Abstract
Emergence and spread of Plasmodium falciparum resistance to artemisinin-based combination therapies (ACT) is a major challenge for Greater Mekong Subregion countries in their goal to eliminate malaria by 2030. Tools to efficiently monitor drug resistance beyond resource-demanding therapeutic efficacy studies are necessary. A custom multiplex amplicon sequencing assay based on Illumina technology was designed to target the marker of partial resistance to artemisinin (K13), five candidate modulators of artemisinin resistance, the marker of resistance to chloroquine (crt), and four neutral microsatellite loci. The assay was used to genotype 635 P. falciparum-positive blood samples collected across seven provinces of Vietnam and one of Cambodia between 2000 and 2016. Markers of resistance to artemisinin partner-drugs piperaquine (copy number of plasmepsin-2) and mefloquine (copy number of multidrug-resistance 1) were determined by qPCR. Parasite population structure was further assessed using a 101-SNP barcode. Validated mutations of artemisinin partial resistance in K13 were found in 48.1% of samples, first detection was in 2000, and by 2015 prevalence overcame > 50% in Central Highlands and Binh Phuoc province. K13-C580Y variant became predominant country-wide, quickly replacing an outbreak of K13-I543T in Central Highlands. Mutations in candidate artemisinin resistance modulator genes paralleled the trends of K13 mutants, whereas resistance to piperaquine and mefloquine remained low (≈ 10%) by 2015–2016. Genomic tools applied to malaria surveillance generate comprehensive information on dynamics of drug resistance and population structure and reflect drug efficacy profiles from in vivo studies.
Subject terms: Malaria, Parasite genomics, Epidemiology
Introduction
Artemisinin derivatives (ART) administered in combination with a partner antimalarial drug (ART-based combination therapies, ACT) are the current first-line treatment for uncomplicated Plasmodium falciparum malaria worldwide1. However, the high-rate of failures after ACT treatment reported in the Greater Mekong Subregion (GMS) challenges malaria elimination in the region and poses a potential threat for malaria control if it expands to other endemic areas2. Monitoring drug resistance is thus of paramount importance for National Malaria Control Programs worldwide. The gold standard for drug resistance surveillance is standardized therapeutic efficacy clinical studies, which are costly and logistically challenging in countries with low endemicity -or in those moving towards elimination-, due to the difficulty to enroll sufficient cases in the study period2,3. With advances in both elucidation of molecular markers of resistance and access to sequencing technology, genomic tools offer the possibility to increase geographical scale and sample size of surveillance studies without the complex conditions required for clinical trials, as well as detect early signs of antimalarial resistance4–7. In this article, we present the use of a custom multiplex amplicon sequencing approach for surveillance of resistance to drugs in ACT formulations and chloroquine in a collection of P. falciparum samples from different locations in Vietnam spanning 17 years.
Reduced susceptibility of P. falciparum to ART was first reported in Western Cambodia in 2008 and has since spread across GMS8–10. It is characterized by delayed clearance of parasites from peripheral blood (i.e. remaining parasitemia 72 h after treatment initiation or a parasite clearance half-life > 5 h) and therefore represents a partial resistance, mainly affecting the ring-stage parasite forms circulating in peripheral blood. Ten non-synonymous mutations in the propeller domain of kelch13 gene (K13) have been validated as markers of artemisinin partial resistance (ART-R) according to World Health Organization (WHO; namely, F446L, N458Y, M476I, Y493H, R539T, I543T, P553L, R561H, P574L and C580Y)2,11. Independent emergence of C580Y and R561H has also occurred in Central America12,13 and South-Saharan Africa14–16, respectively. In GMS, K13 mutants are more frequently associated with parasite genetic backgrounds containing mutation V127M in apicoplast ribosomal protein S10 (arps10-V127M), D193Y in ferredoxin (fd-D193Y), T484I in multidrug-resistance protein 2 (mdr2-T484I), and N326S in chloroquine resistance transporter (crt-N326S)17–19. Although efficacy of common ACT formulations -such as dihydroartemisinin-piperaquine (DHA-PPQ) or artesunate-mefloquine (AS-MQ)-remains high in most of the countries where ART-R has been reported, co-emergence of resistance to the ACT-partner drug is a threat for current treatment policies. Resistance to PPQ is associated with multiple gene copies in the plasmepsin-2 (pm2) and -3 gene cluster20,21, but also with mutation E415G in exonuclease (exo415)21 and variants H97Y, F145I, and I218F in crt10,22–24. Indeed, the expansion of a multidrug resistant lineage combining K13-C580Y with pm2 amplification10,25 (KEL1/PLA1) was likely responsible for high treatment failure rates to DHA-PPQ in Cambodia and Vietnam26,27. Resistance to mefloquine is associated with multiple copies of mdr1 gene28, and double K13/mdr1 mutants have been linked to treatment failure increases in Thailand and Myanmar29.
Chloroquine is not used anymore for the treatment of P. falciparum but is still widely used in the GMS as first-line treatment for uncomplicated malaria caused by Plasmodium vivax. Resistance to chloroquine in P. falciparum is attributed to K76T codon change in crt together with mutations in nearby codons 73–76, forming the resistant haplotype CVI[E/D]T30, which remains highly prevalent in the GMS6,31. Strains with CVI[E/D]T background accompanied by mutations in mdr1 such as Y184F, can modulate susceptibility of parasites to PPQ, mefloquine or lumefantrine31.
Vietnam aims to eliminate malaria by 203032. Malaria cases have decreased by 90% between 2000 and 2019—despite occasional resurgences33,34, and currently ≈ 80% of all cases occur in forested areas of Central Highlands provinces, populated by ethnic minorities with cross-border mobility with Cambodia35. Controlling the expansion of drug resistant lineages is critical to sustain gains in malaria burden reduction. In this study, we developed a custom multiplex amplicon sequencing approach for high-throughput screening of resistance markers for surveillance purposes. The assay, combined with qPCR CNV analysis and data from in vivo therapeutic efficacy trials, was used to investigate the history of emergence and dynamics of resistance to ACT in Vietnam, using samples from eight malaria-endemic provinces collected during a 17-year period (2000–2016).
