Abstract
In this prospective study, the use of a culture-enhanced PCR assay for the detection of Mycoplasma pneumoniae, followed by hybridization with a specific probe (MP-HPCR) or without hybridization (MP-PCR), and the use of a nested PCR (MP-NPCR) were evaluated. Clinical samples (190 specimens) from 190 patients with respiratory complaints were incubated in culture broth overnight and then subjected to PCR. The results of the PCR were compared to those obtained by culture, the direct antigen test, and serologic testing by microparticle agglutination and by immunoblotting in unclear cases. The sensitivities were 19 CFU for MP-PCR, 1.9 CFU for MP-HPCR, and 0.019 CFU for MP-NPCR. PCR amplification of the β-globin gene was possible in 98% of cases: after dilution of the β-globin-negative samples, all samples were reactive. Correlation between negative MP-NPCR results and negative serology results was found in 89% of cases; a positive correlation was found with 10% of the patients. Samples from three immunocompromised patients were MP-NPCR positive but serologically negative. High respiratory colonization by M. pneumoniae (>105 CFU/ml) in patients with acute respiratory disease could be detected by culture, MP-PCR, and MP-NPCR. These results indicate that MP-PCR and MP-NPCR are reliable methods for the detection of M. pneumoniae in respiratory tract samples of patients with respiratory complaints.
Mycoplasma pneumoniae is a common respiratory tract pathogen causing pharyngitis, tracheobronchitis, or pneumonia (5). At present, laboratory diagnosis of M. pneumoniae infections relies on conventional serological methods. The complement fixation test is the most widely used method for M. pneumoniae antibody detection but lacks specificity due to cross-reactivity of the antigen preparations. The commercially available microparticle agglutination test (MAG test) and the enzyme-linked immunosorbent assay (ELISA) are more specific and sensitive, but they require paired sera for diagnosis (13). A recently developed Western immunoblot technique for the P1 antigen, which is a major virulence factor of M. pneumoniae, has high specificity and sensitivity and allows discrimination between colonization with less-virulent P1 protein-negative M. pneumoniae cells and colonization with virulent P1 protein-positive M. pneumoniae strains, but it is offered by only a few laboratories (14).
Culture methods are relatively insensitive and time-consuming, requiring up to 3 weeks for signal detection (15). More-rapid tests such as the direct antigen assay (4, 16) or hybridization with specific DNA probes (24) have good specificities but low sensitivity. Recently developed PCR techniques (2, 6, 12, 18, 22) show high specificity and sensitivity. Detection of the amplification product is usually performed by hybridization with a specific probe, which is very time-consuming (18).
A rapid alternative method for sensitive detection of M. pneumoniae DNA is a two-step PCR (nested PCR) (NPCR). In this prospective study, we evaluated the use of direct M. pneumoniae PCR with hybridization (MP-HPCR) or without hybridization (MP-PCR) and M. pneumoniae NPCR (MP-NPCR) to detect M. pneumoniae in 190 clinical samples obtained from 190 patients. The results were compared with those obtained by culture, the direct antigen test, and serological testing, including Western immunoblotting, for about 20% of the samples obtained from patients with severe problems.
MATERIALS AND METHODS
Patients.
Clinical specimens were routinely obtained from patients admitted with acute respiratory complaints to the Department of Internal Medicine of the Ludwig-Maximilians-University of Munich or to the Children’s Hospital Munich-Schwabing. One hundred ninety patients were divided into three groups according to their clinical status: group I (n = 90) consisted of immunocompromised patients with respiratory complaints after organ or bone marrow transplantation (mean age, 25 ± 5.0 years), group II (n = 50) were adults with acute respiratory tract disease (mean age, 44 ± 12.6 years), and group III (n = 50) comprised children with lower respiratory tract infections (mean age, 8 ± 4.3 years). A total of 190 samples (50 tracheal aspirates and 140 nasopharyngeal aspirates) obtained in 2 ml of Hayflick broth as the transport medium were examined. Samples of acute-phase sera were taken 1 day and 5 to 7 days after the onset of disease, and samples of convalescent-phase sera were taken 20 to 30 days after the onset of disease in a follow-up visit.
Cultures.
