1 and thereby enhances the ApoA-I-mediated release of cholesterol from macrophages. Biochim Biophys Acta. 2004;1636(1):69–76. doi: 10.1016/j.bbalip.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 58.Rodriguez A, Usher DC. Anti-atherogenic effects of the acyl-CoA:cholesterol acyltransferase inhibitor, avasimibe (CI-1011), in cultured primary human macrophages. Atherosclerosis. 2002;161(1):45–54. doi: 10.1016/S0021-9150(01)00620-7. [DOI] [PubMed] [Google Scholar]
- 59.Kellner-Weibel G, Luke SJ, Rothblat GH. Cytotoxic cellular cholesterol is selectively removed by apoA-I via ABCA1. Atherosclerosis. 2003;171(2):235–243. doi: 10.1016/j.atherosclerosis.2003.08.024. [DOI] [PubMed] [Google Scholar]
- 60.Huang ZH, Lin CY, Oram JF, Mazzone T. Sterol efflux mediated by endogenous macrophage ApoE expression is independent of ABCA1. Arterioscler Thromb Vasc Biol. 2001;21(12):2019–2025. doi: 10.1161/hq1201.100242. [DOI] [PubMed] [Google Scholar]
- 61.An S, Jang YS, Park JS, Kwon BM, Paik YK, Jeong TS. Inhibition of acyl-coenzyme A:cholesterol acyltransferase stimulates cholesterol efflux from macrophages and stimulates farnesoid X receptor in hepatocytes. Exp Mol Med. 2008;40(4):407–417. doi: 10.3858/emm.2008.40.4.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamauchi Y, Chang CC, Hayashi M, Abe-Dohmae S, Reid PC, Chang TY, et al. Intracellular cholesterol mobilization involved in the ABCA1/apolipoprotein-mediated assembly of high density lipoprotein in fibroblasts. J Lipid Res. 2004;45(10):1943–1951. doi: 10.1194/jlr.M400264-JLR200. [DOI] [PubMed] [Google Scholar]
- 63.Schmitz G, Niemann R, Brennhausen B, Krause R, Assmann G. Regulation of high density lipoprotein receptors in cultured macrophages: role of acyl-CoA:cholesterol acyltransferase. EMBO J. 1985;4(11):2773–2779. doi: 10.1002/j.1460-2075.1985.tb04003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stahlberg D, Angelin B, Einarsson K. Age-related changes in the metabolism of cholesterol in rat liver microsomes. Lipids. 1991;26(5):349–352. doi: 10.1007/BF02537197. [DOI] [PubMed] [Google Scholar]
- 65.Geltinger F, Tevini J, Briza P, Geiser A, Bischof J, Richter K, et al. The transfer of specific mitochondrial lipids and proteins to lipid droplets contributes to proteostasis upon stress and aging in the eukaryotic model system Saccharomyces cerevisiae. Geroscience. 2020;42(1):19–38. doi: 10.1007/s11357-019-00103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Beas AO, Gordon PB, Prentiss CL, Olsen CP, Kukurugya MA, Bennett BD, et al. Independent regulation of age associated fat accumulation and longevity. Nat Commun. 2020;11(1):2790. doi: 10.1038/s41467-020-16358-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sienski G, Narayan P, Bonner JM, Kory N, Boland S, Arczewska AA, et al. APOE4 disrupts intracellular lipid homeostasis in human iPSC-derived glia. Sci Transl Med. 2021;13(583). [DOI] [PMC free article] [PubMed]
- 68.Farmer BC, Kluemper J, Johnson LA. Apolipoprotein E4 alters astrocyte fatty acid metabolism and lipid droplet formation. Cells. 2019;8(2). [DOI] [PMC free article] [PubMed]
- 69.Chang TY, Chang CCY, Harned TC, De La Torre AL, Lee J, Huynh TN, et al. Blocking cholesterol storage to treat Alzheimer's disease. Explor Neuroprotective Ther. 2021;1(3):173–184. doi: 10.37349/ent.2021.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rubio JSA, Cedillo TJ. ACAT1 as a therapeutic target and its genetic relationship with Alzheimer's disease. Curr Alzheimer Res. 2019. [DOI] [PubMed]