Methods
Sample collection and preparation
Blood samples with confirmed or suspected P. falciparum infection were selected from NIMPE/NMCP routine dried blood spot (DBS) collections at malaria sentinel sites across Vietnam, or from research studies conducted by ITM and NIMPE between September 2000 to September 2016 (DBS, whole blood microtainers or DNA aliquots at − 20 °C)36–40. A total of 946 candidate samples were identified. Samples with unclear information on location and/or date of collection and those with a reported parasite density of < 250 parasites/μl by microscopy were excluded (see Supplementary Fig. S1). DNA was extracted from three 5 mm punches of DBS or from 200 μl whole blood using the FavorPrep™ 96-well Genomic DNA kit (Favorgen) following manufacturer’s instructions, and a final elution in 200 μl nuclease-free water. Samples without confirmed P. falciparum diagnosis were screened by varATS-qPCR41, and excluded if they tested negative or if Ct > 35. Samples with sufficient DBS left -or DNA in case DBS was not available- were shipped for external genotyping at Wellcome Sanger Institute (Cambridge, UK; see Supplementary Methods) as part of MalariaGEN SpotMalaria Project (https://www.malariagen.net/projects/SpotMalaria; see Supplementary Fig. S1).
Control DNAs were obtained from P. falciparum lab strains 3D7 (reference wild-type strain, maintained in continuous culture at ITM), Dd2 (MRA-150, BEI Resources, NIAID, NIH, contributed by David Walliker), CamWT_C580Y (MRA-1251, BEI Resources, NIAID, NIH, contributed by David A. Fidock) and IPC4912 (MRA-1241, BEI Resources, NIAID, NIH, contributed by Didier Ménard), and from non-infected human white blood cells.
Multiplex amplicon sequencing
Detailed procedures for parasite genotyping using next-generation sequencing are provided in Supplementary Methods, including a step-by-step lab protocol. Briefly, DNA samples were first enriched for P. falciparum DNA using selective whole genome amplification (sWGA) adapted from Oyola et al.42. Selective WGA products were purified using AMPure XP magnetic beads (Beckman Coulter) and quantified using a Qubit fluorometer (Invitrogen).
The custom oligonucleotide probe panel was designed for the sequencing chemistry of Illumina’s TruSeq Custom Amplicon (pfTSCA) and included 21 amplicons targeting 11 different genes: K13 as the marker of ART-R (full-length gene), crt codons 72–76 as the marker of chloroquine resistance43, mdr1 codon Y184F as a modulator of resistance to multiple antimalarials31,43, candidate genetic modulators of ART-R arps10-V127M, fd-D193Y, crt-N326S, MAL10: 688956 and MAL13/RAD5-homolog-S1158A18,44–46, and four microsatellites for parasite genetic diversity (poly-α, ARAII, TA81 and pk2; see Supplementary Table S1, Supplementary Fig S2). Libraries were prepared from sWGA amplified DNA using TruSeq Custom Amplicon Low Input Library Prep Kit (Illumina), following manufacturer’s guidelines for 96 samples (see Supplementary Methods). All libraries were quantified using KAPA kit for LightCycler 480 (Roche) before pooling. Library denaturation and sequencing was conducted at Centre for Medical Genetics (University of Antwerp, Edegem, Belgium), using a MiSeq instrument and MiSeq Reagent Kit v2 (Illumina) for paired-end sequencing of 2 × 150 bp reads.
Sequence data for 101-SNP barcode47 was generated at Wellcome Sanger Institute and provided as nucleotide sequences within a Genetic Report Card (see Supplementary Methods).
Sequence data analysis
Demultiplexing, alignment and variant calling were performed using TruSeq Amplicon Workflow in MiSeq Reporter software (Illumina), with P. falciparum 3D7 build 29 (PlasmoDB) as reference genomce and a variant filter quality cut-off score of 30. A read depth cut-off was set as the maximum number of mapped reads for each amplicon found in negative controls. Mutations were only reported if detected in at least two samples, or if the read count for the alternative allele was above the read cut-off for that amplicon. Loci with missing calls in more than 50% of all samples and controls were excluded. Haplotypes were built from calls with a ≥ 75% within-sample allele frequency to minimize risk of confounding by complex infections. Two amplicons of the panel (k13.i and MAL10) were sequenced in less than 50% of positive controls and were excluded from further analysis (see Supplementary Table S2). Median read depth of positive controls for the remaining valid amplicons was 7074× (medians range 34–27,805×; Supplementary Table S2). The minimum proportion for minor allele calling was estimated using parasite mixes and set at 0.2 for all markers (see Supplementary Methods, Supplementary Fig. S3).
Complexity of infection (COI; i.e. the estimated number of genetically distinct parasites within the infection) was determined from both microsatellites and SNP barcodes (see Supplementary Methods). Infections were categorized as single clone (COI = 1) vs. multiple clone infections (COI ≥ 2), if 2 microsatellite alleles were found for one or multiple microsatellite markers. COI from SNP barcodes was determined using The Real McCOIL program48. Metrics of genetic diversity He (expected heterozygosity) and FST (genetic differentiation) were calculated using the R packages adegenet and diveRsity, respectively49,50. Clustering analysis was performed using both unsupervised (Principal Component Analysis, PCA) and supervised population analysis (Discriminant Analysis of Principal Components, DAPC; see Supplementary Methods). Both microsatellite data from pfTSCA and the SNP barcode was combined to achieve the highest possible resolution.
Copy number of pm2 and mdr1 by qPCR
Copy number of plasmepsin-2 (pm2) and mdr1 were determined by qPCR using the original DNA eluate. Reactions were set-up in a LightCycler480 (Roche) with Power SYBR Green master mix (ThermoFisher), using previously published primers and ubiquitin conjugated enzyme (uce) as reference gene20,51. Amplification efficiencies were calculated using a 7-point tenfold dilution of genomic DNA from 3D7 (94.1% for pm2, 93.9% for mdr1 and 92% for uce). Copy numbers were calculated using ddCt method and 3D7 as calibrator sample (one copy of both pm2 and mdr1). Samples with CNV > 1.5 were considered gene amplifications.
Definitions and statistical analysis
Samples were grouped by region and year of collection for analytical purposes. Three regions were defined based on the geographical location and epidemiological characteristics of each province (see Supplementary Table S3). In brief, Region 1 included Central provinces, characterized by high endemicity, high Day 3 positivity after ACT but moderate treatment failure rates; Region 2 included provinces in South-Central Coast with low malaria risk, moderate Day 3 positivity and low treatment failure rates (i.e., < 10%); finally, Region 3 corresponded to Binh Phuoc province, accounting for the highest Day 3 positivity and treatment failure rates in the country. By time of collection, samples before 2010 was divided in two groups, 2000–2005 and 2006–2010, whereas those > 2010 were divided in three biannual groups (2011–2012, 2013–2014 and 2015–2016) for increased resolution on recent changes in circulating markers. Validated markers of partial artemisinin resistance consisted of 10 mutations in K13 according to WHO list from 20202. Differences in allele frequency by time or region were evaluated using Chi2 or Fisher’s exact test.
