Abstract
Dengue virus serotype 4 (DENV-4) has been the rarest circulating serotype in Malaysia, resulting in it being an understudied area. A recent observation from institutional surveillance data indicated a rapid increase in DENV-4-infected cases. The present study aimed to investigate the resurgence of DENV-4 in relation to the demographic, clinical and genomic profiles of 75 retrospective dengue samples. First, the demographic and clinical profiles obtained between 2017 and July 2022 were statistically assessed. Samples with good quality were subjected to full genome sequencing on the Illumina Next Seq 500 platform and the genome data were analysed for the presence of mutations. The effect of the mutations of interest was studied via an in silico computational approach using SWISS-MODEL and AlphaFold2 programs. The predominance of DENV-4 was discovered from 2021 to 2022, with a prevalence of 64.3% (n = 9/14) and 89.2% (n = 33/37), respectively. Two clades with a genetic divergence of 2.8% were observed within the dominant genotype IIa. The majority of DENV-4-infected patients presented with gastrointestinal symptoms, such as vomiting (46.7%), persistent diarrhoea (30.7%) and abdominal pain (13.3%). Two mutations, His50Tyr and Pro144Ser, located at the wing domain of the NS1 protein were discovered to be unique to the recently sequenced DENV-4.
Keywords: dengue virus serotype 4, genomics, mutation, clinical presentation
1. Introduction
The perpetual dengue endemic in Malaysia has been the result of a constant increase in cases in a 12-decade-long (1902–2022) journey of battling dengue infections. Seemingly, an interesting pattern of dengue serotype circulation during an outbreak period has been observed, whereby major outbreaks are likely to follow switches in DENV serotypes. In other words, a predominantly circulating dengue serotype before an outbreak is replaced by another serotype that persists towards the end of the outbreak. Once the number of cases depreciates, the persistent serotype is again replaced by another serotype. The reason for the occurrence of this pattern is unclear, thus generating an epidemiological gap that needs to be addressed.
The first isolated dengue serotype in Malaysia was DENV-1, which was identified in 1954 from a minor outbreak of dengue fever in a local school in the Federal Territory of Kuala Lumpur [1]. Later, three other serotypes were discovered, indicating the circulation of a total of four serotypes in Malaysia [2]. DENV-4 marked its predominance in early 1967–1969. The beginning of the 1970s coincided with the theory of serotype shift when DENV-4 was replaced by DENV-2 as the predominant serotype. This seemed to have suppressed the existence of DENV-4 in Malaysia, as it has accounted for less than 5% of all DENV isolated in Malaysia since then [3].
As a result, DENV-4 has been an understudied serotype, characterised by the scarcity of recently published data. The available data describing the genomic evolution of DENV-4 in Malaysia is also limited. The emergence of DENV-4 genotype IIa in Malaysia was reported for the first time in 2001 [4]. This serotype was also documented in East Malaysia (Sandakan) in 2016 [5]. However, no further genomic characterization is currently available.
A recent observation from institutional surveillance data indicated a rapid upsurge in DENV-4-infected cases from 2021–2022, indicating a switch from DENV-2. Although the precise method or cause of the phenomena of serotype switching is yet unknown, there is a possibility that this was brought on by fitness selection, in which viruses continuously mutate to produce fit progeny that allow them to become more dominant than the others. This is similar to the pattern shown by the most recent epidemic of SARS-CoV-2. Using this information as a foundation, we set out to look into the clinical and demographic features of DENV-4 circulation in Malaysia from 2017 to mid-2022 and to examine the genome of the currently circulating DENV-4. We also intended to perform in silico prediction of the effect of any mutations identified in this study to elucidate the link between fitness and transmission of DENV-4.
2. Materials and Methods
2.1. Study Subjects
RNA extracted from 75 archived clinical samples that were confirmed to be DENV-4 positive by multiplex real-time RT-PCR [6] were used in this study. These samples were referred to the virology laboratory in the Institute for Medical Research, Malaysia, by sentinel hospitals for dengue molecular diagnostics from 2017 to July 2022. The sample types included serum, plasma, cerebrospinal fluid and liver biopsies.
2.2. Analysis of Clinical and Demographic Features
Clinical, demographic and laboratory information corresponding to the DENV-4 cases were retrieved from medical notes and our laboratory database. The data were tabulated according to categorical variables, including gender, location, antibody status and a spectrum of clinical manifestations and continuous variables, such as age and day of fever. The categorical data were described in proportions (percentage), while the continuous variables were reported as mean and standard deviation (SD).
2.3. Viral RNA Extraction and Full Genome Sequencing
Viral RNA extraction was performed using a QIAamp Mini Viral RNA Extraction kit (Qiagen, Germantown, MD, USA) according to the manufacturer’s instruction. The samples were subsequently subjected to DNAse treatment using a Turbo DNA-free kit (Invitrogen, Waltham, MA, USA) to eliminate any host genome interference. Full genome sequencing was performed for the samples, with a cycle threshold (Ct) of less than 30. The library was prepared using a TruSeq Stranded Total RNA kit (Illumina, San Diego, CA, USA) according to kit manual. The final library was assessed for concentration using a Qubit Fluorometer 2.0 (Invitrogen, Waltham, MA, USA). Sequencing was performed on a NextSeq 500 (Illumina, San Diego, CA, USA), with a high output run and a final loading concentration of 1.4pM.
2.4. Genomic Data Analysis
High-quality sequencing reads for the study subjects were obtained through BBDuk (BBTools version 38.57) trimming and filtering, after which they were subjected to assembly using MEGAHIT 1.2.8. [7]. A sequence similarity search for DENV-specific contig filtering was performed using BLASTN (version 2.9.0+) while full genome sequences were generated in FASTA format. Moreover, full genome sequences of the reference DENV-4 strains were obtained from the National Centre for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov/nuccore (accessed on 4 January 2023)). Multiple sequence alignments of the DENV-4 full genomes were executed using the MEGA-X [8] tool, and the genome sequences were studied for the presence of unique mutations. A phylogenetic tree was constructed using the Neighbor Joining algorithm. Subsequently, pairwise genetic distance analysis was performed to estimate the genetic diversity between clades and sub-clades. Additionally, the SimPlot (v3.5.1) [9] tool was utilised to check for inter- and intra-genotype recombination signals of DENV-4.
