Abstract
Gene editing strategies are attractive for treating genetic pulmonary diseases such as cystic fibrosis (CF). However, challenges have included the development of safe and effective vector systems for gene editing of airway epithelia and model systems to report their efficiency and durability. The domestic ferret (Mustela putorius furo) has a high degree of conservation in lung cellular anatomy with humans, and has served as an excellent model for many types of lung diseases, including CF. In this study, we evaluated the efficiency of amphiphilic shuttle peptide S10 for protein delivery and gene editing using SpCas9, and AsCas12a (Cpf1) ribonucleoproteins (RNPs). These approaches were evaluated in proliferating ferret airway basal cells, polarized airway epithelia in vitro, and lungs in vivo, by accessing the editing efficiency using reporter ferrets and measuring indels at the ferret CFTR locus. Our results demonstrate that shuttle peptides efficiently enable delivery of reporter proteins/peptides and gene editing SpCas9 or Cpf1 RNP complexes to ferret airway epithelial cells in vitro and in vivo. We measured S10 delivery efficiency of green fluorescent protein (GFP)-nuclear localization signal (NLS) protein or SpCas9 RNP into ferret airway basal cells and fully differentiated ciliated and nonciliated epithelial cells in vitro. In vitro and in vivo gene editing efficiencies were determined by Cas/LoxP-gRNA RNP-mediated conversion of a ROSA-TG Cre recombinase reporter using transgenic primary cells and ferrets. S10/Cas9 RNP was more effective, relative to S10/Cpf1 RNP at gene editing of the ROSA-TG locus. Intratracheal lung delivery of the S10 shuttle combined with GFP-NLS protein or D-Retro-Inverso (DRI)-NLS peptide demonstrated efficiencies of protein delivery that were ∼3-fold or 14-fold greater, respectively, than the efficiency of gene editing at the ROSA-TG locus using S10/Cas9/LoxP-gRNA. Cpf1 RNPs was less effective than SpCas9 at gene editing of LoxP locus. These data demonstrate the feasibility of shuttle peptide delivery of Cas RNPs to the ferret airways and the potential utility for developing ex vivo stem cell-based and in vivo gene editing therapies for genetic pulmonary diseases such as CF.
Keywords: ferret, cell-penetrating peptides, CRISPR, genome editing, airway epithelia
INTRODUCTION
The discovery and refinement of clustered regularly interspaced short palindromic repeats (CRISPR)/Cas systems have had an incredible impact on the gene editing field.1,2 Examples of ex vivo cell-based therapies using gene editing afford efficient methods of transiently delivering gene editing cargo to stem cells in the absence of an immune response.3 However, challenges remain in the development of safe and efficient methods for delivering in vivo the CRISPR/Cas components into target cells of an organ.4,5 Given preexisting humoral and cellular immune response to Cas9 in most individuals,6 transient expression of gene editing components using RNA or protein delivery has immunologic advantages over vectors that may give prolonged expression of these bacterial derived foreign proteins. Strategies for efficient in vivo delivery of CRISPR/Cas elements are thus crucial for genome editing applications.
Technically, the CRISPR-Cas system can be introduced into cells using a DNA- or mRNA-based vector expressing the Cas nuclease and its single-guide RNA (sgRNA) or preloaded ribonucleoprotein (RNP) containing Cas protein and sgRNA. Each approach has unique advantages and disadvantages,5 which can be delivered to cells using three main categories of technologies that apply physical, chemical, or biological methods.7 The physical approaches include electroporation and microinjection, while the chemical methods rely on synthesized materials, such as liposomes and lipid nanoparticles (LNPs). The biological delivery methods include viral vectors and cell-penetrating peptides (CPPs).8
Viral delivery represents the most investigated approach for CRISPR/Cas9-mediated genome editing with adeno-associated virus being one of the dominant vector systems.9,10 Such approaches have been efficiently applied for genome editing in mouse airway epithelial cells in vivo11 and ferret airway epithelia in vitro.12 However, the tissue-specific tropism, immunogenicity, and potential genotoxicity caused by genomic integration of viral genomes present challenges for this genome editing approach. The development of nonviral systems to deliver CRISPR/Cas elements has been pursued as alternatives, but they showed less efficient than virus-mediated approaches.
With respect to potential off-target DNA breaks and host immune responses to Cas nuclease expression, an approach with short-term expression of functional nucleases, such as mRNA and RNP, is superior to nucleases expressed by DNA-based vectors that give prolonged expression. To this end, the delivery of Cas nuclease mRNA or protein is a “hit-and-run” strategy that reduces the above potential off-target risks and host immune responses.
Delivery of recombinant Cas nuclease in the form of an RNP complex has advantages over mRNA since the delivered complex is immediately functional without the need for mRNA translation and RNP complex formation and the transient half-life of the protein also theoretically reduces off-target effects.13,14 However, the delivery efficiency of RNPs into targeted cells can be lower, particularly following in vivo delivery to lung epithelia due to physicochemical barriers that protect the airways,15–17 macromolecular size and instability of RNPs,18 the lack of effective delivery carriers, and the difficulty of in vivo electroporation.19–21
A number of methods, including nucleofection,22 LNP,23 and CPPs,24–26 have been developed for delivery of Cas RNPs. Among these delivery methods, the CPPs, a class of short peptides capable of mediating molecule internalization to across cell membranes, have shown promise in the delivery of nucleic acids, proteins, and Cas RNPs into various cell types in vitro and in vivo.27–31 Furthermore, recently developed amphiphilic shuttle peptides have demonstrated great promises in efficient delivery of Cas RNPs into multiple cell types for genome editing of different genes, including difficult to transfect airway epithelial cells for CFTR gene editing in vitro and in vivo.27,28,32
In this study, we investigated the efficiency of delivering fluorescent recombinant protein, synthetic peptide, and Cas RNPs mediated by amphiphilic peptides in airway epithelia of ferrets, a species that has produced excellent genetic models of pulmonary diseases, including cystic fibrosis (CF),33–36 chronic obstructive pulmonary disease-related chronic bronchitis and emphysema,37 obliterative bronchiolitis in lung transplantation,38,39 and alpha-1 antitrypsin deficiency.40 Our results demonstrate that amphiphilic shuttle peptides were able to deliver proteins, peptides, and Cas RNPs to ferret airway epithelial cells to facilitate genome editing in vitro and in vivo.
MATERIALS AND METHODS
Peptides, proteins, and gRNAs
Amphiphilic shuttle peptides, nuclear targeted Cy5 conjugated D-Retro-Inverso (DRI)-nuclear localization signal (NLS) (DRI-NLS-Cy5) fluorescent peptide, green fluorescent protein (GFP)-NLS, CRISPR nuclease SpCas9, and AsCas12a (Cpf1) were provided or purchased by Feldan Therapeutics (Quebec, QC, Canada) as previously described.27,28 The shuttle peptides and GFP-NLS, DRI-NLS-Cy5, or RNPs of gRNA/Cas protein complex were thawed and formulated before the experiment.
Ferret airway epithelial cell cultures
Primary trachea (TR) and intralobar (ILB) airway epithelial cells isolated from nontransgenic and Cre/LoxP reporter transgenic ferrets (ROSA-TG)41 were propagated in PneumaCult EX-plus complete medium (Cat No. 05041; STEMCELL, Vancouver, BC, Canada). Passage 4–6 basal epithelial cells were seeded on collagen-coated 96-well plate and cultured in PneumaCult EX-plus medium, or seeded on Corning transwell membranes for differentiation into polarized epithelia with PneumaCult air-liquid interface (ALI) medium at an ALI state as previously described.12,42,43
Formulations of shuttle peptide-peptide, shuttle peptide-protein, and shuttle peptide-Cas RNP
Shuttle peptides were diluted in PBS to achieve final concentrations (5–20 μM) before addition of peptide (DRI-NLS-Cy5) (10 μM), protein cargo (GFP-NLS) (10 μM), or Cas RNP. The Cas RNP was prepared by mixing the Cas nuclease-NLS protein (SpCas9, 2.5 μM and AsCas12a [Cpf1], 1.33 μM) and the sgRNA (gRNA) (2.0 μM) targeting the LoxP sequence, or the mixture of SpCas9 (1.25 μM) and the sgRNAs targeting LoxP (1.0 μM) and SpCas9 1.25 μM and the sgRNAs targeting the ferret CFTR-G551 locus (1.0 μM) for simultaneous editing (Supplementary Table S1), in PBS and incubating at room temperature (RT) for 20–30 min.