Ethics statement
Blood samples were collected after written informed consent was obtained from the patient or their parents/guardian. Ethical approvals including secondary use of samples were obtained from ethics committees at National Institute of Malariology Parasitology and Entomology (NIMPE, Hanoi) and Ministry of Health (Hanoi) in Vietnam; and from Institute of Tropical Medicine (ITM, Antwerp) and Antwerp University Hospital (UZA, Antwerp) in Belgium. All procedures involving human subjects were performed in accordance with the Declaration of Helsinki. Clinical trials contributing with samples to the present analysis were registered at ClinicalTrials.gov under identifiers NCT01775592 and NCT02604966.
Results
Sample selection and performance of pfTSCA
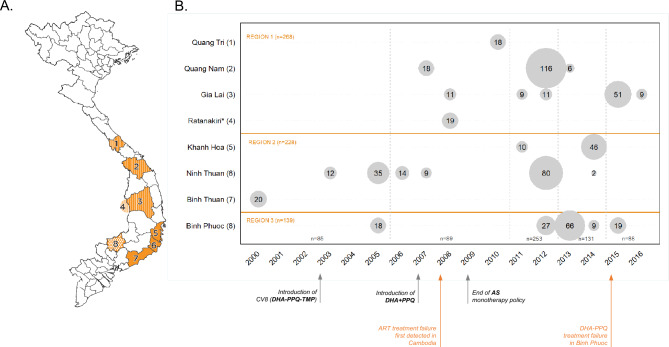
Out of the 946 samples available, 635 (67%) had confirmed P. falciparum infection and complete records on location and time and were processed in pfTSCA workflow (see Supplementary Fig. S1). Fresh DNA extractions were performed on 545 samples originally collected as DBS (545/635, 85.9%), and on 60 samples originally collected as blood in EDTA Vacutainer tubes (60/635, 9.4%); the remaining 30 samples (30/635, 4.7%) were frozen aliquots of previously extracted DNA. Samples originated from seven Vietnamese provinces (n = 616) or from an area in the Cambodian province of Ratanakiri within a 6 km radius of Vietnam’s border (n = 19; Fig. 1).
Figure 1.
Origin of the samples. (A) Map of Vietnam with the location of administrative provinces where samples were collected. Three main geographical regions were defined for analytical purposes based on geographical location and malaria epidemiology criteria (‘Region 1’, stripes; ‘Region 2’, plain orange; ‘Region 3’, dots). Note that Ratanakiri is an administrative province of Cambodia. (B) Detail on sample size per location and year. Circles are proportional to sample size. Time groups used for analysis are indicated by vertical dashed lines. The bottom panel indicates relevant events regarding antimalarial treatment policy in Vietnam and Greater Mekong Subregion. This map was developed for the purpose of this article using QGIS 3.10.
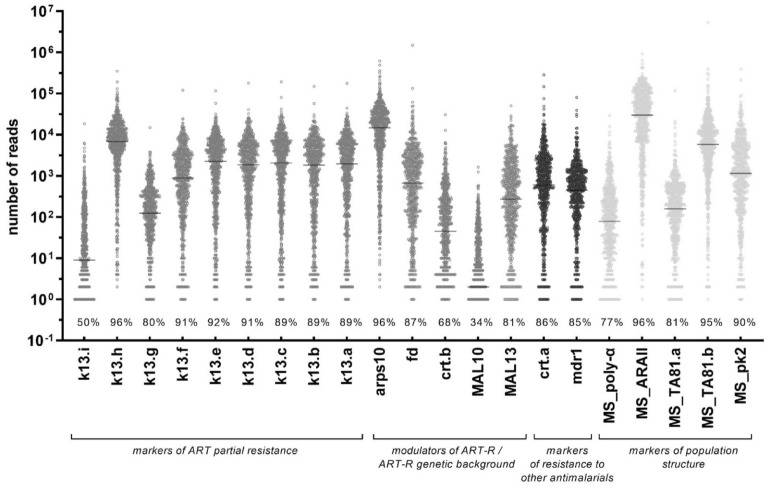
The mean number of samples with valid read counts for amplicons in pfTSCA was 86.0% (means range 68.7–98.1%), and the median read depth was 1561x (medians range 142–36,281×; Fig. 2, Supplementary Table S4). The number of sequenced amplicons per sample increased with parasite density, irrespective of quantification method (microscopy or qPCR) or collection method (DBS or as whole blood pellet from venipuncture, see Supplementary Fig. S4). External genotyping of 529 samples conducted at Wellcome Sanger Institute showed an agreement of > 95.6% with pfTSCA results for variant detection at any frequency, and > 94.1% for the identification of the major allele (see Supplementary Methods, Supplementary Table S5, Supplementary Data S1).
Figure 2.
Read depth for amplicons in the pfTSCA assay. The name of each amplicon corresponds to the gene target followed by a letter in genes that require multiple amplicons to cover the target region. Horizontal lines indicate median read count. Values (%) indicate the proportion of samples with reads above the read depth cut-off for each amplicon. MS, microsatellite.
Genetic markers of resistance to ACT in Vietnam (2000–2016)
Markers of ART-R
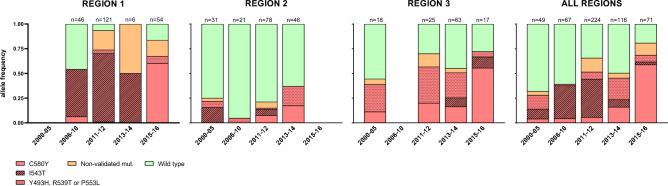
A total of 13 different non-synonymous K13-allelic variants were identified in 327 samples (327/610, 53.6%). K13 SNPs included ART-R validated mutations Y493H, R539T, I543T, P553L and C580Y as well as previously unreported codon changes G484V and P443L (see Supplementary Table S6, Supplementary Data S1). Among samples with the whole K13 propeller domain sequence (i.e., valid sequences for amplicons from k13.f to k13.a; n = 526), 56.5% (296/526) carried a non-synonymous mutation and in 48.1% (252/526) this was a validated marker of ART-R (Fig. 3). ART-R validated mutations increased from 28.6% in the 2000–2005 period to 69% in 2015–2016 period (p < 0.001, Chi2 test), and was highest in Region 1 followed by Region 3/Binh Phuoc (red-colored bars in Fig. 3). Frequency of K13 validated mutations was lowest in Region 2 for all time periods, with provincial prevalence of 10.5% (13/124) in Ninh Thuan (data until 2012) and 38.3% (18/47) in Khanh Hoa (data until 2014). Most common ART-R mutations were I543T (first detected in 2 samples from Binh Thuan in year 2000 and the predominant mutation until 2013), and C580Y (first detected in 2005 and the predominant across all regions by 2015; Fig. 3, Supplementary Data S1).
Figure 3.