2.5. Modelling of DENV-4 Wild Type (WT) and the Mutant of Interest
The effects of mutations in DENV-4 NS1 protein structure identified in this study were observed through computational modelling. The X-ray crystal structure of DENV-4 NS1 is currently not available; therefore, two separate programs, SWISS-MODEL [10] and AlphaFold2 [11], were used to model the DENV-4 protein structure containing the mutation of interest in order to obtain the consensus. The X-ray crystal structure of DENV-2 NS1 was retrieved from the Protein Databank (RCSB-PDB) (PBD ID: 4O6B) and used as a template to build the wild type (WT) and mutant DENV4 NS1 model. Once the WT and mutant DENV-4 protein models were generated, energy minimisations were performed using the Swiss-PDB viewer [12] program to remove unwanted contacts. The GROMOS96 force field was used to assign atomic charges to all residues. Furthermore, for structure prediction in the AlphaFold2 program, ColabFold was used (https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/AlphaFold2.ipynb#scrollTo=kOblAo-xetgx (accessed on 30 January 2023)) by applying the following setting: template_mode: pdb70, model type: AlphaFold2-multimer-v2. The WT and mutant DENV-4 protein models were then visualised using PyMol software [13]. The qualities of the predicted structure models produced by both programs were validated using ProCheck [14], ProSA-web [15] and ERRAT [16] programs to determine the stereo-chemical quality and overall quality of the protein model.
2.6. Structure Stability and Hydrogen Bond Analyses
The effects of the mutations were studied through structure stability and hydrogen bond analyses. Additionally, the thermodynamic stability of the WT and mutant DENV-4 protein models was analysed using FoldX [17]. The change in free energy (kcal/mol) was calculated and interpreted as follows: >3 kcal/mol was regarded as severely destabilising, 1–3 kcal/mol as destabilising and <1 kcal/mol as benign [18]. Meanwhile, hydrogen bond interactions between the native and neighbouring residues were analysed using Chimera software [19]. The relax hydrogen bond constraint was set to be within 0.4 Å, with a maximum angle of 30 degrees.
3. Results
3.1. Trend of DENV-4 Circulation in Malaysia
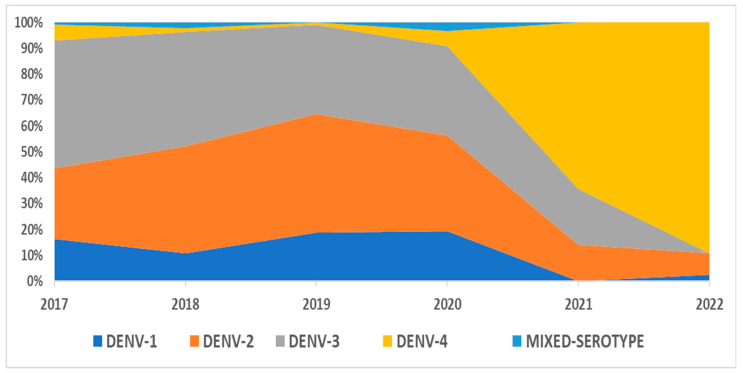
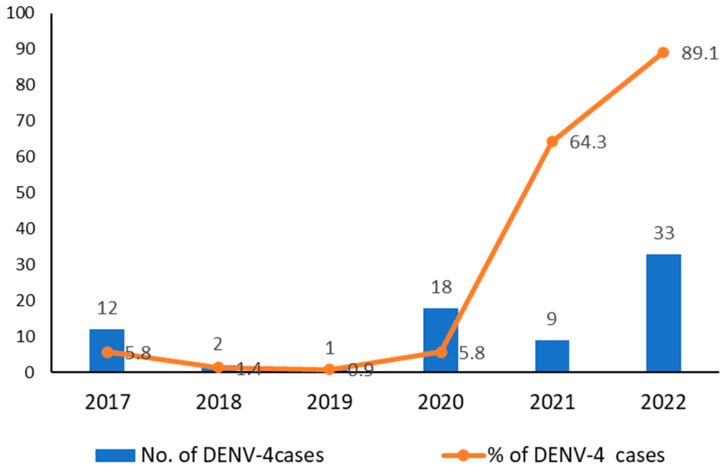
A total of 828 dengue-positive samples, confirmed by Real-Time rt-PCR, from 2017–2020 were further classified into serotypes, as shown in Table 1. DENV-3 and DENV-2 were the predominantly circulating serotypes in 2017–2018 and 2019–2020, respectively, after which, a serotype shift was observed from 2021 onwards, with DENV-4 becoming the major circulating serotype (Figure 1). The breakdown of serotypes by states is provided in Supplement File S1. The prevalence of DENV-4 serotypes over the past 6 years, from 2017 to 2022, ranged from 0.9% in 2019 to 89.2% in 2022. Moreover, an upward trend in the DENV-4 serotype has been observed since the beginning of the COVID-19 pandemic in year 2020, as shown in Figure 2.
Table 1.
Circulating dengue serotypes in Malaysia from 2017 to 2022.
| Year | Number of Dengue Positive Cases Confirmed by Real-Time rt-PCR |
DENV-1 n (%) |
DENV-2 n (%) |
DENV-3 n (%) |
DENV-4 n (%) |
Mixed Serotype n (%) |
|---|---|---|---|---|---|---|
| 2017 | 206 | 34 (16.5) | 56 (27.2) | 102 (49.5) | 12 (5.8) | 2 (1.0) |
| 2018 | 138 | 15 (10.9) | 57 (41.3) | 61 (44.2) | 2 (1.4) | 3 (2.2) |
| 2019 | 116 | 22 (19.0) | 53 (45.7) | 40 (34.4) | 1 (0.9) | 0 (0.0) |
| 2020 | 310 | 60 (19.4) | 115 (37.1) | 107 (34.5) | 18 (5.8) | 10 (3.2) |
| 2021 | 14 | 0 (0.0) | 2 (14.2) | 3 (21.4) | 9 (64.3) | 0 (0.0) |
| 2022 | 37 | 1(2.7) | 3 (8.1) | 0 (0.0) | 33 (89.2) | 0 (0.0) |
Figure 1.
Dengue serotype distribution in Malaysia from 2017 to 2022.
Figure 2.
DENV-4 serotype distribution in Malaysia from 2017 to 2022.
3.2. Demographic and Clinical Features of Patients Infected with DENV-4
For the overall study period (2017–2022), 75 samples positive for the DENV-4 serotype were identified, wherein the age of patients ranged from 2–80 years, with a mean of 36.1. The majority of the patients were over 18 years of age (n = 62, 82.7%), males (n = 42, 56.0%) and hailed from the central region of peninsular Malaysia (n = 37, 49.3%) (Table 2). Blood samples were collected from the patients from Day 1 to Day 12 of fever (mean = 4.3). An equal proportion of the cases (32.0%) presented with primary and secondary infections, as determined by IgM and IgG detection via dengue rapid test kits. Among the DENV-4-infected cases, 32%, 40.0% and 25.3% were diagnosed as dengue without warning signs, dengue with warning signs and severe dengue, respectively, while 2.7% succumbed to the disease (Table 3).
Table 2.
