Abstract
Acinetobacter baumannii is one of the significant healthcare-associated meningitis agents characterized by multidrug resistance and a high mortality risk. Thirty-seven A. baumannii strains were isolated from thirty-seven patients of Moscow neuro-ICU with meningitis in 2013–2020. The death rate was 37.8%. Strain susceptibility to antimicrobials was determined on the Vitek-2 instrument. Whole-genome sequencing was conducted using Illumina technology; the sequence types (ST), capsular types (KL), lipooligosaccharide outer core locus (OCL), antimicrobial resistance genes, and virulence genes were identified. The prevalent ST was ST2, belonging to the international clone IC2, and rarer, ST1, ST19, ST45, ST78, ST106, and ST400, with prevalence of KL9 and OCL1. Twenty-nine strains belonged to multidrug-resistant (MDR) and eight extensively drug-resistant (XDR) categories. Genes conferring resistance to beta-lactams (blaPER, blaGES, blaADC, blaCARB, blaCTX-M, blaTEM, and blaOXA-types), aminoglycosides (aac, aad, ant, aph, and arm), tetracyclines (tet), macrolides (msr and mph), phenicols (cml, cat, and flo), sulfonamides (dfr and sul), rifampin (arr), and antiseptics (qac) were identified. Virulence genes of nine groups (Adherence, Biofilm formation, Enzymes, Immune evasion, Iron uptake, Regulation, Serum resistance, Stress adaptation, and Antiphagocytosis) were detected. The study highlights the heterogeneity in genetic clones, antimicrobial resistance, and virulence genes variability among the agents of A. baumannii meningitis, with the prevalence of the dominant international clone IC2.
Keywords: Acinetobacter baumannii, meningitis, ICU, MDR, XDR, virulence genes, biofilm
1. Introduction
Carbapenem-resistant Acinetobacter baumannii (CRAB) has recently been labeled as a “critical” pathogen, i.e., constituting a significant global human health risk, in the World Health Organization’s list of antimicrobial-resistant bacteria intended to guide the research and development of new effective antibiotic treatments [1]. The recent COVID-19 pandemic was associated with an increasing risk of hospital-acquired ventilator-associated secondary infections, for which A. baumannii was a leading cause [2]. A. baumannii has been frequently reported as the causative pathogen of nosocomial infections, such as ventilator-associated pneumonia, bloodstream infection, urinary tract infection, wound infection, and meningitis. Nosocomial meningitis is a potentially life-threatening central nervous system infection associated with increased mortality, morbidity, and hospital costs; A. baumannii accounted for more than 25% of all pathogens isolated from cerebrospinal fluid (CSF) [3]. The increasing prevalence of CRAB has become a serious clinical problem in many countries because the mortality rate of CRAB meningitis has been reported to exceed 70% [4,5].
The predominance of a few successful multidrug-resistant lineages worldwide underlines the importance of elucidating the epidemiology of A. baumannii isolates in single hospitals, at a country-wide level, and on a global scale. The population structure of A. baumannii is composed of A. baumannii strains assigned to 176 different sequence types (STs) that can be grouped into 20 clonal complexes (CCs) according to Pasteur’s Multilocus sequence typing (MLST) scheme [6]. Three A. baumannii international clones (ICs) have been determined as prevalent: IC I (includes ST1, ST7, ST8, ST19, and ST20); IC II (includes ST2, ST45, ST47, and ST59); IC III (includes ST3, ST13, and ST14). In Europe, IC I and IC II cause the vast majority of infections, and IC II was reported in most cases. IC I, IC II, and IC III were characterized by increasing resistance to clinically significant antimicrobials [7,8]. The emergent IC VI (ST78) strains belonged to the CRAB and presented high biofilm-forming ability, increased resistance to drying, and an enhanced capacity for host cell adhesion/invasion, favoring its diffusion and persistence, and have been identified in European countries (Italy, Russia, Greece, and Germany), Asia (Kuwait), and North and South America (the USA and French Guiana) [9,10,11]. Moreover, the new sequence type ST400 is not related to those of the main clonal complexes known to have spread worldwide [12].
A. baumannii carbapenem resistance is mainly associated with the production of carbapenemases, and more rarely, with the efflux pumps and with the reduction or inactivation of porins expression [13]. The most common A. baumannii carbapenemases are OXA-23-like, OXA-24/40-like, and OXA-58-like enzymes [14]. The prevalence of OXA-23-like-producers is attributed to the spread of successful global clones such as GC1 and GC2 [15]. Moreover, A. baumannii resistomes contain other groups of beta-lactamases, e.g., the extended-spectrum beta-lactamases (ESBLs) SHV-5, TEM-92, CTX-M-2, CTX-M-15, PER-1, PER-2, PER-7, VEB-1, and GES-14, narrow-spectrum beta-lactamases (NSBLs) TEM-1 and SCO-1, and AmpC beta-lactamases ADC-30, ADC-56, etc. [16]. A. baumannii is a life-threatening problem not only because of multidrug resistance but also its ability to evade the host immune response, to survive under stressful environmental conditions, its resistance to disinfectants, and its ability to form complex structures of biofilms. It was shown that there are several master regulators of antimicrobial resistance, stress response, and virulence in A. baumannii, such as GigAB and DksA [17,18].
The virulence factors of A. baumannii cells include (i) the adherence due to Acinetobacter trimeric autotransporter Ata and Type IV pili TFP; (ii) the effector delivery system including type II and VI secretion systems T2SS and T6SS, as well as T6SS effectors Tse1, Tse2, Tse3, and Tse4; (iii) the exotoxins, e.g., phospholipases C and D; (iv) the exoenzyme, coagulation targeting metalloendopeptidase CpaA; (v) the immune modulation due to capsule, lipopolysaccharide LPS, outer membrane protein A OmpA, and penicillin-binding protein PbpG; (vi) the biofilm formation associated with efflux pump AdeFGH, biofilm-associated protein Bap, fimbriae Csu, poly-(1,6)-N-acetyl glucosamine PNAG, and the autoinducer–receptor mechanism for quorum sensing; and (vii) the nutritional/metabolic factors such as acinetobactin and Heme uptake system [19].
This study analyzed the antimicrobial susceptibility, biofilm formation, and virulence factors, as well as their genetic determinants in whole-genome sequences, of 37 A. baumannii strains that caused meningitis in 37 patients (mortality rate 37.8%) in Moscow Neurological intensive care unit in 2013 to 2020. Seven sequence types were identified in this study using the Pasteur MLST scheme: ST1, ST2, ST19, ST45, ST78, ST106, and ST400. The most prevalent (23/37) was ST2, which was associated with 12 of 14 patient deaths. The results are significant for expanding our knowledge about the molecular epidemiology of the nosocomial A. baumannii in critical care units in Russia. The resistome and virulome analysis provides the data that can be used for the development of control strategies against A. baumannii infections and novel antimicrobial and anti-virulence targets for detection systems and treatment.
2. Materials and Methods
2.1. Research, Bioethical Requirements, and Patients
This study was a retrospective cohort observation analytical study. Under the requirements of the Russian Federation Bioethical Committee, each patient signed informed voluntary consent to treatment and laboratory examination. The study did not contain the personal data of patients, such as the name, date of birth, address, and disease history. The study was a retrospective observational study in the neuro-ICU department in a specialized Neurosurgical Hospital in Moscow, Russia, with 300 beds that care for approximately 8000 patients per year, 95% of whom undergo surgery [20].
2.2. Bacterial Isolates and Growing
A. baumannii isolates (n = 37) were collected from clinical samples (cerebrospinal fluid) of the patients (n = 37) in the neuro-ICU in the period from January 2013 to September 2020. Bacterial identification was performed using a MALDI-TOF Biotyper (Bruker Daltonics, Bremen, Germany) instrument. Bacterial isolates were grown at 37 °C on Nutrient Medium No. 1 (SRCAMB, Obolensk, Moscow region, Russia), Luria–Bertani broth (Difco Laboratories, Detroit, MI, USA), and Muller–Hinton broth (Himedia, Mumbai, Maharashtra, India). Bacterial isolates were stored in 15% glycerol at minus 80 °C.
2.3. Antimicrobial Susceptibility
Susceptibility to 23 antimicrobials (AMs) of 11 functional groups—aminoglycosides (gentamicin, tobramycin, amikacin, netilmicin); carbapenems (imipenem, meropenem), penicillin (ampicillin); penicillin/beta-lactam inhibitors (ampicillin–sulbactam, piperacillin/tazobactam); cephalosporins (cefuroxime, cefoxitin, cefotaxime, ceftazidime, ceftriaxone, cefepime, cefoperazone/sulbactam); fluoroquinolones (ciprofloxacin); sulfonamides (trimethoprim–sulfamethoxazole); phenicols (chloramphenicol); tetracyclines (tetracycline, tigecycline); nitrofurans (nitrofurantoin); and polymyxins (colistin)—were determined using Vitek-2 Compact instrument (BioMerieux, Paris, France). The results were interpreted according to the European Committee on Antimicrobial Susceptibility Testing, Version 13.0, 2023 (http://www.eucast.org, access date: 21 March 2023). Reference strains Escherichia coli ATCC 25922 and ATCC 35218 were used as quality controls. The isolates were categorized as multidrug-resistant (MDR), non-susceptible to ≥1 agent in ≥3 antimicrobial groups; and extensively drug-resistant (XDR), non-susceptible to ≥1 agent in all but ≤2 groups according to the criteria proposed by Magiorakos et al. [21].
Minimal inhibitory concentrations (MICs) of two antiseptics—chlorhexidine digluconate (Sigma-Aldrich, Saint Louis, MO, USA) and benzalkonium chloride (Sigma-Aldrich, Saint Louis, MO, USA)—were determined via the broth microdilution assay on the 96-well plates. Bacterial growth in the wells was detected via the xMARK microplate spectrophotometer (Bio-Rad Laboratories, Inc., Watford, UK). MIC values were compared with antiseptic concentrations recommended for use in ICU: 500 mg/L chlorhexidine digluconate; 10,000 mg/L benzalkonium chloride [22].
2.4. Biofilm Formation
The biofilm formation ability of A. baumannii isolates was determined via polystyrene tube assay based on the crystal violet staining method [23]. Briefly, the wells of a 96-well plate containing 0.1 mL of Luria broth (Thermo Fisher Scientific, Waltham, MA, USA) were inoculated with 100 µL of overnight bacterial culture in concentration 1 × 107 CFU/mL and incubated at 37 °C for ~20 h. The liquid media was discarded, and the adherent cells were washed twice with 0.2 mL of phosphate-buffered saline (PBS) and stained with 0.1% solution of crystal violet for 10 min. The stain was discarded, and the cells were washed three times with 0.2 mL of PBS. The stain was eluted from the adherent cells via 0.2 mL of 96% ethanol for 5 min and transferred in. The absorbance of the eluted stain was measured at 600 nm using the xMark microplate spectrophotometer (Bio-Rad, Hercules, CA, USA). The results were divided into four categories according to the sample optical densities (ODs) in comparison with the control optical densities (ODc): strong biofilm producer (4 × ODc < ODs); medium biofilm producer (2 × ODc < ODs ≤ 4 × ODc); weak biofilm producer (ODc < ODs ≤ 2 × ODc); and non-biofilm producer (ODs ≤ ODc [24].