- 71.Shibuya Y, Chang CC, Huang LH, Bryleva EY, Chang TY. Inhibiting ACAT1/SOAT1 in microglia stimulates autophagy-mediated lysosomal proteolysis and increases Abeta1-42 clearance. J Neurosci. 2014;34(43):14484–14501. doi: 10.1523/JNEUROSCI.2567-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shibuya Y, Chang CC, Chang TY. ACAT1/SOAT1 as a therapeutic target for Alzheimer's disease. Future Med Chem. 2015;7(18):2451–2467. doi: 10.4155/fmc.15.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bryleva EY, Rogers MA, Chang CC, Buen F, Harris BT, Rousselet E, et al. ACAT1 gene ablation increases 24(S)-hydroxycholesterol content in the brain and ameliorates amyloid pathology in mice with AD. Proc Natl Acad Sci U S A. 2010;107(7):3081–3086. doi: 10.1073/pnas.0913828107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Murphy SR, Chang CC, Dogbevia G, Bryleva EY, Bowen Z, Hasan MT, et al. Acat1 knockdown gene therapy decreases amyloid-beta in a mouse model of Alzheimer's disease. Mol Ther. 2013;21(8):1497–1506. doi: 10.1038/mt.2013.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hutter-Paier B, Huttunen HJ, Puglielli L, Eckman CB, Kim DY, Hofmeister A, et al. The ACAT inhibitor CP-113,818 markedly reduces amyloid pathology in a mouse model of Alzheimer's disease. Neuron. 2004;44(2):227–238. doi: 10.1016/j.neuron.2004.08.043. [DOI] [PubMed] [Google Scholar]
- 76.Huttunen HJ, Havas D, Peach C, Barren C, Duller S, Xia W, et al. The acyl-coenzyme A: cholesterol acyltransferase inhibitor CI-1011 reverses diffuse brain amyloid pathology in aged amyloid precursor protein transgenic mice. J Neuropathol Exp Neurol. 2010;69(8):777–788. doi: 10.1097/NEN.0b013e3181e77ed9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bhattacharyya R, Kovacs DM. ACAT inhibition and amyloid beta reduction. Biochim Biophys Acta. 2010;1801(8):960–965. doi: 10.1016/j.bbalip.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rusinol AE, Cui Z, Chen MH, Vance JE. A unique mitochondria-associated membrane fraction from rat liver has a high capacity for lipid synthesis and contains pre-Golgi secretory proteins including nascent lipoproteins. J Biol Chem. 1994;269(44):27494–27502. doi: 10.1016/S0021-9258(18)47012-3. [DOI] [PubMed] [Google Scholar]
- 79.Tai LM, Balu D, Avila-Munoz E, Abdullah L, Thomas R, Collins N, et al. EFAD transgenic mice as a human APOE relevant preclinical model of Alzheimer's disease. J Lipid Res. 2017. [DOI] [PMC free article] [PubMed]
- 80.Trommer BL, Shah C, Yun SH, Gamkrelidze G, Pasternak ES, Ye GL, et al. ApoE isoform affects LTP in human targeted replacement mice. NeuroReport. 2004;15(17):2655–2658. doi: 10.1097/00001756-200412030-00020. [DOI] [PubMed] [Google Scholar]
- 81.Trommer BL, Shah C, Yun SH, Gamkrelidze G, Pasternak ES, Stine WB, et al. ApoE isoform-specific effects on LTP: blockade by oligomeric amyloid-beta1-42. Neurobiol Dis. 2005;18(1):75–82. doi: 10.1016/j.nbd.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 82.Bien-Ly N, Andrews-Zwilling Y, Xu Q, Bernardo A, Wang C, Huang Y. C-terminal-truncated apolipoprotein (apo) E4 inefficiently clears amyloid-beta (abeta) and acts in concert with abeta to elicit neuronal and behavioral deficits in mice. Proc Natl Acad Sci U S A. 2011;108(10):4236–4241. doi: 10.1073/pnas.1018381108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhu Y, Nwabuisi-Heath E, Dumanis SB, Tai LM, Yu C, Rebeck GW, et al. APOE genotype alters glial activation and loss of synaptic markers in mice. Glia. 2012;60(4):559–569. doi: 10.1002/glia.22289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liao F, Zhang TJ, Jiang H, Lefton KB, Robinson GO, Vassar R, et al. Murine versus human apolipoprotein E4: differential facilitation of and co-localization in cerebral amyloid angiopathy and amyloid plaques in APP transgenic mouse models. Acta Neuropathol Commun. 2015;3:70. doi: 10.1186/s40478-015-0250-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fagan AM, Watson M, Parsadanian M, Bales KR, Paul SM, Holtzman DM. Human and murine ApoE markedly alters A beta metabolism before and after plaque formation in a mouse model of Alzheimer's disease. Neurobiol Dis. 2002;9(3):305–318. doi: 10.1006/nbdi.2002.0483. [DOI] [PubMed] [Google Scholar]