For generation of sgRNA duplex for SpCas9 RNP complex, equimolar amounts of CRISPR RNA (crRNA) and trans-activating CRISPR RNA (tracrRNA) were mixed in Duplex Buffer (IDT, Coralville, IA) at final concentrations of 10 μM each, slowly raising the temperature to 95°C, and then cooling the samples to RT in stepwise manner (for annealing) at a rate of −5°C/min before mixing with SpCas9 (2.5 μM).41 The formulation of shuttle peptide-Cas RNP was prepared by mixing equal volumes of Cas RNP complex and PBS-diluted shuttle peptide solution (20 μM). The formulated solution was prepared at the time of delivery and used within 5 min after it was mixed.28
Delivery of reporter protein and Cas RNPs using shuttle peptide formulations in vitro
The shuttle peptide formulations were applied onto ferret airway basal cells and/or polarized (fully differentiated) ferret airway epithelia in vitro. For delivery to proliferation basal cell monolayers and polarized ALI airway epithelia, cells were rinsed twice with PBS and a volume of 50 μL of the shuttle RNP formulation was applied to a well of 96-well plate or the apical surface of 0.6-cm diameter of Transwell membrane and incubated at RT for 15 min before removing. The cells were then rinsed twice with PBS, and incubated in culture media at 37°C, 5% CO2 atmosphere for 30 min to 1 h for protein cargo or peptide cargo delivery, or 2–4 days for genome-editing assays, before they were fixed in 4% paraformaldehyde (PFA) for further analysis.
Delivery of reporter protein/peptide and Cas9 RNP using shuttle peptide formulations to ferret airways in vivo
For in vivo delivery to ferret airways, the formulated shuttle peptide and GFP-NLS, Cas9 RNP, or DRI-NLS-Cy5 fluorescent peptide44 were delivered to airways of 2-week-old ferret kits by intratracheal instillation, or 6–18-month-old adult ferrets, through a laryngotracheal route using MADgic atomization device (MAD720; Teleflex Medical, Morrisville, NC). For in vivo airway delivery, a dose of 1.0 mL/kg body weight formulated shuttle mixture containing GFP-NLS protein, DRI-NLS-Cy5 peptide, or Cas RNP preparations was used.
Animals were anesthetized with ketamine/xylazine by intramuscular injection. Wild-type ferrets were used for transduction of GFP-NLS protein and DRI-NLS-Cy5 peptide, and the efficiency of protein/peptide transduction was evaluated at 1 h after the delivery. ROSA-TG Cre/LoxP reporter transgenic ferrets41 were used for genome editing studies, and the outcome of genome editing was analyzed at 2 weeks after the delivery. The ferret TR and lung tissue were collected, fixed in 4% PFA, and embedded in Tissue-Tek® Optimal Cutting Temperature compound for cryosections. Tissues were sectioned at 6 μm thickness for immunofluorescence analysis by confocal microscopy.
Cell toxicity assay in polarized ferret airway ALI epithelia
The toxicity of shuttle peptides to primary ferret airway epithelial cells was accessed by lactate dehydrogenase (LDH) assay using the CytoSelect™ LDH Cytotoxicity Assay Kit (Cell Biolabs, Inc., San Diego, CA). Polarized primary ferret airway epithelial cells cultured in an ALI state were delivered with the shuttle peptide (S10) alone or co-incubated with proteins (GFP-NLS or RNP) in 50 μL volume for 15 min at RT, and the culture media in the basolateral chamber were collected at 24 h after the delivery. Media collected from untreated cells and cell lysate (1% Triton X-100) served as negative and positive controls for normalization, respectively.
Immunofluorescent localization and morphometric analysis
To immunolocalize the delivered GFP-NLS protein, DRI-NLS-Cy5, or CRISPR/Cas nuclease-mediated genome-edited GFP expression (ROSA-TG ferret) with airway epithelial cell-type specific markers, cryosections of TR and lung tissues were air-dried at RT for 30 min, re-fixed in 4% PFA/PBS for 10 min, and then permeabilized with 0.2% Triton X-100/PBS for 20 min at RT. The sections were incubated with blocking buffer containing 5% donkey serum in PBS for 1 h before being probed with primary antibodies to proteins of interest in dilutant buffer (1% donkey serum, 0.03%Triton X-100, and 1 mM CaCl2 in PBS) at 4°C overnight (Supplementary Table S2).
They were then incubated with appropriate fluorescent dye-conjugated secondary antibodies at RT for 2 h. The slides were extensively washed with PBS for 5 × 5 min, before they were mounted with Aqua-Mount Medium (Richard-Allan Scientific, Kalamazoo, MI). For immunofluorescent staining on whole-mount membranes of ALI cultures, Transwell membranes were fixed in 4% PFA for 30 min at RT and then permeabilized with 0.3% Triton X-100 in PBS for 30 min at RT. Medium containing primary antibody was then applied to the apical surface of the membrane and incubated for 24–48 h at 4°C, followed by incubation with the appropriate secondary antibody at RT for 2 h. Samples were then extensively washed in PBS for 3 × 10 min on a shaker. The stained membranes were cut from the Transwell inserts and mounted on a glass slide using Aqua-Mount.
Nuclei were counterstained with Hoechst 33342 or DAPI (4′,6-diamidino-2-phenylindole) (Invitrogen, Carlsbad, CA). Images were acquired under a Zeiss LSM 880 line-scanning confocal microscope (Carl Zeiss, Germany). All images were captured using the same microscope and identical settings for each condition, and then processed with a FIJI-ImageJ software. Colocalizations of phenotypic markers and transduced protein (GFP-NLS and DRI-NLS-Cy5), or genome-edited cells (membrane-bound GFP) for quantification of colocalized cells were quantified with images collected at 20 × magnification using the Metamorph software multiwavelength cell scoring application. At least five random fields from each section or membrane were acquired, no less than three animals (three sections each) or three ALI membranes were evaluated for each condition. For lung tissues, data were collected from sections containing both large and small ILB airways.
Statistical analysis
Data analyses were performed using the Prism v.9.0 software (GraphPad Software, San Diego, CA). ANOVA with Tukey's multiple comparison test was conducted to analyze differences in mean values between groups. Except where indicated otherwise, data were presented as mean ± standard deviation. p Values of <0.05 were considered statistically significant.
Statement on ethics
The animal protocol used in this study conformed to NIH standards. All protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Iowa. All ferrets were purchased from Marshall Farms (North Rose, NY) and housed in the animal facilitate at the University of Iowa. All animal experiments were performed at the Iowa National Ferret Research and Resource Center (Iowa City, IA).
RESULTS
Amphiphilic shuttle peptides efficiently deliver protein to ferret airway epithelial cells in vitro
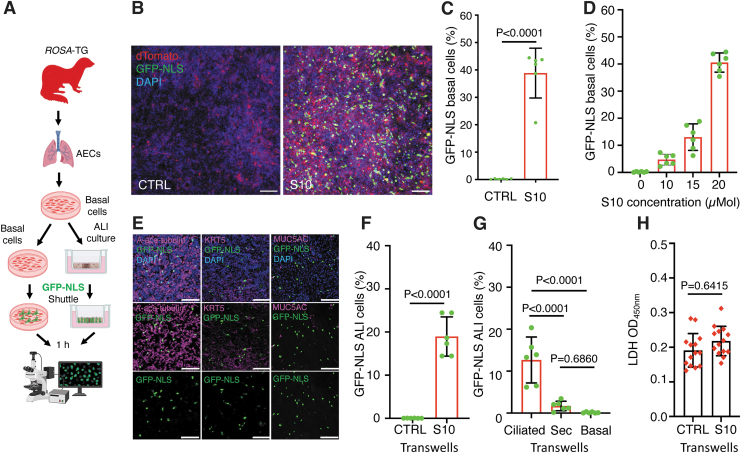
Previous proof-of-concept studies with engineered amphiphilic shuttle peptides have demonstrated the efficient delivery of proteins and Cas RNPs into multiple cell types,32 including difficult to transfect primary natural killer cells,27 polarized human airway epithelial cells in vitro, and mouse airway epithelial cells in vivo.28 To extend potential applications of amphiphilic shuttle peptide to genome editing in the lung of larger animal models, we tested the efficiency of 10 to 20 μMol of S10 shuttle peptide, one of the tested peptides with most potential for protein delivery to mouse airway epithelia.28 S10 is a synthetic amphiphilic peptide (KWKLARAFARAIKKLGGSGGGSYARALRRQARTG) engineered and optimized from the combination of an endosomal leakage domain (ELD highlighted in italics) with a CPP (CPP highlighted in bold) joined by a glycine-serine linker.