Demographic features of DENV-4 cases in Malaysia from 2017 to 2022.
| Variable | Overall (n = 75) |
|
|---|---|---|
| % | ||
| Age: | ||
| Min–Max | 2–80 | |
| Mean (SD): | 36.1 (19.2) | |
| ≤18 years | 13 | 17.3 |
| >18 years | 62 | 82.7 |
| Gender: | ||
| Male | 42 | 56.0 |
| Female | 33 | 44.0 |
| State: | ||
| Johor | 14 | 18.7 |
| Kedah | 17 | 22.7 |
| Kuala Lumpur | 3 | 4.0 |
| Melaka | 2 | 2.7 |
| Perak | 5 | 6.7 |
| Selangor | 34 | 45.3 |
| Region: | ||
| South (Johor, Melaka) |
16 | 21.3 |
| Central (Kuala Lumpur, Selangor) |
37 | 49.3 |
| North (Perak, Kedah) |
22 | 29.3 |
Table 3.
Laboratory and diagnostic characteristics of DENV-4 cases in Malaysia from 2017 to 2022.
| Variable | Overall | |
|---|---|---|
| n | % | |
| Day of fever: | ||
| Min–Max | 1–12 | |
| Mean (SD) | 4.3 (2.1) | |
| Antibody status: | ||
| Primary infection | 24 | 32.0 |
| Secondary infection | 24 | 32.0 |
| NA | 27 | 35.0 |
| Diagnosis: | ||
| Dengue without warning signs | 24 | 32.0 |
| Dengue with warning signs | 30 | 40.0 |
| Severe dengue | 19 | 25.3 |
| Fatal | 2 | 2.7 |
NA = Data not available for either one or more components (IgM, IgG or NS1).
The most frequently reported warning signs for DENV-4 infection were vomiting (46.7%), followed by persistent diarrhoea (30.7%), lethargy (21.3%) and abdominal pain (13.3%). The remaining warning signs were observed less frequently (<10%). The most frequently reported non-warning signs were myalgia (18.7%), arthralgia (16%) and cough (10.7%). Furthermore, transaminitis and hemoconcentration were the most frequently reported dengue complications, at 16.0% and 13.3%, respectively (Table 4).
Table 4.
Clinical manifestations of DENV-4 cases (n = 75) in Malaysia from 2017 to 2022.
| Clinical Presentation | All (n = 75) |
|---|---|
| Warning signs: | |
| Persistent vomiting | 35 (46.7) |
| Persistent diarrhoea | 23 (30.7) |
| Lethargy | 16 (21.3) |
| Abdominal pain | 10 (13.3) |
| Gum bleeding | 5 (6.7) |
| Occult bleeding | 7 (9.3) |
| Epigastric pain | 4 (5.3) |
| Other bleeding | 2 (2.7) |
| Hemoconcentration | 10 (13.3) |
| Non-warning signs: | |
| Myalgia | 14 (18.7) |
| Arthralgia | 12 (16.0) |
| Cough | 8 (10.7) |
| Chills/Rigor | 7 (9.3) |
| Headache | 6 (8.0) |
| Rash | 2 (2.7) |
| Retro-orbital pain | 2 (2.7) |
| Severe dengue: | |
| Plasma leakage | 5 (6.7) |
| Compensated shock | 6 (8.0) |
| Decompensated shock | 5 (6.7) |
| Complications: | |
| Acute kidney injury | 2 (2.7) |
| Transaminitis | 12 (16.0) |
| Encephalitis | 3 (4.0) |
| Hepatitis | 1 (1.3) |
| Multiorgan failure | 1 (1.3) |
| Myocarditis | 3 (4.0) |
| Respiratory distress | 1 (1.3) |
3.3. Genomic Characteristics of DENV-4
A total of 14 full genomes of DENV-4 were successfully sequenced in this study. This included five cases from 2020, three cases from 2021 and six cases from 2022. Phylogenetic analysis revealed that all 14 sequences belonged to DENV-4 genotype IIa. In addition, two distinct clades were observed within the genotype IIa cluster (Figure 3). Intraclade pairwise distance showed a 2.8% (2.8% ± 0.16%) genetic divergence between clades 1 and 2 of the Malaysian DENV-4 genotype IIa, although no recombination was observed. Furthermore, interclade analysis revealed minimal diversities of 0.4 ± 0.04% within clade 1 and 0.9 ± 0.07% within clade 2.
Figure 3.
Phylogeny of the DENV-4 full genome sequences. The sequences in this study are colour-coded and can be identified as OPxxxxxx.
Notably, a possible geographical link was also observed between clades 1 and 2. A mixture of both clades of DENV-4 (n = 7) was found in the central region of Malaysia, consisting of states such as Selangor and the Federal Territory of Kuala Lumpur. Among samples successfully sequenced, clade 1 was largely detected in Kedah (n = 5). On the other hand, clade 2 was detected in Johor (n = 2) (Figure 3).
Two non-synonymous mutations, His50Tyr and Pro144Ser, were found in the NS1 gene, unique to the Malaysian DENV-4 genome sequences obtained in this study, while the other genes were well conserved. The mutant 50Y was detected in an overall prevalence of 57.1% of the sequenced cases in 2020 (n = 1), 2021 (n = 2) and 2022 (n = 5), while mutant 144S was detected with a prevalence of 64.3% in 2020 (n = 2), 2021 (n = 2) and 2022 (n = 5), as shown in Table 5. The impact of these mutations on the clinical presentations was not seen.
Table 5.
NS1 mutation mapping for the recently sequenced DENV-4 genotype II in comparison with reference sequences.
| Sample ID | Country | Case Presentation |
Collection Year | NS1 Mutations | Clade | ||
|---|---|---|---|---|---|---|---|
| His50Tyr | Pro144Ser | ||||||
| 1 | *OP600497 | Malaysia | DF without WS | 2020 | ● | ● | 1 |
| 2 | *OP600506 | Malaysia | DF with WS | 2020 | 2 | ||
| 3 | *OP600478 | Malaysia | DF with WS | 2020 | 2 | ||
| 4 | *OP600495 | Malaysia | DF with WS | 2020 | ● | 1 | |
| 5 | *OP600507 | Malaysia | DF with WS | 2020 | 2 | ||
| 6 | *OP600496 | Malaysia | Severe Dengue | 2021 | 2 | ||
| 7 | *OP600508 | Malaysia | Severe Dengue | 2021 | ● | ● | 1 |
| 8 | *OP600498 | Malaysia | Severe Dengue | 2021 | ● | ● | 1 |
| 9 | *OP600499 | Malaysia | DF with WS | 2022 | ● | ● | 1 |
| 10 | *OP600509 | Malaysia | Severe Dengue | 2022 | ● | 2 | |
| 11 | *OP600510 | Malaysia | DF with WS | 2022 | ● | ● | 1 |
| 12 | *OP600511 | Malaysia | DF without WS | 2022 | ● | ● | 1 |
| 13 | *OP600512 | Malaysia | DF with WS | 2022 | ● | 1 | |
| 14 | *OP604360 | Malaysia | DF with WS | 2022 | ● | ● | 1 |
| 15 | MH888334.1 | Malaysia | NA | 2013 | |||
| 16 | MH051734.1 | Malaysia | NA | 2014 | |||
| 17 | JX024757.1 | Singapore | NA | 2010 | |||
| 18 | MW301595.1 | Malaysia | NA | 2013 | |||
| 19 | KC762695.1 | Indonesia | NA | 2007 | |||
| 20 | MH823210.1 | Indonesia | NA | 2014 | |||
| 21 | MH382789.1 | Papua NG | NA | 2016 | |||
| 22 | KJ579245.1 | Brazil | NA | 2012 | |||
| 23 | KY474335.1 | Ecuador | NA | 2014 | |||
DF = Dengue fever, WS = Warning sign, NA = Data not available, * = Samples sequenced in this study, • = mutation detected.