2.5. Whole-Genome Sequencing, Assembly, and Annotation
DNA isolation was performed via the CTAB method [25]. WGS was carried out using Nextera DNA Library Preparation Kit (Illumina, San Diego, CA, USA) and MiSeq Reagent Kits v3 (Illumina, San Diego, CA, USA) for platform Illumina MiSeq (Illumina, San Diego, CA, USA). Moreover, WGS was performed on MGI (MGI Tech, Shenzhen, China) platform using MGIEasy FS DNA Library Prep Kit MGI-Seq (MGI Tech, Shenzhen, China) and 2000RS High-throughput sequencing kit PE200 (MGI Tech, Shenzhen, China). The assemblies of the genomes were obtained using Unicycler v. 0.4.7 software (The University of Melbourne, Victoria, Australia) with default settings that included primary filtering and quality control [26]. Annotation was carried out via NCBI Prokaryotic Genome Annotation Pipeline (PGAP) v. 6.4 and v. 6.5 (National Center for Biotechnology Information, Bethesda, MD, USA) [27].
2.6. Multilocus Sequence Typing, KL-Typing, and OCL-Typing
Sequence Types were identified using the MLST resource of the Center for Genomic Epidemiology (CGE) with a 95% threshold for minimum identity and 60% minimum coverage (http://www.genomicepidemiology.org/, access date: 20 March 2023). A. baumannii capsular types (KL) and lipooligosaccharide outer core locus (OCL) were identified using Kaptive software v.2.0.7 (https://www.microbiology research.org/content/journal/mgen/, access date: 30 March 2023) [28].
2.7. Antimicrobial Resistance and Virulence Genes
Antimicrobial resistance genes were recognized using ResFinder and KmerResistance resources of CGE with a 90% identity threshold and 10% threshold for depth corr. (http://www.genomicepidemiology.org/, access date: 20 March 2023). The virulence factor database (VFDB) online resource was used to identify virulence genes within the genomes with e-value cut-off 1 × 10−5, identity > 60%, and coverage > 90% (http://www.mgc.ac.cn/VFs/, access date: 20 March 2023) [29].
2.8. Phylogenetic Analysis
The phylogenetic tree of A. baumannii whole genomes was constructed with core SNPs identified via WOMBAC (https://github.com/tseemann/wombac, access date: 20 March 2023) (https://github.com/tseemann/wombac, access date: 20 March 2023) and visualized with SplitsTree4 (https://github.com/husonlab/splitstree4, access date: 20 March 2023) using the neighbor-joining (NJ) method [30].
2.9. Nucleotide Sequences Accession Numbers
The complete sequences of 37 isolates were deposited under BioProject number PRJNA269675: JAROBQ000000000, JASKJB000000000, JASKJA000000000, JAROBM000000000, JAROBU000000000, JASKIZ000000000, JAROBS000000000, JAROBO000000000, JAROBG000000000, JASKIY000000000, JASKIX000000000, JASKIW000000000, JASKIV000000000, JASKIU000000000, JAROBR000000000, JAROBT000000000, JASKIT000000000, JAROBB000000000, JAROBI000000000, JASKIR000000000, JASKIQ000000000, JASKIP000000000, JASKIO000000000, JASKIN000000000, JASKIM000000000, JASKIL000000000, JASKIK000000000, JASKIJ000000000, JASKII000000000, JASKIH000000000, JASKIG000000000, JASKIF000000000, JASKIE000000000, JASKID000000000, JASKIC000000000, JASKIB000000000.
3. Results
3.1. Patients and Clinical Data
During the period from January 2013 to Sepember 2020, 37 cases of A. baumannii meningitis (MG) were detected in a Moscow neurosurgery ICU in the patients after neurosurgery operations. Among them were 25 males of 12–75 years (median 46.7) and 12 females of 2–65 years (median 44.8). The diagnoses of the patients were oncological (n = 25), traumatic brain injury (n = 8), and disorders of the cerebral circulation including ruptured aneurysms (n = 4). Meningitis was detected in 37 patients; moreover, 19 respiratory tract infections (RTIs), 7 surgical site infections (SSIs), 5 gastrointestinal dysfunctions (GD), 5 urinary tract infections (UTIs), and 1 blood infection (BI) were detected in combinations from one (MG) to four (MG + RTI + SSI + GD or MG + RTI + UTI + GD) infections. Infections were located in the brain and other body sites simultaneously. The stays in the ICU were 8–229 days (median 67.6 days). The death rate was estimated as 37.8% (14/37 patients) (Table 1).
Table 1.
Patient information.
| Case | Gender, Age | Date | Patient’s Diagnosis | Infections | Stay in the ICU, Days | Outcome |
|---|---|---|---|---|---|---|
| #1 | M, 16 | 15 January 2013 | Medulloblastoma of the fourth ventricle | MG, RTI, SSI, GD | 186 | Survival |
| #2 | M, 46 | 27 February 2013 | Anaplastic astrocytoma | MG, RTI, UTI | 24 | Died |
| #3 | M, 50 | 4 February 2014 | Cholesteatoma | MG, BI, GD | 184 | Survival |
| #4 | F, 65 | 25 June 2014 | Suprasellar craniopharyngioma | MG | 31 | Died |
| #5 | F, 24 | 5 November 2014 | Anterior cerebral artery aneurysm | MG | 62 | Survival |
| #6 | F, 52 | 19 January 2015 | Craniofacial tumor | MG, SSI, UTI | 34 | Died |
| #7 | M, 48 | 4 February 2015 | Pilocytic astrocytoma | MG, RTI | 43 | Survival |
| #8 | M, 33 | 29 September 2015 | Traumatic brain injury | MG, RTI, SSI | 45 | Survival |
| #9 | F, 2 | 13 November 2015 | Cerebellar astrocytoma | MG, RTI, SSI | 62 | Survival |
| #10 | M, 57 | 7 December 2015 | Giant cell tumor of the middle cranial fosses | MG | 15 | Survival |
| #11 | M, 44 | 9 December 2015 | Anterior cerebral artery aneurysm | MG, RTI | 8 | Died |
| #12 | M, 43 | 18 May 2016 | Traumatic brain injury | MG | 15 | Survival |
| #13 | M, 43 | 15 June 2016 | Cerebellum tumor | MG, RTI, SSI | 63 | Died |
| #14 | M, 12 | 16 June 2016 | Giant craniopharyngioma | MG | 62 | Survival |
| #15 | M, 28 | 1 August 2016 | Atypical parasagittal meningioma | MG, SSI | 49 | Survival |
| #16 | F, 60 | 2 August 2016 | Giant olfactory meningioma | MG | 229 | Died |
| #17 | M, 75 | 16 August 2016 | Ependymoma | MG, RTI, UTI | 136 | Died |
| #18 | M, 35 | 12 September 2016 | Glioblastoma | MG | 98 | Died |
| #19 | F, 57 | 10 October 2016 | Traumatic brain injury | MG, RTI, UTI | 14 | Survival |
| #20 | M, 67 | 29 November 2016 | Sub-ependymoma | MG | 31 | Survival |
| #21 | M, 61 | 4 September 2017 | Craniopharyngioma | MG | 133 | Survival |
| #22 | F, 61 | 12 December 2017 | Hemangiopericytoma | MG | 24 | Survival |
| #23 | F, 36 | 15 January 2018 | Giant meningioma | MG, RTI, SSI | 65 | Died |
| #24 | M, 55 | 21 February 2018 | Hemangioblastoma | MG | 46 | Survival |
| #25 | M, 57 | 20 June 2018 | Gunshot wound head trauma | MG, RTI | 181 | Survival |
| #26 | F, 58 | 19 September 2018 | Glioma | MG | 52 | Died |
| #27 | M, 43 | 1 October 2018 | Hemangioblastoma of the cerebellum | MG, RTI | 53 | Survival |
| #28 | M, 53 | 23 October 2018 | Traumatic brain injury | MG, RTI | 58 | Survival |
| #29 | M, 53 | 12 November 2018 | Traumatic brain injury | MG, RTI, UTI, GD | 103 | Survival |
| #30 | M, 61 | 3 December 2018 | Post-hemorrhagic hydrocephalus | MG, RTI, GD | 19 | Died |
| #31 | M, 64 | 15 May 2019 | Skull base chordoma | MG, RTI | 25 | Died |
| #32 | F, 25 | 20 May 2019 | Traumatic brain injury | MG, RTI | 52 | Survival |
| #33 | M, 26 | 18 July 2019 | Traumatic brain injury | MG, RTI | 45 | Survival |
| #34 | M, 51 | 6 August 2019 | Ruptured middle cerebral artery aneurysms | MG | 26 | Survival |
| #35 | M, 47 | 27 January 2020 | Giant skull base chordoma | MG | 89 | Died |
| #36 | F, 52 | 28 August 2020 | Syringomyelia of the cervical spinal cord | MG, RTI | 71 | Died |
| #37 | F, 45 | 8 September 2020 | Hemangiopericytoma | MG, GD | 69 | Survival |
Note: M, Male; F, Female; MG, Meningitis; RTI, Respiratory Tract Infection; SSI, Surgical Site Infections; BI, Blood Infection; GD, Gastrointestinal Dysfunction; UTI, Urinary Tract Infection.
3.2. Sequence Types, Capsular Polysaccharide Locus Types, and Lipooligosaccharide Outer Core Locus Types
Sequence Type (ST), Capsular Polysaccharide Locus (KL) Type, and Lipooligosaccharide Outer Core Locus (OCL) Type were identified using whole genomes of A. baumannii strains. Seven STs were revealed: ST1 (n = 3); ST2 (n = 15); ST19 (n = 3); ST45 (n = 2); ST78 (n = 2); ST106 (n = 1); and ST400 (n = 1). The prevalent ST2 belonging to the pandemic lineage, named “international clone IC2”, was associated in our study with 12 deaths. Thirteen KLs were identified. The prevalent KL was KL9 (n = 14), following KL2 (n = 3), KL3 (n = 3), KL4 (n = 2), KL15 (n = 2), KL17 (n = 1), KL49 (n = 2), KL77 (n = 1), KL91 (n = 3), KL104 (n = 1), KL165 (n = 1), KL213 (n = 2), and KL235 (n = 2). The major OCL was OCL1 (n = 33), rarer–OCL5 (n = 3), and OCL6 (n = 1) (Table 2).
Table 2.
Characteristics of A. baumannii strains caused meningitis in neuro-ICU in 2013–2020.
