Abstract
Three Mycobacterium genavense strains and three American Type Culture Collection reference strains each of Mycobacterium fortuitum, Mycobacterium simiae, and Mycobacterium tuberculosis were subcultured onto Mycobacteria 7H11 agar (Difco Laboratories, Detroit, Mich.) supplemented with mycobactin J (Allied Laboratories, Fayette, Mo.). After 4 weeks of incubation at 37°C in 10% CO2, the cultures were analyzed by gas-liquid chromatography (GLC) for their fatty acids and mycolic acid cleavage products. M. fortuitum was clearly differentiated from M. genavense by the presence of the specific marker 2-methyloctadecenoic acid in M. fortuitum and by the ratio of tetracosanoic acid to hexacosanoic acid. This ratio was <1 for M. genavense and >3 for M. fortuitum. M. fortuitum also contained docosanoic acid, which was not detected in M. genavense. M. genavense, M. simiae, and M. tuberculosis, which have similar GLC profiles, were also differentiated from each other by the presence of either cis-10-hexadecenoic acid or cis-11-hexadecenoic acid and by tetradecanoic acid content.
Mycobacterium genavense, which has been isolated repeatedly from patients with AIDS (1, 6, 8, 11, 13, 15), has also been recognized as a pathogen in pet animals such as birds (14) and dogs (9). This organism fails to grow on solid media (8) except when subcultured onto Mycobacteria 7H11 agar (Difco Laboratories, Detroit, Mich.) supplemented with mycobactin J (Allied Laboratories, Fayette, Mo.) (6). Amplification of the 16S rRNA fragment from the nucleic acid extract of M. genavense has revealed a unique sequence which is most closely related to that of Mycobacterium simiae but not to that of Mycobacterium fortuitum (1). Recently, DNA homology studies of M. genavense isolates have indicated that they belong to a single and unique DNA homology group (16).
The results of thin-layer chromatographic mycolic acid studies have revealed that M. genavense is related to M. simiae and Mycobacterium malmoense (8). Study of the mycolic acids of this organism by high-performance liquid chromatography has indicated that it can be recognized by the absence of a terminal peak in the third cluster (1, 17). However, gas-liquid chromatographic (GLC) fatty acid analysis with the Microbial Identification System misidentified 14 of 17 M. genavense isolates as M. fortuitum (6).
In this study, we analyzed the fatty acids as well as the mycolic acid cleavage products (MACP) of M. genavense, M. fortuitum, M. simiae, and Mycobacterium tuberculosis by GLC. With the additional fatty acid and MACP chromatographic information, M. genavense was clearly differentiated from M. fortuitum and other closely related species.
Three M. genavense strains (isolated relatively rarely from clinical specimens) were studied. One was isolated from an AIDS patient at the Ottawa General Hospital (3), and two were received from the Laboratoire de Sante Publique du Quebec. Three American Type Culture Collection reference strains each of M. fortuitum (ATCC 6842, 9820, and 49403), M. simiae (ATCC 25273, 25275, and 15080), and M. tuberculosis (ATCC 27294, 25177, and 35800) were also examined. These M. genavense, M. fortuitum, M. simiae, and M. tuberculosis strains were subcultured onto Mycobacteria 7H11 agar slants supplemented with 2 μg of mycobactin J per ml. After 4 weeks of incubation at 37°C in 10% CO2, the bacterial cells were harvested and transferred to 2-ml microcentrifuge tubes containing pH 6.8 buffer. The cells were washed by centrifugation in a Micro Centrifuge (VWR Canlab, Missisauga, Ontario, Canada) at 9,000 rpm for 10 min. The wash step was repeated twice, and the bacterial pellets were then transferred to Teflon-lined screw-capped tubes for derivatization as described by Luquin et al. (10).
In brief, the cells were added to 1 ml of a mixture of methanol, toluene, and H2SO4, vortexed, and heated at 80°C for 16 h before being extracted twice with 2 ml of hexane. The hexane extracts were mixed with an equal amount of pH 11 phosphate buffer. After the aqueous layers were allowed to separate, the hexane layers were transferred to v-bottomed vials and evaporated to 0.1 ml under a gentle stream of nitrogen gas. The final samples were analyzed by GLC or stored at −20°C.