The result showed that the S10 peptide was able to mediate GFP protein delivery to ferret basal cells, with the concentration of 20 μM yielding 40.5% cell delivery in the absence of obvious toxicity (Fig. 1A–D and data not shown). Next, we sought to examine whether S10 enabled efficient delivery of protein into differentiated ferret airway epithelia grown at an ALI (Fig. 1A). The S10 shuttle delivered GFP into multiple epithelial cell types, including ciliated cells and secretory cells, as determined by immunolocalization of GFP and cell-specific markers α-tubulin (ciliated cell) and MUC5AC (secretory cell), although the efficiency was lower than in monolayer proliferating basal cells (Fig. 1E, F).
Figure 1.
Shuttle peptide-mediated GFP delivery to ferret airway epithelial cells in vitro. (A) Schematic showing the experimental workflow for the delivery of GFP protein to ferret airway epithelial cells with S10 shuttle peptide. Primary airway epithelial cells were isolated from ferret TRs and propagated in PneumaCult Ex Plus medium. Basal cells at passage 4–6 were either used directly for delivery studies as submerged monolayer culture or further polarized on Transwell membranes at ALI. The delivery mix of shuttle peptide/nuclear-targeted GFP protein (GFP-NLS) was applied on the basal cells or apical surface of ALI epithelia at RT for 15 min and then rinsed in PBS. Cultures were returned to the incubator for an additional hour before being evaluated for GFP-positive cells. (B, C) Representative images (B) and efficiency (C) of GFP-NLS (10 μM) delivery in ferret airway basal cells by S10 shuttle peptide at a concentration of 20 μM. (D) Dose-dependent S10 shuttle-mediated delivery of GFP-NLS (10 μM) to ferret airway basal cells at S10 concentrations of 0–20 μM. (E) Representative images of GFP immunolocalization with specific markers (magenta) of ciliated cells (α-tubulin, left panel), basal cells (KRT5, middle panel), or secretory (Sec) cells (MUC5AC, right panel) in polarized ferret ALI cultures delivered with GFP-NLS (10 μM) with the S10 shuttle peptide (20 μM). (F) The efficiency of S10-mediated delivery in ALI cultures quantified as the percentage of DAPI nuclei positive for GFP-NLS. (G) S10 shuttle-mediated delivery of GFP-NLS in ciliated cells, secretory cells (Sec), and basal cells. (H) In vitro toxicity of S10/GFP-NLS (20 μM/10 μM) complexes on polarized ferret airway epithelia at 24 h postdelivery. Cytotoxicity was assessed by LDH released into basolateral compartment and compared to undelivered cultures. Nuclei were counterstained with DAPI in B and E. Bars equal 100 μm in B and E. Data in C, D, F, G, and H represent the mean ± SD from three independent replicated experiments (N = 6 ALI cultures). Significant differences were determined evaluated by one-way ANOVA followed by Tukey's multiple comparisons test (G) or two-tailed Student's t-test (F, H). ALI, air-liquid interface; GFP-NLS, green fluorescent protein-nuclear localization signal; LDH, lactate dehydrogenase; RT, room temperature; SD, standard deviation; TR, trachea.
Ciliated cells were the major cell population delivered (12.7%) (Fig. 1G), while only a small fraction of MUC5AC-positive secretory cells were delivered (1.7%) (Fig. 1G). Only rare KRT5-positive basal cells were apically delivered in ferret ALI cultures (Fig. 1G). The S10 shuttle/GFP protein showed undetectable toxicity to ferret airway epithelia, as accessed by the LDH assay (Fig. 1H).
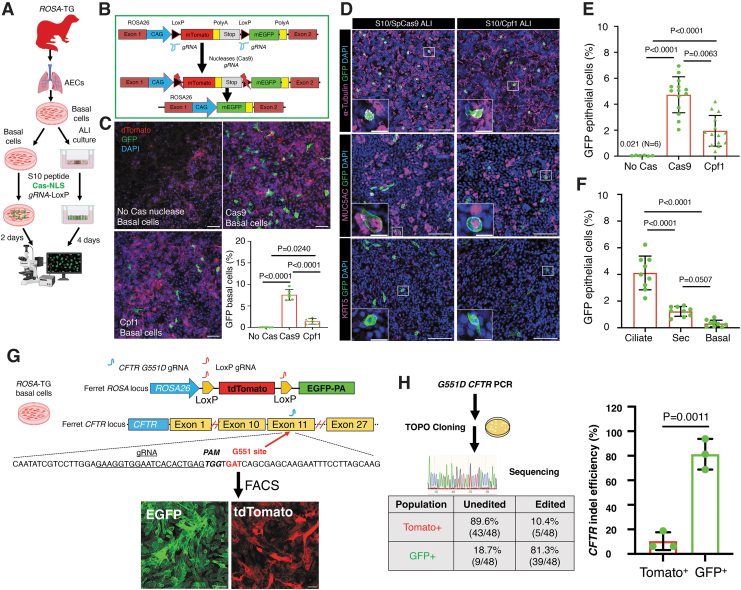
S10 shuttle peptide enables delivery of Cas RNPs to ferret airway epithelia in vitro
To test the potential of shuttle peptides to deliver Cas RNPs capable of gene editing in ferret airway epithelial cells, RNPs comprising gRNA to LoxP sites and Cas9 or Cpf1 (AsCas12a) were delivered by S10 shuttle to airway basal cells and differentiated ALI cultures derived from ROSA-TG Cre/LoxP reporter ferrets (Fig. 2A). This transgenic model expresses a floxed membrane-bound Tomato (mT) that switches to express membrane-bound EGFP (mG) in the presence of Cre or Cas/LoxP-gRNA RNP (Fig. 2B).41 The S10 shuttle delivery of SpCas9 RNP or Cpf1 RNP to ferret airway basal cells resulted in genome editing efficiencies of 7.57% and 1.41%, respectively, as accessed by EGFP conversion at ROSA locus (Fig. 2C).
Figure 2.
S10 shuttle peptides deliver SpCas9 and Cpf1 ribonucleoproteins to ferret airway epithelial cells in vitro. (A) Schematic showing the experimental workflow for delivery of ribonucleoproteins (RNPs) to ferret basal cells and differentiated airway epithelium using the S10 shuttle peptide. Primary ROSA-TG Cre reporter ferret airway basal cells and polarized ALI cultures were delivered with the indicated Cas RNPs (SpCas9, 2.5 μM; Cpf1, 1.33 μM) containing LoxP-gRNA (2.0 μM) and S10 peptide (20 μM) for 15 min. Cultures were then washed and cultured for an additional 2 days (basal cells) or 4 days (ALI cultures) before being evaluated for genome editing by accessing EGFP+ cells. (B) Schematic illustration of the strategy for Cas nuclease RNP-mediated genome editing of LoxP sites at the ROSA-TG reporter locus in ferret airway epithelial cells. Cells were delivered with the S10 shuttle peptides complexed with Cas RNP and LoxP-gRNAs. Cas cleavage at the LoxP sites leads to the conversion of genome-edited cells from Tomato+ to EGFP+. (C) S10/Cas RNP delivery to ferret airway basal cells. Basal cells were either mock delivered with no Cas protein (top left) or delivered with SpCas9 (top right) or Cpf1 (bottom left). The genome editing efficiency was accessed as the percentage of EGFP+ cells from N = 6 donor ferrets (bottom right panel). (D) S10/Cas RNP delivery to differentiated ferret airway epithelia in ALI cultures. Representative images of immunolocalization for EGFP protein and cell-type markers for ciliated cells (α-tubulin, top panel), secretory (Sec) cells (MUC5AC, middle panel), and basal cells (KRT5, bottom panel) in epithelia delivered with S10/SpCas9 (left panel) and S10/Cpf1 (right panel) RNPs. Insets show magnifications of the boxed area in the corresponding image. (E) The efficiency of genome editing was accessed as the percentage of EGFP+ cells in ALI epithelia delivered with S10/SpCas9 and S10/Cpf1 RNPs (N = 14 ALI cultures derived from three donors). (F) The portion of genome-edited epithelial cell types in ferret ALI epithelia delivered with S10/SpCas9 (N = 9 ALI cultures derived from three donors). (G, H) Efficiency of S10-delivered CRISPR/Cas9-mediated genome editing at the CFTR-G551 locus (Gene ID: 101672484) in ferret ROSA-TG airway basal cells. ROSA-TG reporter ferret basal cells were transfected with RNP containing Cas9 protein and sgRNAs to LoxP and the CFTR-G551 locus. Following transfection, the LoxP sgRNA directs Cas9-mediated cleavage at the LoxP sites resulting in conversation of Tomato to EGFP expression. Indels at the CFTR loci were enriched by fluorescence-activated cell sorting (FACS) of EGFP+ cells (G). The CFTR indel efficiency was assessed by TOPO cloning and Sanger sequencing gave a percentage of clones carrying CFTR indel sequence(s) in Tomato+ cells (lacking LoxP-mediated conversion) and EGFP+ cells (including LoxP-mediated conversion) (H). Upper left panel: schematic workflow for the TOPO cloning assay. Lower left panel: the average indel efficiency of three donor ferrets assessed by Sanger sequencing is presented in a table. Right panel: the indel efficiency of three donors was plotted in bar graphs (N = 3). Nuclei were counterstained with DAPI in C and D. Bars equal 100 μm in C and D; 20 μm in insets of D. Data in C, E, F, and H represent the mean ± SD from three independent replicated experiments. Statistical significance was evaluated by one-way ANOVA followed by Tukey's multiple comparisons test, or two-tailed Student's t-test (H). FACS, fluorescence-activated cell sorting; RNPs, ribonucleoproteins.