| Case | Strain | ST | KL | OCL | AMR Phenotype | AM gr. | ARP |
|---|---|---|---|---|---|---|---|
| #1 | B-8162 | 2 | 9 | 1 | AG, CAR, FQN, CEF, SUL, PEN, PEN-IN, TET, NIT | 9 | XDR |
| #2 | B-9371 | 2 | 9 | 1 | AG, CAR, FQN, CEF, SUL, PEN, PEN-IN, NIT | 8 | XDR |
| #3 | B-9374 | 2 | 9 | 1 | AG, CAR, FQN, CEF, PEN, TET, CM, NIT | 8 | MDR |
| #4 | B-8161 | 2 | 9 | 1 | AG, CAR, FQN, CEF, SUL, PEN, PEN-IN, TET, CM, NIT | 10 | XDR |
| #5 | B-9378 | 45 | 77 | 1 | AG, CAR, FQN, CEF, PEN, PEN-IN, CM, NIT | 8 | MDR |
| #6 | B-9379 | 2 | 9 | 1 | AG, CAR, FQN, CEF, SUL, PEN, TET, CM, NIT | 9 | MDR |
| #7 | B-9380 | 2 | 2 | 1 | AG, FQN, CEF, PEN, PEN-IN, TET, CM, NIT | 8 | MDR |
| #8 | B-9381 | 2 | 9 | 1 | AG, CAR, FQN, CEF, PEN, PEN-IN, CM, NIT | 8 | MDR |
| #9 | B-9387 | 1 | 17 | 1 | AG, CAR, FQN, CEF, PEN, TET, CM, NIT | 8 | XDR |
| #10 | B-9389 | 2 | 9 | 1 | AG, CAR, FQN, CEF, SUL, PEN, CM, NIT | 8 | MDR |
| #11 | B-10836 | 2 | 9 | 1 | AG, CAR, FQN, CEF, PEN, PEN-IN, CM, NIT | 8 | MDR |
| #12 | B-9390 | 78 | 49 | 1 | AG, CAR, FQN, CEF, SUL, PEN, PEN-IN, CM, NIT | 9 | MDR |
| #13 | B-9391 | 2 | 9 | 1 | AG, CAR, FQN, CEF, SUL, PEN, TET, CM, NIT | 9 | XDR |
| #14 | B-9395 | 2 | 165 | 1 | AG, FQN, CEF, PEN, PEN-IN, CM, NIT | 7 | MDR |
| #15 | B-9397 | 2 | 213 | 1 | AG, FQN, CEF, PEN, PEN-IN, POL, NIT | 7 | MDR |
| #16 | B-9398 | 2 | 2 | 1 | AG, FQN, CEF, SUL, PEN, PEN-IN, POL, NIT | 8 | MDR |
| #17 | B-8158 | 45 | 9 | 1 | AG, FQN, CEF, PEN, POL, CM | 6 | MDR |
| #18 | B-9399 | 2 | 213 | 1 | AG, FQN, CEF, PEN, PEN-IN, TET, CM, NIT | 8 | MDR |
| #19 | B-9396 | 1 | 4 | 1 | AG, FQN, CEF, SUL, PEN, PEN-IN, CM, NIT | 8 | MDR |
| #20 | B-9400 | 1 | 4 | 1 | AG, FQN, CEF, SUL, PEN, PEN-IN, CM, NIT | 8 | MDR |
| #21 | B-9401 | 19 | 91 | 5 | AG, FQN, CEF, SUL, PEN, PEN-IN, CM, NIT | 8 | MDR |
| #22 | B-9402 | 2 | 3 | 1 | AG, CAR, FQN, CEF, SUL, PEN, PEN-IN, TET, CM, NIT | 10 | XDR |
| #23 | B-9403 | 2 | 9 | 1 | AG, CAR, FQN, CEF, PEN, CM, NIT | 7 | MDR |
| #24 | B-9404 | 106 | 104 | 6 | CEF, SUL, PEN, PEN-IN, CM, NIT | 6 | MDR |
| #25 | B-9405 | 2 | 235 | 1 | AG, CAR, FQN, CEF, SUL, PEN, PEN-IN | 7 | MDR |
| #26 | B-9406 | 400 | 15 | 1 | AG, FQN, CEF, SUL, PEN-IN | 6 | MDR |
| #27 | B-9407 | 2 | 235 | 1 | AG, CAR, FQN, CEF, SUL, PEN, PEN-IN | 7 | MDR |
| #28 | B-9408 | 78 | 3 | 1 | CAR, FQN, CEF, SUL, PEN, PEN-IN | 6 | MDR |
| #29 | B-9409 | 19 | 91 | 5 | CAR, FQN, CEF, SUL, PEN, PEN-IN | 6 | MDR |
| #30 | B-9410 | 2 | 9 | 1 | AG, CAR, FQN, CEF, SUL, PEN, PEN-IN, TET | 8 | XDR |
| #31 | B-9411 | 2 | 3 | 1 | AG, CAR, FQN, CEF, SUL, PEN, PEN-IN, TET | 8 | XDR |
| #32 | B-9412 | 2 | 9 | 1 | CAR, FQN, CEF, PEN, PEN-IN, NIT | 6 | MDR |
| #33 | B-9413 | 78 | 15 | 1 | CAR, FQN, CEF, SUL, PEN, PEN-IN | 6 | MDR |
| #34 | B-9414 | 19 | 91 | 5 | AG, FQN, CEF, SUL, PEN, PEN-IN | 6 | MDR |
| #35 | B-9415 | 2 | 2 | 1 | AG, CAR, FQN, CEF, SUL PEN, | 6 | MDR |
| #36 | B-9416 | 2 | 49 | 1 | AG, CAR, FQN, CEF, SUL, PEN, POL | 7 | MDR |
| #37 | B-9417 | 2 | 9 | 1 | AG, CAR, FQN, CEF, SUL, PEN, PEN-IN | 7 | MDR |
Note: ST, sequence type; CL, capsular locus type; OCL, lipooligosaccharide outer core locus type; AMR, antimicrobial resistance phenotype; AM gr., number of antimicrobial groups (resistance); ARP, antimicrobial resistance patterns [21]; AG, aminoglycosides; CAR, carbapenems; FQN, fluoroquinolones; CEF, cephalosporins; SUL, sulfonamides; PEN, penicillin; PEN-IN, penicillin/beta-lactam inhibitors; TET, tetracyclines; CM, phenicols (chloramphenicol); POL, polymyxins; NIT, nitrofurans; MDR, multidrug resistant; XDR, extensively drug resistant.
3.3. Antimicrobial Susceptibility
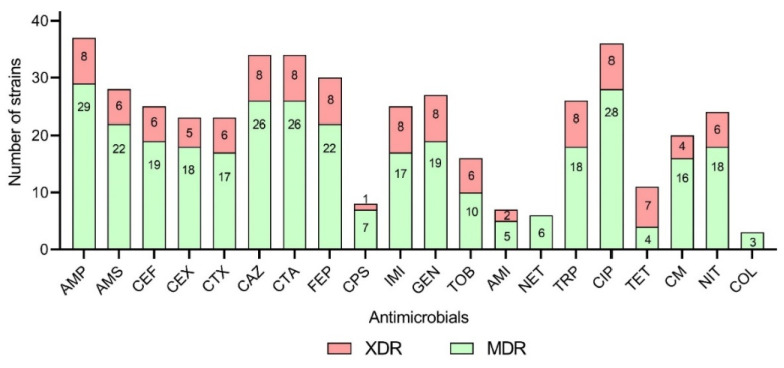
All A. baumannii strains in this study were attributed to two antimicrobial resistance patterns: MDR (n = 29) and XDR (n = 8), according to Magiorakos et al.’s criterium [21]. Major strains were resistant to penicillin and cephalosporins (n = 37), fluoroquinolones (n = 36), aminoglycosides (n = 32), penicillin/beta-lactam inhibitors (n = 28), sulfonamides (n = 26), carbapenems (n = 25), and chloramphenicol (n = 20). At the same time, dominant strains were susceptible to polymyxins (n = 34) and tetracyclines (n = 26) (Figure 1).
Figure 1.
The number of A. baumannii strains of MDR and XDR phenotypes, resistant to antibiotics: AMP, ampicillin; AMS, ampicillin–sulbactam; CEF, cefuroxime; CEX, cefoxitin; CTX, cefotaxime; CAZ, ceftazidime; CTA, ceftriaxone; FEP, cefepime; CPS, cefoperazone/sulbactam; IMI, imipenem; GEN, gentamicin; TOB, tobramycin; AMI, amikacin; NET, netilmicin; TRP, trimethoprim–sulfamethoxazole; CIP, ciprofloxacin; TET, tetracycline; CM, chloramphenicol; NIT, nitrofurantoin; COL, colistin.
Nine strains were resistant to six antimicrobial groups, seven to seven groups, fifteen to eight groups, four to nine groups, and two to ten antimicrobial groups simultaneously. The phenotypic diversity of antibiotic resistance among the strains was very high—23 variants of AMR phenotype were revealed. Carbapenem-resistant strains represented ~68% of strains (Table 2).
It was shown that planktonic cells of all A. baumannii strains were sensitive to chlorhexidine digluconate and benzalkonium chloride, with MICs 8–64 mg/L and 4–8 mg/L, respectively. Such levels of susceptibility to antiseptics are within their concentrations recommended for use in ICUs (http://dezreestr.ru/, access date 30 March 2023): 500 mg/L for chlorhexidine digluconate; 10,000 mg/L for benzalkonium chloride (Table 3).
Table 3.
Susceptibility of A. baumannii planktonic cells to antiseptics.
| Strain | MIC, mg/L | Strain | MIC, mg/L | ||
|---|---|---|---|---|---|
| CHD | BZK | CHD | BZK | ||
| B-9387 | 32 | 8 | B-9389 | 32 | 8 |
| B-9396 | 64 | 8 | B-9395 | 32 | 8 |
| B-9400 | 64 | 8 | B-9397 | 32 | 8 |
| B-9371 | 32 | 8 | B-9402 | 32 | 4 |
| B-8161 | 32 | 8 | B-9405 | 16 | 4 |
| B-9379 | 32 | 8 | B-9407 | 32 | 8 |
| B-9391 | 32 | 8 | B-9412 | 32 | 8 |
| B-9398 | 32 | 8 | B-9417 | 32 | 8 |
| B-9399 | 32 | 8 | B-9401 | 32 | 8 |
| B-9403 | 32 | 8 | B-9409 | 16 | 8 |
| B-9410 | 32 | 8 | B-9414 | 16 | 4 |
| B-9411 | 16 | 4 | B-8158 | 16 | 8 |
| B-9415 | 8 | 4 | B-9378 | 16 | 8 |
| B-9416 | 32 | 8 | B-9390 | 16 | 4 |
| B-10836 | 32 | 8 | B-9408 | 32 | 8 |
| B-8162 | 32 | 8 | B-9413 | 16 | 4 |
| B-9374 | 32 | 8 | B-9404 | 16 | 4 |
| B-9380 | 32 | 8 | B-9406 | 32 | 8 |
| B-9381 | 32 | 8 | |||
Note: CHD, chlorhexidine digluconate; BZK, benzalkonium chloride.
3.4. Biofilm Formation
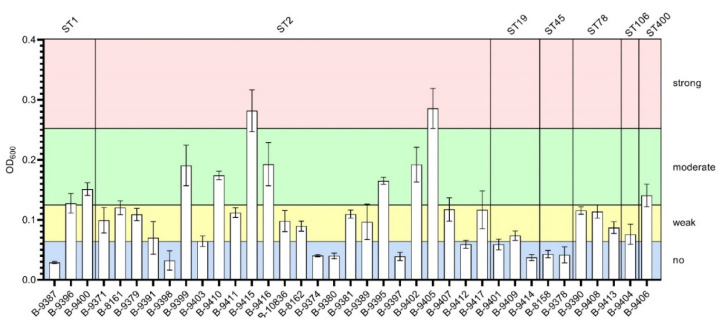
A. baumannii strains were divided into four groups according to their ability for biofilm (BF) formation: strong BF (n = 2); moderate BF (n = 8); weak BF (n = 17); and no BF (n = 10). Notably, strong BF formation was detected for the strains of ST2 only, and moderate BF formation for the strains of ST2, ST1, and ST400 (Figure 2).
Figure 2.
Biofilm formation ability for A. baumannii strains causing meningitis in Moscow neuro-ICU in 2013–2020.
3.5. Whole-Genome Analysis
The complete genome assemblies of 37 A. baumannii strains contained 3553–3891 coding genes, 48–108 pseudogenes, and 69–71 RNA genes. The GC content of the genomes varied from 39.1 to 42.9% (Table 4).
Table 4.
