Abstract
This report describes the detection of mutations in the pol gene of human immunodeficiency virus type 1 associated with resistance to zidovudine, didanosine, and lamivudine by genotyping by an oligonucleotide ligation assay specific codons in the pol gene amplified by PCR. Our studies demonstrate the sensitivity, simplicity, and specificity of this genotyping system.
Ligation assays have several features that make them ideally suited for typing point mutations in the human immunodeficiency virus type 1 (HIV-1) genome, including their specificity, sensitivity, and compatibility with DNA amplification by PCR. Ligation assays are based on the covalent joining of two adjacent oligonucleotide probes (usually 20-mers) when they are hybridized to a DNA sample, usually a PCR product. The specificity of the ligation between two oligonucleotide primers is regulated by three factors: (i) the specificity of hybridization of the oligonucleotide primers to their complementary sequences on the template, (ii) the need for these primers to hybridize in a head-to-tail (5′→3′) orientation on the template, and (iii) the fact that the oligonucleotides must have perfect base pairing with the target at their junction (7). These characteristics allow nonstringent annealing conditions to be used without compromising specificity. The ligation primers can also be modified with reporters by labelling the primers with a hapten such as digoxigenin or fluorescein, or the primers can be used to capture the reaction product on a streptavidin-coated plate by adding a biotin moiety. Applied in this format, the results of the assay are simple to interpret and can be classified as positive or negative for the reporters marking the wild-type or mutant allele (13). In this report, we describe the development of a high-throughput microtiter-based ligation system that types mutations in the pol gene associated with HIV-1 resistance to zidovudine (ZDV), didanosine (ddI), and lamivudine.
RNA was obtained by silica extraction (1) of plasma or was obtained from cerebrospinal fluid with the QIAmp Viral RNA kit (QIAGEN, Inc., Chatsworth, Calif.) from patients consenting to genotypic evaluation of their HIV-1 isolates. Procedures approved by the Children’s Hospital and Medical Center Institutional Review Board were followed. cDNA was synthesized by using SuperScript II RNase H reverse transcriptase (Gibco BRL, Gaithersburg, Md.), 10 μl of extracted RNA, and 20 pmol of the oligonucleotide primer RT2 (5′-GTATGTCATTGACAGTCCAGC). Peripheral blood mononuclear cells were lysed (14) to a concentration of 1.5 × 105 cells/10 μl. DNA or cDNA was amplified by nested PCR (outer pair, RT1 [5′-GTTGACTCAGATTGGTTGCAC] and RT2 [5′-GTATGTCATTGACAGTCCAGC]; inner pair, RT3 [5′-TATCAGGATGGAGTTCATAAC] and RT4 [5′-GGATGGCCCAAAAGTTAAAC]). The amplicon was a 665-bp region encoding amino acids 17 to 237 of the HIV-1 pol gene. The PCR conditions were as follows: 94°C for 5 min followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min. A final extension step at 72°C was carried out for 4 min. The PCR product was visualized on a 3% agarose gel with ethidium bromide staining.
Oligonucleotide primers for ligation detection were assembled by standard phosphoramidite chemistry on an Applied Biosystems (Foster City, Calif.) 394 DNA synthesizer. Ligation primers specific for wild-type or mutant codon sequences (Table 1) were modified with a 5′-aminohexylphosphate linker (Aminolink2; Applied Biosystems), and following deprotection, digoxigenin or fluorescein was added to the 5′ end by using N-hydroxysuccinimide esters for these primers (Boehringer Mannheim, Indianapolis, Ind.) (7). After modification, the primers were purified by reverse-phase high-performance liquid chromatography. Joining primers complementary to common sequences adjacent to both wild-type and mutant codons were synthesized on a 3′-Biotin-ON CPG Column (Clontech, Palo Alto, Calif.) and were chemically phosphorylated with 5′-Phosphate-ON (Applied Biosystems).
TABLE 1.