Similar to that seen in basal cells, the S10 peptide also enabled RNP delivery into differentiated ferret airway cultures and yielded genome editing in multiple epithelial cell types, including ciliated cells and secretory cells, as ascertained by immunolocalization with cell-specific markers (Fig. 2D). The overall genome editing efficiencies were greater for Cas9 RNP (4.73%) than for Cpf1 RNP (1.95%) in ferret ALI cultures (Fig. 2E). Among the Cas9 RNP genome-edited cell types, 4.11% ± 1.27% ciliated cells and 1.23% ± 0.37% secretory cells were genome edited, and basal cells were rarely genome edited in ALI cultures (Fig. 2F).
Next, we investigated if the gene editing efficiency at ROSA-TG Cre/LoxP reporter locus is a good indicator of editing at locus of interest, such as ferret CFTR gene (Gene ID: 101672484). Cas9 RNPs containing gRNAs targeting LoxP and the ferret CFTR-G551 locus were co-delivered using S10 peptide into ferret basal cells containing ROSA-TG Cre/LoxP reporter. Cells with biallelic LoxP-targeted EGFP conversion or nonconverted Tomato+ cells were isolated by flow cytometry. The ferret CFTR-G551 locus was amplified by PCR in EGFP+ and Tomato+ cell populations and the indel frequency was determined by TOPO cloning followed by Sanger sequencing.
We detected in isolated EGFP+ and Tomato+ cells indel efficiencies of 81.3% and 10.4%, respectively (Fig. 2H and Supplementary Fig. S1), while the percentage of clones with unedited target sequence was 18.7% and 89.6% in EGFP+ and Tomato+ cells, respectively (Fig. 2H and Supplementary Fig. S1). These results suggest that the editing of ROSA-TG Cre/LoxP reporter shows good correlation with ferret CFTR editing using S10 peptide.
S10 shuttle peptide delivers protein and peptide into ferret airway epithelia in vivo
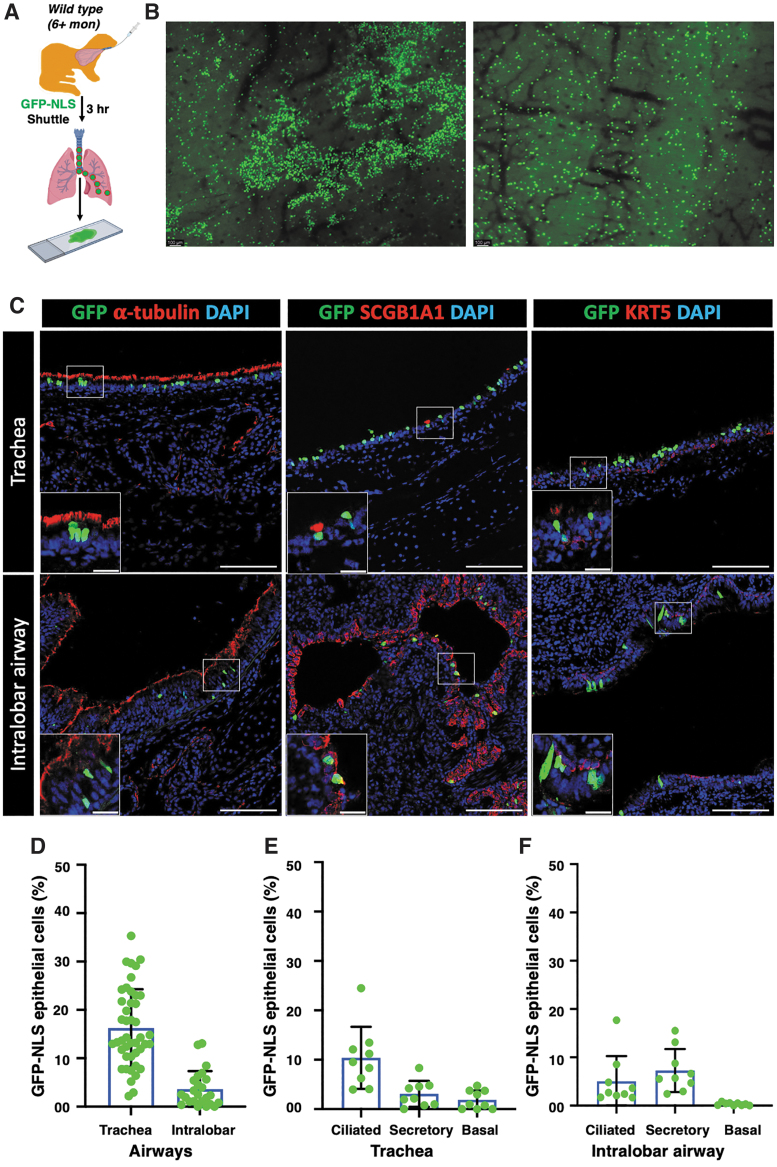
The S10 peptide shuttle has been demonstrated to efficiently deliver GFP and RNPs to mouse airways in vivo.28 To examine the efficiency for deliver to ferret airways, a single dose of S10/GFP-NLS or S10/DRI-NLS-Cy5 was intratracheally administered to adult ferrets and 2-week-old ferret kits, and the distribution of nuclear-targeted GFP or DRI-NLS-Cy5 in airway epithelia was evaluated.
First, S10/GFP-NLS was intratracheally administered to adult ferrets (Fig. 3A) and evaluated at 3 h postdelivery. En face epifluorescence images of the tracheal surface epithelium showed widespread GFP signal in adult ferrets (Fig. 3B). Immunolocalization further confirmed that GFP colocalized with markers of ciliated (α-tubulin), secretory (SCGB1A1), and basal (KRT5) cells, the three main epithelial cell types in the TR and ILB airway epithelia (Fig. 3C). Quantitative analysis demonstrated more GFP+ cells in tracheal epithelia (16.2%) compared to that of ILB airway epithelia (3.6%) (Fig. 3D). In the adult TR, GFP+ cells were observed in 10.4% of ciliated cells, 3.1% of secretory cells, and 1.9% of basal cells (Fig. 3E). By contrast, in the ILB airways, GFP delivery of secretory cells (7.3%) was greater compared with ciliated cells (5.0%) and basal cells were infrequently delivered (0.31%) (Fig. 3F).
Figure 3.