Frequency of K13 mutations as markers of artemisinin partial resistance in Vietnam (2000–2016). Bar charts indicate the percentage of samples with K13 validated mutations (red), non-validated mutations after codon 440 (orange), and wild-type parasites (green). Specific codon changes for validated mutations are indicated with different fill patterns. Data is shown by region and years. Region 1: Quang Tri, Quang Nam, Gia Lai and Ratanakiri (Cambodia) provinces; Region 2: Khanh Hoa, Ninh Thuan, and Binh Thuan provinces: Region 3: Binh Phuoc province.
Genetic background of ART-R
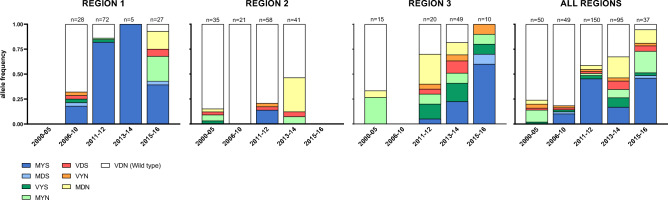
Three out of the four major mutations previously associated with a parasite genetic background (PGB) of ART-R in the GMS were successfully genotyped in the multiplex pfTSCA assay (arps10-V127M, fd-D193Y and crt-N326S; but not mdr2-T484I). A steady increase in frequency of mutations at all three PGB loci was observed in the study period (Fig. 4, Supplementary Fig. S5). The most common haplotype was the arps10/fd/crt triple mutant MYS, and it was significantly associated with presence of K13 mutations (p < 0.001). Parasites with mutations at one or more of PGB markers represented > 75% of the parasite population in Region 1 (Central Highland) by 2011. On the contrary, the frequency of PGB mutants was low in Region 2 (coastal provinces). The MAL13/RAD5-homolog-S1158A allele, a marker previously associated with both delayed clearance and increased survival in RSA44–46 was almost fixed at 99.0% (511/516; see Supplementary Data S1).
Figure 4.
Frequency of ART-R genetic background alleles in Vietnam (2000–2016). Bar charts show the frequency of haplotypes constructed from the combination of arps10-V127M, fd-D193Y and crt-N326S. Reference haplotype VDN is shown in white and mutant haplotypes at either one, two or all three positions are colored. Data is shown by region and years. Region 1: Quang Tri, Quang Nam, Gia Lai and Ratanakiri (Cambodia) provinces; Region 2: Khanh Hoa, Ninh Thuan, and Binh Thuan provinces: Region 3: Binh Phuoc province.
Markers of resistance to partner drugs piperaquine and mefloquine
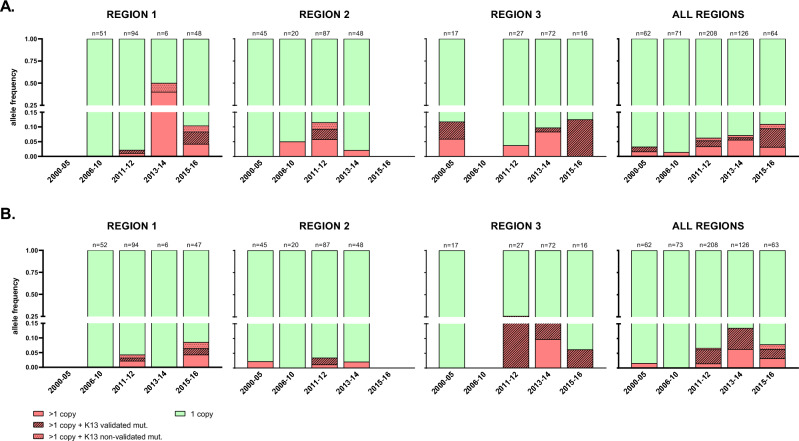
Resistance to the artemisinin partner drug PPQ was primarily assessed based on gene copy numbers of pm2. Overall prevalence of pm2 multiple copies was 6% (32/531), increasing from 3.2% (2/62) in 2000–2005 to 10.9% (7/64) in 2015–2016 (p = 0–161, Fisher’s Exact; Fig. 5A). Of note, 31.2% (10/32) of all parasites with pm2 amplifications also carried K13 ART-R validated SNP, a proportion that was notably higher in the last period (2015–2016; Fig. 5A). Three mutations in crt gene (T93S, H97Y and M343L), which were found to emerge in GMS associated to PPQ resistant phenotypes and ART-R genetic backgrounds10,24, were also covered by crt.a and crt.b amplicons in the pfTSCA multiplex. Both T93S and H97Y were detected but in only one sample each (Region 3, year 2014) and therefore not considered for prevalence calculations.
Figure 5.
Frequency of pm2 and mdr1 multiple copy numbers in Vietnam (2000–2016). Bar charts indicate the percentage of samples with multiple copies (red) or single copies (green) of pm2 (marker of piperaquine resistance; A) and mdr1 (marker of mefloquine resistance; B). Samples with multiple copies also carrying K13 mutations are indicated with different fill patterns. Data is shown by region and years. Note that the y axis in both (A,B) is split in two segments (from 0 to 0.15 and from 0.25 to 1) to improve visualization of low frequencies. Region 1: Quang Tri, Quang Nam, Gia Lai and Ratanakiri (Cambodia) provinces; Region 2: Khanh Hoa, Ninh Thuan, and Binh Thuan provinces: Region 3: Binh Phuoc province.
Resistance to mefloquine was determined based on copy number of mdr1. Parasites with multiple mdr1 copies were found in 6.9% (37/532) of the samples tested. The frequency was highest in Region 3/Binh Phuoc in 2011–2012 (35%) but decreased in 2015–2016 (Fig. 5B). A total of 56.8% (21/37) of samples with mdr1 amplifications carried a K13 ART-R validated SNP. Multiple gene copies at both pm2 and mdr1 were found in 0.8% of the samples (4/530), all four collected after 2012. One sample carried polymorphisms for both K13 (Y511H), pm2 and mdr1 (Region 1, year 2015).
Association between markers of resistance to ACT and in vivo treatment outcomes
The study included Day 0 samples (n = 118) from two clinical trials that assessed efficacy of DHA-PPQ for uncomplicated malaria, both of them conducted in Region 2 (one in Quang Nam province 2012–2013 and one in Gia Lai 2015–2016)36,37. The presence of mutations in markers or modulators of DHA-PPQ resistance was analyzed relative to in vivo parasite clearance and parasite positivity at Day 3 after treatment initiation. Infections with K13 non-synonymous mutations had a significantly slower PC1/2 (6.6 h [IQR 4.7–7.9]) as compared to K13 wild-type infections (3.8 h [3.1–4.0], p < 0.001), either for ART-R validated or non-validated mutations (p < 0.001, Mann–Whitney U test; see Supplementary Fig. S6). Prevalence of validated K13 mutants was 89% (33/37) in Day 3-positive patients and 74% in Day 3-negative (46/62; p = 0.119, see Supplementary Table S7). The sample with triple mutations in K13 (Y511H) + pm2 + mdr1 was part of the Gia Lai trial and showed adequate clearance of infection by Day 3 after DHA-PPQ treatment37.