3.4. DENV-4 NS1 Wild Type (WT) and Mutant Models
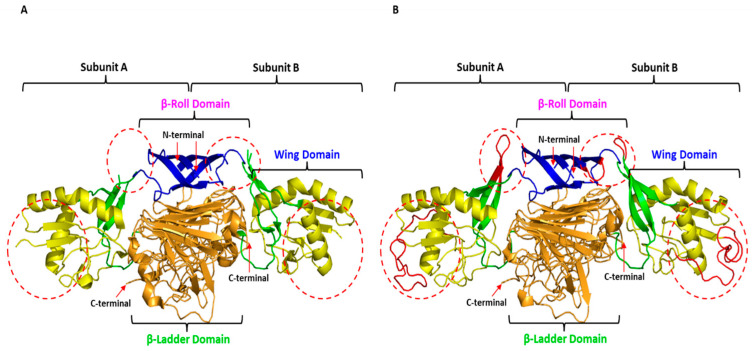
The mutation of interest was identified in the NS1 gene of DENV-4. Since the X-ray crystal structure of DENV-4 NS1 is currently not available, that of DENV-2 NS1 was retrieved from the Protein Databank (RCSB-PDB) (PBD ID: 4O6B) [20] and used as a template to build the WT and mutant DENV-4 models. However, this X-ray crystal structure, with a resolution of 3.0 Å, consisted of missing residues at each of the two domains. Therefore, the missing residues were added using the SWISS-MODEL program. The unresolved regions in the two domains—β-roll (amino acids 7 to 11) and wing (amino acids 108 to 128) of the DENV-2 NS1 structure were successfully constructed in the DENV-4 NS1 structure, as depicted in Figure 4. The quality of predictions by the WT and mutant DENV-4 NS1 models revealed that both structures exhibited good quality and were within the acceptance range (Table 6).
Figure 4.
The homo-dimer X-ray crystal structure of DENV-2 NS1 (PDB ID: 4O6B) and wild type of the DENV-4 NS1 structure model: (A). X-ray crystal structure of DENV-2 NS1 with missing residue (red circle); (B). Complete wild type DENV-4 NS1 structure model developed based on the X-ray crystal structure of DENV-2 NS1. Blue colour represents the β-roll domain, yellow indicates the wing domain, green shows the connector sub-domain and orange signifies the central β-ladder domain. The red dotted circle represents unresolved regions (amino acids 7 to 11 and amino acids 108 to 128).
Table 6.
Quality assessment of the predicted WT and mutant DENV-4 NS1 structures using SWISS-MODEL and AlphaFold2.
| Program | WT/ Mutation |
Procheck (Ramachandran Plot Statistic (%)) |
ProSA | Errat (%) | |||
|---|---|---|---|---|---|---|---|
| Most Favoured |
Additionally Allowed | Generously Allowed | Disallowed | Z-Score | Overall Quality Factor Score | ||
| SWISS-MODEL | WT | 85.6 | 13.1 | 0.5 | 0.8 | −6.67 | 91.2442 |
| p.His50Tyr | 87.1 | 11.7 | 0.7 | 0.5 | −6.7 | 88.2083 | |
| p.Pro144Ser | 87.1 | 11.7 | 0.7 | 0.5 | −6.71 | 88.2083 | |
| AlphaFold2 | WT | 90.4 | 9.3 | 0 | 0.3 | −6.95 | 87.1681 |
| p.His50Tyr | 90.4 | 9.3 | 0 | 0.3 | −7 | 86.9822 | |
| p.Pro144Ser | 90.4 | 9.2 | 0 | 0.3 | −7.04 | 87.1681 | |
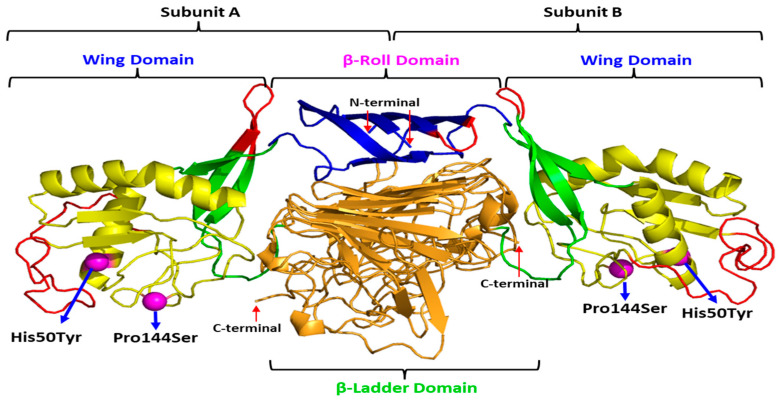
Figure 5 depicts the position of the mutations in the DENV-4 NS1 homo-dimer structure. The mutations are mapped to subunits A and B of the homo-dimer DENV-4 NS1 structure model. Both mutations, His50Tyr and Pro144Ser, are located in the wing domain. The His50Tyr is located at α-helix 1, while the Pro144Ser is observed at the loop after β-sheet 5.
Figure 5.
Mutations mapped onto subunits A and B of the homo-dimer wild type DENV-4 NS1.
3.5. Mutation Effect
The effect of the mutation on the stability of NS1 is presented in Table 7. The stability of both mutant models, which were developed using Swiss Model and Alphafold, was assessed using FoldX. The prediction produced by FoldX was consistent for both mutant models. The His50Tyr mutant was predicted to be benign while the Pro144Ser mutation was predicted to be destabilizing based on the following criteria; ΔΔG value > 3 kcal/mol is severely destabilizing, 1–3 kcal/mol is destabilizing and <1 kcal/mol is neutral or benign.
Table 7.