S10 shuttle peptide-mediated GFP protein delivery to adult ferret airway epithelia in vivo. (A) Schematic showing the experimental workflow for the delivery of GFP protein to ferret airways with S10 shuttle peptide. The GFP-NLS protein (10 μM) was delivered with S10 (20 μM) peptide intratracheally to wild-type ferret airways at a dose of 1.0 mL/kg body weight. The delivery efficiency was evaluated at 3 h postdelivery. (B) Representative immunofluorescent images showing nuclear GFP expression in the surface airway epithelium of whole mount TRs following S10/GFP-NLS delivery. (C) Representative immunofluorescent images of tissue sections showing GFP protein with cell-type markers for ciliated cells (α-tubulin), secretory cells (SCGB1A1), and basal cells (KRT5) in ferret tracheal epithelia (top panels) and ILB airway epithelia (bottom panels) following S10/GFP-NLS delivery. Insets show magnifications of the boxed area in the corresponding image. (D) The efficiency of S10/GFP-NLS delivery in tracheal and ILB airway epithelium. (E, F) The frequencies of S10/GFP-NLS delivery of the indicated epithelial cell types in ferret TR (E) and ILB airways (F). Nuclei were counterstained with DAPI in C. Bars equal 100 μm (C) and 20 μm (C insets). Data in D–F represent the mean ± SD (N = 9–44 randomly quantified fields, from three ferrets). Individual count areas are represented by distinct data points in bars of each graph. ILB, intralobar.
A comparable efficiency of S10-mediated GFP delivery was also observed in 2-week-old ferret kits with 15.8% of the tracheal and 10.6% of the ILB GFP+ airway epithelial cells (Supplementary Fig. S2A–C). Cell-type distribution of delivered cells in the TR of kits was also similar to that in adult ferrets, with 11.0% of ciliated cells, 5.8% of secretory cells, and 0.39% of basal cells being GFP+ (Supplementary Fig. S2D).
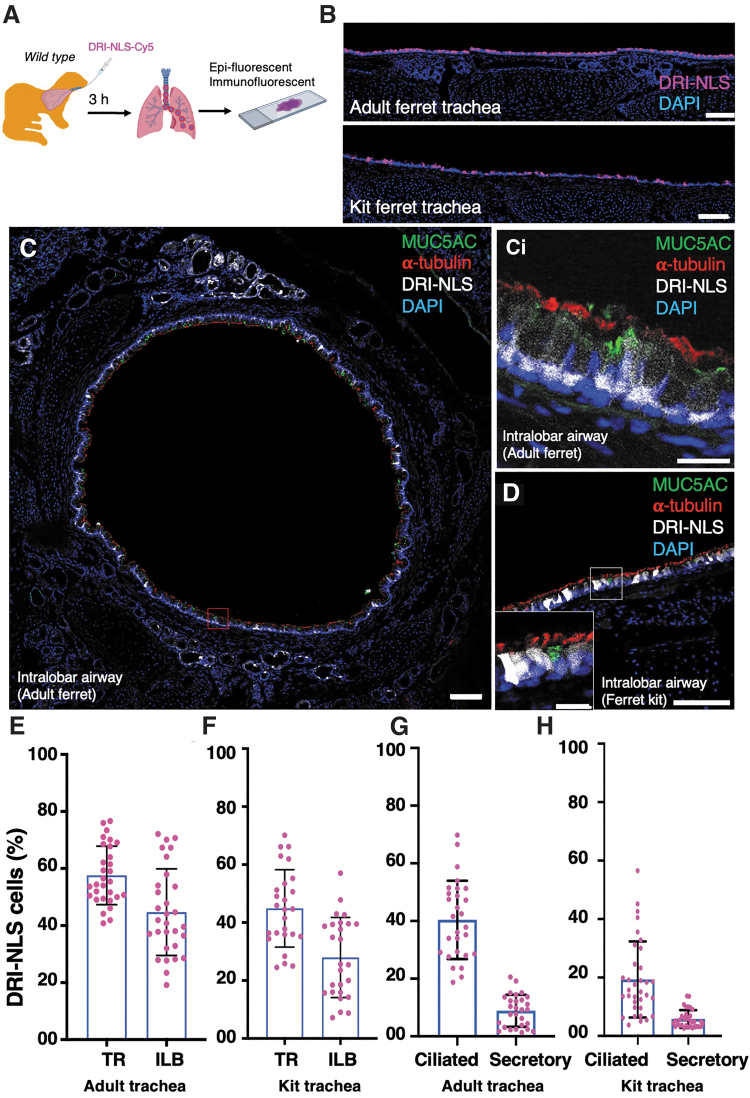
The S10 shuttle also enabled efficient delivery of the peptide cargo DRI-NLS-Cy5 into airway epithelial cells of both adult and kit ferret airways at levels several fold higher than with GFP protein (Fig. 4A–D). The DRI-NLS-Cy5 is a Cy5-labeled synthetic peptide composed of an NLS sequence made of d-amino acids in a retro-inverso sequence to conserve the amino acid residue disposition of the NLS. DRI-NLS peptide delivery was greatest in the TR versus ILB airways in both adult (57.59% vs. 44.71%) and kit (44.89% vs. 27.95%) ferrets (Fig. 4E, F). Consistent with the delivery of GFP protein, more ciliated cells were delivered by the fluorescent peptide compared to secretory cells (Fig. 4G, H).
Figure 4.
S10 shuttle delivery of DRI-NLS-Cy5 peptide to ferret airway epithelia in vivo. (A) Schematic showing the experimental workflow for the S10-mediated delivery of Alexa Fluor Cy5 conjugated DRI-NLS (DRI-NLS-Cy5) peptide into ferret airways. The DRI-NLS-Cy5 peptide (10 μM) was delivered with S10 (20 μM) intratracheally to wild-type ferret airways at a dose of 1.0 mL/kg body weight. The delivery efficiency was evaluated at 30 min postdelivery. (B) Representative fluorescent images of tissue sections showing DRI-NLS peptide (magenta) in adult (top panel) and kit (bottom) ferret tracheal epithelia. (C, D) Representative immunofluorescent image colocalization of DRI-NLS-Cy5 peptide (gray) with cellular markers for ciliated cells (α-tubulin, red) and goblet cells (MUC5AC, green) in ILB airway epithelia of adult (C, ci) and kit (D) ferret lungs. (ci) Inset shows magnification of the red boxed area in (C). (E, F) The efficiency of S10/DRI-NLS peptide delivery to adult (E) and kit (F) TR. (G, H) The frequencies of S10/DRI-NLS peptide delivery to the indicated epithelial cell types in adult (G) and kit (H) TR. Bars equal 100 μm (B–D) and 20 μm (inset ci). Data in (E–H) represent the mean ± SD (N = 26–34 randomly quantified fields, from three ferrets per group). Individual count areas are represented by distinct data points in bars of each graph.
Amphiphilic peptide shuttle delivers Cas9 RNP to airway epithelia for genome editing in ferrets
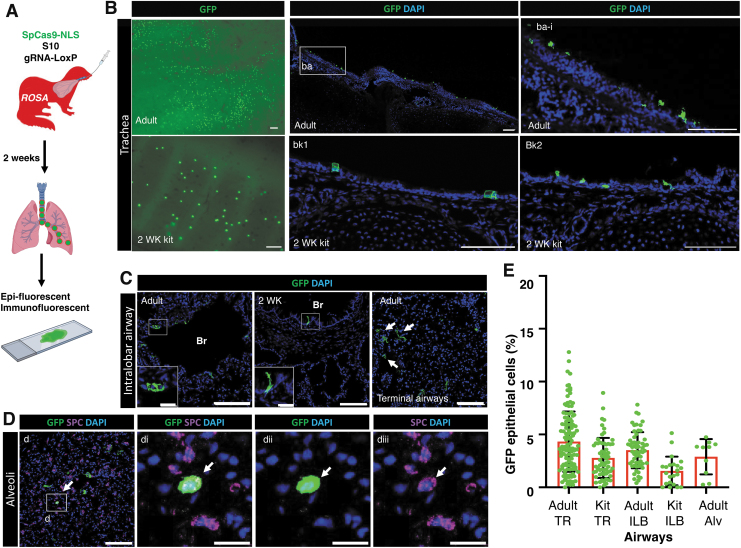
We next investigated the efficiency of genome editing mediated by S10 shuttle-delivered SpCas9 RNP in ferret airways in vivo using ROSA-TG adult ferrets and 2-week-old kits.41 Intratracheal delivery of S10/Cas9 RNP complexes to ROSA-TG ferrets led to the conversion of membrane-bound Tomato to EGFP expression (Fig. 5A). At 2 weeks postdelivery of S10/Cas9 RNP to adult and kit ferrets, the frequency of EGFP+ cells in the surface airway epithelia of the TR and ILB conducting airways ranged from 1.6% to 4.3% of cells (Fig. 5B–E). Of note, genome-edited EGFP cells were also observed in the terminal airway epithelia and alveoli (Fig. 5C), including alveolar type II cells that expressed cell-type marker surfactant protein C (Fig. 5D).