- 86.Fryer JD, Simmons K, Parsadanian M, Bales KR, Paul SM, Sullivan PM, et al. Human apolipoprotein E4 alters the amyloid-beta 40:42 ratio and promotes the formation of cerebral amyloid angiopathy in an amyloid precursor protein transgenic model. J Neurosci. 2005;25(11):2803–2810. doi: 10.1523/JNEUROSCI.5170-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Oddo S, Caccamo A, Cheng D, LaFerla FM. Genetically altering abeta distribution from the brain to the vasculature ameliorates tau pathology. Brain Pathol. 2009;19(3):421–430. doi: 10.1111/j.1750-3639.2008.00194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Balu D, Karstens AJ, Loukenas E, Maldonado Weng J, Valencia-Olvera AC, LaDu MJ. The role of APOE in transgenic mouse models of AD. Neurosci Lett. 2019:134285. [DOI] [PMC free article] [PubMed]
- 89.White JA, Manelli AM, Holmberg KH, Van Eldik LJ, LaDu MJ. Differential effects of oligomeric and fibrillar amyloid-beta1-42 on astrocyte-mediated inflammation. Neurobiol Dis. 2005;18(3):459–465. doi: 10.1016/j.nbd.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 90.Nwabuisi-Heath E, Rebeck GW, Ladu MJ, Yu C. ApoE4 delays dendritic spine formation during neuron development and accelerates loss of mature spines in vitro. ASN Neuro. 2014;6(1):e00134. doi: 10.1042/AN20130043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Youmans KL, Tai LM, Kanekiyo T, Stine WB, Jr, Michon SC, Nwabuisi-Heath E, et al. Intraneuronal abeta detection in 5xFAD mice by a new abeta-specific antibody. Mol Neurodegener. 2012;7(1):8. doi: 10.1186/1750-1326-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu DS, Pan XD, Zhang J, Shen H, Collins NC, Cole AM, et al. APOE4 enhances age-dependent decline in cognitive function by down-regulating an NMDA receptor pathway in EFAD-Tg mice. Mol Neurodegener. 2015;10:7. doi: 10.1186/s13024-015-0002-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Thomas R, Morris AWJ, Tai LM. Epidermal growth factor prevents APOE4-induced cognitive and cerebrovascular deficits in female mice. Heliyon. 2017;3(6):e00319. doi: 10.1016/j.heliyon.2017.e00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Baldi E, Efoudebe M, Lorenzini CA, Bucherelli C. Spatial navigation in the Morris water maze: working and long lasting reference memories. Neurosci Lett. 2005;378(3):176–180. doi: 10.1016/j.neulet.2004.12.029. [DOI] [PubMed] [Google Scholar]
- 95.Kim S, Kim N, Park S, Jeon Y, Lee J, Yoo SJ, et al. Tanycytic TSPO inhibition induces lipophagy to regulate lipid metabolism and improve energy balance. Autophagy. 2020;16(7):1200–1220. doi: 10.1080/15548627.2019.1659616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rodriguez GA, Tai LM, LaDu MJ, Rebeck GW. Human APOE4 increases microglia reactivity at abeta plaques in a mouse model of abeta deposition. J Neuroinflammation. 2014;11(1):111. doi: 10.1186/1742-2094-11-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Esteban-Salan M, Guimon-Bardesi A, de La Viuda-Unzueta JM, Azcarate-Ania MN, Pascual-Usandizaga P, Amoroto-Del-Rio E. Analytical and clinical evaluation of two homogeneous assays for LDL-cholesterol in hyperlipidemic patients. Clin Chem. 2000;46(8 Pt 1):1121–1131. doi: 10.1093/clinchem/46.8.1121. [DOI] [PubMed] [Google Scholar]
