Abstract
Neurodegeneration and cerebrovascular disease share an underlying microvascular dysfunction that may be remedied by selective transgene delivery. To date, limited options exist in which cellular components of the brain vasculature can be effectively targeted by viral vector therapeutics. In this study, we characterize the first engineered adeno-associated virus (AAV) capsid mediating high transduction of cerebral vascular pericytes and smooth muscle cells (SMCs). We performed two rounds of in vivo selection with an AAV capsid scaffold displaying a heptamer peptide library to isolate capsids that traffic to the brain after intravenous delivery. One identified capsid, termed AAV-PR, demonstrated high transduction of the brain vasculature, in contrast to the parental capsid, AAV9, which transduces mainly neurons and astrocytes. Further analysis using tissue clearing, volumetric rendering, and colocalization revealed that AAV-PR enabled high transduction of cerebral pericytes located on small-caliber vessels and SMCs in the larger arterioles and penetrating pial arteries. Analysis of tissues in the periphery indicated that AAV-PR also transduced SMCs in large vessels associated with the systemic vasculature. AAV-PR was also able to transduce primary human brain pericytes with higher efficiency than AAV9. Compared with previously published AAV capsids tropisms, AAV-PR represents the first capsid to allow for effective transduction of brain pericytes and SMCs and offers the possibility of genetically modulating these cell types in the context of neurodegeneration and other neurological diseases.
Keywords: adeno-associated virus vector, pericytes, smooth muscle cells
INTRODUCTION
Many common neurodegenerative diseases involve the brain vasculature, including small vessel disease, Alzheimer's disease, Parkinson's disease, multiple sclerosis, and amyotrophic lateral sclerosis.1 Among diseases with the most severe phenotypes are monogenic causes of cerebrovascular disease affecting small vessel and/or arterial function that lead to white matter degeneration, stroke, and death in childhood.2–4
Gene therapy using adeno-associated virus (AAV) vectors has the potential to treat diseases that affect the vasculature. The gold standard in central nervous system (CNS) gene therapy, AAV9, which is FDA approved for treatment of spinal muscular atrophy, mainly transduces neurons and astrocytes with minimal vasculature transduction.5 Thus, for therapies targeting components of the vasculature, other vectors are needed.
In recent years, several groups have engineered and selected AAV capsids capable of transducing the vasculature.6–9 While these capsids are very useful for transgene expression in endothelial cells, none has reported transduction of other components of the cerebral vasculature, specifically smooth muscle cells (SMCs) and pericytes. Both cell types are affected in a variety of diseases. For example, pericyte degeneration has been implicated as a factor in the development of Alzheimer's disease10 and Cerebral Autosomal Dominant Arteriopathy with Subcortical Ischemic strokes and Leukoencephalopathy (CADASIL), the most common familial stroke disorder; and ACTA2 mutations in SMC lead to a devastating multisystemic smooth muscle dysfunction syndrome and childhood strokes.4,11 Additionally, neuroinflammation, including HIV infection, can lead to a loss of pericytes.12,13 Therefore, developing new gene delivery systems that can target these cell types would have important implications for therapeutic development and studies of basic biology.
In this study, through an in vivo selection process in mice using an AAV9 peptide display library, we discovered a new capsid, called AAV-PR, which displays high tropism for the CNS vasculature after systemic delivery. AAV-PR transduces several cell types, including pericytes and SMCs, which has not been effectively targeted by prior AAV capsids.
MATERIALS AND METHODS
Animals
All animal experiments were approved by the Massachusetts General Hospital Subcommittee on Research Animal Care following guidelines set forth by the National Institutes of Health Guide for the Care and Use of Laboratory Animals. We used adult age (8- to 10-week-old) Ai9 [B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J, Strain No. 007909], C57BL/6J (Strain No. 000664), and BALB/cJ (Strain No. 000651) mice all from The Jackson Laboratory (Bar Harbor, ME).
Primary human pericyte cell culture, AAV transduction, and immunostaining
Adult primary human pericytes were isolated from microvessels present in resected brain tissue (from healthy areas and not from the focal resection) of patients undergoing surgery for the treatment of intractable epilepsy, supplied by Michael Bernas and Dr. Marlys Witte (University of Arizona, Tucson, AZ). The tissue was mechanically fragmented and size filtered using polyester meshes.14 The resulting microvessel fragments were placed onto type-I collagen-coated flasks to allow cells to proliferate. From the resulting mixed culture of brain endothelial cells and pericytes, cells were detached by ethylenediaminetetraacetic acid and sorted using flow cytometry (as described previously15). Cells were sorted to remove the endothelial cells, and the remaining cells (pericytes) were placed back into culture.
Pericytes were plated on collagen type I, Rat Tail (50 μg/mL; Corning, Corning, NY)-coated tissue culture flasks in premedium; Dulbecco's modified Eagle's medium/F-12 media supplemented with 10% heat inactivated fetal bovine serum (Fisher Scientific), heparin (1 mg/mL; Thomas Scientific), cell growth supplement (endothelial cell growth supplement; Fisher Scientific), amphotericin B (2.5 μg/mL; Invitrogen™), penicillin (100 U/mL; Invitrogen), and streptomycin (100 μg/mL; Invitrogen). Characterization by immunofluorescence staining of these donor cells indicated that >90% expressed protein markers such as α-SMC actin and platelet-derived growth factor receptor (PDGFR)-β, used to commonly identify pericytes.
For transduction with AAV, the pericytes (passages below 6) were plated on to collagen type I, Rat Tail (50 μg/mL; Corning)-coated six-well plates at 1 × 106 cells/mL. Then, cells were rinsed with 1 × phosphate-buffered saline (PBS) and maintained in postmedium for 24 h to remove heparin from the culture. Next, AAV9-sc-CBA-DsRed or AAV-PR-sc-CBA-DsRed (both at 2 × 105 vg/cell) were suspended in postmedium and incubated with pericytes for 24 h. The medium was then changed to remove unbound virus and the pericyte cultures were rinsed with 1 × PBS. Pericytes were incubated in postmedium at least for 3 days to allow for DsRed expression. Before imaging, pericytes were rinsed with 1 × PBS, fixed with 4% ultrapure paraformaldehyde (PFA) for 30 min, and permeabilized with 0.1% Triton X-100 in PBS. The cells were incubated with primary antibodies for 1 h at room temperature. Secondary antibodies anti-Rb Alexa Fluor-488 (Invitrogen) or anti-Mouse Fluor-594 were used at 1:200 for 1 h. Cells were then washed and mounted with ProLong Antifade Reagent without 4′,6-diamidino-2-phenylindole (DAPI).
Images of the DsRed expression of monolayers exposed to AAV-PR and AAV9 were captured at 10 × magnification using the EVOS imaging system (Thermo Fisher). All the images were taken with same settings (i.e., exposure time). Primary antibodies and dilutions used included rabbit polyclonal antibodies to DsRed (1:200; Rockland) and mouse monoclonal to α-smooth muscle actin (α-SMA, 1:200; Abcam). Image analysis to determine transduction efficiency was performed with the AIVIA imaging software (Leica) using the pixel classifier and particle counting functions.
AAV-PR selection and identification
We performed an in vivo selection with an AAV peptide display library to isolate AAV capsids, which could transduce brain after systemic injection. To do this we used our previously described iTransduce library16 (AAV-CBA-Cre-p41-Cap), which combines a 7-mer peptide display library with a Cre recombinase cassette to couple transgene expression in mice, which express a Cre-sensitive fluorescent reporter (Ai9 mice) with rescued peptide encoding sequences. We initially set out with the goal of isolating capsids capable of transducing myeloid-derived cells (e.g., microglia) in the brain. We performed two rounds of selection with the capsid library. For the first round of selection, 1.27 × 1011 vector genomes (vg) of the library was injected intravenously in one adult male and one female Ai9 mouse. Three weeks postinjection, DNA was extracted from the brain tissue, and the CAP region containing the 21-mer randomized inserts was amplified before recloning it back into the AAV plasmid backbone and repackaging for the second round of selection (“brain-enriched capsid library”).
For the second round, we used the Cre-cassette to isolate transduction-competent capsids. Two Ai9 females and one male were injected through the tail vein with 1.91 × 1010, respectively, of the rescued and repackaged library. After 3 weeks, mice were killed, brain cells dissociated, and cells isolated using anti-CD11b/magnetic beads (magnetic activated cell sorting; Miltenyi). Next, cells were flow sorted into tdTomato+ and tdTomato− fractions. DNA was isolated from each cell pellet as described for the first round and polymerase chain reaction (PCR) amplification of the peptide insert-encoding region submitted for next-generation sequencing (NGS). From the NGS data we selected capsid candidates that had a high read frequency in the tdTomato+ fraction. DNA was isolated from each cell pellet using the Pico Pure™ DNA Extraction Kit (Thermo Fisher Scientific) and we PCR amplified the cap region containing the insert to determine the peptide profile by NGS. From the NGS results, four capsid clones were chosen to evaluate in a pilot study in mice.