From each specimen, a 0.2-ml volume was inoculated into 1.8 ml of Hayflick broth with glucose (7) and 0.02 ml was cultured onto Hayflick agar plates (37°C, 5% CO2) and incubated for 3 weeks. Colonies on plates were identified by indirect immunofluorescence (23), and positive broths were verified by various methods: subculture onto agar plates, a direct antigen test (0.2 ml) (Virion, Würzburg, Germany), and detection of glass-adherent M. pneumoniae cells by phase-contrast microscopy (4). For quantitative determination, five 10-fold dilutions (5 × 0.2 ml in 0.8 ml of Hayflick broth) were prepared, 0.02 ml of each dilution was cultured onto a Hayflick agar plate, colonies were identified and counted, and the number of CFU/ml was calculated.
For PCR, 0.2 ml of each specimen was incubated in 3.8 ml of Hayflick broth overnight, 0.25 ml of this dilution was extracted, and a 5-μl volume was subjected to MP-PCR the next morning. MP-PCR-negative samples were subjected to MP-NPCR by using 5 μl of a 1:10 dilution of MP-PCR product. To evaluate the specificity of MP-NPCR, M. genitalium G37c, M. salivarium A889, M. orale T519, and 80 bacterial strains cultured from clinical specimens were subjected to MP-PCR, MP-HPCR, and MP-NPCR. For evaluation of the sensitivity, the reference strain M. pneumoniae FH was used.
Direct antigen test.
The direct antigen test, a species-specific capture ELISA for direct detection of M. pneumoniae antigen (Virion), is based on monoclonal antibodies directed to the P1 protein (8). The test was performed by using 0.2 ml of Hayflick broth within 4 h after it changed color to yellow according to the manufacturer’s recommendations.
DNA extraction.
To compare the efficacies of DNA extraction by cell lysis with proteinase K without further nucleic acid purification and of DNA extraction after lysis by phenol-chloroform followed by ethanol precipitation, a 10-fold dilution series of M. pneumoniae FH ranging from 10−1 to 10−6 CFU/ml was prepared and 0.25 ml of each dilution was subjected to both extraction methods.
For DNA extraction by proteinase K treatment, samples were prepared as described for ureaplasmas by other authors (1, 3). Briefly, 250 μl of each diluted sample was centrifuged (12,000 × g for 20 min at 4°C). The pellet was resuspended in 50 μl of solution A (10 mM Tris-HCl, pH 8.3, containing 100 mM KCl and 2.5 mM MgCl2) and an equal volume of solution B (solution A supplemented with 1% Tween 20, 1% Triton X-100, and 5 mg of proteinase K per ml). After incubation for 1 h at 60°C and heating to 95°C for 10 min, a 5-μl volume was used for PCR. In the second extraction protocol, 0.3 ml of phenol was added after proteinase K treatment and extraction was continued as described by other authors (23). Finally, a 5-μl volume was used for PCR. For clinical samples (0.25 ml), only the first extraction method was used and a 5-μl volume was subjected to PCR.
PCR.
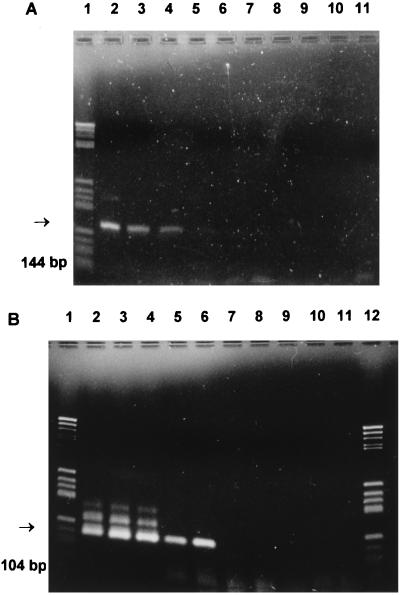
Prior to M. pneumoniae amplification, each DNA sample was tested for its ability to be amplified with β-globin-specific primers KM 38 and KM 39 (21). A positive signal was defined by a 262-bp fragment visualized on an ethidium bromide-stained agarose gel. Negative samples were diluted (1:50 with sterile water), and β-globin PCR was repeated. One hundred samples (50 tracheal and 50 nasopharyngeal aspirates) were tested directly from specimens and, in parallel, from culture-enhanced overnight samples, and for the remaining 90 specimens only culture-enhanced overnight samples were used. Primer set MP-1 (5′-GAA GCT TAT GGT ACA GGT TGG-3′) and MP-2 (5′-ATT ACC ATC CTT GTT GTA AGG-3′) (MWG, Biotech, Ebersberg, Germany) was used for M. pneumoniae-specific amplification as described by Bernet et al. (2). The reaction volumes for the first and second rounds of amplification were 50 μl with 0.1 μM (each) primer, 200 μM (each) deoxynucleoside triphosphate, buffer, and 1 U of AmpliTaq polymerase (Perker-Elmer Cetus). Amplification was carried out for 40 cycles, each consisting of 20 