- 98.Warnick GR, Wood PD. National Cholesterol Education Program recommendations for measurement of high-density lipoprotein cholesterol: executive summary. The National Cholesterol Education Program Working Group on Lipoprotein Measurement. Clin Chem. 1995;41(10):1427–1433. doi: 10.1093/clinchem/41.10.1427. [DOI] [PubMed] [Google Scholar]
- 99.Youmans KL, Leung S, Zhang J, Maus E, Baysac K, Bu G, et al. Amyloid-beta42 alters apolipoprotein E solubility in brains of mice with five familial AD mutations. J Neurosci Methods. 2011;196(1):51–59. doi: 10.1016/j.jneumeth.2010.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Stine WB, Jungbauer L, Yu C, LaDu MJ. Preparing synthetic abeta in different aggregation states. Methods Mol Biol. 2011;670:13–32. doi: 10.1007/978-1-60761-744-0_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tardif JC, Gregoire J, L'Allier PL, Anderson TJ, Bertrand O, Reeves F, et al. Effects of the acyl coenzyme A:cholesterol acyltransferase inhibitor avasimibe on human atherosclerotic lesions. Circulation. 2004;110(21):3372–3377. doi: 10.1161/01.CIR.0000147777.12010.EF. [DOI] [PubMed] [Google Scholar]
- 102.Delsing DJ, Offerman EH, van Duyvenvoorde W, van Der Boom H, de Wit EC, Gijbels MJ, et al. Acyl-CoA:cholesterol acyltransferase inhibitor avasimibe reduces atherosclerosis in addition to its cholesterol-lowering effect in ApoE*3-Leiden mice. Circulation. 2001;103(13):1778–1786. doi: 10.1161/01.CIR.103.13.1778. [DOI] [PubMed] [Google Scholar]
- 103.Zhu Y, Chen CY, Li J, Cheng JX, Jang M, Kim KH. In vitro exploration of ACAT contributions to lipid droplet formation during adipogenesis. J Lipid Res. 2018;59(5):820–829. doi: 10.1194/jlr.M081745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tai LM, Ghura S, Koster KP, Liakaite V, Maienschein-Cline M, Kanabar P, et al. APOE-modulated abeta-induced neuroinflammation in Alzheimer's disease: current landscape, novel data, and future perspective. J Neurochem. 2015;133(4):465–488. doi: 10.1111/jnc.13072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Huttunen HJ, Peach C, Bhattacharyya R, Barren C, Pettingell W, Hutter-Paier B, et al. Inhibition of acyl-coenzyme A: cholesterol acyl transferase modulates amyloid precursor protein trafficking in the early secretory pathway. FASEB J. 2009;23(11):3819–3828. doi: 10.1096/fj.09-134999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Refolo LM, Pappolla MA, LaFrancois J, Malester B, Schmidt SD, Thomas-Bryant T, et al. A cholesterol-lowering drug reduces beta-amyloid pathology in a transgenic mouse model of Alzheimer's disease. Neurobiol Dis. 2001;8(5):890–899. doi: 10.1006/nbdi.2001.0422. [DOI] [PubMed] [Google Scholar]
- 107.Bjorkhem I, Lutjohann D, Diczfalusy U, Stahle L, Ahlborg G, Wahren J. Cholesterol homeostasis in human brain: turnover of 24S-hydroxycholesterol and evidence for a cerebral origin of most of this oxysterol in the circulation. J Lipid Res. 1998;39(8):1594–1600. doi: 10.1016/S0022-2275(20)32188-X. [DOI] [PubMed] [Google Scholar]
- 108.Mast N, Saadane A, Valencia-Olvera A, Constans J, Maxfield E, Arakawa H, et al. Cholesterol-metabolizing enzyme cytochrome P450 46A1 as a pharmacologic target for Alzheimer's disease. Neuropharmacology. 2017;123:465–476. doi: 10.1016/j.neuropharm.2017.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pikuleva IA, Cartier N. Cholesterol hydroxylating cytochrome P450 46A1: from mechanisms of action to clinical applications. Front Aging Neurosci. 2021;13:696778. doi: 10.3389/fnagi.2021.696778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cramer PE, Cirrito JR, Wesson DW, Lee CYD, Karlo JC, Zinn AE, et al. ApoE-directed therapeutics rapidly clear β-amyloid and reverse deficits in AD mouse models. Science. 2012. [DOI] [PMC free article] [PubMed]