AAV-PR capsid construction
To create rep/cap plasmids encoding AAV9 capsids displaying the AAV-PR peptide (PRPPSTH) for production of vectors encoding a transgene of interest (e.g., green fluorescent protein [GFP] or Cre), we digested an AAV9 rep/cap plasmid (pAR9) with BsiWI and BaeI, which removes a fragment flanking the VP3 amino acid 588 site for peptide sequence insertion. Next, we ordered a 997 bp dsDNA fragment from Integrated DNA Technologies (IDT, Coralville, IA), which contains overlapping Gibson homology arms with the BsiWI/BaeI cut AAV9 as well as the 21-mer nucleotide sequence encoding the peptide of interest in frame after amino acid 588 of VP3. Last, we performed Gibson assembly using the Gibson Assembly® Master Mix (NEB, Ipswich, MA) to ligate the peptide-encoding insert into the AAV9 rep/cap plasmid. Next, we transformed NEB 5-alpha competent Escherichia coli (NEB) with 2 μL of the Gibson assembly and plated transformed cells on LB-Amp agar plates.
DNA isolated from selected colonies was sent for Sanger sequencing at the MGH Center for Computational and Integrative Biology DNA Core using a primer that flanked the peptide-encoding region to confirm the correct sequence was inserted.
AAV vector production, purification, and titration
For transgene expression studies with AAV-PR vectors we used the following constructs: (1) AAV expression plasmid, pAAV-CBA-NLS-Cre, a gift from Dr. Miguel Sena-Esteves (UMass Medical Center). This plasmid contains AAV inverted terminal repeats (ITRs) flanking the chicken β-actin (CBA) expression cassette, which consists of: a hybrid CMV-IE enhancer/CBA promoter, SV40 nuclear localization signal (NLS), Cre recombinase cDNA, and a bovine growth hormone (BGH) poly A signal sequence. (2) pAAV-sc-CBA-GFP. This plasmid drives GFP expression under the hybrid CMV immediate-early/CBA promoter (flanked by AAV ITRs), and was kindly provided by Dr. Miguel Sena-Esteves (UMass Medical Center). AAV-sc-CBA-GFP encodes a self-complementary (sc) AAV genome. (3) pAAV-sc-CBA-DsRed. This plasmid replaces the GFP cDNA of AAV-sc-CBA-GFP with DsRed cDNA.
AAV production was performed as previously described.17 Briefly, 293T cells were triple transfected (calcium phosphate method or polyethylenimine) with (1) AAV-PR rep/cap plasmid (2) an adenovirus helper plasmid, pAdΔF6, and (3) ITR-flanked AAV transgene expression plasmid. Cell lysates were harvested 68–72 h posttransfection and purified by ultracentrifugation of an iodixanol density gradient. Iodixanol was removed and buffer exchanged to PBS containing 0.001% Pluronic F68 (Gibco) using 7 kDa molecular weight cutoff Zeba™ desalting columns (Thermo Scientific). Vector was concentrated using Amicon® Ultra-2 100 kDa MWCO ultrafiltration devices (Millipore Sigma). Vector titers in vg/mL were determined by TaqMan quantitative polymerase chain reaction in an ABI Fast 7500 real-time PCR system (Applied Biosystems) using probes and primers to the BGH poly A sequence and interpolated from a standard curve made with a restriction enzyme-linearized AAV plasmid. Vectors were pipetted into single-use aliquots and stored at −80°C until use.
Vector injections in mice
In experiments involving AAV-PR-mediated Cre recombination and tdTomato expression in Ai9 mice, we injected adult (both male and female) Ai9 mice systemically through the lateral tail vein with AAV-PR-CBA-Cre vector. At 3–5 weeks postinjection, animals were killed and brains and other organs processed for cryosectioning and immunofluorescence staining and imaging. In experiments using AAV-PR as a conventional transgene expression vector, male C57BL/6J mice or BALB/c mice were injected systemically through the lateral tail vein with AAV-PR-sc-CBA-GFP. Mice were killed at 2 or 5 weeks (depending on the experiment) postinjection and transcardially perfused with PBS then 4% formaldehyde buffered in PBS.
For intracerebroventricular injection, 8-week-old C57BL/6 mice were anesthetized with 5% isoflurane and placed in a stereotaxic frame. Anesthesia was maintained with 2% isoflurane and body temperature was regulated with a heating pad. An incision was made at midline and a 0.5 mm hole was drilled in the skull at A/P +0.3 mm, M/L +1 mm Bregma. A Hamilton syringe fitted with a 30-gauge beveled needle was loaded with 5 μL AAV-PR-sc-CBA-GFP and was lowered 3 mm into the lateral ventricle. Injections were carried out at 1 μL/min using an injector pump. Five minutes after infusion, the needle was retracted and mice were sutured and returned to their home cages. After a 5-day incubation period, mice were killed through CO2, perfused with PBS and brains were drop-fixed in 4% PFA in PBS for 48 h.
Immunofluorescence staining and microscopy and image analysis
Brain immunostaining and imaging
Once harvested, brains were placed into a 4% PFA solution (Electron Microscopy Sciences, Hatfield, PA) in 1 × PBS (Fisher Scientific) for 24 h at 4°C. Next, brains were removed from the solution and placed into a brain matrix (Zivic Instruments, Pittsburgh, PA) and segmented into 1 mm segments. The brain segments were then placed in a 12-well nontreated tissue culture plate (Corning) and submerged again into 4% PFA for a second 24-h fixation incubation at 4°C. Following fixation, the brain segments were washed five times with 1 × PBS. Samples were then infused in an 4% acrylamide hydrogel solution (Logos Biosystems, Annandale, VA) for 24 h at 4°C in the dark to create polyacrylamide bonds throughout the tissue. Succeeding infusion of the hydrogel, the brain segments were polymerized to hybridization of the hydrogel into the tissue with the X-Clarity™ Polymerization System on a heat block for flat-bottom plates (Logos Biosystems) in the 12-well plate for 3 h at −90 kPa at 37°C.
Once polymerization of the samples was complete, samples were washed three times with 1 × PBS at room temperature on an orbital shaker at 120 RPM to remove residual hydrogel, which can affect proper clearing of the samples. Brain segments were then placed into a holder for 36-1 mm mouse brain slices (C12010; Logos Biosystems) and placed in the X-Clarity Tissue Clearing System II for lipid removal and active tissue clearing. This piece of equipment utilizes an electrophoretic tissue clearing solution (Logos Biosystems) to actively remove lipids from tissue by running a constant current and voltage through the clearing solution, while the machine pumps the solution through the clearing chamber.
Mouse brain segments were cleared for 4 h, 70 RPM, at 37°C, while maintaining 1.2 Å and 60 V consistently through the clearing process. Once clearing had been completed, samples were washed overnight with 1 × PBS at room temperature on an orbital shaker at 120 RPM to remove excess electrophoretic tissue clearing solution from the sample, which can affect antibody staining. Following the overnight wash, the samples appeared opaque white, indicating tissue clearing was successful.
Following tissue clearing, samples were additionally washed three times with 1 × PBS for 5 min each wash on an orbital shaker at 120 RPM, in a 12-well nontreated tissue culture plate for antibody staining. Samples then underwent a 24-h block incubation at 4°C on an orbital shaker at 90 RPM with Invitrogen ReadyProbes™ Mouse-on-Mouse IgG Blocking Solution (Thermo Fisher Scientific), for a final solution volume of 2.5 mL, two drops of the ReadyProbe block in 1 × PBS containing 5% normal donkey serum (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA). After blocking was completed, the samples were washed three times with 1 × PBS for 5 min each wash on an orbital shaker at 120 RPM. Samples were then incubated in primary antibody solution at room temperature for 72 h on an orbital shaker at 120 RPM. The primary antibody solution consisted of the primary antibody diluted in 1 × PBS containing 5% bovine serum albumin (BSA; Fisher Scientific) 0.1% Triton X-100 (Sigma, Burlington, MA), and 0.01% sodium azide (Sigma).