Genetic markers of chloroquine resistance in Vietnam (2000–2016)
The overall frequency of the chloroquine resistant crt haplotypes (CVIET, CVIDT or mixed CVI[E/D]T) was > 52% across all time periods, and steadily increased over time up to 90% (64/71) in 2015–2016 (p < 0.001, chi2). Like markers of resistance to ACT, frequency of the chloroquine resistant haplotypes was lowest in Region 2 (Supplementary Fig. S7).
The mdr1 mutation Y184F was detected in 20.5% (110/536, 20.5%). The frequency of the mutant allele increased over time in Region 1 (from 13.0 to 70.6%; see Supplementary Fig. S8), whereas it remained low and stable in Region 2 (6.2–10.6%). mdr1-Y184F was predominantly found in chloroquine resistant CVI[E/D]T background (n = 97) as compared to parasites with CVMNK (n = 6; p < 0.001, chi2). There was no association between presence of the 184F allele and mdr1 copy number (p = 0.925, Fisher’s exact).
Genetic diversity and population structure
Parasite population genetics metrics were determined from the 4 microsatellite markers in pfTSCA and from a 101-SNP barcode. Out of the 635 samples in the study, 542 (86.8%) returned genetic data for at least one microsatellite locus, and 340 (53.5%) for ≥ 2 loci. SNP barcode data was obtained from 468 out of the 529 samples processed (88.5%).
Complexity of infection (COI)
Based on microsatellite data, 20.5% (111/542) of all samples sequenced were single clone infections, and there was no difference in their prevalence over time (range of single clone = 13.9–27.2%, p = 0.136, Chi2; see Supplementary Table S8). The highest rate of single clone infections was observed for Region 1 in 2011–2012 (35.7%; 41/115). COI estimates from the SNP barcode data indicated that overall prevalence of single clone infections was 92.5% (433/468), remaining at > 75% in all regions and time periods (Supplementary Table S8).
Genetic diversity and genetic differentiation
Overall genetic diversity, measured as expected heterozygosity (He) at four microsatellite loci (poly-α, ARAII, TA81 and pk2) and at 101 SNPs, was stable throughout the studied period (see Supplementary Fig. S9). Microsatellite ARAII had very few different alleles in the overall population, leaving only poly-α, TA81 and pk2 with sufficient variability to be informative for the analysis. The lowest He was found for Region 1 in 2011–2012 (median He by microsatellites = 0.07, p ≤ 0.0004 pairwise comparisons using Wilcoxon rank sum test; median He by 101-SNP barcode = 0.08, p ≤ 0.0004, Wilcoxon rank sum test), but increased in 2015–2016. Parasites in Region 1 in 2011–2012 and 2015–2016 showed great genetic differentiation (FST) both when compared to other regions and to each other (see Supplementary Fig. S10).
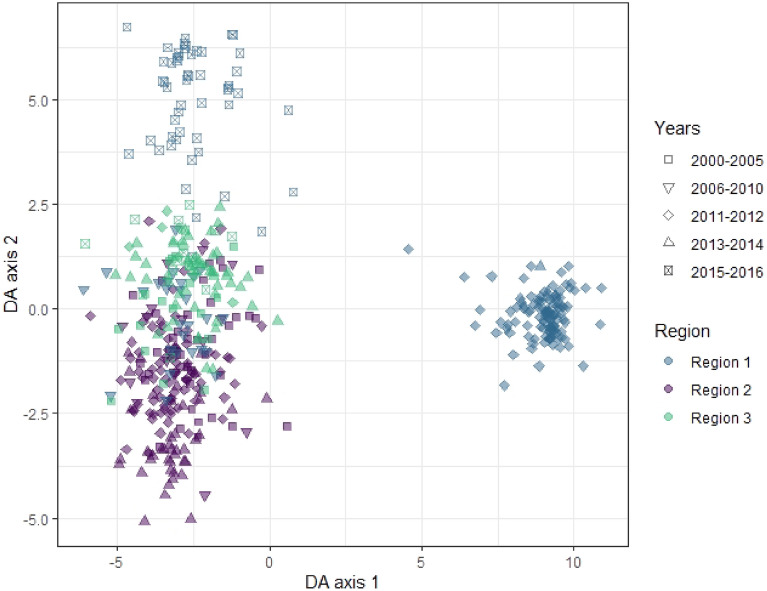
Clustering analysis
Clustering analysis was performed to explore population structure and dynamics over time. Parasites clustered predominantly by geographical region (color), rather than year (shape; see Fig. 6, Supplementary Fig. S11). The parasites from Region 1 were responsible for the majority of the variation along DA 1 and 2, with a very distinct population in 2011–2012 (blue diamonds, Fig. 6). The alleles contributing most to this separation in the DAPC were two SNPs from the barcode (Pf3D7_12_v3_1934745 and Pf3D7_04_v3_891732), followed by the K13 mutations I543T and T474I (see Supplementary Table S9). There was less distinction between samples from Region 2 and 3, as these populations overlapped.
Figure 6.
Discriminant analysis of principal component (DAPC) in samples from Vietnam (2000–2016). DAPC was performed using all markers genotyped in the pfTSCA and the SNP barcode. Scatter plot shows discriminant analysis (DA) eigenvalues 1 and 2, in which populations are differentiated by color (geographical region) and shape (time). Alleles contributing most to the DAPC are listed in Supplementary Table S9.
Discussion
In this study we developed genomic tools based on multiplex amplicon sequencing for the high-throughput molecular surveillance of antimalarial drug resistance. This tool was applied to investigate the prevalence and evolution of molecular markers of P. falciparum resistance to ACT in Vietnam, during the period of ART-R emergence in the GMS (2000–2016). ART-R validated mutations were detected in samples collected as early as year 2000, and steadily increased throughout the studied period, especially in Central Highlands provinces bordering Cambodia where an expansion or potential outbreak of K13-I543T was detected before the K13-C580Y mutation became predominant. Resistance to ACT partner drug PPQ remained moderately low in the surveyed period. Overall, trends in molecular markers coincided with treatment efficacy estimates from in vivo clinical trials.