s at 95°C, 2 min at 63°C, and 1 min at 72°C. For MP-NPCR, primers MUH-1 (5′-TGA CTG GAA GGA TGT TAA GC-3′) and MUH-2 (5′-TTG TAA TCG TCT TTA TTT CG-3′) (MWG, Biotech) were used. Nested amplification was performed by using 5 μl of 1:10-diluted PCR product (5 μl in 45 μl of sterile water) from the first round of amplification under otherwise identical conditions. Positive controls (reference strain M. pneumoniae FH) and negative controls (Hayflick broth, sterile water, master mix, human DNA, and DNA from reference strains M. orale T519, M. salivarium A889, and M. genitalium G37c) were prepared, extracted, and amplified under identical conditions. For MP-NPCR of clinical samples, every fourth sample consisted of a negative control to control cross-contamination. Hayflick broth, which was extracted and subjected to MP-PCR and then to MP-NPCR, served as the negative control. As a positive control for each run, reference strain M. pneumoniae FH was used. The amplified product of the first PCR was a 144-bp DNA fragment of the M. pneumoniae ATPase operon gene (11); the amplicon from the nested PCR had a size of 104 bp (Fig. 1). Both PCR products were analyzed by using 3% agarose gels containing ethidium bromide, initially identified by direct sequence analysis, and subsequently identified by the sizes of the amplicons.
FIG. 1.
Sensitivities of single-step PCR (MP-PCR) (A) and NPCR (MP-NPCR) (B) for M. pneumoniae. Lanes: 1 and 12, size markers; 2, 10−1 dilution of M. pneumoniae FH reference strain; 3, 10−2 dilution; 4, 10−3 dilution; 5, 10−5 dilution; 6, 10−7 dilution; 7, 10−9 dilution; 8, negative control. Lanes 9, 10, and 11 were empty.
Southern blot analysis of PCR products.
Ten-microliter volumes of the PCR products were analyzed on 3% agarose gels. DNA was transferred from the agarose gel to a nylon membrane (Boehringer Mannheim Biochemica, Mannheim, Germany) by standard methods. The internal detection oligonucleotide was labeled with digoxigenin by using commercially available kits (Boehringer Mannheim Biochemica). DNA hybrids were revealed by chemiluminescence (CSPD; Boehringer Mannheim Biochemica).
Serology.
Determination of M. pneumoniae-specific antibodies was performed by using a commercial MAG assay (Serodia-Myco II Kit; Fujirebio, Tokyo, Japan) according to the manufacturer’s recommendations. An antibody titer of ≤1:40 was regarded as indicating a negative result. A fourfold rise in titers of paired sera and titers of ≥1:160 were considered positive results. For all positive titers (about 20%) without a fourfold rise, titers between 1:40 and 1:160, and negative titers from samples from patients for whom PCR was positive, results were confirmed by a Western immunoblot technique as described by Jacobs et al. (14).
RESULTS
Comparison of MP-NPCR with MP-HPCR and MP-PCR.
The sensitivity of the PCR method was tested by using serial dilutions (1:10) of chromosomal M. pneumoniae DNA. Under optimal conditions, primers MP5-1 and MP5-2 (first-step PCR, MP-PCR) detected about 3,000 genome copies, 30 pg of DNA, 19 CFU, or 1.9 × 103 organisms. Sensitivity was increased to 300 genome copies, 3 pg of DNA, 1.9 CFU, or 190 organisms after hybridization (MP-HPCR) and up to 3 to 10 genome copies, 30 to 100 fg of DNA, 0.019 CFU, or 19 organisms by MP-NPCR with primers MUH-1 and MUH-2 (1 CFU = 160 genome copies according to Harris et al. [10] and 1 CFU = 10 to 1,000 organisms according to Razin [19]). There was no amplification of human DNA or DNA from M. salivarium, M. orale, and M. genitalium, confirming the specificity of the primer set MP5-1 and MP5-2 as described by Bernet et al. (2) and of the primer set MUH-1 and MUH-2. In addition, none of the 80 samples containing bacteria other than mycoplasmas was positive by PCR (data not shown). The use of phenol-chloroform-purified DNA was time-consuming and led to a 10-fold decrease in sensitivity. Therefore, we extracted DNA from clinical samples only by cell lysis with proteinase K followed by incubation of the samples at 95°C for 10 min (1, 3).