Succeeding primary incubation, the samples were washed five times over 24 h (roughly every 2–3 h) with 1 × PBS, including an overnight wash at room temperature on an orbital shaker at 120 RPM. Samples were incubated in the secondary antibody solution at room temperature for 48 h, covered from light on an orbital shaker at 120 RPM. The secondary antibody solution consisted of the secondary antibody diluted in 1 × PBS containing 1:1,000 DAPI (Sigma), 2% normal donkey serum, 0.1% Triton X-100, and 0.01% sodium azide. Once secondary antibody incubation is complete, the samples were washed five times over 24 h (roughly every 2–3 h) with 1 × PBS, including and overnight wash at room temperature on an orbital shaker at 120 RPM, while covered from light.
Finally, the samples were rinsed three times with deionized (DI) water for 5 min each wash on an orbital shaker at 120 RPM while covered from light. The DI water was aspirated, and the samples are mounted in X-Clarity Mounting Solution (Logos Biosystems) with an RI = 1.46 and incubated in the dark for 2 h on an orbital shaker at 120 RPM, with enough solution to cover the samples.
Samples were then transferred with the mounting solution to an imaging dish (Thermo Fisher). All samples were imaged with Nikon A1R confocal microscope.
AAV-PR transduction efficiency of pericyte, SMCs, and endothelial cells was performed using the AIVIA imaging analysis software (Leica). Whole hemisphere image stacks were collected (A1R; Nikon) from 2 mm tissue-cleared segments (bregma −1.05). Ten regions of interest at 100 × 100 μm were extracted and processed with the software's machine learning pixel classifier function (for each channel) to eliminate background and isolate tdTomato, endothelial cells (CD31), and pericyte (α-SMA)-positive cells. Next, stacks were processed with the 3D object tracking function to segment and volumetrically render objects ready for object classification and quantitation. CD31+ or α-SMA+ (on small-caliber vessels under 7 μm in diameter) cell/objects were colocalized with tdTomato and measured for the Pearson coefficient. It is important to note how we distinguished pericytes from SMCs given both were identified by α-SMA immunostaining. It is well established that capillary-sized vessels (avg. 7 μm in diameter) are surrounded by pericytes and that large (first-order arterial vessels) are surrounded by SMCs.18,19
Of note, our analysis has focused on capillary and large-caliber vessels, thus avoiding intermediate-sized vessels, which have mural cells with both SMC and pericyte attributes. For transduction efficiency of α-SMA+ SMCs on brain vasculature we focused on first-order arterial vessels. Calculated Pearson coefficients of 0.35 and above were used as the threshold for determining transduction efficiency.
Table 1 displays primary antibodies and Table 2, secondary antibodies, used for whole-tissue immunofluorescent staining at the indicated concentrations.
Table 1.
Primary antibodies used for brain immunostaining
| Antibody | Concentration Used for Immunolabeling | Catalog No. and Vendor |
|---|---|---|
| Goat anti-GFP | 1:1000 | Cat. No. ab5450; Abcam |
| Mouse anti-GFAP | 1:100 | Cat. No. GA5-3670; Cell Signaling Technology |
| Rabbit anti-IBA-1 | 1:50 | Cat. No. 019-19741; Wako |
| Mouse anti-α-SMA | 1:100 | Cat. No. ab7817; Abcam |
| Rabbit anti-CD31 | 1:100 | Cat. No. ab28364; Abcam |
| Rabbit anti-PDGFR-β | 1:100 | Cat. No. ab234965; Abcam |
| Mouse anti-Zo1 | 1:66 | Cat. No. 33-9100; Invitrogen™ |
α-SMA, α-smooth muscle actin; GFAP, glial fibrillary acidic protein; GFP, green fluorescent protein; PDGFR, platelet-derived growth factor receptor.
Table 2.
Secondary antibodies used for brain immunostaining
| Antibody | Concentration Used for Immunolabeling | Catalog No. and Vendor |
|---|---|---|
| Donkey anti-goat IgG (H+L) AF488 highly cross-absorbed | 1:350 | A32814; Invitrogen |
| Donkey anti-rabbit IgG (H+L) AF488 highly cross-absorbed | 1:350 | A21206; Invitrogen |
| Donkey anti-mouse IgG (H+L) AF488 highly cross-absorbed | 1:350 | A21202; Invitrogen |
| Donkey anti-rabbit IgG (H+L) AF647 highly cross-absorbed | 1:350 | A31573; Invitrogen |
| Donkey anti-mouse IgG (H+L) AF647 highly cross-absorbed | 1:350 | A31571; Invitrogen |
Imaging of aorta
Fixed aortas were embedded in optimal cutting temperature (OCT) and cryosectioned at 10 μm. Frozen slides were washed with sterile PBS twice for 2 min followed by antigen retrieval with Borg Decloaker RTU buffer (cat# BD1000G1; BIOCARE Medical). Then slides were washed with PBS twice for 2 min and tissues were blocked with donkey serum at 10% for 1 h followed by incubation overnight at 4°C with anti-α-SMA antibody. Slides were washed with PBS-Tween at 0.1% three times for 3 min each followed by incubation with secondary antibody (1:400) for 1 h at room temperature. Then slides were washed with PBS-Tween at 0.1%, four times for 3 min each and slides were mounted with diamond mounting medium containing DAPI. Intrinsic tdTomato and GFP fluorescence was observed without immunostaining. Slides were visualized with the Leica TCS SP8 confocal microscopy station and micrographs were digitized with the Leica Application Suite X software.
Brain
Forty micrometers of free-floating brain sections were washed 3 × with PBS, permeabilized in 0.01% Phosphate Buffered Saline-Tween (PBST) overnight at 4°C, and then blocked with a 10% donkey serum in PBS overnight at 4°C. Sections were subsequently incubated with primary antibodies made up in blocking solution for 48 h at 4°C on a rocking shaker, washed 3 × with 0.01% PBST, and incubated with secondary antibodies and DAPI made up in blocking solution for 3 h at 4°C. Finally, sections were washed 3 × with 0.01% PBST, 3 × with PBS, and finally mounted on a glass slide and cover slipped for imaging with mounting media. The following primary antibodies were used at the indicated concentrations: Rat anti-CD31 (1:250 dilution, BD550274; BD Biosciences), Rabbit anti-RFP (1:250 dilution, 600401379; Rockland), Alexa Fluor® 488 anti-α-SMA antibody [1A4] (1:250 dilution, SC-32251-AF488; SantaCruz).
Multiple coronal brain sections were acquired from each animal and imaged in its entirety using a TissueFAXS SQL Confocal Slide whole slide Scanner. Representative images were extracted using TissueFAXS SL Viewer software.
Organs
Organs were postfixed in 4% PFA at 4°C overnight. Samples were then washed 3 × with PBS to remove residual PFA. For cryosections, samples were cryopreserved in a 30% sucrose solution at 4°C overnight and then embedded in OCT and cryosectioned at 8 μm. Frozen slides were washed 3 × with PBS followed by antigen retrieval with ice-cold methanol for 15 min. Slides were then washed 3 × with PBS for 5 min each and tissues were blocked with 5% donkey serum with 3% BSA in PBS for 1 h followed by incubation overnight at 4°C with primary antibodies. Slides were washed 3 × with PBS for 5 min each followed by incubation with secondary antibody for 2 h at room temperature. Then slides were washed 3 × with PBS for 5 min each and slides were mounted with mounting medium containing DAPI. Slides were imaged in its entirety using a TissueFAXS SQL Confocal Slide whole slide Scanner. Representative images were extracted using a TissueFAXS SL Viewer software.
Statistics
We used GraphPad Prism 9.0 for PC for statistical analysis. To compare the difference between two means, we used an unpaired two-tailed t-test; p-values <0.05 were accepted as significant. For the comparison of transduction of endothelial cells, pericytes, and SMCs in two strains of mice, we used a two-way ANOVA followed by a Tukey's multiple comparisons test for comparisons of two means.
RESULTS
An engineered peptide displaying AAV9 capsid, AAV-PR, mediates a vasculature-selective transduction phenotype in brain after intravenous delivery
In an earlier in vivo selection with our AAV9 peptide display library (please see Materials and Methods section for details), we identified several candidate 7-mer peptide displaying capsids for evaluation of CNS transduction in mice. We next tested four of these new capsids for transduction of the adult murine brain after systemic delivery. We packaged a single-stranded AAV-CBA-Cre genome into each of the candidate capsids and injected 1012 vg (5 × 1013 vg/kg) into the tail vein of adult Ai9 mice, which have a Cre-sensitive CAG-floxed-STOP-tdTomato reporter in all cells. Any cells, which are successfully transduced by the AAV vector and have Cre-expressed, should result in removal of the floxed-STOP cassette and allow tdTomato expression. Mice were harvested 3 weeks postinjection, brains were sectioned, and tdTomato was detected with immunofluorescence staining. For one capsid, displaying the peptide, PRPPSTH, named AAV-PR, we observed a distinct vasculature-like immunostaining of tdTomato expression throughout the entire brain (Supplementary Fig. S1).