Nucleotide sequences of ligation oligonucleotides used to determine pol genotype at amino acids 41, 70, 74, 184, and 215
| Amino acid | Modification | Genotype detected | Sequence (5′→3′)a |
|---|---|---|---|
| 41 | Digoxigenin | wtb | CA-TTA-GTA-GAA-ATT-TGT-ACA-GAA-A |
| Fluorescein | Mutant A | CA-TTA-GTA-GAA-ATT-TGT-ACA-GAA-T | |
| Fluorescein | Mutant B | CA-TTA-GTA-GAA-ATT-TGT-ACA-GAA-C | |
| Biotin | Common | TG-GAA-AAG-GAA-GGG-AAA-A | |
| 70 | Digoxigenin | wt | CC-ATA-AAG-AAA-AAA-GAC-AGT-ACT-AA |
| Fluorescein | Mutant | CC-ATA-AAG-AAA-AAA-GAC-AGT-ACT-AG | |
| Biotin | Common | A-TGG-AGA-AAA-TTA-GTA-GAT-TTC-AGA | |
| 74 | Digoxigenin | wt | AC-AGT-ACT-AAA-TGG-AGA-AAA-T |
| Fluorescein | Mutant | AC-AGT-ACT-AAA-TGG-AGA-AAA-G | |
| Biotin | Common | TA-GTA-GAT-TTC-AGA-GAA-CTT | |
| 184 | Digoxigenin | wt | AG-ACA-TAG-TTA-TCT-ATC-AAT-AC-A |
| Fluorescein | Mutant | AG-ACA-TAG-TTA-TCT-ATC-AAT-AC-G | |
| Biotin | Common | TG-GAT-GAT-TTG-TAT-GTA-GGA-TC | |
| 215 | Digoxigenin | wt | CAA-CAT-CTG-TTG-AGG-TGG-GGA-TTT-AC |
| Fluorescein | Mutant A | CAA-CAT-CTG-TTG-AGG-TGG-GGA-TTT-TT | |
| Fluorescein | Mutant B | CAA-CAT-CTG-TTG-AGG-TGG-GGA-TTT-TA | |
| Biotin | Common | C-ACA-CCA-GAC-AAA-AAA-CAT-CAG-AA |
Base pairs comprising the codons of interest are in boldface type.
wt, wild type.
For the ligation assay, 2 μl of the pol amplicon, 10 μl of distilled H2O containing 0.1% Triton X-100, 10 μl of a reaction mixture of 20% 10× ligase buffer (200 mM Tris [pH 8.0], 100 mM MgCl2, 10 mM dithiothreitol), 20% 10 mM NAD (Sigma, St. Louis, Mo.), 2.5% 1 M KCl (Sigma), 57.5% distilled H2O containing 0.1% Triton X-100, 3 U of thermostable ligase (Epicentre Technologies, Madison, Wis.), and 3 pmol of each ligation primer (the biotinylated oligonucleotide, the digoxygeninated oligonucleotide, and the fluoresceinated oligonucleotide) were placed into a 96-well V-bottom plate (M. J. Research Inc., Watertown, Mass.). Two mutant genotypes at amino acids 41 and 215 were assayed in separate reactions. Each well was overlaid with 50 μl of mineral oil (Sigma), and 10 cycles (M. J. Research Inc.) of 93°C for 30 s and 37°C for 4 min were performed. Within 10 min of completion of the last cycle, 10 μl of 0.1 M EDTA–0.1% Triton X-100–distilled H2O was added to each well to stop the ligation reaction.