Whole-genome characteristics for A. baumannii strains causing meningitis in Moscow neuro-ICU in 2013–2020.
| Strain | GenBank ID | GC Content, % | Genes (Total) |
CDSs (Total) |
Genes (Coding) |
Genes (RNA) |
rRNA Genes (5S, 16S, 23S) |
tRNAs | ncRNAs | Pseudo Genes |
|---|---|---|---|---|---|---|---|---|---|---|
| B-8162 | JAROBQ000000000 | 39.8 | 3929 | 3858 | 3801 | 71 | 1, 1, 1 | 64 | 4 | 57 |
| B-9371 | JASKJB000000000 | 42.4 | 3722 | 3650 | 3600 | 72 | 0, 2, 3 | 63 | 4 | 50 |
| B-9374 | JASKJA000000000 | 42.4 | 3873 | 3802 | 3745 | 71 | 0, 2, 1 | 64 | 4 | 57 |
| B-8161 | JAROBM000000000 | 39.2 | 3848 | 3778 | 3723 | 70 | 1, 1, 1 | 63 | 4 | 55 |
| B-9378 | JAROBU000000000 | 39.6 | 3707 | 3636 | 3576 | 71 | 1, 1, 1 | 64 | 4 | 60 |
| B-9379 | JASKIZ000000000 | 42.4 | 3704 | 3632 | 3580 | 72 | 1, 2, 2 | 63 | 4 | 52 |
| B-9380 | JAROBS000000000 | 39.9 | 3951 | 3880 | 3829 | 71 | 1, 1, 1 | 64 | 4 | 51 |
| B-9381 | JAROBO000000000 | 39.3 | 3867 | 3797 | 3745 | 70 | 1, 1, 1 | 63 | 4 | 52 |
| B-9387 | JAROBG000000000 | 39.2 | 3877 | 3807 | 3737 | 70 | 1, 1, 1 | 63 | 4 | 70 |
| B-9389 | JASKIY000000000 | 42.2 | 3596 | 3524 | 3475 | 72 | 0, 2, 3 | 63 | 4 | 49 |
| B-9390 | JASKIX000000000 | 42.4 | 3890 | 3820 | 3764 | 70 | 1, 1, 1 | 63 | 4 | 56 |
| B-9391 | JASKIW000000000 | 42.1 | 3679 | 3608 | 3555 | 71 | 0, 2, 2 | 63 | 4 | 53 |
| B-9395 | JASKIV000000000 | 42.9 | 3887 | 3816 | 3758 | 71 | 0, 2, 1 | 64 | 4 | 58 |
| B-9397 | JASKIU000000000 | 42.2 | 3795 | 3725 | 3676 | 70 | 0, 1, 1 | 64 | 4 | 49 |
| B-9398 | JAROBR000000000 | 39.8 | 3965 | 3894 | 3840 | 71 | 1, 1, 1 | 64 | 4 | 54 |
| B-8158 | JAROBT000000000 | 40.3 | 3735 | 3664 | 3605 | 71 | 1, 1, 1 | 64 | 4 | 59 |
| B-9399 | JASKIT000000000 | 42.0 | 3810 | 3741 | 3692 | 69 | 0, 1, 1 | 63 | 4 | 49 |
| B-9396 | JAROBB000000000 | 40.1 | 3855 | 3785 | 3720 | 70 | 1, 1, 1 | 63 | 4 | 65 |
| B-9400 | JAROBI000000000 | 39.8 | 3879 | 3809 | 3744 | 70 | 1, 1, 1 | 63 | 4 | 65 |
| B-9401 | JASKIR000000000 | 40.6 | 3624 | 3553 | 3496 | 71 | 1, 1, 1 | 63 | 5 | 57 |
| B-9402 | JASKIQ000000000 | 39.3 | 3797 | 3726 | 3664 | 71 | 1, 1, 1 | 64 | 5 | 62 |
| B-9403 | JASKIP000000000 | 39.2 | 4019 | 3949 | 3891 | 70 | 1, 1, 1 | 63 | 5 | 58 |
| B-9404 | JASKIO000000000 | 39.7 | 3821 | 3752 | 3644 | 69 | 0, 1, 1 | 63 | 5 | 108 |
| B-9405 | JASKIN000000000 | 39.2 | 3957 | 3887 | 3830 | 70 | 1, 1, 1 | 70 | 5 | 57 |
| B-9406 | JASKIM000000000 | 39.3 | 3803 | 3733 | 3675 | 70 | 1, 1, 1 | 63 | 4 | 58 |
| B-9407 | JASKIL000000000 | 39.3 | 3956 | 3886 | 3830 | 70 | 1, 1, 1 | 63 | 4 | 56 |
| B-9408 | JASKIK000000000 | 39.9 | 3804 | 3733 | 3682 | 71 | 1, 1, 1 | 64 | 4 | 51 |
| B-9409 | JASKIJ000000000 | 39.5 | 3845 | 3775 | 3716 | 70 | 1, 1, 1 | 62 | 5 | 59 |
| B-9410 | JASKII000000000 | 39.4 | 3853 | 3783 | 3728 | 70 | 1, 1, 1 | 63 | 4 | 55 |
| B-9411 | JASKIH000000000 | 38.6 | 3933 | 3862 | 3800 | 71 | 1, 1, 1 | 64 | 4 | 62 |
| B-9412 | JASKIG000000000 | 39.1 | 3654 | 3584 | 3532 | 70 | 1, 1, 1 | 63 | 4 | 52 |
| B-9413 | JASKIF000000000 | 39.4 | 3745 | 3674 | 3626 | 71 | 1, 1, 1 | 64 | 4 | 48 |
| B-9414 | JASKIE000000000 | 39.4 | 3665 | 3593 | 3538 | 72 | 1, 1, 1 | 64 | 5 | 55 |
| B-9415 | JASKID000000000 | 39.4 | 3939 | 3868 | 3813 | 71 | 1, 1, 1 | 64 | 4 | 55 |
| B-9416 | JASKIC000000000 | 39.4 | 3928 | 3859 | 3797 | 69 | 0, 1, 1 | 63 | 4 | 62 |
| B-9417 | JASKIB000000000 | 39.1 | 3743 | 3673 | 3622 | 70 | 1, 1, 1 | 63 | 4 | 51 |
3.6. The Resistome Identification and Analysis
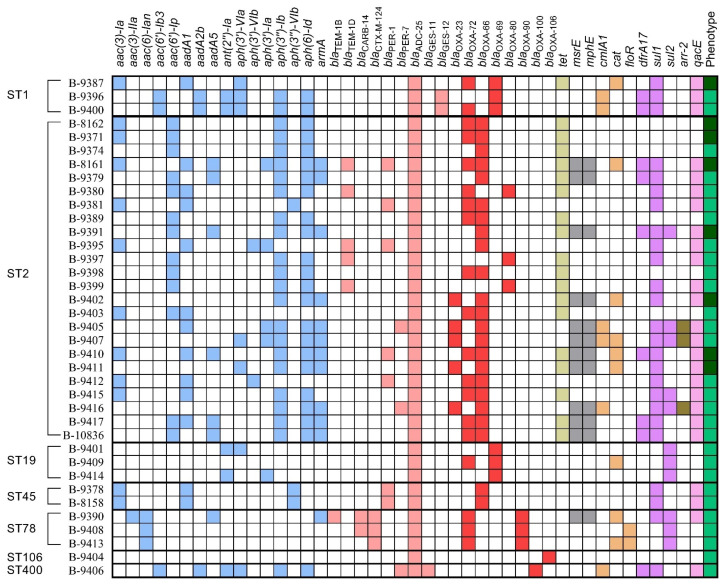
The resistome analysis of 37 A. baumannii genomes showed the presence of a total of 47 types of antimicrobial resistance genes (ARGs) providing resistance to six functional groups of antimicrobials, namely, resistance to aminoglycosides (n = 16), beta-lactams (n = 17), tetracyclines (n = 1), macrolides (n = 2), phenicols (n = 4), sulfonamides (n = 3), rifamycin (n = 1), and antiseptics (n = 1).
Aminoglycoside resistance was associated with aminoglycoside N-acetyltransferase aac (n = 32), aminoglycoside 6-adenylyltransferase aad (n = 23), aminoglycoside nucleotidyltransferase ant (n = 5), aminoglycoside O-phosphotransferase aph (n = 63), and 16S rRNA (guanine(1405)-N(7))-methyltransferase arm (n = 12) genes. The strains of ST1 carried 3–6 ARGs of this group, the strains of ST2 carried 3–7 ARGs each, the strains of ST1 carried 9–2 ARGs each, the strains of ST4 carried 5–3 ARGs each, the strains of ST7 and 8 carried 1–3 ARGs each, the strain of ST400 carried 6 ARGs, and the strain of ST106 had no aminoglycoside resistance genes.
Beta-lactamase ARGs were detected as follows: broad-spectrum class A beta-lactamase blaTEM-1 gene (n = 6), carbenicillin-hydrolyzing class A beta-lactamase blaCARB-14 gene (n = 2), extended-spectrum beta-lactamase (ESBL) of class A blaCTX-M-124 (n = 3), inhibitor-resistant ESBL of class A blaPER-1 (n = 7), and ESBL of class blaPER-7 (n = 2), blaGES-11 (n = 1), and blaGES-12 (n = 2). Acinetobacter-specific ESBL of class C blaADC-25 gene was detected in all 37 strains. Notably, beta-lactamase genes blaTEM-1B, blaCARB-14, and blaCTX-M-124 were identified in the genomes of the strains attributed to ST78 only, while blaTEM-1D gene was identified in the strains of ST2. The blaPER genes were detected in the strains belonging to ST2, ST45, and ST400, whereas blaGES genes were detected in the strains of ST1 and ST400. Carbapenem resistance of the studied strains was supported by eight OXA-type carbapenemase genes: blaOXA-23 (n = 5); blaOXA-72 (n = 19); blaOXA-66 (n = 23); blaOXA-69 (n = 6); blaOXA-80 (n = 3); blaOXA-90 (n = 3); blaOXA-100 (n = 1); and blaOXA-106 (n = 1). Interestingly that blaOXA-23 and blaOXA-80 carbapenemase genes were detected in the strains belonging to ST2 only; blaOXA-66 gene in the strains of ST2 and ST45; blaOXA-69 gene in the strains of ST1 and ST19; blaOXA-90 gene in the strains of ST78; and blaOXA-100 and blaOXA-106 genes—in the stains of ST400 and ST106, respectively.
The tet gene of tetracycline efflux MFS-family transporter was detected in 1 strain of ST1 and 18 strains of ST2. The msrE and mphE genes coding ribosomal protection protein and macrolide 2′-phosphotransferase, respectively, were simultaneously identified in the strains of ST2 (n = 11) and ST78 (n = 1). Regarding phenicol resistance markers, the cat (n = 9) genes encoded subunits of chloramphenicol acetyltransferase, and the cmlA (n = 6) and floR (n = 2) genes encoded chloramphenicol/florfenicol efflux MFS-family transporters. AGRs providing resistance to sulfonamides were detected in the strains of all STs: trimethoprim-resistant dihydrofolate reductase dfrA17 gene (n = 9) and the sulfonamide-resistant dihydropteroate synthase genes sul1 (n = 25) and sul2 (n = 11). Three A. baumannii strains of ST2 carried the gene arr-2, encoding the NAD(+)-rifampin ADP-ribosyltransferase associated with resistance to rifampin. Moreover, the genetic determinant for the antiseptic-resistant qacE gene coding the quaternary ammonium compound efflux SMR-family transporter was detected in the genomes of 25 strains (Figure 3).
Figure 3.
Resistomes of 37 A. baumannii strains causing meningitis in Moscow Neurological ICU in 2013–2020. ARGs of different groups are presented in specific colors. MDR phenotype is designated by the green color, and XDR phenotype is designated by the dark green color.