Thermodynamic stability of the predicted WT and mutant DENV-4 NS1 structures using FoldX.
| Program | Mutation | FoldX (kcal/mol) |
|---|---|---|
| SWISS-MODEL | p.His50Tyr | −0.279206 |
| p.Pro144Ser | 2.9665 | |
| AlphaFold2 | p.His50Tyr | −0.233207 |
| p.Pro144Ser | 3.5151 |
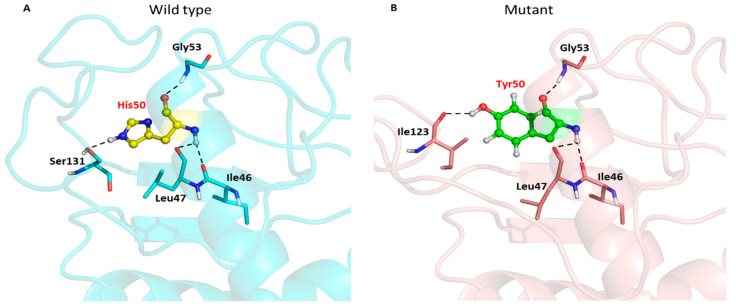
The substitution of histidine to tyrosine caused the loss of the hydrogen bond interaction between the backbone nitrogen atom of histidine and the oxygen atom of the serine side chain at residue 131 (Figure 6A and Table 8). However, the substituted Y50 formed a new hydrogen bond interaction with the oxygen atom of the isoleucine side chain at residue 123, as shown in Figure 6B.
Figure 6.
Hydrogen bond interactions between the wild type (His50) and the mutant (Tyr50) with their neighbouring residues. The wild type residue is presented as the yellow stick and sphere (A), while the mutant residue is presented as the green stick and sphere (B). Hydrogen bonds are indicated by black dotted lines. The DENV-4 NS1 structures are presented in cyan and salmon cartoons, respectively.
Table 8.
Distribution of hydrogen bond interactions in the WT and mutant models.
| WT/ Mutation |
Donor (Amino Acid/Residue/Chain/Atom) |
Acceptor (Amino Acid/Residue/Chain/Atom) |
Hydrogen (Amino Acid/Residue/Chain/Atom) |
Distance (Å) |
|---|---|---|---|---|
| WT_His50 | HIS 50.A (N) | ILE 46.A (O) | HIS 50.A (H) | 2.937 |
| GLY 53.A (N) | HIS 50.A (O) | GLY 53.A (H) | 2.823 | |
| HIS 50.A (N) | LEU 47.A (O) | HIS 50.A (H) | 3.14 | |
| HIS 50.A (NE) | SER131.A (OG) | HIS 50.A (H) | 3.19 | |
| Mut_Tyr50 | TYR 50.A (N) | ILE 46.A (O) | TYR 50.A (H) | 2.844 |
| TYR 50.A (OH) | ILE 123.A (O) | TYR 50.A (HH) | 2.952 | |
| GLY 53.A (N) | TYR 50.A (O) | GLY 53.A (H) | 2.823 | |
| TYR 50.A (N) | LEU 47.A (O) | TYR 50.A (H) | 3.21 | |
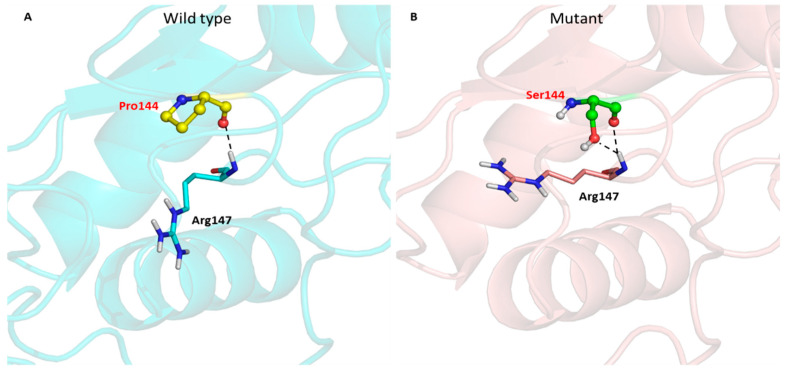
| WT_Pro144 | ARG 147.A (N) | PRO 144.A (O) | ARG 147.A (H) | 2.831 |
| Mut_Ser144 | ARG 147.A (N) | SER 144.A (O) | ARG 147.A (H) | 2.88 |
| ARG 147.A (N) | SER 144.A (OG) | ARG 147.A (H) | 3.418 |
Meanwhile, the substituted Ser144 preserved the hydrogen bond interaction with arginine at residue 147 (Figure 7 and Table 8). This substituted Ser144 even introduced another hydrogen bond interaction between the oxygen atom of the serine side chain and the backbone nitrogen atom of arginine (Figure 7B).
Figure 7.
Hydrogen bond interactions between the wild type (Pro144) and the mutant (Ser144) with their neighbouring residues. The wild type residue is denoted as a yellow stick and sphere (A), while the mutant residue is presented as the green stick and sphere (B). Hydrogen bonds are indicated by black dotted lines. The DENV-4 NS1 structures are presented in cyan and salmon cartoons, respectively.
Next, we analyzed a public dataset of genotype–fitness maps of DENV from a study reported by Dolan et al., 2021 [21] to determine the impact of the mutations observed in our study on virus evolution. The aforementioned study assessed fitness effects for all single-nucleotide variants and determined the beneficial and deleterious effect to the overall fitness of the population. The relative fitness (w) and its 95% confidence interval (CI) (in human cells) for His50Tyr was 0.60 (0.25–0.96) for set A and 0.45 (0.28–0.58) for set B and for Pro144Ser was 0.32 (0.13–0.53) for set A and 0.27 (0.18–0.45) for set B. Set A and B are independent passage lineage for each cell line to represent technical replicates. In mosquito cells, the w and CI for His50Tyr was 0.92 (0.16–1.92) for set A and 0.28 (0.19–0.44) for set B and for Pro144Ser was 0.18 (0.08–0.25) for set A and 0.15 (0.01–0.27) for set B. In set A and B, both mutations were interpreted to have deleterious effects on viral fitness in both human and mosquito cells.
4. Discussion
This study focuses on the circulating trend of DENV-4 in Malaysia from 2017 to 2022 to elucidate possible changes in clinicodemographic and genome profiles at points where a significant predominance was observed. National surveillance data on circulating dengue serotypes in Malaysia from 1990–2014 revealed a consistent predominance of other serotypes, such as DENV-1, 2 and 3, as compared to DENV-4 [22]. The data also showed that DENV-4 retained its existence at the background level, except for a minor increase during 2011–2012.