- 111.Huttunen HJ, Kovacs DM. ACAT as a drug target for Alzheimer's disease. Neurodegener Dis. 2008;5(3–4):212–214. doi: 10.1159/000113705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shah D, Latif-Hernandez A, De Strooper B, Saito T, Saido T, Verhoye M, et al. Spatial reversal learning defect coincides with hypersynchronous telencephalic BOLD functional connectivity in APP(NL-F/NL-F) knock-in mice. Sci Rep. 2018;8(1):6264. doi: 10.1038/s41598-018-24657-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gulbranson DR, Ho K, Yu GQ, Yu X, Das M, Shao E, et al. Phenotypic differences between the Alzheimer's disease-related hAPP-J20 model and heterozygous Zbtb20 knock-out mice. eNeuro. 2021;8(3). [DOI] [PMC free article] [PubMed]
- 114.Ziegler-Waldkirch S, d'Errico P, Sauer JF, Erny D, Savanthrapadian S, Loreth D, et al. Seed-induced abeta deposition is modulated by microglia under environmental enrichment in a mouse model of Alzheimer's disease. EMBO J. 2018;37(2):167–182. doi: 10.15252/embj.201797021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bouter Y, Kacprowski T, Weissmann R, Dietrich K, Borgers H, Brauss A, et al. Deciphering the molecular profile of plaques, memory decline and neuron loss in two mouse models for Alzheimer's disease by deep sequencing. Front Aging Neurosci. 2014;6:75. doi: 10.3389/fnagi.2014.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lee D, Lee WS, Lim S, Kim YK, Jung HY, Das S, et al. A guanidine-appended scyllo-inositol derivative AAD-66 enhances brain delivery and ameliorates Alzheimer's phenotypes. Sci Rep. 2017;7(1):14125. doi: 10.1038/s41598-017-14559-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tang X, Wu D, Gu LH, Nie BB, Qi XY, Wang YJ, et al. Spatial learning and memory impairments are associated with increased neuronal activity in 5XFAD mouse as measured by manganese-enhanced magnetic resonance imaging. Oncotarget. 2016;7(36):57556–57570. doi: 10.18632/oncotarget.11353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rajavashisth TB, Kaptein JS, Reue KL, Lusis AJ. Evolution of apolipoprotein E: mouse sequence and evidence for an 11-nucleotide ancestral unit. Proc Natl Acad Sci U S A. 1985;82(23):8085–8089. doi: 10.1073/pnas.82.23.8085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nguyen D, Dhanasekaran P, Nickel M, Mizuguchi C, Watanabe M, Saito H, et al. Influence of domain stability on the properties of human apolipoprotein E3 and E4 and mouse apolipoprotein E. Biochemistry. 2014;53(24):4025–4033. doi: 10.1021/bi500340z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hudry E, Dashkoff J, Roe AD, Takeda S, Koffie RM, Hashimoto T, et al. Gene transfer of human apoe isoforms results in differential modulation of amyloid deposition and neurotoxicity in mouse brain. Sci Transl Med. 2013;5(212):212ra161. doi: 10.1126/scitranslmed.3007000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Su YR, Dove DE, Major AS, Hasty AH, Boone B, Linton MF, et al. Reduced ABCA1-mediated cholesterol efflux and accelerated atherosclerosis in apolipoprotein E-deficient mice lacking macrophage-derived ACAT1. Circulation. 2005;111(18):2373–2381. doi: 10.1161/01.CIR.0000164236.19860.13. [DOI] [PubMed] [Google Scholar]
- 122.Hayashi T, Yoshimine N, Naito M, Funaki C, Yamada K, Asai K, et al. HDL does not promote cholesterol efflux from macrophages of hypercholesterolemic rabbit: efflux differences between species. Artery. 1992;19(4):184–198. [PubMed] [Google Scholar]
- 123.Dove DE, Su YR, Zhang W, Jerome WG, Swift LL, Linton MF, et al. ACAT1 deficiency disrupts cholesterol efflux and alters cellular morphology in macrophages. Arterioscler Thromb Vasc Biol. 2005;25(1):128–134. doi: 10.1161/01.ATV.0000148323.94021.e5. [DOI] [PubMed] [Google Scholar]