Figure 5.
S10 shuttle peptide delivery of SpCas9 RNP enables genome editing of ferret airway epithelia in vivo. (A) Schematic showing the experimental workflow for evaluating S10/SpCas9 RNP gene editing efficiency in vivo. ROSA-TG transgenic adult ferrets and 2-week-old kits received S10/SpCas9 RNP intratracheally and genome editing efficiency was evaluated 2 weeks later by accessing the percentage of EGFP+ epithelial cells. Complexes carried SpCas9 RNPs (Cas:gRNA-LoxP; 1.33 μM:2 μM) and S10 peptide (20 μM) at a dose of 1.0 mL/kg body weight. (B) Representative fluorescent images from adult (top panels) and kit (bottom panels) ferrets showing EGFP+ cells in whole mount TRs (left panels) and sections of the TR. Magnification of the boxed region (ba-i) is shown in (ba). (C) Representative fluorescent images of intrapulmonary airways showed genome-edited EGFP+ cells in bronchial (Br) epithelial cells of adult (left panel) and kit (middle panel) lungs, and terminal airways (arrows) of adult ferret lungs. (D) Representative fluorescent images of EGFP+ cells immunolocalized with the alveolar type II cell marker SPC (arrow) in lungs of ferrets delivered with S10/SpCas9 RNP. (di–diii) Enlarged image of boxed area in (D) with dii and diii having dual color channels. (E) The efficiency of genome editing was accessed as the percentage of EGFP+ cells in ferret TR and ILB airway epithelia. Nuclei were counterstained with DAPI in B–D. Bars equal 100 μm (main panels in B–D) and 20 μm (C-insets, D-di–diii). Data in E represent the mean ± SD (N = 10–118 randomly quantified fields, from three ferrets for each group). Individual count areas are represented by distinct data points in bars of each graph. Alv, alveoli; SPC, surfactant protein C.
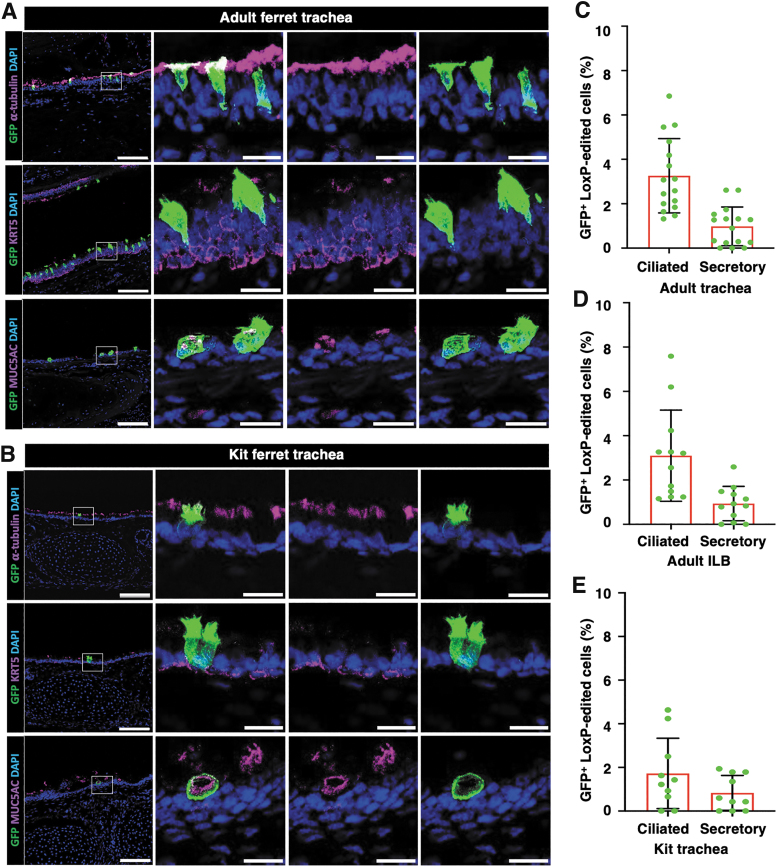
The majority of gene-edited cells were ciliated cells and secretory cells in tracheal epithelia (Fig. 6A, B) and ILB airway epithelia (Supplementary Fig. S3) of both adult and kit ferrets. The average gene editing efficiencies of ciliated cells versus secretory cells were (3.26% vs. 1.00%), (3.10% vs. 0.97%), and (1.77% vs. 0.83%) in ferret epithelia of adult TR (Fig. 6C), adult ILB airways (Fig. 6D), and kit TR (Fig. 6E), respectively. We found no GFP+ genome-edited basal cells (KRT5+) (Fig. 6A, B, and Supplementary Fig. S3A, B).
Figure 6.
S10 shuttle peptide delivery of SpCas9 RNP enables in vivo genome editing of ferret ciliated and secretory cells, but not basal cells. ROSA-TG transgenic adult ferrets and 2-week-old kits received S10/SpCas9 RNP intratracheally and genome editing efficiency was evaluated 2 weeks later by accessing the percentage of EGFP+ epithelial cells. Complexes carried SpCas9 RNPs (Cas:gRNA-LoxP; 1.33 μM:2 μM) and S10 peptide (20 μM) at a dose of 1.0 mL/kg body weight. (A, B) Representative immunofluorescent images of adult (A) and kit (B) tissue sections showing EGFP+ colocalization with cell-type markers for ciliated cells (α-tubulin), secretory cells (MUC5AC), and basal cells (KRT5) in ferret tracheal epithelia following S10/SpCas9 RNP delivery. Insets to the right of the main panel show magnifications of the boxed area (both triple and dual color channels are shown). (C, D) The frequency of genome-edited ciliated cells and secretory cells in adult ferret TR (C) and ILB (D) airways. (E) The frequency of genome-edited ciliated cells and secretory cells in kit ferret TR. Nuclei were counterstained with DAPI in A and B. Bars equal 100 μm (main panels in A, B) and 20 μm (Insets in A, B). Data in C–E represent the mean ± SD (N = 10–16 randomly quantified fields, from three ferrets for each group). Individual count areas are represented by distinct data points in bars of each graph.
DISCUSSION
The domestic ferret has proven to be an excellent model for human lung diseases, including influenza viral infection,45 CF,33,35,36 alpha-1 antitrypsin deficiency40 and chronic obstructive lung disease.37 Thus, the development of gene editing technologies in this species provides a preclinical path for treating inherited lung diseases. In this study, we show that the amphiphilic shuttle peptide S10 is capable of delivering GFP protein, peptide, and Cas RNPs to ferret airway epithelia in vitro and in vivo.
Using the novel ROSA-TG transgenic ferret model, we demonstrate that the S10 shuttle peptide achieves editing efficiency using SpCas9 RNP for gene editing at the ROSA locus in vitro and in vivo, and ferret CFTR gene in vitro. These findings build on the success of amphiphilic peptide for gene editing in human airway epithelia in vitro and mouse lung in vivo.27,28,46 The S10 peptide yielded efficient delivery in this type of cells in a dose-dependent manner with undetectable toxicity (Fig. 1). Together with our results, these studies suggest that shuttle peptides are safe and efficient vectors for developing disease-specific gene editing approaches for lung diseases, such as CF.
In differentiated airway epithelia, both in vitro (Fig. 1) and in vivo (Fig. 3), S10-mediated GFP delivery was approximately half as efficient as delivery to proliferating airway basal cells. Of note, gene editing efficiencies using S10-mediated delivery of SpCas9 RNP at the ROSA locus were fairly similar between differentiated ALI cultures (Fig. 2), and airway epithelium in vivo (Fig. 5).