3.7. The Virulome Identification and Analysis
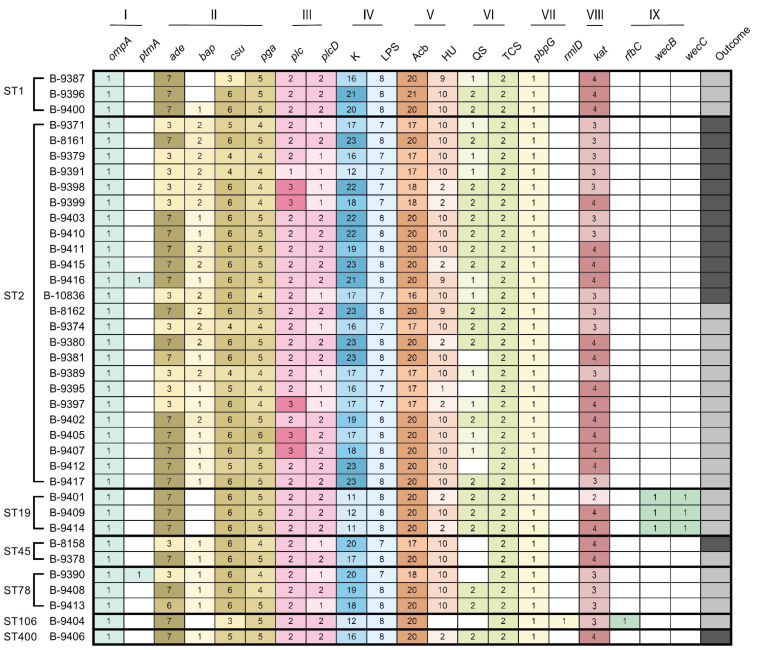
The virulence factor genes identified via the VFDB online resource were classified into nine virulence factor classes: Adherence (outer membrane protein A, ompA, and legionaminic and biosynthesis protein A, ptmA), Biofilm formation (AdeFGH efflux pump, ade, Biofilm-associated protein, bap, Csu pili, csu, PNAG Polysaccharide poly-N-acetylglucosamine, pga), Enzyme (Phospholipase C, plc, Phospholipase D, plcD), Immune evasion (Capsule locus, K, and lipopolysaccharide locus, LPS), Iron uptake (Acinetobactin locus, Acb, Heme utilization, HU), Regulation (Quorum sensing, QS, Two-component system, TCS), Serum resistance (Penicillin-binding protein, pbpG, dTDP-6-deoxy-L-lyxo-4-hexulose reductase, rmlD), Stress adaptation (catalases, kat), and Antiphagocytosis (dTDP-4-dehydrorhamnose 3,5-epimerase, rfbC, UDP-N-acetylglucosamine 2-epimerase (non-hydrolyzing), wecB, UDP-N-acetyl-D-mannosamine dehydrogenase, wecC).
A total of 20 types of putative virulence factor genes (VFGs) were predicted in 37 A. baumannii genomes. The VFG open reading frames (ORFs) associated with Adherence (n = 39), Biofilm formation (n = 537), Enzyme (n = 139), Immune evasion (n = 964), Iron uptake (n = 989), Regulation (n = 124), Serum resistance (n = 32), Stress adaptation (n = 130), and Antiphagocytosis (n = 7) were identified in the genomes (Figure 4).
Figure 4.
Virulomes of 37 A. baumannii strains caused meningitis in Moscow Neurological ICU in 2013–2020. Virulence genes of different groups are presented in specific colors: I, Adherence; II, Biofilm formation; III, Enzyme; IV, Immune evasion; V, Iron uptake; VI, Regulation; VII, Serum resistance; VIII, Stress adaptation; IX, Antiphagocytosis. The color intensity reflects the number of predicted ORFs. The survival outcome is designated by the gray color, death outcome is designated by the dark gray color.
The VFGs identified in all 37 A. baumannii genomes were the following: ompA, ade, csu, pga, plc, plcD, Capsula locus, LPS locus, Acb locus, two-component system genes, pbpG, and kat. At the same time, the ptmA gene encoding legionaminic and biosynthesis protein A was detected in two strains of ST2 and ST78 only. The bap gene encoding biofilm-associated protein was detected in all strains except two strains of ST1 and four strains of ST19 and ST106. The HU locus was obtained in all strains except one of ST106. The abaI inducer and abaR receptor genes associated with QS were detected in 30 strains. The rmlD gene encoding dTDP-6-deoxy-L-lyxo-4-hexulose reductase provides serum resistance, as well as the rfbC gene coding dTDP-4-dehydrorhamnose 3,5-epimerase providing antiphagocytosis, were detected in only one strain of ST106. The wecB and wecC genes encoding UDP-N-acetylglucosamine 2-epimerase (non-hydrolyzing) and UDP-N-acetyl-D-mannosamine dehydrogenase, respectively, providing antiphagocytosis, were identified in the strains of ST19 only. Despite the vast variability of VFGs in the strains under study, the correlation between disease outcomes—patient death or survival—was not obtained.
Upon analyzing Acinetobacter genomes through the VFDB online resource, it was determined that the “katA Catalase (Neisseria)” gene is present in all strains belonging to ST2. This data prompted further examination of kat genes in all studied Acinetobacter genomes. These genes include katA, which encodes a small-subunit mono-functional catalase; katE, the large-subunit mono-functional catalase; katG, the catalase-peroxidase; and katX, a small protein with a catalase-domain. The reference genome of the A. baumannii strain AB5075-UW (CP008706.1) isolated from the tibia/osteomyelitis of the diabetes patient in the USA in 2008 was obtained from GenBank (Table 5).
Table 5.
Analysis of the kat genes in the genomes of A. baumannii strains causing meningitis.
| Strain | ST | katA (AKA32231.1) | katE (AKA32163.1) | katG (AKA33165.1) | katX (AKA31788.1) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ident., % | SNP | SAV | Ident., % | SNP | SAV | Ident., % | SNP | SAV | Ident., % | SNP | SAV | ||
| B-9387 | 1 | 100 | 0 | 0 | 100 | 0 | 0 | 100 | 0 | 0 | 97.85 | 25 | 6 |
| B-9396 | 1 | 100 | 0 | 0 | 100 | 0 | 0 | 100 | 0 | 0 | 100 | 0 | 0 |
| B-9400 | 1 | 100 | 0 | 0 | 100 | 0 | 0 | 100 | 0 | 0 | 100 | 0 | 0 |
| B-9371 | 2 | 97.65 | 25 | 1 | 98.38 | 35 | 3 | 98.89 | 24 | 4 | - | - | - |
| B-8161 | 2 | 97.65 | 25 | 1 | 98.38 | 35 | 3 | 98.89 | 24 | 4 | - | - | - |
| B-9379 | 2 | 97.65 | 25 | 1 | 98.38 | 35 | 3 | 98.89 | 24 | 4 | - | - | - |
| B-9391 | 2 | 97.65 | 25 | 1 | 98.38 | 35 | 3 | 98.89 | 24 | 4 | - | - | - |
| B-9398 | 2 | 97.65 | 25 | 1 | 98.38 | 35 | 3 | 98.89 | 24 | 4 | - | - | - |
| B-9399 | 2 | 97,65 | 25 | 1 | 98.38 | 35 | 3 | 98.89 | 24 | 4 | 97.85 | 25 | 6 |
| B-9403 | 2 | 97.65 | 25 | 1 | 98.38 | 35 | 3 | 98.89 | 24 | 4 | - | - | - |
| B-9410 | 2 | 97.56 | 26 | 1 | 98.38 | 35 | 3 | 98.89 | 24 | 4 | - | - | - |
| B-9411 | 2 | 97.65 | 25 | 1 | 98.38 | 35 | 3 | 98.89 | 24 | 4 | 97.85 | 25 | 6 |
| B-9415 | 2 | 97.65 | 25 | 1 | 98.38 | 35 | 3 | 98.89 | 24 | 4 | 97.85 | 25 | 6 |
| B-9416 | 2 | 97.65 | 25 | 1 | 98.38 | 35 | 3 | 98.89 | 24 | 4 | 97.85 | 25 | 6 |
| B-10836 | 2 | 97.65 | 25 | 1 | 98.38 | 35 | 3 | 98.89 | 24 | 4 | - | - | - |
| B-8162 | 2 | 97.65 | 25 | 1 | 98.38 | 35 | 3 | 98.89 | 24 | 4 | - | - | - |
| B-9374 | 2 | 97.65 | 25 | 1 | 98.38 | 35 | 3 | 98.89 | 24 | 4 | - | - | - |
| B-9380 | 2 | 97.65 | 25 | 1 | 98.38 | 35 | 3 | 98.89 | 24 | 4 | 97.85 | 25 | 6 |
| B-9381 | 2 | 97.65 | 25 | 1 | 98.38 | 35 | 3 | 98.89 | 24 | 4 | 97.85 | 25 | 6 |
| B-9389 | 2 | 97.65 | 25 | 1 | 98.38 | 35 | 3 | 98.89 | 24 | 4 | - | - | - |
| B-9395 | 2 | 97.65 | 25 | 1 | 98.38 | 35 | 3 | 98.89 | 24 | 4 | 97.85 | 25 | 6 |
| B-9397 | 2 | 97.65 | 25 | 1 | 98.38 | 35 | 3 | 98.89 | 24 | 4 | 97.85 | 25 | 6 |
| B-9402 | 2 | 97.65 | 25 | 1 | 98.38 | 35 | 3 | 98.89 | 24 | 4 | 97.85 | 25 | 6 |
| B-9405 | 2 | 97.65 | 25 | 1 | 98.38 | 35 | 3 | 98.89 | 24 | 4 | 97.85 | 25 | 6 |
| B-9407 | 2 | 97.65 | 25 | 1 | 98.38 | 35 | 3 | 98.89 | 24 | 4 | 97.85 | 25 | 6 |
| B-9412 | 2 | 97.65 | 25 | 1 | 98.38 | 35 | 3 | 98.89 | 24 | 4 | 97.85 | 25 | 6 |
| B-9417 | 2 | 97.65 | 25 | 1 | 98.38 | 35 | 3 | 98.89 | 24 | 4 | - | - | - |
| B-9401 | 19 | - | - | - | - | - | - | 100 | 0 | 0 | 100 | 0 | 0 |
| B-9409 | 19 | 100 | 0 | 0 | 100 | 0 | 0 | 100 | 0 | 0 | 100 | 0 | 0 |
| B-9414 | 19 | 100 | 0 | 0 | 100 | 0 | 0 | 100 | 0 | 0 | 100 | 0 | 0 |
| B-8158 | 45 | 97.65 | 25 | 1 | 98.38 | 35 | 3 | 98.89 | 24 | 4 | 97.85 | 25 | 6 |
| B-9378 | 45 | 97.65 | 25 | 1 | 98.36 | 35 | 3 | 98.89 | 24 | 4 | 97.85 | 25 | 6 |
| B-9390 | 78 | 98.03 | 21 | 2 | 98.6 | 30 | 1 | 98.75 | 27 | 4 | pseudo | - | - |
| B-9408 | 78 | 98.03 | 21 | 2 | 98.6 | 30 | 1 | 98.75 | 27 | 4 | pseudo | - | - |
| B-9413 | 78 | 98.03 | 21 | 2 | 98.6 | 30 | 1 | 98.75 | 27 | 4 | pseudo | - | - |
| B-9404 | 106 | pseudo | - | - | 98.83 | 25 | 1 | 98.56 | 31 | 4 | 97.85 | 25 | 6 |
| B-9406 | 400 | 97.84 | 23 | 2 | 98.88 | 24 | 2 | 98.79 | 26 | 2 | 97.85 | 25 | 6 |
Note: ST, sequence type; Ident., identity to the reference gene sequence obtained from GenBank; SNP, single nucleotide polymorphism; SAV, single amino acid variation; “-“, no match; pseudo, pseudogene.
The katA gene with 100% identity compared to the reference (GB accession number AKA32231.1) was detected in three strains of ST1 and two strains of ST19. All strains of ST2 and ST45 carried the katA gene with 25–26 single nucleotide polymorphisms (SNPs), and only one SNP was significant, resulting in the single amino acid variation (SAV), g367c/A123P. The strains of ST78 and ST400 carried the katA gene with 21 and 23 SNPs, respectively, which caused two SAVs, a211t/T71S and g367c/A123P, in the ST78 strains, as well as g367c/A123P and g649t/V217F in the ST400 strain. The katA gene was not detected in strain B-9401 of ST19, while the pseudogene was identified in strain B-9406 of ST400. It should be noted that SAV g367c/A123P was detected in all variants of the katA gene in this study.