The rest of the capsids showed a lower transduction efficiency with many neurons transduced (data not shown). The profile of AAV-PR is in stark contrast to the tropism of parental AAV9-CBA-Cre in adult Ai9 mice, which mediates transduction of mostly astrocytes and neurons.20
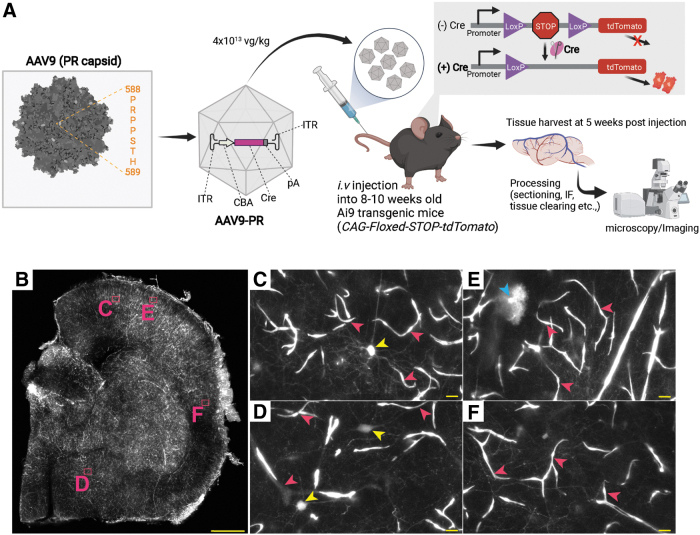
Next, we followed up this pilot experiment by injecting Ai9 mice (n = 2 male, n = 2 females) with AAV-PR-CBA-Cre at a dose of 4 × 1013 vg/kg (∼8 × 1011 vg/mouse) (Fig. 1A). Mouse tissues were harvested at 5 weeks postinjection and similar to the pilot experiment, we observed efficient transduction of the brain vasculature with AAV-PR, as observed by bright tdTomato expression (intrinsic fluorescence without immunostaining, Fig. 1B). No obvious differences in the transduction phenotype/efficiency were observed between male or female mice (data not shown). Higher magnification clearly showed transduction of vascular cells in several regions of the brain (Fig. 1B–F, pink arrowheads). Transduced neurons were confirmed by morphology (Fig. 1D, yellow arrowheads). We also observed transduced cells with “bushy” processes such as those found in basket neurons (Fig. 1E, blue arrowheads).
Figure 1.
AAV-PR-CBA-Cre mediates vasculature-tropic transduction and gene modification in transgenic Ai9 mice (CAG-floxed-STOP-tdTomato). (A) AAV-PR peptide and insertion site. The peptide PRPPSTH is inserted after amino acid 588 of AAV9 VP1. A Cre-recombinase expression cassette is packaged inside AAV-PR and this is administered systemically to Ai9 mice, which contain a floxed-stop-tdTomato cassette in all cells. If AAV-PR mediates Cre expression, the loxP-stop is removed from the genome allowing tdTomato expression. (B) Half brain hemisphere of Ai9 mouse injected i.v. with AAV-PR-CBA-Cre showing bright intrinsic fluorescence (white signal). Letters (C–F) indicate areas of zoomed images in (B). The cells that were transduced had morphology consistent with the vasculature (pink arrowheads), and neurons (yellow and blue arrowheads). Scale bar in (B) = 500 μm. (C–F) = 10 μm. AAV, adeno-associated virus; CBA, hybrid CMV early/chicken β-actin promoter; i.v., intravenous.
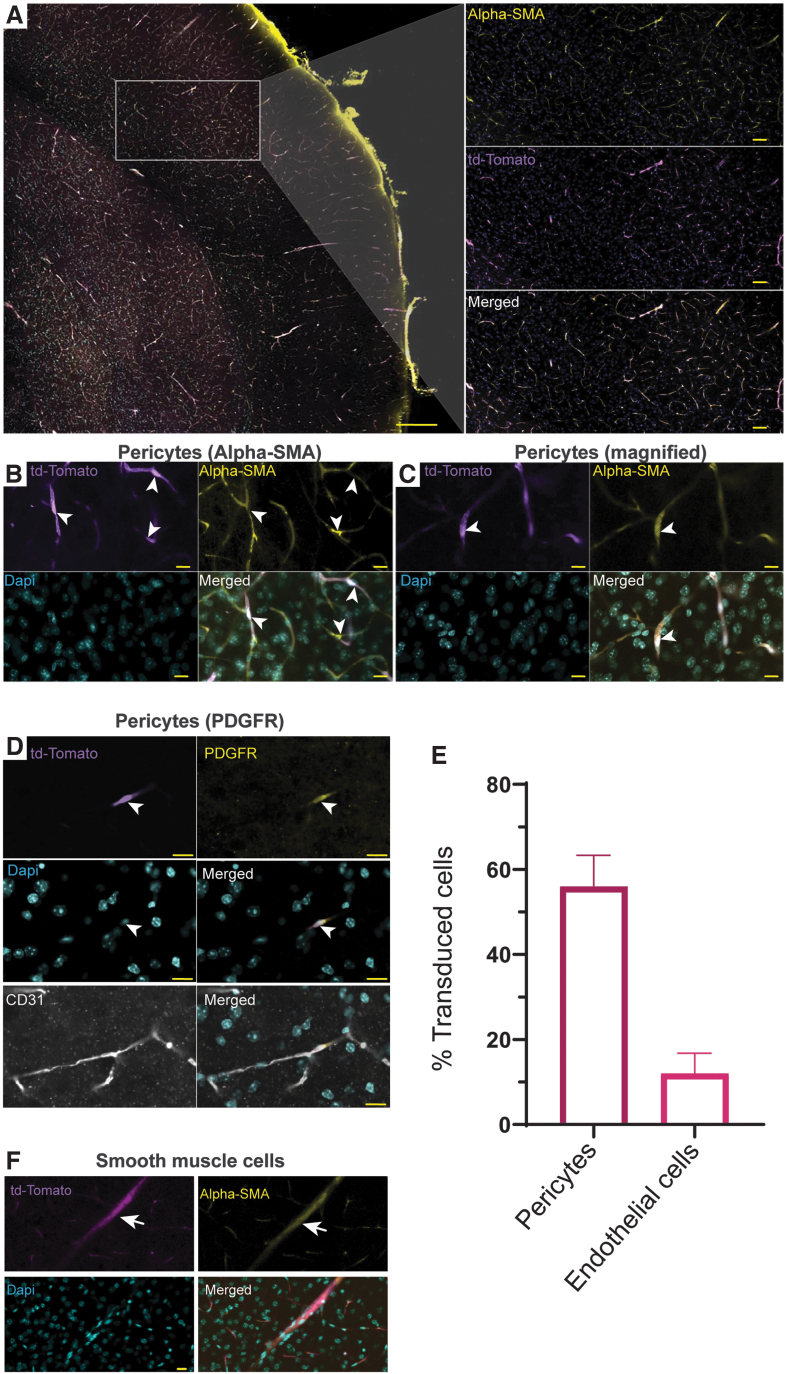
Next, to ascertain the cell types of the vasculature transduced by AAV PR in the brain, we stained with antibodies to proteins on pericytes (α-SMA and PDGFR), and endothelial cells (CD31). Interestingly, we observed colocalization of tdTomato fluorescence with α-SMA (Fig. 2A–C) indicating pericytes were transduced. Further evidence of transduction of pericytes was confirmed with PDGFR colocalization (Fig. 2D). We also immunostained for colocalization image analysis of CD31 and tdTomato, and surprisingly, there was much less than for the pericyte markers (Fig. 2D). Thus, the majority of capillary transduction appeared to be in pericytes. We quantitated the percentage of pericytes and endothelial cells based on the colocalization of tdTomato expression with PDGFR and CD31, respectively. Remarkably, an average of 56% of pericytes were transduced, while only 12% of endothelial cells had tdTomato colocalization (Fig. 2E). We next assessed transduction of vascular SMCs in cerebral arteries stained for α-SMA and we observed tdTomato expression in this cell type (Fig. 2F).
Figure 2.