Gene editing efficiency in differentiated airway epithelium in vitro and in vivo was ∼30% the efficiency observed with GFP delivery. This lower gene editing frequency could be due to the reduced delivery efficiency of the approximately fivefold larger SpCas9 protein and/or the requirement for two simultaneous LoxP breaks required to activate the Cre reporter. Given that S10 peptide delivery of the DRI-NLS peptide in vivo gave rise to the highest percentage of delivered cells (Fig. 4), size of the cargo likely has a significant impact on delivery efficiency. The net negative charge of the gRNA used to form the RNP could also be a major factor influencing the delivery activity of the shuttle peptide, as observed in previous studies.28
Our studies showed that S10/Cas9 RNP yielded more efficient gene editing at LoxP sites than S10/Cpf1 (AsCas12a) RNP in ferret airway basal cells and polarized epithelia (Fig. 2). This finding was the opposite of the previous gene editing studies at the CFTR locus using polarized human airway epithelia, where S10/Cpf1 RNP resulted in a great number of targeted indels in comparison to S10/Cas9 RNP.28 In addition to the difference in intrinsic activities between Cas9 and Cpf1, several reasons may also explain the discrepancy in gene editing efficiency. First the different net negative charge densities of LoxP gRNA for Cas9 and Cpf1 may have influenced the RNP delivery efficiency, despite comparable molecular size of Cas9 (170 kDa) and Cpf1 (156 kDa). For example, the Cas9 utilizes a larger gRNA composed of crRNA and tracrRNA (∼100 nt), while Cpf1 utilizes a shorter ∼42 nt crRNA, which may influence the gene editing efficiency.28,47
Second, the gRNA target sequences to LoxP between these two Cas nucleases may also contributed to the difference in editing efficiencies, as the Cas9 gRNA to LoxP was designed with optimal PAM sequence. Since an optimal Cpf1 PAM at the LoxP site was not present, we used a previously described suboptimal CTTC PAM in gRNA.5,28,48 Third, chromatin conformations adjacent to the target sequence can vary for different genes and/or animal species and may also influence editing outcomes.49–51 In this context, the chromatin structure at the ROSA locus of the ferret genome is likely different from that of CFTR gene in human genome.
Gene editing approaches to treat inherited diseases can provide durable therapy if the genetic alterations occur in stem cells of the target organ. In humans and ferrets, basal cells are the predominant stem cells in the surface airway epithelium of the large conducting cartilaginous airways, whereas club secretory cells in the terminal and respiratory bronchioles are multipotent stem cells in this compartment.15 By contrast, in all levels of mouse ILB airways, the club secretory cells are the main stem cell population,15 emphasizing the need for alternative animal models to translate gene editing approaches to humans.
Our in vitro studies in differentiated ferret airway epithelia and in vivo studies in ferrets demonstrated little to no gene editing in basal cells. Given that basal cells lack an apical membrane, accessibility to S10 shuttle complexes is expected to be very limited. Thus, the application of RNP delivery for stem cell gene editing of the proximal conducting airways will require methods that promote access of complexes to basal cells. One such approach used in virus-mediated transduction includes the use of mild detergent such as lysophosphatidylcholine to temporarily break down tight junctions.52,53
In this study, shuttle peptide and cargo formulations were delivered into adult ferret lungs intratracheally using a MADgic atomization device in proof-of-concept studies. This approach comes with limitations in the distribution of complexes within the lung, as evident by the rather large variation in delivery efficiencies within a given animal. Future studies using co-administered DRI-NLS-Cy5 with RNPs may aid in referencing the amount of fluid delivered to various regions of the lung to obtain a delivery vs gene editing efficiency index. Ultimately, methods of uniform nebulization of shuttle peptides to the entire lung will be required to evaluate the ability of gene editing to reverse or slow disease progression in lung disease models.
Limitations of these studies include the low gene editing frequencies. Overexpression of CFTR in 6–10% of CF airway epithelia has been previously shown to correct chloride transport properties to WT levels.54 However, CFTR gene editing would likely require a greater percentage of cell complementation since endogenous levels of CFTR expression would be the outcome and ciliated cells do not express substantial amount of CFTR protein. Approaches to enhance the genome editing efficiency could include increasing the shuttle concentration, using nebulization formulations for delivery, and repeating dosing.
Although the S10 peptide is not compatible with negatively charged and large DNA template, S10-mediated delivery of Cas nuclease or Cas RNP could be a robust approach for genome and base editing approaches. Such an approach could be translated into genetic therapies for CF lung disease targeting single-base mutations and/or splicing mutations. Furthermore, a genome editing enrichment approach using transgenic ROSA-TG CF ferrets would allow for more accurate quantification of CFTR genome editing by fate mapping transfected cells in vivo using EGFP conversion at the Cre-reporter locus. Using such an approach, methods and formulation improvements could lead to enhanced gene editing efficiencies.
Collectively, we demonstrate that S10 amphiphilic shuttle peptide delivers peptides, proteins, and Cas nuclease RNPs to ferret airway epithelia in vitro and in vivo to achieve gene editing and expand the CRISPR genome editing toolbox for translational medicine in larger animal models. However, many hurdles remain before clinical application of this technology for the delivery of gene editors. Among them are improvements in genome editing efficiency and methods of targeting airway basal stem cells, although the turnover time of normal airway epithelial cells is about 1 year, repeating dosing of Cas RNPs remains a significant safety concern in clinical application. Therefore, genome editing in basal stem cells may be required to improve durability of treatments of pulmonary disease such as CF.
Supplementary Material
AUTHORs' CONTRIBUTIONS
Conceptualization, X.L., J.F.E. and D.G.; methodology, M.L. and J.M.; investigation, M.L., J.M., S.W., and X.L.; data analysis, M.L., J.M., and X.L.; resources, X.C. and D.J.B.; writing—original draft preparation, M.L., J.M., and X.L.; writing—review and editing, D.G., X.C., X.L., and J.F.E.; funding acquisition, J.F.E. All authors have read and agreed to the published version of the article.
INSTITUTIONAL REVIEW BOARD STATEMENT
The animal study protocol was approved by the Institutional Institutional Animal Care and Use Committee of the University of Iowa (UI IACUC# 1071945).
DATA AVAILABILITY STATEMENT
All data generated or analyzed during this study are included in this published article.
AUTHOR DISCLOSURE
D.G., and X.C. are employees of Feldan Therapeutics. D.G. holds equity in Feldan Therapeutics. D.G. has filed patent applications and is inventor of patents related to the Shuttle peptide technology, which are assigned to Feldan Bio Inc. All other authors have nothing to disclose.
FUNDING INFORMATION
NHLBI: R01 HL165404 (to J.F.E.), P01 HL152960 (to J.F.E.), and 75N92019C00010 (to J.F.E.); NIDDK: P30 DK054759 (to J.F.E.); Cystic Fibrosis Foundation ENGELH20XX2 (to J.F.E.); NHLBI-Contract P01 HL158506 to X.L.