The sequence of the katE gene was matched to the reference (GB accession number AKA32163.1). The 100%-identical genes were detected in stains of ST1 and two strains of ST19. All strains attributed to the ST2 and ST45 carried 35 SNPs in this gene, providing three SAVs: a876c/Q292H; c1304t/A435V; and a1610g/N537S. The strains of ST78 carried the katE gene with 30 SNPs compared to the reference, including two nonsynonymous substitutions, g1303a and c1304t, resulting in one SAV A435I. The strains B-9404 of ST106 and B-9406 of ST400 carried 25 and 24 SNPs, respectively, realizing in one (a1820g/N607S) and two (c1304t/A435V and t1939g/L647V) SAVs, respectively. The katE gene was missing in strain B-9401 of ST19. Interestingly, that SAV in position 435 was common for katE genes in this study, two variants of SAVs were detected: A435V and A435I.
The katG gene in 100%-identity (compared to reference AKA33165.1) was detected in the strains of ST1 and ST19. At the same time, all strains attributed to the ST2 and ST45 carried 24 SNPs in this gene, providing four SAVs, namely, c808t/P270S, a1264g/T422A, c1366t/P456S, and g1369a/A457T. Even though the strains of ST78 and ST106 carried different SNPs (27 and 31, respectively) in the katG gene, they have four identical SAVs (c808t/P270S; a1264g/T422A; c1366t/P456S; and a1513c/K505Q). The strain B-9406 belonged to ST400 and carried 26 SNPs in the katG gene providing two SAVs, namely, c808t/P270S and a1264g/T422A. It is noteworthy that two SAVs (c808t/P270S and a1264g/T422A) were common for mutant katG genes in the stains of ST2, ST45, ST78, ST106, and ST400; while the SAV (c1366t/P456S) was frequent in the strains of ST2, ST45, ST78, and ST106; and the SAV (a1513c/K505Q) was prevalent in the strains of ST78 and ST106.
The katX gene was detected in 22 of 37 A. baumannii strains in the study; meanwhile, two strains of ST1 and three strains of ST19 carried this gene with 100%-identity (compared to reference) AKA31788.1. All the katX gene-positive strains carried the same 25 SNPs, resulting in six SAVs (a85g/T29A; g461a/G154D; a694g/T232A; t784a/S262T; a790g/T264A; and t922a-t923c/F308T). The stains attributed to ST78 carried the pseudogene (Table 5).
3.8. Phylogenetic Analysis
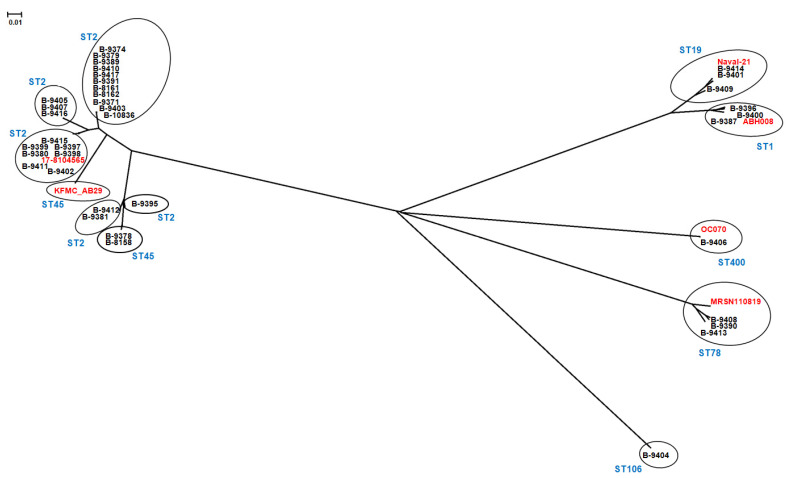
The phylogenetic relationship between 37 A. baumannii genomes determined on the base of the SNPs analysis showed the presence of five main clusters. The first cluster included 24 genome sequences of the strains belonging to ST2 and two strains of ST45; the second cluster included the genomes belonging to ST1 and ST19; while the genomes of the strains belonging to ST78, ST106, and ST400 formed individual clusters each. The phylogenetic analysis also showed that the genomes belonging to ST2 were divided into five sub-clusters, while the genomes of the other STs were combined in a single cluster for a specific type (Figure 5).
Figure 5.
Neighbor-joining phylogenetic tree based on 58,859 core SNPs, showing the phylogenetic relationships between 37 genomes of A. baumannii strains causing meningitis in this study (black color) and 6 A. baumannii genomes accepted as references from the NCBI database (red color): strain ST1 isolate A076 (KT317084.1), Sweden; strain ST2 isolate 17-8104565 (CANDXI000000000.1), United Kingdom; strain ST19 isolate Naval-21 (AMSY00000000.1), USA; strain ST45 isolate KFMC_AB29 (JALGJO000000000.1), Kingdom of Saudi Arabia; strain ST78 isolate 4190 (KT266827.1), Australia; strain ST400 isolate OC070 (CP087298.1), Germany. The scale bar represents a 0.01 base substitution per site.
4. Discussion
The present study characterized 37 Acinetobacter baumannii strains isolated from 37 patients of Moscow neurosurgery ICU with meningitis in 2013–2020, which caused 37.8% of deaths. The overall incidence of nosocomial meningitis in this period was 9.5%; the average proportion of A. baumannii among all nosocomial meningitis causative agents was 22.1%. [31]. Such a high level of mortality is in agreement with reports about A. baumannii as a problematic pathogen to human health due to its high level of drug resistance, increased virulence, and difficulty in treatment, with a high mortality rate (15–73%) because of its multiple drug resistance [32,33].
Seven A. baumannii STs were revealed in the study: ST1, ST2, ST19, ST45, ST78, ST106, and ST400. The prevalent ST2 belonging to the pandemic lineage, named “international clone IC2”, was associated in our study with 12 of 14 deaths. ST2 is a well-known dominant type (59%) among the available complete and draft genomes in the GenBank database. This is also consistent with a large number of previous publications that report outbreaks due to IC2s accounting for the bulk of carbapenem-resistant A. baumannii (CRAB) outbreaks, whilst IC2 strains do not seem to be the dominant type in South American countries, Tunisia, Tanzania, Poland, and Japan [34]. Other A. baumannii strains associated with the death of the patients were ST45, belonging to IC2; and ST400, attributed to the distant phylogenetic group. The last ST was previously rare and described for a nosocomial A. baumannii isolate in Germany [35].
Thirteen KLs were identified in our study; the most prevalent was KL9. In A. baumannii, the capsular polysaccharide is a major virulence factor that is important for survival in vitro and in vivo. Specific capsule types are proven to overwhelm mammalian defenses in vivo, including K2 [36]. To date, more than 128KL gene clusters (KL types) have been identified at the KL in A. baumannii genomes [37]. The major A. baumannii outer core locus (OCL) in our study was OCL1, which includes genes for the synthesis of the variable outer core region. This is in agreement with the data of the Kaptive tool that OCL1 is the most common (~74%) amongst isolates attributed to ST1, ST2, ST3, and ST78 [38].
In this study, a majority of the strains were multidrug-resistant MDR, (78%); the rest were extensively drug-resistant XDR, resistant to 6-10 functional groups of antimicrobials. For comparison, the prevalence of XDR A. baumannii isolates in Bulgarian university hospitals during the period from 2014 to 2016 was reported as 12.4%; during the period between 2018 and 2019, it increased to 78.1% [39,40]. It was reported recently that intracranial infections caused by drug-resistant Gram-negative bacilli, including MDR, XDR, or even pandrug-resistant (PDR) A. baumannii, have very serious consequences [41]. The phenotypic diversity of antibiotic resistance among the strains in our study was very high—23 variants of AMR phenotype were revealed. Carbapenem-resistant strains represented 68%. It is known that CRAB infections are generally nosocomial and frequently occur in the intensive care unit (ICU) for up to 31% around the world with high mortality—greater than 50% in some cases [42]. The carbapenem resistance of the strains in this study was supported by eight blaOXA-type carbapenemase genes with the prevalence of blaOXA-23 and blaOXA-80 genes detected in the strains belonging to ST2 only. This finding is in agreement with reports about blaOXA-type carbapenemase genes associated with members of GC1 and GC2 [34]. Moreover, the genes of broad-spectrum beta-lactamases and ESBLs (blaTEM-1, blaCARB-14, blaCTX-M-124, blaPER-1, blaPER-7, blaGES-11, blaGES-12, and blaADC-25) were detected in our study. Similar sets of beta-lactamase genes were identified in the studies of other authors [43,44].
Additionally, a total of 47 types of antimicrobial resistance genes (ARGs) providing resistance to aminoglycosides, tetracyclines, macrolides, phenicols, sulfonamides, rifamycin, and antiseptics were identified. Aminoglycoside resistance was associated with aac, aad, ant, aph, and arm genes. Recently, many reports have appeared showing that carbapenem resistance in A. baumannii tends to occur in conjunction with aminoglycosides resistance. The co-production of ArmA 16S rRNA methylase and OXA-23 carbapenemase was determined as the most prevalent mechanism conferring MDR in clinical A. baumannii isolates in many countries worldwide, such as Italy, Greece, China, South Korea, India, Yemen, etc. [45]. Among the ARGs providing resistance to other groups of antimicrobials, the tet genes were detected in 19/37 strains in this study, which was approximately the same level as the study in Thailand (56.1%); a similar level of prevalence was also obtained for the arr-2 gene encoding the resistance to rifampin—3/37 strains in our study (9.1%)—in Thailand. Regarding phenicol resistance markers—cat, cmlA, and floR genes—17/37 strains carried these determinants in our study, which was higher than in the Thailand study (8.1%). As well as the prevalence of AGRs providing resistance to sulfonamides, sul1 and sul2 genes were detected in 25/37 strains and 11/37 strains, respectively, in our study, compared to 46.1% and 13.6% in Thailand. In contrast, the msrE and mphE genes providing the macrolide resistance were less common in our study (12/37 strains) than in Thailand (79.6%) [46]. Importantly, three colistin-resistant strains were identified among A. baumannii causing meningitis, all of which were attributed to ST2 as in the previously described colistin-resistant isolates collected from the urinary tract, respiratory tract, skin, soft tissue, and blood of patients in some Russian regions in 2013–2014 [47].
Moreover, the genetic determinant for the antiseptic-resistant qacE gene was detected in the genomes of 25/37 strains in our study, which was higher than the previously published prevalence of this gene in A. baumannii isolates collected from urine, pus, blood, and endotracheal fluid/aspirate in Malaysia (28%). However, the phenotypic resistance of strains in our study was lower (MIC = 4–8 mg/L of benzalkonium chloride; and MIC = 8–64 of chlorhexidine gluconate) than in the Malaysia study (from 12.5 to >50 mg/L and from 25 to >50 mg/L, respectively) [48].