Pericytes are efficiently transduced by AAV-PR after systemic injection. The identity of transduced vascular cells in Ai9 mice injected with AAV-PR-CBA-Cre was determined by immunofluorescence staining. (A) Low magnification image α-SMA staining (yellow) for pericytes and tdTomato (purple). Scale bar = 100 μm. Inset: higher magnification of the cortex showing colocalization. Scale bar = 50 μm (B, C). Higher magnification images of tdTomato/α-SMA colocalization. Scale bar = 10 μm (D). Colocalization of tdTomato with PDGFR-β. Bottom panel shows PDGFR (yellow), tdTomato (purple), DAPI (cyan), and CD31 endothelial cells (white). The colocalization of tdTomato was mainly with PDGFR and not CD31. (B–D) Arrows head point to transduced pericytes. (E) Quantitation of transduction efficiency in pericytes (PDGFR) and endothelial cells (CD31). Colocalization was based on a Pearson correlation of >0.35. (F) Transduction of smooth muscle cells on large caliber vessels. Arrow points to transduced smooth muscle cells. α-SMA, α-smooth muscle actin; DAPI, 4′,6-diamidino-2-phenylindole; PDGFR, platelet-derived growth factor receptor.
This was further confirmed when we stained large vessels for ACTA2 and we observed colocalization with tdTomato expression, and very little tdTomato/CD31 colocalization (Supplementary Fig. S2). Staining for astrocytes (glial fibrillary acidic protein [GFAP]) and microglia (IBA1) revealed no-colocalization with tdTomato expression (Supplementary Fig. S3), including in the cells with “bushy” processes observed in Fig. 1E (data not shown).
Characterization of peripheral tissues and organs transduced by AAV-PR
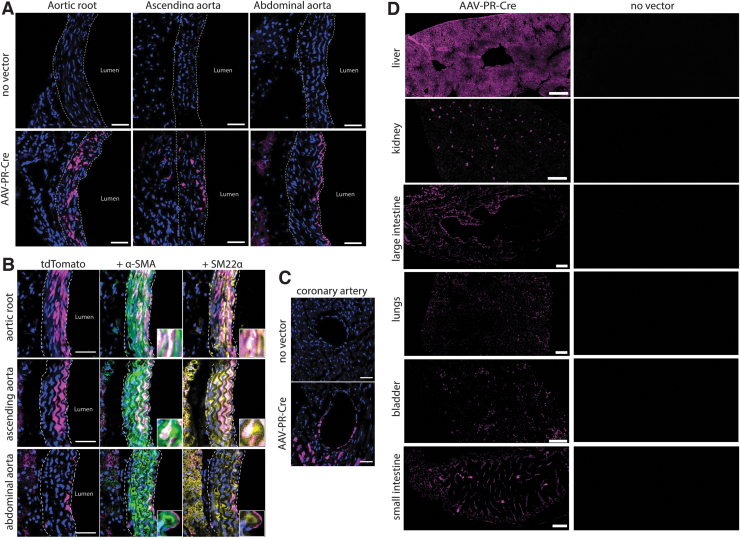
In the same Ai9 mice injected with AAV-PR-CBA-Cre from Figs. 1 and 2, we assessed transduction of peripheral organs and tissues. Due to the transduction of brain vasculature and SMCs, we also assessed transduction of the aorta. Remarkably, tdTomato-positive cells resembling SMCs were seen in the aorta, primarily the aortic valve and root, while no signal was observed in untreated Ai9 mice (Fig. 3A). We confirmed that the AAV-PR-transduced cells were indeed SMCs as they were positive for α-SMA and SM22α (Fig. 3B). We also observed cells lining the coronary artery were transduced by AAV-PR (Fig. 3C).
Figure 3.
AAV-PR transduces aortic smooth muscle cells as well as multiple peripheral tissues. Ai9 mice were left untreated (noninjected) or were injected with AAV-PR-CBA-Cre and 5 weeks later, intrinsic tdTomato fluorescence observed. (A) Intrinsic tdTomato fluorescence (magenta) was readily detected in aorta of injected but not in untreated mice. Scale bar = 30 μm (B). Many of the transduced cells in aorta were smooth muscle cells as they colocalized with α-SMA and SM22α. Scale bar = 30 μm (C). Cell lining the coronary artery were transduced by AAV-PR. Scale bar = 100 μm (D). tdTomato expression in peripheral tissues of Ai9 mice injected with AAV-PR-CBA-Cre injected (left panels) and noninjected (right panels). Scale bars are as follows: liver, kidney = 500 μm; lungs, bladder, small intestine = 200 μm; large intestine = 100 μm.
Liver is efficiently transduced by many AAV capsids after systemic dosing, and not surprisingly AAV-PR transduced hepatocytes with high efficiency (Fig. 3D). In lung, bladder, kidney, esophagus, and small intestine, robust intrinsic tdTomato expression was observed in parenchymal vessels, including glomeruli, alveolar, and submucosa capillaries and arteries that surrounded or colocalized with CD31. In addition, a number of histiocyte-like CD31-positive cells in the kidney medulla and submucosa of gastrointestinal organs and skeletal myocytes were transduced by AAV-PR (Fig. 3D and Supplementary Fig. S4).
AAV-PR transduces brain pericytes using a conventional “gene addition” transgene expression cassette
The impressive transduction efficiency of the vasculature by AAV-PR-CBA-Cre in the Ai9 (Floxed-stop-tdTomato) transgenic mouse is a model of genetic editing, as Cre expression leads to permanent tdTomato expression after Cre-mediate excision of the transcription stop signal. To assess whether AAV-PR would function in a more traditional “gene addition” transgene expression strategy, we packaged a sc AAV genome, which encodes GFP cDNA.
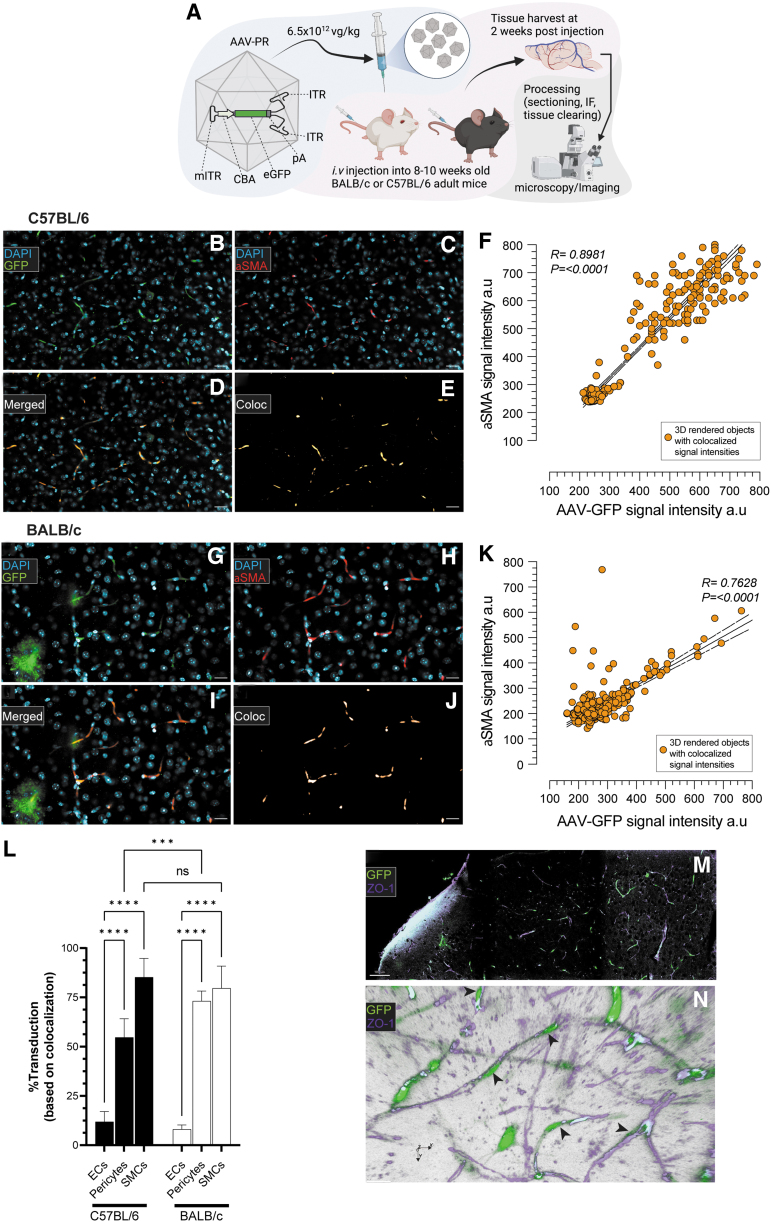
We tested transduction of AAV-PR in the two most widely used strains of mice, C57BL/6 and BALB/c, to observe if the capsid's vascular tropism translated across these two strains. Adult mice (n = 3/strain) were injected systemically with 1.2 × 1011 vg/mouse of AAV-PR-sc-CBA-GFP, which on a dose/weight was 6.5 × 1012 vg/kg for C57BL/6 and 4.8 × 1012 vg/kg for BALB/c and harvested 2 weeks later. Brains were cleared and imaged for GFP expression as well as α-SMA-positive pericytes (Fig. 4A). Similar to the gene editing experiment with AAV-PR-CBA-Cre in Ai9-floxed-STOP-tdTomato mice, a vascular transduction phenotype was detected upon the expression of GFP in the brain of AAV-PR-sc-CBA-GFP-injected C57BL/6 mice (Fig. 4B).