Samples were transferred to a 96-well flat-bottom plate (Falcon) previously coated with streptavidin (50 μl of a solution of 25 μg/μl per well overnight at 4°C) and were blocked with 200 μl of bovine serum albumin (BSA) blocking buffer (0.5% BSA [Sigma] in 1× phosphate-buffered saline) per well at room temperature (RT) for 30 min to eliminate nonspecific binding. The biotinylated end of the ligation product was allowed to be captured by the streptavidin plate for 1 h at RT. The plates were washed twice with NaOH wash (0.01 N NaOH, 0.05% Tween 20) and twice with Tris wash (0.1 M Tris [pH 7.5], 0.15 M NaCl, 0.05% Tween 20). Forty microliters of a 1:1,000 dilution of both anti-digoxigenin-peroxidase Fab fragments (Boehringer Mannheim) and anti-fluorescein-alkaline phosphatase Fab fragments (Boehringer Mannheim) in BSA blocking buffer was added to each well. The plates were incubated for 30 min at RT and were then washed six times with Tris wash. Twenty-five microliters of alkaline phosphatase (AP) substrate (GibcoBRL) was added to each well and the plates were incubated for 10 min at RT, after which 25 μl of amplifier (GibcoBRL) was added to each well and the plates were incubated in the dark at RT for a maximum of 10 min or until color from the AP substrate-amplifier reaction was full. Spectrophotometric absorbances were read at 490 nm by an ELISA plate reader (Dynatech, Chantilly, Va.). The plates were then washed six times with Tris wash, after which 50 μl of 3,3′,5,5′-tetramethylbenzidine (TMB) substrate (Sigma) was added to each well. The plates were incubated at RT for a maximum of 10 min or until color from the peroxidase-TMB reaction was complete, and then 50 μl of 0.3 M sulfuric acid was added to each well. Spectrophotometric absorbances were read at 450 nm. Absorbances were blanked against wells containing distilled H2O in lieu of the PCR product. HIV-1-infected 8E5 cells (3) were used as a wild-type control, and HIV-1RTMDR1/MT-2 (8) and sequenced HIV-1 isolates from patient specimens were used as mutant controls. The PCR products of plasma HIV-1 from 10 pregnant women treated with ZDV or ddI, or both, were assayed by the oligonucleotide ligation assay (OLA), and for comparison, plus- and minus-strand sequences were obtained (ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction Kit; Applied Biosystems) by using an Applied Biosystems 373 Stretch Automated Sequencer after purification of the amplicons (Wizard DNA Clean-Up System; Promega).
There was no overlap in the ranges of optical density readings for positive controls or for patient specimens and the negative controls. A random selection of 100 negative wells containing either water or PCR-amplified pol from patient specimens had a range of optical absorbances of 0.000 to 0.020, with a mean of 0.002. One hundred randomly selected wells positive for wild-type virus had a range of optical absorbances of from 0.088 to 1.45, and 100 wells positive for mutant virus had a range of from 0.041 to 0.471.
Among the 40 codons (codons 41, 70, 74, and 215 for each of 10 patients) from pregnant women’s plasma examined by both OLA and direct sequencing, 19 typed as mutant codons. It is worth noting that for 8 of the 19 codons typed as containing only mutant sequences by sequence analysis, OLA also detected wild-type sequences. These results reflect the higher sensitivity of OLA compared to that of direct sequence analysis (unpublished data).
To further determine the sensitivity of OLA in discriminating wild-type and mutant virus genotypes, serial dilutions and reciprocal mixtures of lysates of cells infected with HIV-1 containing either mutant or wild-type sequences at the pol codons of interest were evaluated. PCR products from laboratory virus strains and patient lysates with sequences representing those of all genotypes whose codons 41, 70, 74, and 215 could be assessed by this system were diluted so that 100, 10, 1, 0.1, or 0.01 ng of amplicon was typed by OLA. As little as 0.01 ng of each of the amplicons of mutant B codon 41, wild-type codon 70, wild-type codon 74, and mutant B codon 215 could be detected by OLA, while 0.1 ng of each of the amplicons of wild-type codon 41 and mutant codon 74 was detected. One nanogram of each of the amplicons of mutant A codon 41, mutant codon 70, and wild-type codon 215 was detected by OLA, while the amplicon of mutant A codon 215 was not detected at levels below 10 ng. Similar findings were obtained when reciprocal mixtures of the patient cell lysates at 5, 12, 25, and 50% were tested with a total of 6 ng of DNA/reaction. Wild-type codons 41, 70, 74, and 215 were all detected at a level of 5% in the appropriate mutant genome background, and mutant B codon 41 was detected at 5% in wild-type codon 41 background. Mutant codon 70, mutant codon 74, and mutant B codon 215 were all detected at 12% in a background of the corresponding wild-type genome. Mutant A codon 41 and mutant A codon 215 were detected at intermediate levels (between 25 and 50%) in a background of the corresponding wild-type genome.