PCR of clinical samples versus culture and serology.
Internal inhibition of the PCR was controlled for 100 specimens and the corresponding overnight cultures by amplification of a 262-bp β-globin gene fragment. Ten of the undiluted specimens (five tracheal aspirates and five nasopharyngeal aspirates), but only one of the overnight cultures, were initially β-globin PCR negative but yielded a positive signal after dilution (1:50 in sterile water). Consequently, PCR for the remaining half of the samples was performed solely with overnight cultures, and only two specimens (2%) were initially β-globin PCR negative.
In order to avoid false-positive results (19), we routinely coamplified samples known to be negative (Hayflick broth only) in large numbers (25% of all samples in every run). MP-NPCR was considered valid when all negative controls were negative in both rounds of PCR. Specimens giving false-positive results, which were obtained with about 0.5% of all samples, were retested and only included when the repeated control tests were all negative.
Among the 190 patients, 20 (11%) were found to be positive by MP-NPCR, 16 (8%) were found to be positive by MP-HPCR, and 15 (8%) were positive by MP-PCR. M. pneumoniae was detected in 8 patients (4%) by culture and in 11 patients (6%) by the direct antigen test. In addition, 17 patients (9%) were confirmed to be serologically positive by the MAG test or by immunoblot (Table 1).
TABLE 1.
Correlation among results obtained by amplification of M. pneumoniae DNA, culture, direct antigen test, and serology (immunoblot)
| Group (n) | Materiala | No. (%) of patients positive by:
|
|||||
|---|---|---|---|---|---|---|---|
| NPCR | HPCR | PCR | AGb | Culture | Serology | ||
| I (90) | NPA | 6 | 3 | 3 | 1 | 0 | 3 |
| II (50) | TA | 6 | 6 | 6 | 5 | 4 | 6 |
| III (50) | NPA | 8 | 7 | 6 | 5 | 4 | 8 |
| Total (190) | 20 (11) | 16 (8) | 15 (8) | 11 (6) | 8 (4) | 17 (9) | |
NPA, nasopharyngeal aspirates; TA, tracheal aspirates.
AG, direct antigen test.
Corresponding negative serology and MP-NPCR results were obtained for 170 patients (89%), and corresponding positive results were obtained for 17 patients (10%) (Table 2); with respect to culture, serology, and MP-NPCR, corresponding negative results were obtained for 170 patients (89%) and corresponding positive results were obtained for only 8 patients (4%; data not shown in Table 2).
TABLE 2.
Correlation between results obtained by amplification of M. pneumoniae DNA (NPCR and PCR) and serology (immunoblot)
| Group (n) | No. of patients with indicated resulta by:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| NPCR/Serology
|
PCR/Serology
|
|||||||
| +/+ | 0/0 | +/0 | 0/+ | +/+ | 0/0 | +/0 | 0/+ | |
| I (90) | 3 | 84 | 3 | 0 | 3 | 87 | 0 | 0 |
| II (50) | 6 | 44 | 0 | 0 | 6 | 44 | 0 | 0 |
| III (50) | 8 | 42 | 0 | 0 | 6 | 42 | 0 | 2 |
| Total (190) | 17 | 170 | 3 | 0 | 15 | 173 | 0 | 2 |
+, positive result; 0, negative result; +/+, correlation of two test systems.
Besides being tested for M. pneumoniae, all patients were also tested for other respiratory pathogens. Thirty percent of the patients in group I were positive for one of the following: Legionella pneumophila (two patients), Streptococcus pneumoniae (three patients), Haemophilus influenzae (three patients), Aspergillus fumigatus (two patients), Pneumocystis carinii (three patients), cytomegalovirus (nine patients), and Herpes simplex virus (five patients). In group II, 32% had either Staphylococcus aureus (two patients), S. pneumoniae (eight patients), H. influenzae (five patients), or Klebsiella pneumoniae (one patient). In group III, 32% were positive for one of the following: S. aureus (one patient), S. pneumoniae (one patient), H. influenzae (three patients), Moraxella catarrhalis (two patients), Herpes simplex virus (one patient), and Respiratory syncytial virus (eight patients).