Figure 4.
AAV-PR transduces pericytes and smooth muscle cells in brain vasculature of C57BL/6 and BALB/c mice using a “gene addition” transgene expression cassette. (A) Overview of experiment. AAV-PR capsid packaged a sc cassette encoding GFP under the CBA promoter (AAV-PR-sc-CBA-GFP) was injected through the tail vein into adult C57BL/6 mice and BALB/c mice and 2 weeks later mice were sacrificed and processed for imaging of GFP fluorescence. (B) Transduction of α-SMA-positive pericytes in C57BL/6 mice. Scale bar = 25 μm. (C) Pericyte staining on small caliber vessels detected with α-SMA staining. (D) Merge of GFP (green) with α-SMA (red). (E) Rendering of GFP/α-SMA colocalization. (F) Correlation of α-SMA fluorescence signal intensity versus GFP signal intensity. (G–K) The same depiction as for C57BL/6, except in BALB/c mice. (L) Quantitation of endothelial cells, pericytes, and smooth muscle cells on large caliber vessels in both strains of mice. Values of significance indicated by asterisks compare transduction to endothelial cells. ****p < 0.0001; ***p = 0.0001; ns = not statistically significant. (M, N) An independent experiment was performed in C57BL/6 mice injected with AAV-PR-sc-CBA-GFP (6 × 1012 vg/kg) and 5 weeks later, brains were imaged for GFP (green) and ZO-1 to label endothelium (magenta). (M) Low magnification image of brain showing GFP cells transduced by AAV-PR and the endothelium labeled with ZO-1. Scale bar = 100 μm. (N) Volumetric rendering of stacked images shows spatial location of GFP-positive cells with pericyte morphology (black arrowheads) interacting with the ZO-1-positive cerebrum endothelium. GFP, green fluorescent protein; ITR, wild-type inverted terminal repeats; mITR, mutant inverted terminal repeats; pA, poly A signal sequence; sc, self-complementary; vg, vector genomes.
Brains were costained with α-SMA to label pericytes on capillaries (Fig. 4C). In line with the prior data with AAV-PR-Cre, we observed GFP signal colabeling with the pericyte marker α-SMA (Fig. 4D–F). Similarly, AAV-PR transduced pericytes in BALB/c mice (Fig. 4G–K). Next, we performed quantitation of transduction efficiency of AAV-PR for endothelial cells (CD31), pericytes (α-SMA, small-caliber vessels), and SMCs (α-SMA, large-caliber vessels), in both strains of mice. Similar to the experiment with AAV-PR-CBA-Cre in Ai9 mice, AAV-PR-sc-CBA-GFP transduced a high percentage of pericytes in the brain, which was 54.7% in C57BL/6 and 73.1% for BALB/c, with BALB/c group being significantly higher than C57BL/6 group (Fig. 4L). Quantifying AAV-PR mediated transduction of SMCs at first-order large vessels revealed 85.2% for C57BL/6 and 79.7% for BALC/c (Fig. 4L). Transduction of endothelial cells was significantly lower than either pericytes or SMCs for both mouse strains (11.9% for C57BL/6 and 8.1% for BALB/c) (Fig. 4L).
In an independent experiment, adult C57BL/6 mice were injected systemically with 1.2 × 1011 vg/mouse (6.5 × 1012 vg/kg) of AAV-PR-sc-CBA-GFP and harvested 5 weeks later. Brains were cleared and imaged for GFP expression and costained with ZO-1, which labels the intracellular tight-junction protein on endothelial cells. We observed GFP signal in cells with pericyte morphology that were wrapped around the ZO-1 signal (Fig. 4M). Three-dimensional rendering of the brain images clearly showed GFP-positive cells interacting closely with the ZO-1-labeled endothelium (Fig. 4N).
We also looked for SMC transduction in the aorta in these mice. We observed SMC transduction with AAV-PR-sc-CBA-GFP in both strains of mice, although it appeared to be much lower efficiency than with AAV-PR-CBA-Cre injection in Ai9 transgenic mice (Supplementary Fig. S5).
Cell type tropism of AAV-PR in the brain is dependent on route of administration
To understand if AAV-PR pericytes and SMC transduction are driven by the intravenous route of administration, we tested intracerebroventricular injection (lateral ventricle) of AAV-PR-sc-CBA-GFP in adult C57BL/6 mice. Five days later, mice were harvested and cryosection of brain imaged for GFP expression. We chose a shorter-in-life duration to speed up the experimental timeframe, as we have previously found that intracranial injections generally yield fast transgene expression kinetics with AAV vectors likely owing to higher vector genome concentrations versus systemic injections (unpublished observations). Transduction was not widespread and was confined mainly to cells immediately surrounding the subventricular zone of the injected lateral ventricle (Supplementary Fig. S6). The morphology of the cells (round cell bodies with uni or bipolar fine projections) did not appear to be associated with the vasculature and were most consistent with neural and progenitor cells. This suggests that AAV-PR's pericyte and SMC tropism is driven by its ability to penetrate the endothelium after intravascular delivery.
AAV-PR transduces primary human pericyte cultures
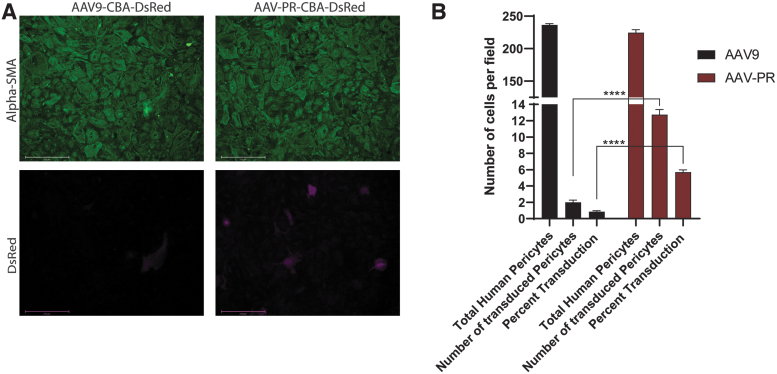
Since AAV-PR demonstrated a remarkable ability to transduce pericytes after systemic injection in vivo in mice, we were curious to test whether this capsid could mediate transduction of primary human pericytes in culture. Human pericyte cultures were transduced with 2 × 105 vg/cell of either AAV9-sc-CBA-DsRed or AAV-PR-sc-CBA-DsRed vectors and 3 days later, cells were examined for α-SMA expression to confirm the purity of the cultures as well as DsRed to observe AAV-mediated transgene expression. We found that although both capsids could transduce human pericytes, the transduction efficiency was 6.7-fold higher (p < 0.001) for AAV-PR (4.5–6.63% efficiency) compared with AAV9 (0.41–1.3% efficiency) (Fig. 5A, B).
Figure 5.
AAV-PR transduces primary human pericytes in culture. Human pericytes were transduced with equal doses of either AAV9 or AAV-PR both packaging a sc AAV-CBA-DsRed genome. Three days posttransduction cells were examined for α-SMA to determine culture purity and DsRed for transgene expression. (A) Representative images of transduced cells with either vector. DsRed fluorescence is shown in magenta. Scale bars = 275 μm. (B) Quantitation results from particle counting image analysis of transduced (DsRed positive) positive pericytes for AAV9 and AAV-PR. Shown are the number of cells/field counted, the number of DsRed-positive cells/field counted and the percentage of total cells expressing DsRed. Performed in triplicate, n = 8 averaged fields of view, ****p < 0.0001 by Student's unpaired t-test.