On the basis of the accuracy and sensitivity of this approach, we examined pol codons 41, 70, 74, 184, and 215 (a total of 700 codons) from viruses in 175 patient specimens using OLA. For 97.9% of the codons, the codons were clearly found to have wild-type or mutant sequences by OLA. Fifteen of the 700 codons (2.1%) examined failed to be genotyped by OLA; i.e., the color representing neither the wild-type nor the mutant appeared. Although PCR failure can lead to a negative OLA result, our samples with a negative result by OLA had amplicons detected by agarose gel electrophoresis. The sequences of these 15 codons obtained directly (Table 2) revealed alternative mutations in the pol gene which were no longer complementary to the OLA primers. For 13 of the 15 specimens negative by OLA, mutations in the pol gene were found within two bases of the ligation site. The sequences from the remaining two negative specimens had two or more base changes located three to five bases from the ligation site.
TABLE 2.
Nucleotide sequences of codons of isolates from patient specimens which were indeterminant by OLA, including 10 bases on either side of the ligation site
| Patient sample no. | Results of direct sequencing (5′→3′)a | pol amino acid |
|---|---|---|
| 1 | AC-AGT-GAT-AA*A-TGG-AGA-AAA | K70 |
| 2 | TGT-ACA-GAG-A*TG-GAA-AAG-GA | M41 |
| 2 | AC-AGT-GAC-AG*A-TGG-AGA-AAA | K70R |
| 2 | TGG-AGA-AAA-A*TA-GTA-GAT-TT | L74I |
| 3 | AC-AGT-ACT-GG*A-TGG-AGA-AAA | K70G |
| 4 | GG-GGA-TTT-TG*C-ACA-CCA-GAC | T215C |
| 5 | TGT-ACA-GAG-A*TG-GAA-AAG-GA | M41 |
| 6 | AC-AGT-ACT-AA*G-TGG-AGA-AAA | K70 |
| 7 | TGG-AGA-AAG-T*TA-GTA-GAT-TT | L74 |
| 8 | TGT-ACA-GAT-A/C*TG-GAA-AAG-GA | M41M/L |
| 8 | GG-GGA-TTT-AC*T-ACA-CCA-GA | T215 |
| 9 | TGT-ACA-GAG-A*TG-GAA-AAG-GA | M41 |
| 10 | TGG-AGA-AAA-A*TA-GTA-GAT-TT | L741 |
| 11 | TGT-ACA-GAT-A*TG-GAA-AAG-GA | M41 |
| 11 | GG-GGA-TTT-A/GT*T-ACA-CCA-GA | T215F/L |
Boldface type indicates base changes which were not complementary to the ligation oligonucleotides. *, ligation site. The codon of interest is underlined.
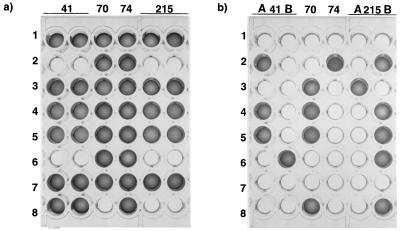
Ligation assays have a number of advantages in genotyping mutations within the HIV pol gene compared to other phenotypic (2, 5) or genotypic (6, 9, 12) approaches. First, because of their highly specific nature, ligation assays yield clear positive or negative outcomes that are easy to interpret visually (Fig. 1) or that can be interpreted by a spectrophotometer and computer program with samples on a microtiter plate (10, 13). In the past, we used a solid-phase ligase-based detection reaction (LDR) in which one of the oligonucleotides was covalently coupled to latex beads (4). Visual scoring of the LDR was on occasion blotchy and resulted in an element of subjectivity in reading the results of LDR.