DISCUSSION
The prospective study reported here describes the use of PCR (MP-PCR) and NPCR (MP-NPCR) for rapid diagnosis of M. pneumoniae infections. It differs from other studies in several points. First, the PCR procedure is performed not directly on the specimens but on culture-enhanced Hayflick broth, into which specimens are inoculated immediately after reception and incubated overnight. By this step, PCR-inhibitory substances, which were initially observed in 10% of the specimens as demonstrated by a directly performed β-globin PCR, are diluted and internal inhibition is reduced to 2%, a rate which is clearly lower than those described by other authors (18, 20, 22, 23).
As a second modification, preparation of samples was simplified by replacing the time-consuming phenol-chloroform extraction by treatment with proteinase K (1, 3). This proved to be a fast method which did not result in loss of DNA (23). For detection of low concentrations of mycoplasmas, two-step PCR (MP-NPCR) was used instead of a time-consuming hybridization protocol (2 versus 4 days). By this method, the detection limit ranges from 3 to 10 genome copies, 30 to 100 fg of DNA, or 0.019 CFU. Sensitivity is 10-fold better than that for single-step PCR followed by hybridization and exceeds that for antigen capture enzyme immunoassay or culture by 104- to 105-fold.
However, increase in sensitivity was accompanied by an enhanced risk of contamination and false-positive MP-NPCR results. In preliminary assays (data not shown), contamination, detected by positive results after the second amplification step with randomly incorporated negative samples, probably caused by carryover (12), occurred in 10% of all samples. By strictly following the guidelines for the general handling of the PCR procedure (17), such as separation of rooms, boards, and lab benches, we minimized contamination to 0.5% in this study. Another approach to establishing a more sensitive test is to examine genes that occur in multiple copies, such as rRNA genes (23, 24). Such a protocol may also provide greater sensitivity, without the additional work and enhanced risk of contamination associated with NPCR. However, a disadvantage of this method is the need for purification of RNA, making it less suitable for routine procedures (12).
In this study, the best correlation between culture, serology, and PCR results was observed among patients with current infections of the lower respiratory tract (groups II and III). In accordance with data reported in other publications (19), these patients revealed high respiratory colonization levels (>105 CFU/ml) and, therefore, a high probability for positive culture, antigen detection (2), and positive immune response by microparticle agglutination. The difficulty is not in the detection of mycoplasmas but in the reliable confirmation of a rapid diagnosis, which is important for early and adequate antibiotic therapy. Consequently, due to its short detection time of 2 days, MP-PCR should be preferentially used. An ultrasensitive test like MP-NPCR is not necessary.
In contrast, among immunocompromised patients interpretation of positive NPCR results was, with exception of one case, more difficult. In the one exception, the patient had a manifest mycoplasma infection associated with positive PCR results (MP-NPCR, MP-HPCR, and MP-PCR), a positive antigen assay, and a good antibody response by the MAG assay. The other five patients with a positive MP-NPCR result gave negative results by culture and had no serological response by the MAG test. Two of them had had M. pneumoniae infections 9 months previously (elevated antibody titer of >1:160). They revealed a weak immunoglobulin G (IgG) response by immunoblot at admission, but single-step PCRs (MP-HPCR and MP-PCR) were negative. This might reflect a recent mycoplasma infection with persistence of a very low concentration of M. pneumoniae DNA in the respiratory tract. The differences between positive results by MP-NPCR and negative serology for the remaining three patients may be explained in several ways: first, false-positive PCR results; second, lack of immunological response in immunocompromised patients (22); third, persistence of mycoplasmas after a recent infection (24); and fourth, a mild infection without immunological response. The possibility of false-positive PCR results seems very unlikely, because PCR was positive in two independent assays which were performed separately from daily diagnostic assays and in different rooms, with all possible precautions being taken. The hypothesis of the presence of seronegative immunocompromised patients could be more relevant (22); however, we could not distinguish whether PCR detected a mild acute infection, a carrier state (9), or the persistence of mycoplasmas in the respiratory tract after a recent infection (24).