- 124.Shafaati M, Olin M, Bavner A, Pettersson H, Rozell B, Meaney S, et al. Enhanced production of 24S-hydroxycholesterol is not sufficient to drive liver X receptor target genes in vivo. J Intern Med. 2011;270(4):377–387. doi: 10.1111/j.1365-2796.2011.02389.x. [DOI] [PubMed] [Google Scholar]
- 125.Reverter M, Rentero C, de Muga SV, Alvarez-Guaita A, Mulay V, Cairns R, et al. Cholesterol transport from late endosomes to the Golgi regulates t-SNARE trafficking, assembly, and function. Mol Biol Cell. 2011;22(21):4108–4123. doi: 10.1091/mbc.e11-04-0332r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lim WL, Lam SM, Shui G, Mondal A, Ong D, Duan X, et al. Effects of a high-fat, high-cholesterol diet on brain lipid profiles in apolipoprotein E epsilon3 and epsilon4 knock-in mice. Neurobiol Aging. 2013;34(9):2217–2224. doi: 10.1016/j.neurobiolaging.2013.03.012. [DOI] [PubMed] [Google Scholar]
- 127.Mahley RW. Central nervous system lipoproteins: ApoE and regulation of cholesterol metabolism. Arterioscler Thromb Vasc Biol. 2016;36(7):1305–1315. doi: 10.1161/ATVBAHA.116.307023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhao J, Davis MD, Martens YA, Shinohara M, Graff-Radford NR, Younkin SG, et al. APOE epsilon4/epsilon4 diminishes neurotrophic function of human iPSC-derived astrocytes. Hum Mol Genet. 2017;26(14):2690–2700. doi: 10.1093/hmg/ddx155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rawat V, Wang S, Sima J, Bar R, Liraz O, Gundimeda U, et al. ApoE4 alters ABCA1 membrane trafficking in astrocytes. J Neurosci. 2019;39(48):9611–9622. doi: 10.1523/JNEUROSCI.1400-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lindner K, Beckenbauer K, van Ek LC, Titeca K, de Leeuw SM, Awwad K, et al. Isoform- and cell-state-specific lipidation of ApoE in astrocytes. Cell Rep. 2022;38(9):110435. doi: 10.1016/j.celrep.2022.110435. [DOI] [PubMed] [Google Scholar]
- 131.Bhattacharyya R, Barren C, Kovacs DM. Palmitoylation of amyloid precursor protein regulates amyloidogenic processing in lipid rafts. J Neurosci. 2013;33(27):11169–11183. doi: 10.1523/JNEUROSCI.4704-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Simonovitch S, Schmukler E, Bespalko A, Iram T, Frenkel D, Holtzman DM, et al. Impaired autophagy in APOE4 astrocytes. J Alzheimers Dis. 2016;51(3):915–927. doi: 10.3233/JAD-151101. [DOI] [PubMed] [Google Scholar]
- 133.Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH, et al. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer's disease. J Neurosci. 2008;28(27):6926–6937. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nixon RA. Autophagy, amyloidogenesis and Alzheimer disease. J Cell Sci. 2007;120(Pt 23):4081–4091. doi: 10.1242/jcs.019265. [DOI] [PubMed] [Google Scholar]
- 135.Llaverias G, Laguna JC, Alegret M. Pharmacology of the ACAT inhibitor avasimibe (CI-1011) Cardiovasc Drug Rev. 2003;21(1):33–50. doi: 10.1111/j.1527-3466.2003.tb00104.x. [DOI] [PubMed] [Google Scholar]
- 136.Robertson DG, Breider MA, Milad MA. Preclinical safety evaluation of avasimibe in beagle dogs: an ACAT inhibitor with minimal adrenal effects. Toxicol Sci. 2001;59(2):324–334. doi: 10.1093/toxsci/59.2.324. [DOI] [PubMed] [Google Scholar]
- 137.Zabielska J, Sledzinski T, Stelmanska E. Acyl-coenzyme a: cholesterol acyltransferase inhibition in cancer treatment. Anticancer Res. 2019;39(7):3385–3394. doi: 10.21873/anticanres.13482. [DOI] [PubMed] [Google Scholar]
- 138.Qi G, Mi Y, Shi X, Gu H, Brinton RD, Yin F. ApoE4 impairs neuron-astrocyte coupling of fatty acid metabolism. Cell Rep. 2021;34(1):108572. doi: 10.1016/j.celrep.2020.108572. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, GRJT, upon reasonable request.