SUPPLEMENTARY MATERIAL
REFERENCES
- 1. Cring MR, Sheffield VC. Gene therapy and gene correction: Targets, progress, and challenges for treating human diseases. Gene Ther 2022;29:3–12. [DOI] [PubMed] [Google Scholar]
- 2. Sayed N, Allawadhi P, Khurana A, et al. Gene therapy: Comprehensive overview and therapeutic applications. Life Sci 2022;294:120375. [DOI] [PubMed] [Google Scholar]
- 3. Li Y, Glass Z, Huang M, et al. Ex vivo cell-based CRISPR/Cas9 genome editing for therapeutic applications. Biomaterials 2020;234:119711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lohia A, Sahel DK, Salman M, et al. Delivery strategies for CRISPR/Cas genome editing tool for retinal dystrophies: Challenges and opportunities. Asian J Pharm Sci 2022;17:153–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Taha EA, Lee J, Hotta A. Delivery of CRISPR-Cas tools for in vivo genome editing therapy: Trends and challenges. J Control Release 2022;342:345–361. [DOI] [PubMed] [Google Scholar]
- 6. Ferdosi SR, Ewaisha R, Moghadam F, et al. Multifunctional CRISPR-Cas9 with engineered immunosilenced human T cell epitopes. Nat Commun 2019;10:1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Behr M, Zhou J, Xu B, et al. In vivo delivery of CRISPR-Cas9 therapeutics: Progress and challenges. Acta Pharm Sin B 2021;11:2150–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yang W, Yan J, Zhuang P, et al. Progress of delivery methods for CRISPR-Cas9. Expert Opin Drug Deliv 2022;19:913–926. [DOI] [PubMed] [Google Scholar]
- 9. Asmamaw Mengstie M. Viral vectors for the in vivo delivery of CRISPR components: Advances and challenges. Front Bioeng Biotechnol 2022;10:895713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yin H, Song CQ, Dorkin JR, et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol 2016;34:328–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liang SQ, Walkey CJ, Martinez AE, et al. AAV5 delivery of CRISPR-Cas9 supports effective genome editing in mouse lung airway. Mol Ther 2022;30:238–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yan Z, Vorhies K, Feng Z, et al. Recombinant adeno-associated virus-mediated editing of the G551D cystic fibrosis transmembrane conductance regulator mutation in ferret airway basal cells. Hum Gene Ther 2022;33:1023–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu X, Zhao Z, Wu F, et al. Tailoring hyperbranched poly(beta-amino ester) as a robust and universal platform for cytosolic protein delivery. Adv Mater 2022;34:e2108116. [DOI] [PubMed] [Google Scholar]
- 14. Krishnamurthy S, Traore S, Cooney AL, et al. Functional correction of CFTR mutations in human airway epithelial cells using adenine base editors. Nucleic Acids Res 2021;49:10558–10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu X, Driskell RR, Engelhardt JF. Stem cells in the lung. Methods Enzymol 2006;419:285–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wan T, Ping Y. Delivery of genome-editing biomacromolecules for treatment of lung genetic disorders. Adv Drug Deliv Rev 2021;168:196–216. [DOI] [PubMed] [Google Scholar]
- 17. McLachlan G, Alton E, Boyd AC, et al. Progress in respiratory gene therapy. Hum Gene Ther 2022;33:893–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Park J, Choe S. DNA-free genome editing with preassembled CRISPR/Cas9 ribonucleoproteins in plants. Transgenic Res 2019;28:61–64. [DOI] [PubMed] [Google Scholar]
- 19. Genish I, Gabay B, Ruban A, et al. Electroporation-based proteome sampling ex vivo enables the detection of brain melanoma protein signatures in a location proximate to visible tumor margins. PLoS One 2022;17:e0265866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Haddadzadegan S, Dorkoosh F, Bernkop-Schnurch A. Oral delivery of therapeutic peptides and proteins: Technology landscape of lipid-based nanocarriers. Adv Drug Deliv Rev 2022;182:114097. [DOI] [PubMed] [Google Scholar]
- 21. Banskota S, Raguram A, Suh S, et al. Engineered virus-like particles for efficient in vivo delivery of therapeutic proteins. Cell 2022;185:250–265.e216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Petkovic I, Bischof J, Kocher T, et al. COL17A1 editing via homology-directed repair in junctional epidermolysis bullosa. Front Med (Lausanne) 2022;9:976604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mirjalili Mohanna SZ, Djaksigulova D, Hill AM, et al. LNP-mediated delivery of CRISPR RNP for wide-spread in vivo genome editing in mouse cornea. J Control Release 2022;350:401–413. [DOI] [PubMed] [Google Scholar]
- 24. Kumari A, Pal S, Betsy Resma G, et al. Surface-engineered mucus penetrating nucleic acid delivery systems with cell penetrating peptides for the lungs. Mol Pharm 2022;19:1309–1324. [DOI] [PubMed] [Google Scholar]
- 25. Lostale-Seijo I, Louzao I, Juanes M, et al. Peptide/Cas9 nanostructures for ribonucleoprotein cell membrane transport and gene edition. Chem Sci 2017;8:7923–7931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ramakrishna S, Kwaku Dad AB, Beloor Jet al. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res 2014;24:1020–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Del'Guidice T, Lepetit-Stoffaes JP, Bordeleau LJ, et al. Membrane permeabilizing amphiphilic peptide delivers recombinant transcription factor and CRISPR-Cas9/Cpf1 ribonucleoproteins in hard-to-modify cells. PLoS One 2018;13:e0195558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Krishnamurthy S, Wohlford-Lenane C, Kandimalla S, et al. Engineered amphiphilic peptides enable delivery of proteins and CRISPR-associated nucleases to airway epithelia. Nat Commun 2019;10:4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Morris MC, Depollier J, Mery J, et al. A peptide carrier for the delivery of biologically active proteins into mammalian cells. Nat Biotechnol 2001;19:1173–1176. [DOI] [PubMed] [Google Scholar]
- 30. Nakase I, Kobayashi S, Futaki S. Endosome-disruptive peptides for improving cytosolic delivery of bioactive macromolecules. Biopolymers 2010;94:763–770. [DOI] [PubMed] [Google Scholar]
- 31. Trofimenko E, Grasso G, Heulot M, et al. Genetic, cellular, and structural characterization of the membrane potential-dependent cell-penetrating peptide translocation pore. Elife 2021;10:e69832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guay D, Del'Guidice T, Lepetit-Stoffaes JP. Rationally-Designed Synthetic Peptide Shuttle Agents for Delivering Polypeptide Cargos from an Extracellular Space to the Cytosol and/or Nucleus of a Target Eukaryotic Cell, Uses Thereof, Methods and Kits Relating to Same. US Patent 9, 267, ed. United State of America; 2017. [Google Scholar]
- 33. Rosen BH, Chanson M, Gawenis LR, et al. Animal and model systems for studying cystic fibrosis. J Cyst Fibros 2018;17:S28–S34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sun X, Sui H, Fisher JT, et al. Disease phenotype of a ferret CFTR-knockout model of cystic fibrosis. J Clin Invest 2010;120:3149–3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sun X, Yi Y, Yan Z, et al. In utero and postnatal VX-770 administration rescues multiorgan disease in a ferret model of cystic fibrosis. Sci Transl Med 2019;11:eaau7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sun X, Olivier AK, Liang B, et al. Lung phenotype of juvenile and adult cystic fibrosis transmembrane conductance regulator-knockout ferrets. Am J Respir Cell Mol Biol 2014;50:502–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Raju SV, Kim H, Byzek SA, et al. A ferret model of COPD-related chronic bronchitis. JCI Insight 2016;1:e87536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sui H, Olivier AK, Klesney-Tait JA, et al. Ferret lung transplant: an orthotopic model of obliterative bronchiolitis. Am J Transplant 2013;13:467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Swatek AM, Lynch TJ, Crooke AK, et al. Depletion of airway submucosal glands and TP63(+)KRT5(+) basal cells in obliterative bronchiolitis. Am J Respir Crit Care Med 2018;197:1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. He N, Liu X, Vegter AR et al. Ferret models of alpha-1 antitrypsin deficiency develop lung and liver disease. JCI Insight 2022;7:e143004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yu M, Sun X, Tyler SR, et al. Highly efficient transgenesis in ferrets using CRISPR/Cas9-mediated homology-independent insertion at the ROSA26 locus. Sci Rep 2019;9:1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu X, Luo M, Guo C, et al. Comparative biology of rAAV transduction in ferret, pig and human airway epithelia. Gene Ther 2007;14:1543–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. O'Malley Y, Rotti PG, Thornell IM, et al. Development of a polarized pancreatic ductular cell epithelium for physiological studies. J Appl Physiol (1985) 2018;125:97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pandey P, Nguyen N, Hansmann UHE. d-Retro inverso amylin and the stability of amylin fibrils. J Chem Theory Comput 2020;16:5358–5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Belser JA, Lau EHY, Barclay W, et al. Robustness of the ferret model for influenza risk assessment studies: A cross-laboratory exercise. mBio 2022;13:e0117422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Suresh B, Ramakrishna S, Kim H. Cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA for genome editing. Methods Mol Biol 2017;1507:81–94. [DOI] [PubMed] [Google Scholar]
- 47. Park HM, Liu H, Wu J, et al. Extension of the crRNA enhances Cpf1 gene editing in vitro and in vivo. Nat Commun 2018;9:3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yamano T, Zetsche B, Ishitani R, et al. Structural basis for the canonical and non-canonical PAM recognition by CRISPR-Cpf1. Mol Cell 2017;67:633–645.e633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sun X, Li Z, Yi Y, et al. Chromatin configurations in the ferret germinal vesicle that reflect developmental competence for in vitro maturation. Reprod Domest Anim 2009;44:320–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chechik L, Martin O, Soutoglou E. Genome editing fidelity in the context of DNA sequence and chromatin structure. Front Cell Dev Biol 2020;8:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Janssen JM, Chen X, Liu J, et al. The Chromatin structure of CRISPR-Cas9 target DNA controls the balance between mutagenic and homology-directed gene-editing events. Mol Ther Nucleic Acids 2019;16:141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. McCarron A, Cmielewski P, Drysdale V, et al. Effective viral-mediated lung gene therapy: is airway surface preparation necessary? Gene Ther 2023;30:469–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. McCarron A, Farrow N, Cmielewski P, et al. Breaching the delivery barrier: Chemical and physical airway epithelium disruption strategies for enhancing lentiviral-mediated gene therapy. Front Pharmacol 2021;12:669635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Johnson LG, Olsen JC, Sarkadi B, et al. Efficiency of gene transfer for restoration of normal airway epithelial function in cystic fibrosis. Nat Genet 1992;2:21–25. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.