The use of genomic tools to support malaria control and elimination is currently a growing area of research and is already part of NMCP malaria surveillance activities and programmatic-decision making in some countries5,6,52,53. Multiplex amplicon sequencing assays, such as the pfTSCA described here or the more recent AmpliSeq technology7, allow for rapid analysis of multiple markers and samples in a < 1 week-protocol with semi-automated laboratory and analysis procedures, which highly increases the potential for integration into pathogen surveillance systems of reference laboratories in endemic countries. Additional advantages of the approach are its high sensitivity and low DNA input and the combination of both drug resistance markers with direct programmatic relevance, drug resistance markers under research, and markers of population genetics to better understand dynamics and evolution of resistance. Moreover, the flexible custom designs can potentially incorporate additional targets for other surveillance purposes: hrp2 and hrp3 genes to monitor deletions associated with false negative results in HRP2-based rapid diagnostic tests, circumsporozoite protein gene (csp) elements present in RTS,S vaccine construct, other polymorphic antigens used to characterize recurrences in therapeutic efficacy trials, or SNP barcodes with regional and country-level resolution7.
The analysis of a large retrospective collection of blood samples from Vietnam showed that the overall proportion of infections with K13 validated mutations was high (48.1%), with notable changes between time periods and regions surveyed. Whereas in the Central Highlands (Region 1) K13 validated mutations represented > 50% of infections since 2006, they remained a minority in coastal areas (Region 2), in agreement with the different treatment efficacy profiles reported in these two areas (see Supplementary Table S3). The type of K13 variants detected changed over time, the most notable change being that from I543T predominance to C580Y predominance in the Central Highlands between 2014 and 2015. In this region, C580Y was also reported as the most common K13 variant in the 2017–2019 period6. P553L was the third most frequent K13 codon change; it was detected in provinces belonging to all three regions analyzed (Gia Lai, Khanh Hoa and Binh Phuoc) in agreement with findings from previous studies46,54, and frequencies were stable throughout the studied years. Variant I543T was present in all regions, and its earliest detection was in 53% of the Cambodian samples collected in 2008. However, most I543T mutant parasites were from Quang Nam, likely due to an outbreak in 2011–2012, evidenced by the lowest level of genetic diversity and high genetic differentiation in Region 1 for this time period. Interestingly, another retrospective study conducted in the neighboring province of Gia Lai in the same time period did not report cases of I543T, suggesting the expansion of this variant was localized to some districts55.
Parasites resistant to PPQ and mefloquine, partner drugs in most of the ACT formulations used in the GMS, were uncommon, as indicated by low frequencies of copy number amplifications in pm2 and mdr1 genes. A notable exception was Binh Phuoc province in 2012, where > 20% of samples carried multiple mdr1 copies in their genomes. The presence of CNV at these loci was frequently accompanied by presence of K13 ART-R markers, but the combination of both pm2 and mdr1 CNV in the same sample (i.e., PPQ + MQ resistance) was rare. Another study with samples collected immediately after 2016 reported much higher prevalence of PPQ resistant parasites6, what may be attributable to a rapid spread of lineages with pm2 duplication, or to differences in genetic profiles of parasites sampled form different districts. With regards to new emerging crt mutations, reported by Hamilton et al. at frequencies of 11–21% in Vietnam in 2016–201710, they were only partially targeted in our assay; mutants were rare, indicating these variants likely emerged in Vietnam after 2016.
Despite declining malaria transmission in recent years, as measured by traditional epidemiological indicators34,56, we did not observe a decrease in COI or genetic diversity measures during the surveyed time period. On the contrary, genetic diversity in Region 1 and 3 (derived from both SNP barcode and microsatellites) increased from 2000–2012 to 2015–2016, what may reflect evolutionary processes in the opposite direction of transmission intensity. In these areas, the frequency of parasites carrying the C580Y variant in a diverse PGB background also increased, which might suggest a population expansion after initial bottleneck events. On the other hand, genetic diversity remained stable over time in coastal provinces of Region 2, where levels of drug resistance were relatively low.
This retrospective study presents some limitations that should be considered in data interpretation. First, the convenient sampling approach included all available samples for a given province and year, resulting in unbalanced sample sizes for different temporal and geographical origins. The larger sample size for Region 1 might have resulted in higher resolution for this region as compared to others, but also allowed to establish associations between molecular and in vivo treatment efficacy data for these provinces. Future routine molecular surveillance strategies aiming at programmatic decision-making will require harmonized sampling strategies to avoid interpretation biases. Second, the molecular panel missed some loci of interest in the context of GMS countries, either due to incompatibility of primers in the in silico design (e.g. exo415 and mdr2-T484I) or because they had not been described at the time of the assay validation (e.g. KEL1 lineage SNP or crt variants conferring resistance to PPQ10). Third, the number of microsatellite markers for genetic diversity was insufficient to uncover parasite population structuring at the geographical scale needed, although in this case resolution increased by using the 101-SNP barcode data in population genetic analysis. An additional factor limiting optimal microsatellite allele calling was the use of algorithms for tandem repeat calling in NGS data. These algorithms are generally designed for diploid organisms and not samples of unknown ploidy (as it is often encountered in malaria complex infections), and require high and balanced allele depth, which resulted in missing microsatellite alleles in the present dataset. Our team has currently transitioned to AmpliSeq® technology (Illumina) and added SNP barcodes in panels applied to recent surveillance studies7,57.
In conclusion, multiplexed targeted amplicon sequencing combined with adequate sampling strategies are a powerful tool for routine malaria molecular surveillance. The application of pfTSCA assay to a retrospective collection of samples from Vietnam allowed to characterize in detail the increase in P. falciparum ART-R observed in the past two decades, and to detect regional and temporal changes in the parasite population. Population genetic analysis requires higher resolution markers like SNP-barcodes and cannot rely only on a reduced number of microsatellites.
Supplementary Information
Acknowledgements
The authors sincerely thank all individuals who provided blood samples for malaria research studies included in this work. They are especially grateful to the team at the Centre of Medical Genetics, University of Antwerp, for hosting the next generation sequencing facilities and contributing to MiSeq data analysis. They would also like to thank Dr. Harvie Portugaliza for support in sample processing, and Dr. Cristina Ariani and Dr. Dirk Schaerlaeckens for advice on experimental procedures.
Author contributions
E.R.V. and A.R.U. conceived and designed the study. E.R.V., P.G. and D.C. performed the experiments. N.V.H., A.E., B.Q.P., N.X.X. and A.R.U. obtained scientific and ethical approvals. N.V.H., B.Q.P., A.E. and N.X.X. coordinated field work activities and collected clinical data. E.R.V., J.H.K., P.G., H.I. and P.M. analyzed the genetic data. E.R.V., J.H.K. and A.R.U. wrote the manuscript. All authors have revised and approved the final submitted version of the manuscript.
Funding
The study was supported by Belgium Development Cooperation (DGD) under the Framework Agreement Programs between DGD and ITM (FA3-III Vietnam 2014–2016 and FA4 Vietnam 2017–2021). This publication uses data from the MalariaGEN SpotMalaria Project, coordinated by the MalariaGEN Resource Centre supported by Wellcome (098051, 090770); we thank staff of Wellcome Sanger Institute (Sample Management, Genotyping, Sequencing and Informatics teams) for their contribution.