Nine groups of virulence factor genes (VFGs) were identified in A. baumannii in this study, including a total of 20 types of VFGs containing the specific numbers of the open reading frames (ORFs): adherence (n=39); biofilm formation (n = 537); enzyme (n = 139); immune evasion (n = 964); iron uptake (n = 989); regulation (n = 124); serum resistance (n = 32); stress adaptation (n = 130); and antiphagocytosis (n = 7). The adherence group was presented by the ompA gene detected in all strains. A significantly increasing expression of the ompA gene in persisters was recently shown, which proposes ompA as a potential target for drug development against A. baumannii persisters [49]. Additionally, the ptmA gene was detected in two strains of ST2 and ST7; this is in agreement with the opinion that this virulence factor is quite rare [50]. Several genes involved in the biofilm formation of A. baumannii were identified in the strains in this study, such as adeFGH efflux pump genes, bap gene, csu locus, and pga locus. However, the capacity to form biofilms varied among the strains, with the majority of the isolates forming either moderate (8/37) or weak (17/37) biofilms and only a minority (2/37) forming strong biofilms. This is in agreement with other studies aimed at the evaluation of the distribution of VFGs involved in biofilm formation in multi-drug-resistant A. baumannii [51]. The plc and plcD genes contributing to pathogenesis by aiding in the lysis of host cells to facilitate bacterial invasion were also detected in all studied A. baumannii strains. The major differences between the strains were observed in the capsular export region, where the number of genes varied from 11 to 23. These findings were in agreement with recently published data [52]. A number of putative genes associated with acinetobactin biosynthesis were detected in all A. baumannii strains in our study and varied from 16 to 21. In contrast, the heme utilization locus was obtained in all strains except one of ST106. Surprisingly, only this strain of ST106 contained the additional gene rmlD involved in serum resistance, as well as the rfbC gene providing antiphagocytosis activity [53]. Surprisingly, the VFDB online resource revealed “katA Catalase (Neisseria)” in all A. baumannii strains belonging to ST2. A further comprehensive examination of kat genes showed that the strains carried the katA (35/37), katE (36/37), katG (37/37), and katX (23/37) genes. This finding is consistent with current knowledge of the significance of katE and katG genes in H2O2 resistance, as well as the lesser influence of katA and katX genes on peroxide resistance [54]. It should be noted that putative KatE was different in the strains of ST2 and ST45 (Q292H, A435V, and N537S), ST78 (A435I), ST106 (N607S), and ST400 (A435V and L647V) compared to the KatE of ST1 strains. At the same time, putative KatG varied in the strains of ST2 and ST45 (P270S, T422A, P456S, and A457T), ST78 and ST106 (P270S, T422A, P456S, and K505Q), and ST400 (P270S and T422A) compared to ST1 and ST19. This observation demonstrates the ongoing evolution of these virulence factor determinants. The wide variety of the virulence factors identified in the Acinetobacter strains that caused meningitis in neuro-intensive care patients indicates the plasticity of the pathogen genome. This is consistent with the previously described diverse interaction between different pathogenicity factors, such as iron acquisition, cell adherence, cell motility, biofilm formation, and antibiotic susceptibility [55].
In conclusion, this study provides a detailed account of the genetic determinants associated with multidrug resistance and virulence in clinically significant A. baumannii isolates from the patients of a Moscow neurosurgery ICU with meningitis. The sequence type ST2 was the prevalent genetic line in this cohort. Numerous variants of antimicrobial resistance genes and virulence gene sets were revealed in the genomes of meningitis agents. At the same time, the correlation between STs and their molecular–genetic characteristics was obtained. This observation could form the basis for the future design of novel diagnostic and treatment strategies. We propose to continue this study by focusing on the further investigation of antimicrobial resistance islands, plasmid compositions, the localization of the genetic mobile elements, etc.
Acknowledgments
We acknowledge Alexander Bogun for administrative and technical support of the whole-genome sequencing.
Author Contributions
Conceptualization, N.K.F., O.N.E. and I.A.S.; methodology, N.K.F., E.I.A., M.V.F. and O.N.E.; software, A.A.K., A.A.S. and M.V.F.; validation, E.I.A., A.A.K. and I.A.S.; formal analysis, A.A.K., O.N.E. and I.A.S.; investigation, E.I.A., T.S.N., A.A.S., A.D.F. and G.N.F.; resources, N.K.F., A.A.K. and O.N.E.; data curation, E.I.A., T.S.N., A.A.S., M.V.F. and G.N.F.; writing—original draft preparation, N.K.F., E.I.A., A.A.K. and M.V.F.; writing—review and editing, N.K.F.; visualization, A.D.F., A.A.S. and T.S.N.; supervision, N.K.F., O.N.E. and I.A.S.; project administration, N.K.F. and O.N.E.; funding acquisition, N.K.F. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
The following whole-genome sequences were deposited in the GenBank database: JAROBQ000000000, JASKJB000000000, JASKJA000000000, JAROBM000000000, JAROBU000000000, JASKIZ000000000, JAROBS000000000, JAROBO000000000, JAROBG000000000, JASKIY000000000, JASKIX000000000, JASKIW000000000, JASKIV000000000, JASKIU000000000, JAROBR000000000, JAROBT000000000, JASKIT000000000, JAROBB000000000, JAROBI000000000, JASKIR000000000, JASKIQ000000000, JASKIP000000000, JASKIO000000000, JASKIN000000000, JASKIM000000000, JASKIL000000000, JASKIK000000000, JASKIJ000000000, JASKII000000000, JASKIH000000000, JASKIG000000000, JASKIF000000000, JASKIE000000000, JASKID000000000, JASKIC000000000, JASKIB000000000.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by the Ministry of Science and Higher Education of the Russian Federation, grant number 075-15-2019-1671 (agreement dated 31 October 2019).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D.L., Pulcini C., Kahlmeter G., Kluytmans J., Carmeli Y., et al. WHO pathogens priority list working group. discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018;18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 2.Lescure F.X., Bouadma L., Nguyen D., Parisey M., Wicky P.H., Behillil S., Gaymard A., Bouscambert-Duchamp M., Donati F., Le Hingrat Q., et al. Clinical and virological data of the first cases of COVID-19 in Europe: A case series. Lancet Infect. Dis. 2020;20:697–706. doi: 10.1016/S1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen T., Fu Y., Hua X., Xu Q., Lan P., Jiang Y., Yu Y., Zhou Z. Acinetobacter baumannii strains isolated from cerebrospinal fluid (CSF) and bloodstream analysed by cgMLST: The dominance of clonal complex CC92 in CSF infections. Int. J. Antimicrob. Agents. 2021;58:106404. doi: 10.1016/j.ijantimicag.2021.106404. [DOI] [PubMed] [Google Scholar]
- 4.Tuon F.F., Penteado-Filho S.R., Amarante D., Andrade M.A., Borba L.A. Mortality rate in patients with nosocomial Acinetobacter meningitis from a Brazilian hospital. Braz. J. Infect. Dis. 2010;14:437–440. doi: 10.1016/S1413-8670(10)70090-8. [DOI] [PubMed] [Google Scholar]
- 5.Chusri S., Sakarunchai I., Kositpantawong N., Panthuwong S., Santimaleeworagun W., Pattharachayakul S., Singkhamanan K., Doi Y. Outcomes of adjunctive therapy with intrathecal or intraventricular administration of colistin fo.; post-neurosurgical meningitis and ventriculitis due to carbapenem-resistant Acinetobacter baumannii. Int. J. Antimicrob. Agents. 2018;51:646–650. doi: 10.1016/j.ijantimicag.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Jolley K.A., Bray J.E., Maiden M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lopez-Otsoa F., Gallego L., Towner K.J., Tysall L., Woodford N., Livermore D.M. Endemic carbapenem resistance associated with OXA-40 carbapenemase among Acinetobacter baumannii isolates from a hospital in northern Spain. J. Clin. Microbiol. 2002;40:4741–4743. doi: 10.1128/JCM.40.12.4741-4743.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dijkshoorn L., Aucken H., Gerner-Smidt P., Janssen P., Kaufmann M.E., Garaizar J., Ursing J., Pitt T.L. Comparison of outbreak and nonoutbreak Acinetobacter baumannii strains by genotypic and phenotypic methods. J. Clin. Microbiol. 1996;34:1519–1525. doi: 10.1128/jcm.34.6.1519-1525.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zarrilli R., Pournaras S., Giannouli M., Tsakris A. Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int. J. Antimicrob. Agents. 2013;41:11–19. doi: 10.1016/j.ijantimicag.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 10.Giannouli M., Antunes L.C., Marchetti V., Triassi M., Visca P., Zarrilli R. Virulencerelated traits of epidemic Acinetobacter baumannii strains belonging to the international clonal lineages I–III and to the emerging genotypes ST25 and ST78. BMC Infect. Dis. 2013;13:282. doi: 10.1186/1471-2334-13-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayanskiy N., Chebotar I., Alyabieva N., Kryzhanovskaya O., Savinova T., Turenok A., Bocharova Y., Lazareva A., Polikarpova S., Karaseva O. Emergence of the uncommon clone ST944/ST78 carrying blaOXA-40-like and blaCTX-M-like genes among carbapenem-nonsusceptible Acinetobacter baumannii in Moscow, Russia. Microb. Drug Resist. 2017;23:864–870. doi: 10.1089/mdr.2016.0302. [DOI] [PubMed] [Google Scholar]
- 12.Cayô R., Rodrigues-Costa F., Matos A.P., Carvalhaes C.G., Jové T., Gales A.C. Identification of a new integron harboring bla(IMP-10) in carbapenem-resistant Acinetobacter baumannii clinical isolates. Antimicrob. Agents Chemother. 2015;59:3687–3689. doi: 10.1128/AAC.04991-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poirel L., Nordmann P. Carbapenem resistance in Acinetobacter baumannii: Mechanisms and epidemiology. Clin. Microbiol. Infect. 2006;12:826–836. doi: 10.1111/j.1469-0691.2006.01456.x. [DOI] [PubMed] [Google Scholar]
- 14.Zarrilli R., Bagattini M., Migliaccio A., Esposito E.P., Triassi M. Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii in Italy. Ann. Ig. 2021;33:401–409. doi: 10.7416/ai.2020.2395. [DOI] [PubMed] [Google Scholar]
- 15.Wohlfarth E., Kresken M., Higgins P.G., Stefanik D., Wille J., Hafner D., Körber-Irrgang B., Seifert H., Study Group “Antimicrobial Resistance” of the Paul-Ehrlich-Society for Infection Therapy The evolution of carbapenem resistance determinants and major epidemiological lineages among carbapenem-resistant Acinetobacter baumannii isolates in Germany, 2010–2019. Int. J. Antimicrob. Agents. 2022;60:106689. doi: 10.1016/j.ijantimicag.2022.106689. [DOI] [PubMed] [Google Scholar]