DISCUSSION
Developing AAV vectors, which can deliver genes more efficiently and specifically to the vasculature, has great promise to treat diseases that affect the cells that form the cerebrovascular system. In this study, we found that the AAV9-based capsid, AAV-PR, is highly efficient at mediating transduction of both brain pericytes and SMCs. AAV9 capsid crosses the blood–brain barrier and while it can transduce the brain endothelium, it is with relatively low efficiency.5
In the past decade, there has been considerable progress in the development of AAV capsids that mediate transduction of the vasculature. For example, the capsid AAV2-BR1 was identified through an in vivo selection with an AAV2-based peptide display library.7 This capsid (NRGTEWD peptide) mediates efficient transduction of endothelial cells of the brain vasculature, with significant detargeting from peripheral organs such as the liver. The same group identified another AAV2 capsid variant with the peptide ESGHGYF after an in vivo selection directed at the lung vasculature.8 Similar to BR1, AAV2-ESGHGYF transduced endothelial cells (although transduction was localized to the lung).
Ravindra Kumar et al. developed the PHP.V1 capsid using the CREATE system.9 This capsid, with peptide TALKPFL, was able to transduce both cortical astrocytes as well as endothelial cells of the brain. More recently, Krolak et al. derived an AAV9 capsid, called AAV-BI30 displaying the peptide, NNSTRGG, after selection in vivo in mice and in vitro in human and mouse brain microvascular endothelial cells (BMVECs).6 AAV-BI30 was able to efficiently transduce endothelial cells of the murine brain, as well as human and mouse BMVECs.
While AAV-PR can transduce endothelial cells to some extent, what profoundly sets it apart from these prior capsids, is that it can transduce both pericytes and SMCs after systemic injection. As a variety of disease affect these two cell types, AAV-PR should be a useful tool to modulate these cells at the genetic level, for both basic research and preclinical therapeutic development. It should be noted that an engineered capsid, AAV2.5, has been shown to transduce SMC in injured aortas of rats, although the route of delivery was local through clamping the aorta and instilling virus locally for 30 min.21 It may be interesting to try local delivery of AAV-PR to the aorta for more selective SMC transduction.
In addition to the high level of vascular transduction, we observed transduced cells with a phenotype resembling basket neurons (Fig. 1E). We were unable to identify the precise identity of this cell type, as it did not stain positive for either GFAP or Iba1. It will be interesting in future work to determine its phenotype in case it turns out to be a cell type of interest to genetically modify with this capsid. AAV-PR could also transduce other cells in the body after systemic injection (e.g., hepatocytes), and in future applications the transcriptional activity can be controlled using selective promoters/and or liver detargeting using specific microRNA seed sequences in the AAV transgene cassette.22 For example, the PDGFR-β promoter has been cloned for both human and mouse.23,24 Testing different regions of the promoter in the context of an AAV transgene expression cassette may allow further restriction of transduction to pericytes.
We tested the transduction of AAV-PR using two transduction models. In the first model we used the ultrasensitive Cre/lox system to model a genome editing strategy. In this system (Figs. 1–3), transduction resulting from even transient or low levels of transgene expression by AAV can result in permanent genetic modification (similar to gene editing strategies with Cas9/gRNA).25 Using the AAV-PR-CBA-Cre (single-stranded genome) in Cre-sensitive reporter mice, we found highly efficient transduction of pericytes and SMCs at clinically relevant doses (4 × 1013 vg/kg).
In the second transduction system, we modeled a traditional gene addition strategy, AAV genomes encoding GFP cDNA. Because observing transduction events by AAV is limited by the sensitivity of the expression cassette and reporter molecule, we opted to use a self-complementary genome to maximize transgene expression. Again, in this system, we observed efficient transduction of brain pericytes and SMCs (Fig. 4). Transduction of SMC cells in the aorta was observed with AAV-PR-sc-CBA-GFP, although it appeared to be much lower than in the AAV-PR-Cre/Ai9 system.
Prior work has shown that many AAV genomes do not form stable dsDNA forms, resulting in transient transgene expression.26 Thus, AAV-PR may be able to efficiently enter and transiently transduce SMC, but some AAV genomes may be lost. Other explanations, could be transcriptional shutdown in these cells, which will be the focus of future work. At any rate, these data in both transduction models suggest that AAV-PR may be useful in gene editing strategies as well as gene addition/gene knockdown studies targeted to the vasculature, particularly in the brain, although aorta SMC transduction may be best suited for transient expression strategies, such as CRISPR/Cas9 gene editing.
We also observed that AAV-PR requires systemic injection to efficiently transduce the vasculature, including pericytes. This was determined by injecting AAV-PR-sc-CBA-GFP directly into the lateral ventricle of adult mice. Transduced cells were restricted to the subventricular zone immediately surrounding the ventricle and did not appear to be of vascular origin (i.e., they were not pericytes) (Supplementary Fig. S5). This is not surprising in hindsight as AAV-PR was selected from a library that was administered intravenously. Therefore, this capsid likely has the unique ability to penetrate past the endothelium, but unlike AAV9, AAV-PR mainly transduces the cells intimately in contact with the endothelium (SMC and pericytes). It will be interesting in future research to determine if AAV-PR binds to a unique receptor on the surface of pericytes and SMC or if it is more related to postentry transduction events in pericytes that differ from AAV9.
It is important to note that the initial selection with the AAV9 peptide display library that identified AAV-PR had the goal of deriving AAV capsids that could transduce microglia. To drive the selection, we recovered AAV genomes from cells that were pulled down with anti-CD11b beads. The target cells were intended to be microglia/brain macrophage as they express CD11b on their surface. However, when AAV-PR was tested individually after systemic injection, it did not transduce microglia.
One explanation is that the promoter for the transgene expression cassette, CBA, is not optimized for microglia-specific transcription, as it has recently been shown that even AAV9 can transduce microglia in the context of specific promoters and regulatory elements.27 Another potential explanation for AAV-PR pericyte tropism, is that the CD11b isolation may inadvertently have isolated pericytes, which coisolated with CD11b-positive cells. Support for this is a recent study, which showed that a subpopulation of microglia, termed pericyte-associated microglia are in close proximity to pericytes, as the name implies.28
A more speculative explanation would be that the anti-CD11b-conjugated beads pulled down pericytes themselves as pericytes have been reported to express CD11b on their surface in vivo,29,30 and in some reports, pericytes can differentiate into CD11b-positive microglia-like cells.31,32 The results of the selection demonstrate that obtaining the desired traits from a peptide library of capsids can be challenging and needs to be carefully optimized, especially for cell types that are very recalcitrant to AAV transduction. In any case, the pericyte tropism obtained by AAV-PR was very useful and unprecedented and is the reason we chose to pursue this capsid for the current study.
Virus vector transduction efficiency in a given cell type can vary greatly between in vivo systems and cultured cells, owing to differences in cell biology as well as biophysical factors. There are several examples of transduction efficiency and cell-type tropism differences with AAV vectors in primary human cultured cells and in vivo in mice33 and between mice and larger animals, including nonhuman primates (NHPs).34 For example, Nonnenmacher et al. identified novel AAV capsids with high transduction efficiency of murine brain after systemic injection in mice compared with AAV9. The group then tested these same capsids to bind cultured murine BMVECs, and while some capsids had an increased ability to bind to these cells compared with AAV9, many did not.35 This is just one of several examples that shows the difficulty in predicting how a given capsid will perform in vivo based on in vitro results and vice versa. Interestingly, we found that AAV-PR transduced primary cultured human pericytes at a higher efficiency than AAV9, although at a relatively low level.
While these results are encouraging, a more accurate measure of clinical potential of AAV9 for transduction of human vasculature after systemic delivery will be determined in ongoing NHP experiments, as they are currently the most complex biological system with highest genetic similarity to humans. In the gene addition experiment (Fig. 4), we demonstrate AAV-PR's tropism for SMC and pericytes extends to the two most widely used mouse strains, C57BL/6 and BALB/c (Fig. 4). Thus, the results in mice with AAV-PR will allow the study of pericyte and SMC biology as well as testing of new therapeutics in animal models of human disease.
Supplementary Material
ACKNOWLEDGMENTS
For generation of the artwork in the Figures, BioRender software was utilized for some of the objects. For the AAV capsid cartoons, we used the Protein Data Bank (RCSB PDB) AAV9 capsid structure PDB ID: http://doi.org/10.2210/pdb7MT0/pdb36 We exported the AAV9 structure in 3D viewer into a drawing program for custom coloring.
DATA AVAILABILITY STATEMENT
The AAV rep/cap plasmid containing the AAV-PR capsid sequence is available upon completion of a standard Material Transfer Agreement with The Massachusetts General Hospital. Any other raw data that support the findings of this study are available from the corresponding author. The AAV-PR rep/cap plasmid is available by request from Addgene (plasmid #197565).