Among children with acute respiratory infections in group III, diagnosis was easy, except for those who were treated with erythromycin. In these cases, cultures and MAG assays with acute-phase sera gave negative results (antibody titer, <1:160) and there was not a fourfold increase in convalescent-phase sera; only immunoblots revealed weak IgG and often strong IgA or IgM responses. These cases demonstrate the limitation of conventional serology and confirm the necessity of more-specific assays such as the immunoblot. Additionally, the use of more-sensitive nucleic acid tests like NPCR may be helpful.
During the period studied, the incidence of M. pneumoniae (11%) was in accordance with that described in other publications (12) but differed considerably among age groups and types of clinical disease. Whereas respiratory syncytial virus is more prevalent in very young children, M. pneumoniae infections occur predominantly in older children or in young adults. Among the elderly, typical respiratory pathogens such as pneumococci or H. influenzae are more frequently isolated, and among immunocompromised patients, endogenous microorganisms are prominent.
In conclusion, conventional PCR and two-step PCR (NPCR) performed with culture-enhanced overnight samples proved to be reliable methods for detecting M. pneumoniae rapidly and specifically in clinical specimens. For routine diagnosis of acute respiratory disease we prefer conventional serology, culture, and conventional PCR without hybridization. For pretreated or immunocompromised patients, the more sensitive two-step PCR (NPCR) method and immunoblotting may be helpful.
REFERENCES
- 1.Abele-Horn M, Wolff C, Dressel P, Zimmermann A, Vahlensieck W, Pfaff F, Ruckdeschel G. Polymerase chain reaction versus culture for detection of Ureaplasma urealyticum and Mycoplasma hominis in the urogenital tract of adults and the respiratory tract of newborns. Eur J Clin Microbiol Infect Dis. 1996;15:595–598. doi: 10.1007/BF01709369. [DOI] [PubMed] [Google Scholar]
- 2.Bernet C, Garret M, Barbeyrac B, Bonnet J. Detection of Mycoplasma pneumoniae by using the polymerase chain reaction. J Clin Microbiol. 1889;27:2492–2496. doi: 10.1128/jcm.27.11.2492-2496.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanchard A, Hentschel J, Duffy L, Baldus K. Detection of Ureaplasma urealyticum by polymerase chain reaction in the urogenital tract of adults, in amniotic fluid, and in the respiratory tract of newborns. Clin Infect Dis. 1993;17:S148–S153. doi: 10.1093/clinids/17.supplement_1.s148. [DOI] [PubMed] [Google Scholar]
- 4.Bredt W, Lam W, Berger J. Evaluation of a microscopy method for rapid detection and identification of Mycoplasma pneumoniae. J Clin Microbiol. 1975;2:541–545. doi: 10.1128/jcm.2.6.541-545.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clyde, W. A. 1993. Clinical overview of typical Mycoplasma pneumoniae infections. Clin. Infect. Dis. 17(Suppl. 1):S32–S37. [PubMed]
- 6.De Barbeyrac, B., C. Bernet-Poggi, F. Fébrer, H. Renaudin, M. Dupont, and C. Bébéar. 1993. Detection of Mycoplasma pneumoniae and Mycoplasma genitalium in clinical samples by polymerase chain reaction. Clin. Infect. Dis. 17(Suppl. 1):S83–S89. [DOI] [PubMed]
- 7.Deutsche Gesellschaft für Hygiene und Mikrobiologie. Isolierung und Identifizierung von Mykoplasmen und Chlamydien. Zentralbl Bakteriol Hyg A. 1989;270:470–486. [PubMed] [Google Scholar]
- 8.Gerstenecker B, Jacobs E. Development of a capture-ELISA for the specific detection of M. pneumoniae in patient material. In: Kahane I, Adoni A, editors. Rapid diagnosis of mycoplasmas. New York, N.Y: Plenum Press; 1993. pp. 152–159. [Google Scholar]
- 9.Gnarpe J, Lundbäck A, Sündelöf B, Gnarpe H. Prevalence of M. pneumoniae in subjectively healthy individuals. Scand J Infect Dis. 1992;24:161–164. doi: 10.3109/00365549209052607. [DOI] [PubMed] [Google Scholar]