- 16.Kyriakidis I., Vasileiou E., Pana Z.D., Tragiannidis A. Acinetobacter baumannii antibiotic resistance mechanisms. Pathogens. 2021;10:373. doi: 10.3390/pathogens10030373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gebhardt M.J., Shuman H.A. GigA and GigB are master regulators of antibiotic resistance, stress responses, and virulence in Acinetobacter baumannii. J. Bacteriol. 2017;199:e00066-17. doi: 10.1128/JB.00066-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maharjan R.P., Sullivan G.J., Adams F.G., Shah B.S., Hawkey J., Delgado N., Semenec L., Dinh H., Li L., Short F.L., et al. DksA is a conserved master regulator of stress response in Acinetobacter baumannii. Nucleic Acids Res. 2023;9:gkad341. doi: 10.1093/nar/gkad341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu B., Zheng D., Zhou S., Chen L., Yang J. VFDB 2022: A general classification scheme for bacterial virulence factors. Nucleic Acids Res. 2022;50:D912–D917. doi: 10.1093/nar/gkab1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ershova K., Savin I., Kurdyumova N., Wong D., Danilov G., Shifrin M., Alexandrova I., Sokolova E., Fursova N., Zelman V., et al. Implementing an infection control and prevention program decreases the incidence of healthcare-associated infections and antibiotic resistance in a Russian neuro-ICU. Antimicrob. Resist. Infect. Control. 2018;7:94. doi: 10.1186/s13756-018-0383-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magiorakos A.-P., Srinivasan A., Carey R.B., Carmeli Y., Falagas M.E., Giske C., Harbarth S., Hindler J.F., Kahlmeter G., Olsson-Liljequist B., et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 22.Babalska Z.Ł., Korbecka-Paczkowska M., Karpiński T.M. Wound antiseptics and European Guidelines for antiseptic application in wound treatment. Pharmaceuticals. 2021;14:1253. doi: 10.3390/ph14121253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vijayakumar S., Rajenderan S., Laishram S., Anandan S., Balaji V., Biswas I. Biofilm formation and motility depend on the nature of the Acinetobacter baumannii clinical isolates. Front. Public. Health. 2016;4:105. doi: 10.3389/fpubh.2016.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stepanović S., Vuković D., Hola V., Di Bonaventura G., Djukić S., Cirković I., Ruzicka F. Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. Apmis. 2007;115:8918–8999. doi: 10.1111/j.1600-0463.2007.apm_630.x. [DOI] [PubMed] [Google Scholar]
- 25.Wilson K. Preparation of genomic DNA from bacteria. Curr. Protoc. Mol. Biol. 2001;56:2–4. doi: 10.1002/0471142727.mb0204s56. [DOI] [PubMed] [Google Scholar]
- 26.Wick R.R., Judd L.M., Gorrie C.L., Holt K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tatusova T., DiCuccio M., Badretdin A., Chetvernin V., Nawrocki E.P., Zaslavsky L., Lomsadze A., Pruitt K.D., Borodovsky M., Ostell J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wyres K.L., Cahill S.M., Holt K.E., Hall R.M., Kenyon J.J. Identification of Acinetobacter baumannii loci for capsular polysaccharide (KL) and lipooligosaccharide outer core (OCL) synthesis in genome assemblies using curated reference databases compatible with Kaptive. Microb. Genom. 2020;6:e000339. doi: 10.1099/mgen.0.000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen L., Yang J., Yu J., Yao Z., Sun L., Shen Y., Jin Q. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2005;33:D325–D328. doi: 10.1093/nar/gki008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huson D.H., Bryant D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 31.Kurdyumova N.V., Savin I.A., Ershova O.N., Aleksandrova I.A., Sazykina S.J., Gadjieva O.A., Danilov G.V., Shifrin M.A., Fursova N.K. Nosocomial meningitis caused by Acinetobacter baumannii in ICU patients. Rus. J. Anaesthesiol. Reanimatol. (Anesteziologiya I Reanimatologiya) 2019;4:43–49. doi: 10.17116/anaesthesiology201904143. [DOI] [Google Scholar]
- 32.Zheng G., Wang S., Lv H., Zhang G. Nomogram analysis of clinical characteristics and mortality risk factor of non-fermentative gram-negative bacteria-induced post-neurosurgical meningitis. Infect. Drug Resist. 2022;15:6379–6389. doi: 10.2147/IDR.S385502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Panic H., Gjurasin B., Santini M., Kutlesa M., Papic N. Etiology and outcomes of healthcare-associated meningitis and ventriculitis-a single center cohort study. Infect. Dis. Rep. 2022;14:420–427. doi: 10.3390/idr14030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamidian M., Nigro S.J. Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii. Microb. Genom. 2019;5:e000306. doi: 10.1099/mgen.0.000306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eigenbrod T., Reuter S., Gross A., Kocer K., Günther F., Zimmermann S., Heeg K., Mutters N.T., Nurjadi D. Molecular characterization of carbapenem-resistant Acinetobacter baumannii using WGS revealed missed transmission events in Germany from 2012–15. J. Antimicrob. Chemother. 2019;74:3473–3480. doi: 10.1093/jac/dkz360. [DOI] [PubMed] [Google Scholar]
- 36.Yang J.L., Yang C.J., Chuang Y.C., Sheng W.H., Chen Y.C., Chang S.C. Association of capsu-lar polysaccharide locus 2 with prognosis of Acinetobacter baumannii bacteraemia. Emerg. Microbes Infect. 2022;11:83–90. doi: 10.1080/22221751.2021.2011624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arbatsky N.P., Kasimova A.A., Shashkov A.S., Shneider M.M., Popova A.V., Shagin D.A., Shelenkov A.A., Mikhailova Y.V., Yanushevich Y.G., Azizov I.S., et al. Structure of the K128 capsular polysaccharide produced by Acinetobacter baumannii. Carbohydr. Res. 2019;485:107814. doi: 10.1016/j.carres.2019.107814. [DOI] [PubMed] [Google Scholar]
- 38.Sorbello B.M., Cahill S.M., Kenyon J.J. Identification of further variation at the lipooligosac-charide outer core locus in Acinetobacter baumannii genomes and extension of the OCL reference sequence database for Kaptive. Microb. Genom. 2023;9:001042. doi: 10.1099/mgen.0.001042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stratev A., Tanova R., Dimov S., Mitov I., Strateva T. Clonal spread of carbapenem-resistant Acinetobacter baumannii isolates among Bulgarian critically ill patients undergoing renal replacement therapy (2016–2018) Infect. Dis. 2020;52:430–433. doi: 10.1080/23744235.2020.1725622. [DOI] [PubMed] [Google Scholar]
- 40.Strateva T.V., Sirakov I., Stoeva T.J., Stratev A., Peykov S. Phenotypic and molecular characteristics of carbapenem-resistant Acinetobacter baumannii isolates from Bulgarian Intensive Care Unit patients. Microorganisms. 2023;11:875. doi: 10.3390/microorganisms11040875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Munari M., Franzoi F., Sergi M., De Cassai A., Geraldini F., Grandis M., Caravello M., Boscolo A., Navalesi P. Extensively drug-resistant and multidrug-resistant gram-negative pathogens in the neurocritical intensive care unit. Acta Neurochir. 2022;164:859–865. doi: 10.1007/s00701-020-04611-3. [DOI] [PubMed] [Google Scholar]
- 42.Ayobami O., Willrich N., Harder T., Okeke I.N., Eckmanns T., Markwart R. The incidence and prevalence of hospital-acquired (carbapenem-resistant) Acinetobacter baumannii in Europe, Eastern Mediterranean and Africa: A systematic review and meta-analysis. Emerg. Microbes Infect. 2019;8:1747–1759. doi: 10.1080/22221751.2019.1698273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hamed S.M., Hussein A.F.A., Al-Agamy M.H., Radwan H.H., Zafer M.M. Genetic configuration of genomic resistance islands in Acinetobacter baumannii clinical isolates from Egypt. Front. Microbiol. 2022;13:878912. doi: 10.3389/fmicb.2022.878912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farzana R., Swedberg G., Giske C.G., Hasan B. Molecular and genetic characterization of emerging carbapenemase-producing Acinetobacter baumannii strains from patients and hospital environments in Bangladesh. Infect. Prev. Pract. 2022;4:100215. doi: 10.1016/j.infpip.2022.100215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen M., Luan G., Wang Y., Chang Y., Zhang C., Yang J., Deng S., Ling B., Jia X. Coexistence of blaOXA-23 with armA in quinolone-resistant Acinetobacter baumannii from a Chinese university hospital. Diagn. Microbiol. Infect. Dis. 2016;84:230–231. doi: 10.1016/j.diagmicrobio.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 46.Chukamnerd A., Singkhamanan K., Chongsuvivatwong V., Palittapongarnpim P., Doi Y., Pomwised R., Sakunrang C., Jeenkeawpiam K., Yingkajorn M., Chusri S., et al. Whole-genome analysis of carbapenem-resistant Acinetobacter baumannii from clinical isolates in Southern Thailand. Comput. Struct. Biotechnol. J. 2022;20:545–558. doi: 10.1016/j.csbj.2021.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sheck E.A., Edelstein M.V., Sukhorukova M.V., Ivanchik N.V., Skleenova E.Y., Dekhnich A.V., Azizov I.S., Kozlov R.S. Epidemiology and genetic diversity of colistin nonsusceptible nosocomial Acinetobacter baumannii strains from Russia for 2013–2014. Can. J. Infect. Dis. Med. Microbiol. 2017;2017:1839190. doi: 10.1155/2017/1839190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nor A’shimi M.H., Alattraqchi A.G., Mohd Rani F., Rahman N.I.A., Ismail S., Abdullah F.H., Othman N., Cleary D.W., Clarke S.C., Yeo C.C. Biocide susceptibilities and biofilm-forming capacities of Acinetobacter baumannii clinical isolates from Malaysia. J. Infect. Dev. Ctries. 2019;13:626–633. doi: 10.3855/jidc.11455. [DOI] [PubMed] [Google Scholar]
- 49.Schmitt B.L., Leal B.F., Leyser M., de Barros M.P., Trentin D.S., Ferreira C.A.S., de Oliveira S.D. Increased ompW and ompA expression and higher virulence of Acinetobacter baumannii persister cells. BMC Microbiol. 2023;23:157. doi: 10.1186/s12866-023-02904-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones C.L., Clancy M., Honnold C., Singh S., Snesrud E., Onmus-Leone F., McGann P., Ong A.C., Kwak Y., Waterman P., et al. Fatal outbreak of an emerging clone of extensively drug-resistant Acinetobacter baumannii with enhanced virulence. Clin. Infect. Dis. 2015;61:145–154. doi: 10.1093/cid/civ225. [DOI] [PubMed] [Google Scholar]
- 51.Thummeepak R., Kongthai P., Leungtongkam U., Sitthisak S. Distribution of virulence genes involved in biofilm formation in multi-drug resistant Acinetobacter baumannii clinical isolates. Int. Microbiol. 2016;19:121–129. doi: 10.2436/20.1501.01.270. [DOI] [PubMed] [Google Scholar]
- 52.Gheorghe I., Barbu I.C., Surleac M., Sârbu I., Popa L.I., Paraschiv S., Feng Y., Lazăr V., Chifiriuc M.C., Oţelea D., et al. Subtypes, resistance and virulence platforms in extended-drug resistant Acinetobacter baumannii Romanian isolates. Sci. Rep. 2021;11:13288. doi: 10.1038/s41598-021-92590-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Law S.K.K., Tan H.S. The role of quorum sensing, biofilm formation, and iron acquisition as key virulence mechanisms in Acinetobacter baumannii and the corresponding anti-virulence strategies. Microbiol. Res. 2022;260:127032. doi: 10.1016/j.micres.2022.127032. [DOI] [PubMed] [Google Scholar]
- 54.Harrison A., Fernando D.M., Mason K.M., Santana E., Loewen P.C., Kumar A., Liu Y. KatG and KatE confer Acinetobacter resistance to hydrogen peroxide but sensitize bacteria to killing by phagocytic respiratory burst. Life Sci. 2016;148:31–40. doi: 10.1016/j.lfs.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cook-Libin S., Sykes E.M.E., Kornelsen V., Kumar A. Iron acquisition mechanisms and their role in the virulence of Acinetobacter baumannii. Infect. Immun. 2022;90:e0022322. doi: 10.1128/iai.00223-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The following whole-genome sequences were deposited in the GenBank database: JAROBQ000000000, JASKJB000000000, JASKJA000000000, JAROBM000000000, JAROBU000000000, JASKIZ000000000, JAROBS000000000, JAROBO000000000, JAROBG000000000, JASKIY000000000, JASKIX000000000, JASKIW000000000, JASKIV000000000, JASKIU000000000, JAROBR000000000, JAROBT000000000, JASKIT000000000, JAROBB000000000, JAROBI000000000, JASKIR000000000, JASKIQ000000000, JASKIP000000000, JASKIO000000000, JASKIN000000000, JASKIM000000000, JASKIL000000000, JASKIK000000000, JASKIJ000000000, JASKII000000000, JASKIH000000000, JASKIG000000000, JASKIF000000000, JASKIE000000000, JASKID000000000, JASKIC000000000, JASKIB000000000.