Data availability
The datasets (fastq files) generated and/or analyzed during the current study are available in the NCBI Sequence Read Archive (SRA) repository under BioProject accession number PRJNA957102, and individual library accession numbers are listed in the Supplementary Data S1 file. Variant call format (vcf) files are available from the corresponding author upon request.
Competing interests
The authors declare no competing interests.
Footnotes
The original online version of this Article was revised: The original version of this Article contained an error in the Data availability section, where the SRA Bioproject accession number for DNA sequences was incorrect. Full information regarding the corrections made can be found in the Correction for this Article.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
10/11/2023
A Correction to this paper has been published: 10.1038/s41598-023-43996-w
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-40935-7.
References
- 1.World Health Organization. Guidelines for malaria. Global Malaria Program (2021).
- 2.World Health Organization. Report on Antimalarial Drug Efficacy, Resistance and Response: 10 Years of Surveillance (2010–2019) (2020).
- 3.World Health Organization. Methods for Surveillance of Antimalarial Drug Efficacy (2009).
- 4.Ippolito MM, Moser KA, Kabuya J-BB, Cunningham C, Juliano JJ. Antimalarial drug resistance and implications for the WHO global technical strategy. Curr. Epidemiol. Rep. 2021;8:46–62. doi: 10.1007/s40471-021-00266-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noviyanti R, et al. Implementing parasite genotyping into national surveillance frameworks: Feedback from control programmes and researchers in the Asia-Pacific region. Malar. J. 2020;19:271. doi: 10.1186/s12936-020-03330-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacob CG, et al. Genetic surveillance in the Greater Mekong subregion and South Asia to support malaria control and elimination. Elife. 2021;10:e62997. doi: 10.7554/eLife.62997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kattenberg JH, et al. Malaria molecular surveillance in the Peruvian Amazon with a novel highly multiplexed Plasmodium falciparum AmpliSeq Assay. Microbiol. Spectr. 2023;11:e0096022. doi: 10.1128/spectrum.00960-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noedl H, et al. Evidence of artemisinin-resistant malaria in western Cambodia. N. Engl. J. Med. 2008;359:2619–2620. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- 9.Ashley EA, et al. Spread of artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 2014;371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamilton WL, et al. Evolution and expansion of multidrug-resistant malaria in southeast Asia: A genomic epidemiology study. Lancet Infect. Dis. 2019;19:943–951. doi: 10.1016/S1473-3099(19)30392-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ariey F, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2013;505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chenet SM, et al. Independent emergence of the Plasmodium falciparum Kelch Propeller domain mutant allele C580Y in Guyana. J. Infect. Dis. 2016;213:1472–1475. doi: 10.1093/infdis/jiv752. [DOI] [PubMed] [Google Scholar]
- 13.Mathieu LC, et al. Local emergence in Amazonia of Plasmodium falciparum k13 C580Y mutants associated with in vitro artemisinin resistance. Elife. 2020;9:1–21. doi: 10.7554/eLife.51015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uwimana A, et al. Association of Plasmodium falciparum kelch13 R561H genotypes with delayed parasite clearance in Rwanda: An open-label, single-arm, multicentre, therapeutic efficacy study. Lancet Infect. Dis. 2021;21:1120–1128. doi: 10.1016/S1473-3099(21)00142-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balikagala B, et al. Evidence of artemisinin-resistant malaria in Africa. N. Engl. J. Med. 2021;385:1163–1171. doi: 10.1056/NEJMoa2101746. [DOI] [PubMed] [Google Scholar]
- 16.Moser KA, et al. Describing the current status of Plasmodium falciparum population structure and drug resistance within mainland Tanzania using molecular inversion probes. Mol. Ecol. 2021;30:100–113. doi: 10.1111/mec.15706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Straimer J, et al. Drug resistance. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science. 2015;347:428–431. doi: 10.1126/science.1260867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miotto O, et al. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat. Genet. 2015;47:226–234. doi: 10.1038/ng.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MalariaGEN Plasmodium falciparum Community Project Genomic epidemiology of artemisinin resistant malaria. Elife. 2016;5:8714. doi: 10.7554/eLife.08714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Witkowski B, et al. A surrogate marker of piperaquine-resistant Plasmodium falciparum malaria: A phenotype–genotype association study. Lancet Infect. Dis. 2017;17:174–183. doi: 10.1016/S1473-3099(16)30415-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amato R, et al. Genetic markers associated with dihydroartemisinin–piperaquine failure in Plasmodium falciparum malaria in Cambodia: A genotype–phenotype association study. Lancet Infect. Dis. 2017;17:164–173. doi: 10.1016/S1473-3099(16)30409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Pluijm RW, et al. Determinants of dihydroartemisinin-piperaquine treatment failure in Plasmodium falciparum malaria in Cambodia, Thailand, and Vietnam: A prospective clinical, pharmacological, and genetic study. Lancet Infect. Dis. 2019;19:952–961. doi: 10.1016/S1473-3099(19)30391-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dhingra SK, Small-Saunders JL, Ménard D, Fidock DA. Plasmodium falciparum resistance to piperaquine driven by PfCRT. Lancet Infect. Dis. 2019;19:1168–1169. doi: 10.1016/S1473-3099(19)30543-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ross LS, et al. Emerging Southeast Asian PfCRT mutations confer Plasmodium falciparum resistance to the first-line antimalarial piperaquine. Nat. Commun. 2018;9:3314. doi: 10.1038/s41467-018-05652-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imwong M, Hien TT, Thuy-Nhien NT, Dondorp AM, White NJ. Spread of a single multidrug resistant malaria parasite lineage (PfPailin) to Vietnam. Lancet Infect. Dis. 2017;17:1022–1023. doi: 10.1016/S1473-3099(17)30524-8. [DOI] [PubMed] [Google Scholar]
- 26.Leang R, et al. Evidence of Plasmodium falciparum malaria multidrug resistance to artemisinin and piperaquine in Western Cambodia: Dihydroartemisinin–piperaquine open-label multicenter clinical assessment. Antimicrob. Agents Chemother. 2015;59:4719–4726. doi: 10.1128/AAC.00835-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phuc BQ, et al. Treatment failure of dihydroartemisinin/piperaquine for Plasmodium falciparum Malaria, Vietnam. Emerg. Infect. Dis. 2017;23:715–717. doi: 10.3201/eid2304.161872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Price R, Uhlemann A, Brockman A. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–447. doi: 10.1016/S0140-6736(04)16767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phyo AP, et al. Declining efficacy of artemisinin combination therapy against P. falciparum malaria on the Thai-Myanmar border (2003–2013): The role of parasite genetic factors. Clin. Infect. Dis. 2016;63:784–791. doi: 10.1093/cid/ciw388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fidock DA, et al. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol. Cell. 2000;6:861–871. doi: 10.1016/S1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Veiga MI, et al. Globally prevalent PfMDR1 mutations modulate Plasmodium falciparum susceptibility to artemisinin-based combination therapies. Nat. Commun. 2016;7:11553. doi: 10.1038/ncomms11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaehler N, et al. Prospects and strategies for malaria elimination in the Greater Mekong Sub-region: A qualitative study. Malar. J. 2019;18:1–13. doi: 10.1186/s12936-019-2835-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kattenberg JH, et al. Characterization of Plasmodium falciparum and Plasmodium vivax recent exposure in an area of significantly decreased transmission intensity in Central Vietnam. Malar. J. 2018;17:1. doi: 10.1186/s12936-018-2326-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.National Institute of Malariology Entomology and Parasitology. Malaria Report 2018 (2019).