- 10.Harris R, Marmion B P, Varkansis G, Kok P W, Lunn B, Martin J. Laboratory diagnosis of Mycoplasma pneumoniae infection. 2. Comparison of methods for the direct detection of specific antigen or nucleic acid sequences in respiratory exudates. Epidemiol Infect. 1988;101:685–694. doi: 10.1017/s0950268800029563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hilbert H, Himmelreich R, Plagens H, Herrmann R. Sequence analysis of 56 kbp from the genome of the bacterium Mycoplasma pneumoniae comprising the dnaA region, the atp operon and a cluster of ribosomal protein genes. Nucleic Acids Res. 1996;24:628–639. doi: 10.1093/nar/24.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ieven M, Ursi D, Van Bever H, Quint W, Niesters H G M, Goossens H. Detection of Mycoplasma pneumoniae by two polymerase chain reactions and role of M. pneumoniae in acute respiratory tract infections in pedriatric patients. J Infect Dis. 1996;173:1445–1452. doi: 10.1093/infdis/173.6.1445. [DOI] [PubMed] [Google Scholar]
- 13.Jacobs, E. 1993. Serological diagnosis of Mycoplasma pneumoniae infections: a critical review of current procedures. Clin. Infect. Dis. 17(Suppl. 1):S79–S82. [DOI] [PubMed]
- 14.Jacobs E, Bennewitz A, Bredt W. Reaction pattern of human anti-Mycoplasma pneumoniae antibodies in enzyme-linked immunosorbent assays and immunoblotting. J Clin Microbiol. 1986;23:517–522. doi: 10.1128/jcm.23.3.517-522.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kenny G E, Kaiser C G, Cooney M K. Diagnosis of Mycoplasma pneumoniae pneumonia: sensitivities and specificities of serology with lipid antigen and isolation of the organism on soy peptone medium for identification of infections. J Clin Microbiol. 1990;28:2087–2093. doi: 10.1128/jcm.28.9.2087-2093.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kok T W, Varkanis G, Marmion B P, Martin J, Esterman A. Laboratory diagnosis of Mycoplasma pneumoniae infection. Direct detection of antigen in respiratory exudates by enzyme immunoassay. Epidemiol Infect. 1988;101:669–684. doi: 10.1017/s0950268800029551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwok S, Higuchi R. Avoiding false positives with PCR. Nature. 1989;339:237–238. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- 18.Lüneberg E, Jensen J S, Frosch M. Detection of Mycoplasma pneumoniae by polymerase chain reaction and nonradioactive hybridization in microtiter plates. J Clin Microbiol. 1993;31:1088–1094. doi: 10.1128/jcm.31.5.1088-1094.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Razin S. DNA probes and PCR in diagnosis of mycoplasma infections. Mol Cell Probes. 1994;8:497–511. doi: 10.1006/mcpr.1994.1071. [DOI] [PubMed] [Google Scholar]
- 20.Reznikov M, Blackmore T K, Finlay-Jones J J, Gordon D L. Comparison of nasopharyngeal aspirates and throat swab specimens in a polymerase chain reaction-based test for Mycoplasma pneumoniae. Eur J Clin Microbiol Infect Dis. 1995;14:58–61. doi: 10.1007/BF02112622. [DOI] [PubMed] [Google Scholar]
- 21.Saiki, R. K., D. H. Gelfand, S. Stoffel, S. J. Scharf, R. Higuchi, G. T. Horn, K. B. Mullis, and H. Ehrlich. Primer directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239:487–491. [DOI] [PubMed]
- 22.Skakni L, Sardet A, Just J, Landman-Parker J, Costil J, Moniot-Ville N, Bricout F, Garbarg-Chenon A. Detection of Mycoplasma pneumoniae in clinical samples from pedriatric patients by polymerase chain reaction. J Clin Microbiol. 1992;30:2638–2643. doi: 10.1128/jcm.30.10.2638-2643.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tjhie J H T, Van Kuppefeld F J M, Rossendaal R. Direct PCR enables detection of Mycoplasma pneumoniae in patients with respiratory tract infections. J Clin Microbiol. 1994;32:11–16. doi: 10.1128/jcm.32.1.11-16.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williamson J, Marmion B P, Worswick D A, Kok T W, Tannock G, Herd R, Harris J. Laboratory diagnosis of Mycoplasma pneumoniae infection. Antigen capture and PCR-gene amplification for detection of the mycoplasma: problems of clinical correlation. Epidemiol Infect. 1992;109:519–537. doi: 10.1017/s0950268800050512. [DOI] [PMC free article] [PubMed] [Google Scholar]