The findings from our study demonstrated an unprecedented predominance of DENV-4 from 2021 until 2022, while DENV-2 and DENV-3 were observed to be the major serotypes prevalent prior to these periods. A review of the most recent dengue outbreaks in neighbouring countries revealed a diverse occurrence. Singapore reported a predominance of DENV-3 from 2020–2022 [23,24], while DENV-4 had the lowest prevalence. Meanwhile, Indonesia documented a striking predominance of the DENV-4 genotype IIa during an outbreak in East Jawa from 2019–2020, with the majority of the cases presenting mild infection [25]. Additionally, the Philippines reported the emergence of the DENV-4 genotype IIa for the first time in 2015–2017 [26], whereas a national epidemic declared in 2019 was caused by DENV-3 [27]. The scenario of DENV-4 incidence in the Philippines coincided with Malaysia. In the past, DENV-4 was the least common serotype in the Philippines, where one outbreak was noted in 1964. However, a dramatic rise in DENV-4 infection cases from 2015 to 2018 was reported, which prompted a molecular epidemiological investigation. It was also determined that DENV-4 has been evolving quickly in recent years, with a significant genotype turnover to genotype IIa and the ensuing disappearance of genotype I. It was suggested that this genotype may possess higher fitness than previous strains or enhanced ability to replicate or transmit at the population level in the Philippines. This emphasises how important the genetic evolution of DENV is to establishing and maintaining transmission.
The majority of the DENV-4-infected cases during the period of our study were identified in male adults residing in the central region of peninsular Malaysia, namely the Federal Territory of Kuala Lumpur. The clinical presentations mostly indicated dengue with warning signs, such as persistent vomiting, diarrhoea, lethargy and abdominal pain. A handful of studies have shown that dengue symptoms are serotype specific [28,29]. DENV-4-infected patients commonly manifested respiratory and cutaneous symptoms [30]. Another study also reported a high likelihood of DENV-4 patients exhibiting rashes, in addition to headache and nausea [31]. In contrast, our study demonstrated a high incidence of gastrointestinal manifestations in DENV-4 infection, consistent with a study in Brazil that reported vomiting and abdominal pain as the two most frequent symptoms [32]. Gastrointestinal manifestation can be a medical concern when patients advance to bleeding and require the need for intensive care. According to one study [33], gastrointestinal bleeding complicated 4.4% of dengue infections, and according to another [34], a vast majority of patients experiencing gastrointestinal symptoms required hospitalisation. Therefore, it is critical to recognise the significance of gastrointestinal symptoms in dengue patients as soon as possible for a number of reasons, including the potential financial impact on patients and their families and the possibility that early diagnosis could prevent these patients from being admitted to the intensive care unit [35].
Similar to the findings of the present study, which showed infrequent severe cases compared to non-severe dengue, DENV-4 has rarely been related to severe clinical presentations [36]. One study, however, revealed a high prevalence of severe haemorrhagic manifestations [37]. In our study, both primary and secondary infections were identified as occurring in equal frequency during the 6-year period of analysis. However, limitations pertaining to missing antibody detection results for a number of cases were also observed. Since the data collection process was performed for surveillance purposes, it relied on the precision of the involved healthcare workers when documenting clinical information on patient forms.
In the present study, the DENV-4 full genome sequences were successfully obtained from 14 cases (average Ct 23.0 ± 5.0) between 2020 and 2022. The attempt to sequence the remaining samples was not fruitful, owing to the relatively low viral load (average Ct 31.0 ± 7.0), insufficient original samples to repeat (n = 10) and possible degradation of the RNA due to long duration of storage despite maintenance of an appropriate temperature. This resulted in small sample size and can be regarded as a limitation of this study. Nevertheless, the contribution of this study to the literature comes from its provision of more genome data, since only two Malaysian DENV-4 full genome sequences were found to be available in the NCBI database (MH888334 and MH051734) over the past years. The majority of the sequence submissions were, in fact, partial genomes, which hampered in-depth comparisons with the DENV-4 genome data prior to 2020.
The genome investigation of DENV-4 revealed the dominance of a mono-genotype and the localization of genotype IIa in Malaysia. This genotype which was first discovered in the country in 2001, was reported to confer a unique mutation at amino acid position 120 in the E gene, characterised by a substitution of serine (S) with leucine (L) [4]. The Ser120Leu mutation was found to have been well conserved in all 14 DENV-4 genomes that were sequenced in our study. A phylogenetic tree, constructed using the whole genome, revealed the existence of two clades of genotype IIa, along with a clear geographical segregation between these clades. Both clades had a distribution of cases from the central region of Malaysia, which then diverged to cases from the northern and southern regions, suggesting the possibility of the central region being the hub for the recent DENV-4 outbreak before its spread. In addition, clade 1 exhibited a stronger founder effect due to its relatively limited divergence, thus creating a high chance of widespread DENV-4 genotype IIa in the northern state of Kedah in the near future. However, this finding should be interpreted cautiously due to small sample size.
Two novel mutations in the NS1 gene from the cases in 2020–2022 were discovered to be unique to the Malaysian DENV-4 strains. Substitutions of histidine to tyrosine at amino acid position 50 of the NS1 protein and from proline to serine at position 144 were observed. As evident in this study, compared to the strains isolated in 2021–2022, the majority of DENV-4 from the year 2020 did not confer these mutations. Interestingly, this finding corresponds to the spike of DENV-4 cases from 2021 onwards. It is noteworthy that these mutations did not have a concluding pattern on the clinical outcomes of the patients. This warrants further study on the association of His50Tyr and Pro144Ser with disease severity.
The high sequence conservation of NS1 between DENV-2 and DENV-4 (>70%) (data not shown) allowed us to produce a high-quality model of mutant DENV-4. Even though protein structure analysis revealed some important characteristics of the mutations, such as its location and potential atomic interactions, the results of thermodynamic stability analysis was inconclusive. In our analysis, we found that His50Tyr mutant was predicted to be stable while the Pro144Ser mutation was predicted to be unstable. However, the positions of the mutations could influence this result because His50Tyr was located on stable alpha helices while Pro144Ser was located on a flexible loop region. FoldX is more accurate on mutations located on rigid structures and less accurate on mutations on flexible loops/disordered regions. This led us to explore the impact of these mutations on viral fitness based on a public dataset.
The NS1 protein is an enigmatic component of DENV that plays a significant role in viral replication and virion production [38]. The impact of NS1 mutations has been reported. Mutants, such as Val236Ala in DENV-2 or Trp68➔stop codon in DENV-3, were associated with decreased NS1 production and secretion, resulting in ELISA-negative diagnoses [39]. Another mutation, Thr164Ser in DENV-2 was found to decrease virus production but increased production of secreted NS1, leading to greater production of proinflammatory cytokines such as IL-1 and TNFα [40]. Pairwise growth competition assay showed that Thr164Ser has reduced fitness compared to wild-type virus [40]. The reduction in viral fitness due to mutations was a common phenomenon as it was estimated that DENV genomes have a 40–50% probability to acquire a deleterious mutation but only a 0.2–0.3% probability to acquire a beneficial mutation per replication cycle [21]. However, the consequence of having a reduced viral fitness on dengue transmission remained unclear.