FIG. 1.
OLA plate demonstrating the detection of wild-type genotypes (a) and mutant genotypes (b) from randomly selected patients receiving nucleosides. Some wells contain mixtures of genotypes (e.g., row 2, codon 74; row 4, codons 41, 70, and 215), while others reveal only wild-type (e.g., rows 1 and 7, all codons) or mutant (e.g., rows 2 and 6, codons 41 and 215) genotypes. Positive results were easily determined visually by the development of a colored product. The digoxigenin reporter (wild type) was identified by a specific antibody coupled with horseradish peroxidase, which resulted in a brilliant blue color (a). After washing this microtiter plate, the presence of the fluorescein reporter (mutant) was assessed by the production of a deep magenta color due to the alkaline phosphatase labelling of the antibodies to fluorescein (b). The absence of either genotype was also quite clear, because no color was produced in the wells (a and b). A and B, mutants A and B.
Although the highly specific nature of OLA may lead to completely negative assays for both wild-type and mutant pol alleles, this is rare, occurring in approximately 2% of the codons typed and is indicative of the presence of alternative mutations in the joining or priming regions for the ligation reaction. Therefore, specimens containing sequences which are amplified in PCR assays but which are negative for wild-type and mutant sequences by OLA should be investigated by sequence analysis.
It is worth noting that as more pol mutations are identified and associated with drug failure and phenotypic resistance, it is simple to establish new OLAs for these variants. Once the appropriate oligonucleotide primers are synthesized, the new primers can be incorporated into plate assays used for other pol variants because of the use of nonstringent assay conditions.
Another attribute of OLA is its sensitivity. In some cases, OLA was able to accurately detect genotypes when less than 0.01 ng of PCR-amplified DNA was present. Even with the variability in the sensitivity of OLA among the codons tested, the least sensitive reaction required only 1 to 10 ng of amplified DNA, whereas direct sequencing requires 30 to 90 ng of PCR product. Furthermore, it is difficult to detect the presence of mixtures of wild-type and mutant virus sequences by fluorescence-based sequencing, which has a sensitivity of polymorphism detection of about 25% (11), while the analysis of mixtures of wild-type and mutant virus in samples by OLA is well documented in this and previous (13) studies. Two factors contribute to the excellent sensitivity of the OLA format. First, the ligation reaction itself can be cycled by using a thermostable ligase to generate additional ligated products. Second, an amplification step is associated with the enzyme-linked immunosorbent assay (ELISA)-based phase of detection of the specific hapten reporters in the ligated products.
Lastly, the OLA adapts well to a clinical format. Allowing for controls, up to 65 codons can be screened in one OLA on one 96-well ELISA plate in less than 4 h after the completion of PCR amplification. Throughput can be even higher with the use of automation for the processing of large numbers of microtiter plates (13).
In summary, the high specificity, sensitivity, and objectivity, the ease of use, the adaptability, and the high throughput make OLA a useful clinical tool for screening HIV-1 for the pol genotypes associated with resistance to antiretroviral agents.
Acknowledgments
This project was supported in part by NIH U01 AI32910 and the Foster Foundation, and technology development was supported in part by an NSF grant (DIR 8809710).