- 35.Canavati SE, et al. Targeting high risk forest goers for malaria elimination: A novel approach for investigating forest malaria to inform program intervention in Vietnam. BMC Infect. Dis. 2020;20:8. doi: 10.1186/s12879-020-05476-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thriemer K, et al. Delayed parasite clearance after treatment with dihydroartemisinin–piperaquine in Plasmodium falciparum malaria patients in central Vietnam. Antimicrob. Agents Chemother. 2014;58:7049–7055. doi: 10.1128/AAC.02746-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rovira-Vallbona E, et al. Efficacy of dihydroartemisinin/piperaquine and artesunate monotherapy for the treatment of uncomplicated Plasmodium falciparum malaria in Central Vietnam. J. Antimicrob. Chemother. 2020;75:2272–2281. doi: 10.1093/jac/dkaa172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thang ND, et al. Rapid decrease of malaria morbidity following the introduction of community-based monitoring in a rural area of central Vietnam. Malar. J. 2009;8:3. doi: 10.1186/1475-2875-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Erhart A, et al. Epidemiology of forest malaria in central Vietnam: A large scale cross-sectional survey. Malar. J. 2005;4:58. doi: 10.1186/1475-2875-4-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Erhart A, et al. Forest malaria in Vietnam: A challenge for control. Am. J. Trop. Med. Hyg. 2004;70:110–118. doi: 10.4269/ajtmh.2004.70.110. [DOI] [PubMed] [Google Scholar]
- 41.Natama HM, et al. Diagnosing congenital malaria in a high-transmission setting: Clinical relevance and usefulness of P. falciparum HRP2-based testing. Sci. Rep. 2017;7:2080. doi: 10.1038/s41598-017-02173-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oyola SO, et al. Whole genome sequencing of Plasmodium falciparum from dried blood spots using selective whole genome amplification. Malar. J. 2016;15:597. doi: 10.1186/s12936-016-1641-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Srimuang K, et al. Analysis of anti-malarial resistance markers in pfmdr1 and pfcrt across Southeast Asia in the tracking resistance to artemisinin collaboration. Malar. J. 2016;15:1–12. doi: 10.1186/s12936-016-1598-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spring MD, et al. Dihydroartemisinin-piperaquine failure associated with a triple mutant including kelch13 C580Y in Cambodia: An observational cohort study. Lancet Infect. Dis. 2015;15:683–691. doi: 10.1016/S1473-3099(15)70049-6. [DOI] [PubMed] [Google Scholar]
- 45.Takala-Harrison S, et al. Genetic loci associated with delayed clearance of Plasmodium falciparum following artemisinin treatment in Southeast Asia. Proc. Natl. Acad. Sci. U.S.A. 2012;110:240–245. doi: 10.1073/pnas.1211205110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takala-Harrison S, et al. Independent emergence of artemisinin resistance mutations among Plasmodium falciparum in Southeast Asia. J. Infect. Dis. 2015;211:670–679. doi: 10.1093/infdis/jiu491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baniecki ML, et al. Development of a single nucleotide polymorphism barcode to genotype Plasmodium vivax infections. PLoS Negl. Trop. Dis. 2015;9:e0003539. doi: 10.1371/journal.pntd.0003539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang H-H, et al. THE REAL McCOIL: A method for the concurrent estimation of the complexity of infection and SNP allele frequency for malaria parasites. PLoS Comput. Biol. 2017;13:e1005348. doi: 10.1371/journal.pcbi.1005348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jombart T. adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- 50.Keenan K, Mcginnity P, Cross TF, Crozier WW, Prodöhl PA. diveRsity: An R package for the estimation and exploration of population genetics parameters and their associated errors. Methods Ecol. Evol. 2013;4:782–788. doi: 10.1111/2041-210X.12067. [DOI] [Google Scholar]
- 51.Joice R, et al. Plasmodium falciparum transmission stages accumulate in the human bone marrow. Sci. Transl. Med. 2014;6:244. doi: 10.1126/scitranslmed.3008882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mayor A, et al. Prospective surveillance study to detect antimalarial drug resistance, gene deletions of diagnostic relevance and genetic diversity of Plasmodium falciparum in Mozambique: Protocol. BMJ Open. 2022;12:e063456. doi: 10.1136/bmjopen-2022-063456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dalmat R, Naughton B, Kwan-Gett TS, Slyker J, Stuckey EM. Use cases for genetic epidemiology in malaria elimination. Malar. J. 2019;18:1–11. doi: 10.1186/s12936-019-2784-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maeno Y, et al. Detection of the Plasmodium falciparum Kelch-13 gene P553L mutation in sporozoites isolated from mosquito salivary glands in South-Central Vietnam. Parasit. Vectors. 2017;10:1–5. doi: 10.1186/s13071-017-2247-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thuy-Nhien N, et al. K13 propeller mutations in Plasmodium falciparum populations in regions of malaria endemicity in Vietnam from 2009 to 2016. Antimicrob. Agents Chemother. 2017;61:e01578. doi: 10.1128/AAC.01578-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goldlust SM, et al. The decline of malaria in Vietnam, 1991–2014. Malar. J. 2018;17:226. doi: 10.1186/s12936-018-2372-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kattenberg JH, et al. Novel highly-multiplexed AmpliSeq targeted assay for Plasmodium vivax genetic surveillance use cases at multiple geographical scales. Front. Cell Infect. Microbiol. 2022;12:953187. doi: 10.3389/fcimb.2022.953187. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets (fastq files) generated and/or analyzed during the current study are available in the NCBI Sequence Read Archive (SRA) repository under BioProject accession number PRJNA957102, and individual library accession numbers are listed in the Supplementary Data S1 file. Variant call format (vcf) files are available from the corresponding author upon request.