The findings of this study provide valuable information on DENV-4 and its predominant genotype circulation in Malaysia. In addition, this study also provides useful information for consideration during the development of a dengue vaccine. One of the challenges to developing an effective dengue vaccine has been attributed to the characteristics of the four distinct dengue serotypes, wherein the efficacy of a vaccine depends on simultaneously neutralising high-affinity antibodies in all four serotypes. A safe and effective dengue virus vaccine needs to stimulate the neutralisation of antibodies by targeting unique sites on each of the four dengue serotypes [41]. To this end, the genome data of DENV-4 generated in our study would be a tangible reference point.
5. Conclusions
The present study identified DENV-4 as the predominant serotype in Malaysia from 2021 to 2022. Additionally, genotype IIa was observed to have maintained sustained circulation ever since its discovery in 2001. Although there were no significant changes in the clinical outcomes of dengue, unique mutations were discovered among the recently sequenced DENV-4. Both mutations were predicted to have deleterious effects to viral fitness, but the implications especially on dengue transmission remained elusive. Continuous surveillance of DENV is recommended to profile circulating variants in the population such as those identified in this study as well as to identify strains with epidemic potential.
Acknowledgments
The authors would like to thank the Director General of Health, Malaysia, for granting permission to publish this paper.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/tropicalmed8080409/s1, Supplement File S1: Distribution of each serotype by states from 2017-mid 2022.
Author Contributions
Conceptualization, J.S.; methodology, J.S. and E.Z.A.; software, E.Z.A., M.K.N.M.K., K.K.T., S.M.G. and S.S.L.S.; validation, J.S., E.Z.A. and M.K.N.M.K.; formal analysis, J.S., E.Z.A., M.K.N.M.K., K.K.T., S.M.G. and S.S.L.S.; investigation, J.S., M.M.S.Z., N.R., A.H.A., M.N.R. and F.R.; resources, J.S.; data curation, J.S.; writing—original draft preparation, J.S., E.Z.A. and M.K.N.M.K.; writing—review and editing, J.S., S.S.L.S., R.M.Z. and R.T.; supervision, R.M.Z. and R.T.; funding acquisition, J.S. and R.T. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study has been given an expedited ethical approval including a waiver for informed consent by the Medical Research and Ethics Committee (MREC), Ministry of Health Malaysia. All methods were carried out in accordance with Declaration of Helsinki.
Informed Consent Statement
Patient consent was waived for this study due to the use of extracted RNAs from retrospective samples and data were analysed anonymously.
Data Availability Statement
The full genome sequences of DENV-4 generated in this study are available in the NCBI database (accession numbers: OP600478, OP600495-OP600499, OP600506-OP600512). Other data reported in the current study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
The study was funded under the Virology Unit, IMR’s operational budget (OS27403).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Smith C.E.G. Isolation of Three Strains of Type 1 Dengue Virus from a Local Outbreak of the Disease in Malaya. J. Hyg. 1956;54:569–580. doi: 10.1017/S0022172400044843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wallace H.G., Lim T.W., Rudnick A., Knudsen A.B., Cheong W.H., Chew V. Dengue hemorrhagic fever in Malaysia: The 1973 epidemic. Southeast Asian J. Trop. Med. Public Health. 1980;11:1–13. [PubMed] [Google Scholar]
- 3.AbuBakar S., Shafee N. Outlook of dengue in Malaysia: A century later. Malays. J. Pathol. 2002;24:23–27. [PubMed] [Google Scholar]
- 4.AbuBakar S., Wong P.F., Chan Y.F. Emergence of dengue virus type 4 genotype IIA in Malaysia. J. Gen. Virol. 2002;83:2437–2442. doi: 10.1099/0022-1317-83-10-2437. [DOI] [PubMed] [Google Scholar]
- 5.Gintarong T.J., Emran A., Sherin A., Thein T.T., Aung T.S. Circulation of all dengue virus serotypes during dengue outbreak in Sandakan, Sabah, Malaysia (2016) J. Vector Borne Dis. 2008;55:168–171. doi: 10.4103/0972-9062.242566. [DOI] [PubMed] [Google Scholar]
- 6.Johnson B.W., Russell B.J., Lanciotti R.S. Serotype-specific detection of dengue viruses in a fourplex real-time reverse transcriptase PCR assay. J. Clin. Microbiol. 2005;43:4977–4983. doi: 10.1128/JCM.43.10.4977-4983.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li D., Liu C.M., Luo R., Sadakane K., Lam T.W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2019;31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 8.Kumar S., Stecher G., Li M., Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samson S., Lord É., Makarenkov V. SimPlot++: A Python application for representing sequence similarity and detecting recombination. Bioinformatics. 2022;38:3118–3120. doi: 10.1093/bioinformatics/btac287. [DOI] [PubMed] [Google Scholar]
- 10.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mirdita M., Schütze K., Moriwaki Y., Heo L., Ovchinnikov S., Steinegger M. ColabFold: Making protein folding accessible to all. Nat. Methods. 2022;19:679–682. doi: 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johansson M.U., Zoete V., Michielin O., Guex N. Defining and searching for structural motifs using DeepView/Swiss-Pdb Viewer. BMC Bioinform. 2012;13:173. doi: 10.1186/1471-2105-13-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seeliger D., de Groot B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. Comput. Aided Mol. Des. 2010;24:417–422. doi: 10.1007/s10822-010-9352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laskowski R.A., MacArthur M.W., Thornton J.M. PROCHECK: Validation of protein-structure coordinates. Int. Tables Crystallogr. 2012;21:684–687. doi: 10.1107/97809553602060000882. [DOI] [Google Scholar]
- 15.Wiederstein M., Sippl M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colovos C., Yeates T.O. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 1993;2:1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schymkowitz J., Borg J., Stricher F., Nys R., Rousseau F., Serrano L. The FoldX web server: An online force field. Nucleic Acids Res. 2005;33:W382–W388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caswell R.C., Gunning A.C., Owens M.M., Ellard S., Wright C.F. Assessing the clinical utility of protein structural analysis in genomic variant classification: Experiences from a diagnostic laboratory. Genome Med. 2022;14:77. doi: 10.1186/s13073-022-01082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 20.Akey D.L., Brown W.C., Dutta S., Konwerski J., Jose J., Jurkiw T.J., DelProposto J., Ogata C.M., Skiniotis G., Kuhn R.J., et al. Flavivirus NS1 structures reveal surfaces for associations with membranes and the immune system. Science. 2014;343:881–885. doi: 10.1126/science.1247749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dolan P., Taguwa S., Rangel M., Acevedo A., Hagai T., Andino R., Frydman J. Principles of dengue virus evolvability derived from genotype-fitness maps in human and mosquito cells. eLife. 2021;10:e61921. doi: 10.7554/eLife.61921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mudin R.N. Dengue incidence and the prevention and control program in Malaysia. IIUM Med. J. Malays. 2015;14:5–10. doi: 10.31436/imjm.v14i1.447. [DOI] [Google Scholar]