REFERENCES
- 1.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boucher C A B, Keulen W, van Bommel T, Nijuis M, de Jong D, de Jong M D, Schipper P, Back N K T. Human immunodeficiency virus type 1 drug susceptibility determination by using recombinant viruses generated from patient sera tested in a cell-killing assay. Antimicrob Agents Chemother. 1996;40:2404–2409. doi: 10.1128/aac.40.10.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Folks T M, Powell D, Lightfoote M, Koenig S, Fauci A S, Benn S, Rabson A, Daugherty D, Gendelman H E, Hoggan M D, et al. Biological and biochemical characterization of a cloned Leu-3-cell surviving infection with the acquired immune deficiency syndrome retrovirus. J Exp Med. 1986;164:280–290. doi: 10.1084/jem.164.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frenkel L M, Wagner L E N, Atwood S M, Cummins T J, Dewhurst S. Specific, sensitive, and rapid assay for human immunodeficiency virus type 1 pol mutations associated with resistance to zidovudine and didanosine. J Clin Microbiol. 1995;33:342–347. doi: 10.1128/jcm.33.2.342-347.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Japour A J, Mayers D L, Johnson V A, Kuritzkes D R, Beckett L A, Arduino J M, Lane J, Black R J, Reichelderfer P S, D’Aquila R T, Crumpacker C S the RV-43 Study Group; the AIDS Clinical Trials Group Virology Committee Resistance Working Group. Standardized peripheral blood mononuclear cell culture assay for determination of drug susceptibilities of clinical human immunodeficiency virus type 1 isolates. Antimicrob Agents Chemother. 1993;37:1095–1101. doi: 10.1128/aac.37.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaye S, Loveday C, Tedder R S. A microtitre format point mutation assay: application to the detection of drug resistance in human immunodeficiency virus type-1 infected patients treated with zidovudine. J Med Virol. 1992;37:241–246. doi: 10.1002/jmv.1890370402. [DOI] [PubMed] [Google Scholar]
- 7.Landegren U, Kaiser R, Sanders J, Hood L. A ligase-mediated gene detection technique. Science. 1988;241:1077–1080. doi: 10.1126/science.3413476. [DOI] [PubMed] [Google Scholar]
- 8.Larder B A, Kellam P, Kemp S D. Convergent combination therapy can select viable multidrug-resistant HIV-1 in vitro. Nature. 1993;365:451–453. doi: 10.1038/365451a0. [DOI] [PubMed] [Google Scholar]
- 9.Larder B A, Kellam P, Kemp S D. Zidovudine resistance predicted by direct detection of mutations in DNA from HIV-infected lymphocytes. AIDS. 1991;5:137–144. doi: 10.1097/00002030-199102000-00002. [DOI] [PubMed] [Google Scholar]
- 10.Nickerson D A, Kaiser R, Lappin S, Stewart J, Hood L, Landegren U. Automated DNA diagnostics using an ELISA-based oligonucleotide ligation assay. Proc Natl Acad Sci USA. 1990;87:8923–8927. doi: 10.1073/pnas.87.22.8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parker L T, Zakeri H, Deng Q, Spurgeon S, Kwok P-Y, Nickerson D A. AmpliTaq® DNA polymerase, FS dye-terminator sequencing: analysis of peak height patterns. BioTechniques. 1996;21:694–699. doi: 10.2144/96214rr02. [DOI] [PubMed] [Google Scholar]
- 12.Stuyver L, Wyseur A, Rombout A, Louwagie J, Scarcez T, Verhofstede C, Rimland D, Schinazi R, Rossau R. Line probe assay for rapid detection of drug-selected mutations in the human immunodeficiency virus type 1 reverse transcriptase gene. Antimicrob Agents Chemother. 1997;41:284–291. doi: 10.1128/aac.41.2.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tobe V O, Taylor S L, Nickerson D A. Single-well genotyping of diallelic sequence variations by a two-color ELISA-based oligonucleotide ligation assay. Nucleic Acids Res. 1996;24:3728–3732. doi: 10.1093/nar/24.19.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zack J, Haislip A M, Krogstad P, Chen I S Y. Incomplete reverse-transcribed human immunodeficiency virus type 1 genomes in quiescent cells can function as intermediates in the retroviral life cycle. J Virol. 1992;66:1717–1725. doi: 10.1128/jvi.66.3.1717-1725.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]