AUTHORs' CONTRIBUTIONS
C.A.M., E.H., S.H.R., P.L.M., K.S.H., and A.M.A. conceived of the study, performed experiments, analyzed data, and wrote the article. J.F.H., S.M., C.L.C., K.N.U.G.D., R.E.B., and S.D. performed experiments and analyzed data. C.N., D.M.N., D.D.L.C., and J.N. performed experiments.
AUTHOR DISCLOSURE
C.A.M. has a financial interest in Sphere Gene Therapeutics, Inc., Chameleon Biosciences, Inc., and Skylark Bio, Inc., companies developing gene therapy platforms. C.A.M.'s interests were reviewed and are managed by MGH and Mass General Brigham in accordance with their conflict-of-interest policies. C.A.M., P.L.M., E.H., and K.S.H. have a filed patent application surrounding the AAV-PR capsid.
FUNDING INFORMATION
This work was supported by NIH R01 grant DC017117 (to Casey A. Maguire), NIH R01 grant NS117575 (Patricia L. Musolino), NIH DP2 grant DA056172 (to Allison M. Andrews), NIH R01 grant DA052850 (to Servio H. Ramirez), and NIH grant R01AG071567 (Rachel E. Bennett).
SUPPLEMENTARY MATERIAL
REFERENCES
- 1. Sweeney MD, Kisler K, Montagne A, et al. The role of brain vasculature in neurodegenerative disorders. Nat Neurosci 2018;21(10):1318–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Marini S, Anderson CD, Rosand J. Genetics of cerebral small vessel disease. Stroke 2020;51(1):12–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chojdak-Łukasiewicz J, Dziadkowiak E, Budrewicz S. Monogenic causes of strokes. Genes (Basel) 2021;12(12):1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lauer A, Speroni SL, Patel JB, et al. Cerebrovascular disease progression in patients with ACTA2 Arg179 pathogenic variants. Neurology 2021;96(4): e538–e552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Foust KD, Nurre E, Montgomery CL, et al. Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat Biotechnol 2009;27(1):59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Krolak T, Chan KY, Kaplan L, et al. A high-efficiency AAV for endothelial cell transduction throughout the central nervous system. Nat Cardiovasc Res 2022;1(4):389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Körbelin J, Dogbevia G, Michelfelder S, et al. A brain microvasculature endothelial cell-specific viral vector with the potential to treat neurovascular and neurological diseases. EMBO Mol Med 2016;8(6):609–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Körbelin J, Sieber T, Michelfelder S, et al. Pulmonary targeting of adeno-associated viral vectors by next-generation sequencing-guided screening of random capsid displayed peptide libraries. Mol Ther 2016;24(6):1050–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ravindra Kumar S, Miles TF, Chen X, et al. Multiplexed Cre-dependent selection yields systemic AAVs for targeting distinct brain cell types. Nat Methods 2020;17(5):541–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Winkler EA, Sagare AP, Zlokovic BV. The pericyte: A forgotten cell type with important implications for Alzheimer's disease? Brain Pathol 2014;24(4):371–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Milewicz DM, Østergaard JR, Ala-Kokko LM, et al. De novo ACTA2 mutation causes a novel syndrome of multisystemic smooth muscle dysfunction. Am J Med Genet A 2010;152A(10):2437–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Persidsky Y, Hill J, Zhang M, et al. Dysfunction of brain pericytes in chronic neuroinflammation. J Cereb Blood Flow Metab 2016;36(4):794–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hill J, Rom S, Ramirez SH, et al. Emerging roles of pericytes in the regulation of the neurovascular unit in health and disease. J Neuroimmune Pharmacol 2014;9(5):591–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bernas MJ, Cardoso FL, Daley SK, et al. Establishment of primary cultures of human brain microvascular endothelial cells to provide an in vitro cellular model of the blood-brain barrier. Nat Protoc 2010;5(7):1265–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Andrews AM, Lutton EM, Cannella LA, et al. Characterization of human fetal brain endothelial cells reveals barrier properties suitable for in vitro modeling of the BBB with syngenic co-cultures. J Cereb Blood Flow Metab 2018;38(5):888–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hanlon KS, Meltzer JC, Buzhdygan T, et al. Selection of an efficient AAV vector for robust CNS transgene expression. Mol Ther Methods Clin Dev 2019;15:320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ivanchenko MV, Hanlon KS, Devine MK, et al. Preclinical testing of AAV9-PHP.B for transgene expression in the non-human primate cochlea. Hear Res 2020;394:107930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rhodin JA. Ultrastructure of mammalian venous capillaries, venules, and small collecting veins. J Ultrastruct Res 1968;25(5):452–500. [DOI] [PubMed] [Google Scholar]
- 19. Krueger M, Bechmann I. CNS pericytes: Concepts, misconceptions, and a way out. Glia 2010;58(1):1–10. [DOI] [PubMed] [Google Scholar]
- 20. Prabhakar S, Lule S, da Hora CC, et al. AAV9 transduction mediated by systemic delivery of vector via retro-orbital injection in newborn, neonatal and juvenile mice. Exp Anim 2021;70(4):450–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lompré AM, Hadri L, Merlet E, et al. Efficient transduction of vascular smooth muscle cells with a translational AAV2.5 vector: A new perspective for in-stent restenosis gene therapy. Gene Ther 2013;20(9):901–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xie J, Xie Q, Zhang H, et al. MicroRNA-regulated, systemically delivered rAAV9: A step closer to CNS-restricted transgene expression. Mol Ther 2011;19(3):526–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Qin Y, Fortin JS, Tye D, et al. Molecular cloning of the human platelet-derived growth factor receptor beta (PDGFR-beta) promoter and drug targeting of the G-quadruplex-forming region to repress PDGFR-beta expression. Biochemistry 2010;49(19):4208–4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ballagi AE, Ishizaki A, Nehlin JO, et al. Isolation and characterization of the mouse PDGF beta-receptor promoter. Biochem Biophys Res Commun 1995;210(1):165–173. [DOI] [PubMed] [Google Scholar]
- 25. Lang JF, Toulmin SA, Brida KL, et al. Standard screening methods underreport AAV-mediated transduction and gene editing. Nat Commun 2019;10(1):3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang J, Xie J, Lu H, et al. Existence of transient functional double-stranded DNA intermediates during recombinant AAV transduction. Proc Natl Acad Sci U S A 2007;104(32):13104–13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Okada Y, Hosoi N, Matsuzaki Y, et al. Development of microglia-targeting adeno-associated viral vectors as tools to study microglial behavior in vivo. Commun Biol 2022;5(1):1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Morris GP, Foster CP, Courtney JM, et al. Microglia associations with brain pericytes and the vasculature are reduced in Alzheimer's disease. bioRxiv 2022; 2022:2022..08.08.503250. [Google Scholar]
- 29. Balabanov R, Washington R, Wagnerova J, et al. CNS microvascular pericytes express macrophage-like function, cell surface integrin alpha M, and macrophage marker ED-2. Microvasc Res 1996;52(2):127–142. [DOI] [PubMed] [Google Scholar]
- 30. Liu YW, Zhang J, Bi W, et al. Histones of neutrophil extracellular traps induce CD11b expression in brain pericytes via dectin-1 after traumatic brain injury. Neurosci Bull 2022;38(10):1199–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hutter-Schmid B, Humpel C. Primary mouse brain pericytes isolated from transgenic Alzheimer mice spontaneously differentiate into a CD11b(+) microglial-like cell type in vitro. Exp Gerontol 2018;112:30–37. [DOI] [PubMed] [Google Scholar]
- 32. Sakuma R, Kawahara M, Nakano-Doi A, et al. Brain pericytes serve as microglia-generating multipotent vascular stem cells following ischemic stroke. J Neuroinflammation 2016;13(1):57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lisowski L, Dane AP, Chu K, et al. Selection and evaluation of clinically relevant AAV variants in a xenograft liver model. Nature 2014;506(7488):382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hordeaux J, Wang Q, Katz N, et al. The neurotropic properties of AAV-PHP.B are limited to C57BL/6J mice. Mol Ther 2018;26(3):664–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nonnenmacher M, Wang W, Child MA, et al. Rapid evolution of blood-brain-barrier-penetrating AAV capsids by RNA-driven biopanning. Mol Ther Methods Clin Dev 2021;20:366–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Penzes JJ, Chipman P, Bhattacharya N, et al. Adeno-associated virus 9 structural rearrangements induced by endosomal trafficking pH and glycan attachment. J Virol 2021;95(19):e0084321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The AAV rep/cap plasmid containing the AAV-PR capsid sequence is available upon completion of a standard Material Transfer Agreement with The Massachusetts General Hospital. Any other raw data that support the findings of this study are available from the corresponding author. The AAV-PR rep/cap plasmid is available by request from Addgene (plasmid #197565).