- 23.Huang L., Xiao G., Chen H., Niu X., Fu X., Xu H., Xu G., Ma S., Ong J., Ng L.C. Geographical clusters of dengue outbreak in Singapore during the Covid-19 nationwide lockdown of 2020. Sci. Data. 2022;9:547. doi: 10.1038/s41597-022-01666-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheema H.A., Mujtaba R.S., Siddiqui A., Vohra L.I., Shahid A., Shah J., Nashwan A.J., Howard N. Singapore’s Dengue Outbreak Amidst the COVID-19 Pandemic: Challenges, Responses, and Lessons. Infect. Drug Resist. 2023;22:1081–1085. doi: 10.2147/IDR.S397407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aryati A., Wrahatnala B.J., Yohan B., Fanny M., Hakim F.K.N., Sunari E.P., Zuroidah N., Wardhani P., Santoso M.S., Husada D., et al. Dengue virus serotype 4 is responsible for the outbreak of dengue in East Java city of Jember, Indonesia. Viruses. 2020;12:913. doi: 10.3390/v12090913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luz M.A.V., Nabeshima T., Moi M.L., Dimamay M.T.A., Pangilinan L.S., Dimamay M.P.S., Matias R.R., Mapua C.A., Buerano C.C., de Guzman F., et al. An Epidemic of dengue virus serotype-4 during the 2015–2017: The emergence of a novel genotype IIa of DENV-4 in the Philippines. Jpn. J. Infect Dis. 2019;72:413–419. doi: 10.7883/yoken.JJID.2019.208. [DOI] [PubMed] [Google Scholar]
- 27.Dyer O. Dengue: Philippines declares national epidemic as cases surge across South East Asia. BMJ. 2019;366:l5098. doi: 10.1136/bmj.l5098. [DOI] [PubMed] [Google Scholar]
- 28.Tsai J.J., Chan K.S., Chang J.S., Chang K., Lin C.C., Huang J.H., Lin W.R., Chen T.C., Hsieh H.C., Lin S.H., et al. Effect of serotypes on clinical manifestations of dengue fever in adults. J. Microbiol. Immunol. Infect. 2009;42:471–478. [PubMed] [Google Scholar]
- 29.Yung C.F., Lee K.S., Thein T.L., Tan L.K., Gan V.C., Wong J.G.X., Lye D.C., Ng L.C., Leo Y.S. Dengue serotype-specific differences in clinical manifestation, laboratory parameters and risk of severe disease in adults, Singapore. Am. J. Trop. Med. Hyg. 2015;92:999–1005. doi: 10.4269/ajtmh.14-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Halsey E.S., Marks M.A., Gotuzzo E., Fiestas V., Suarez L., Vargas J., Aguayo N., Madrid C., Vimos C., Kochel T.J., et al. Correlation of serotype-specific dengue virus infection with clinical manifestations. PLoS Negl. Trop. Dis. 2012;6:e1638. doi: 10.1371/journal.pntd.0001638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khan E., Prakoso D., Imtiaz K., Malik F., Farooqi J.Q., Long M.T., Barr K.L. The clinical features of co-circulating dengue viruses and the absence of dengue hemorrhagic fever in Pakistan. Front. Public Health. 2020;8:287. doi: 10.3389/fpubh.2020.00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramalho I.L.C., Araújo F.M.C., Cavalcanti L.P.G., Braga D.N.M., Perdigão A.C.B., Santos F.B.D., Nogueira F.B., Escóssia K.N.F.D., Guedes M.I.F. Dengue 4 in Ceará, Brazil: Characterisation of epidemiological and laboratorial aspects and causes of death during the first epidemic in the state. Mem. Inst. Oswaldo Cruz. 2018;113:e180320. doi: 10.1590/0074-02760180320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ooi E.T., Ganesananthan S., Anil R., Kwok F.Y., Sinniah M. Gastrointestinal manifestations of dengue infection in adults. Med. J. Malays. 2008;63:401–405. [PubMed] [Google Scholar]
- 34.Ramos-De La Medina A., Remes-Troche J.M., González-Medina M.F., Anitúa-Valdovinos Mdel M., Cerón T., Zamudio C., Díaz-Vega A. Abdominal and gastrointestinal symptoms of Dengue fever. Analysis of a cohort of 8559 patients. Gastroenterol. Hepatol. 2011;34:243–247. doi: 10.1016/j.gastrohep.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 35.Abrar T., Selamat T., Abbasi A.H., Cheema H.J., Ahmed S., Zaka M. Frequency of Gastrointestinal Signs and Symptoms in Patients with Dengue Fever in Tertiary Care Hospital. Pak. J. Med. Health Sci. 2023;17:721. doi: 10.53350/pjmhs2023172721. [DOI] [Google Scholar]
- 36.Barde P.V., Godbole S., Bharti P.K., Chand G., Agarwal M., Singh N. Detection of dengue virus 4 from central India. Indian. J. Med. Res. 2012;136:491–494. [PMC free article] [PubMed] [Google Scholar]
- 37.Kumaria R. Correlation of disease spectrum among four Dengue serotypes: A five years hospital based study from India. Braz. J. Infect. Dis. 2010;14:141–146. doi: 10.1016/S1413-8670(10)70027-1. [DOI] [PubMed] [Google Scholar]
- 38.Scaturro P., Cortese M., Chatel-Chaix L., Fischl W., Bartenschlager R. Dengue virus non-structural protein 1 modulates infectious particle production via interaction with the structural proteins. PLoS Pathog. 2015;11:e1005277. doi: 10.1371/journal.ppat.1005277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghosh A., Sukla S., Nath H., Saha R., De A., Biswas S. Non-structural protein 1 (NS1) variants from dengue virus clinical samples revealed mutations that influence NS1 production and secretion. Eur. J. Clin. Microbiol. Infect. Dis. 2022;41:803–814. doi: 10.1007/s10096-022-04441-4. [DOI] [PubMed] [Google Scholar]
- 40.Chan K.W.K., Watanabe S., Jin J.Y., Pompon J., Teng D., Alonso S., Vijaykrishna D., Halstead S.B., Marzinek J.K., Bond P.J., et al. A T164S mutation in the dengue virus NS1 protein is associated with greater disease severity in mice. Sci. Transl. Med. 2019;11:eaat7726. doi: 10.1126/scitranslmed.aat7726. [DOI] [PubMed] [Google Scholar]
- 41.Henein S., Adams C., Bonaparte M., Moser J.M., Munteanu A., Baric R., de Silva A.M. Dengue vaccine breakthrough infections reveal properties of neutralizing antibodies linked to protection. J. Clin. Investig. 2021;131:e147066. doi: 10.1172/JCI147066. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The full genome sequences of DENV-4 generated in this study are available in the NCBI database (accession numbers: OP600478, OP600495-OP600499, OP600506-OP600512). Other data reported in the current study are available from the corresponding author upon request.