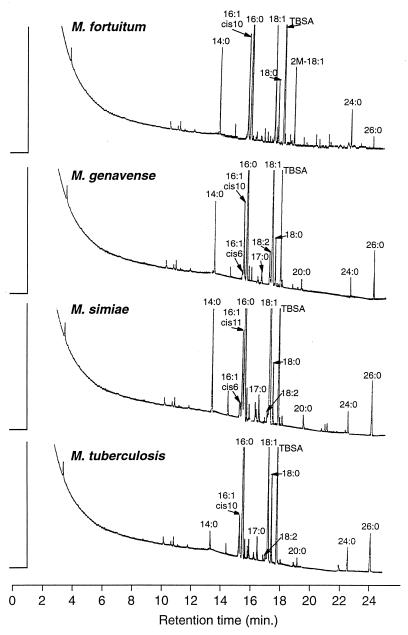
The fatty acids and MACP were analyzed with a model 3700 GLC (Varian, Sunnyvale, Calif.) equipped with a flame ionization detector. The identification of the eluted substances was accomplished by comparing the retention times with those of known standards on both a nonpolar capillary DB-5 column and a polar capillary DB-225 column (J&W Scientific Inc., Folsom, Calif.) (4). The columns were 30 m long with an inner diameter of 0.25 mm and a film thickness of 0.25 μm. For sample analysis, the temperature for both injector and detector was maintained at 300°C. Helium was used as the carrier gas. The initial column temperature of 100°C was increased to 300°C at the rate of 12°C/min. The chromatograms were integrated by a Varian 4400 integrator for identification and quantitation of each eluted substance (5). The fatty acid and MACP standards were supplied by Supelco Inc. (Bellefonte, Pa.), Applied Science Laboratory (Rockwood, Ontario, Canada), Regis Chemical Co. (Morton Grove, Ill.), and Sigma Chemical Co. (St. Louis, Mo.). The percentages of total fatty acids and MACP and the chromatograms of the studied species are shown in Table 1 and Fig. 1. Each of the test strains was regrown and retested in the same lot of medium at least twice. The deviations in the major peaks are less than 2% relative to each test. The percentages of fatty acids shown in Table 1 are mean values and ranges of studied strains of the same Mycobacterium species. Table 1 shows that M. fortuitum could be clearly differentiated from the other strains by the presence of the specific marker 2-methyloctadecenoic acid (2-M-18:1) in M. fortuitum and by the ratio of the percentage of MACP tetracosanoic acid (24:0) to that of hexacosanoic acid (26:0). This ratio was >3 for M. fortuitum and <1 for the other studied mycobacterial species. The presence of 2-M-18:1 in M. fortuitum culture was also reported by Fourche et al. (7). M. fortuitum also contained docosanoic acid (22:0) (3.8%), tetracosenoic acid (24:1) (2.3%), and docosenoic acid (22:1) (0.7%), which were not detected in the other species studied. As a rapid grower, M. fortuitum is able to produce recognizable growth on 5% sheep blood agar (Lowenstein-Jensen [LJ] slant) after 4 days of incubation; the fatty acid profiles of the 4-day-old cultures are identical to those of the 4-week cultures, including the presence of the specific marker 2-M-18:1. The fatty acid profile of M. genavense is in agreement with previous reports (6, 16); the fatty acid profiles reported therein represent only up to eicosanoic acid (20:0) and do not detect the presence of 24:0 and 26:0. The fatty acid and mycolic acid patterns of M. genavense, M. simiae, and M. tuberculosis are similar but are distinguishable from each other. There was 6.9, 7.9, and 6.3% cis-10-hexadecenoic acid (cis-10-16:1) in M. fortuitum, M. genavense, and M. tuberculosis, respectively. This acid was not found in M. simiae. In contrast, 6.3% cis-11 hexadecenoic acid (cis-11-16:1) was detected in M. simiae but not in the other species. The content of tetradecanoic acid (14:0), proposed as the differentiating factor between M. simiae and M. tuberculosis (4), can also be applied to differentiate M. genavense from M. tuberculosis. Fatty acid 14:0 was always <2% for M. tuberculosis and >5% for both M. simiae and M. genavense.
TABLE 1.
Fatty acids and MACP of mycobacterial species
| Organism (no. of strains) | % of fatty acida
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 14:0 | 15:0 | cis-6-16:1 | cis-10-16:1 | cis-11-16:1 | 16:0 | 17:0 | 18:2 | 18:1 | 18:0 | TBSA | 2-M-18:1 | 20:0 | 22:0 | 24:0 | 26:0 | |
| M. fortuitum (3) | 4.3 (4.2–4.3) | 0.4 (0.3–0.5) | 2.7 (2.6–2.7) | 6.9 (6.5–7.3) | 23.6 (22.5–22.4) | 0.7 (0.5–0.9) | 1.0 (0.7–1.2) | 12.2 (12.1–12.3) | 3.7 (3.6–3.7) | 16.3 (15.7–17.3) | 4.3 (3.7–5.2) | 1.8 (1.6–2.0) | 3.8 (3.0–4.2) | 9.4 (9.1–9.8) | 2.7 (2.5–3.0) | |
| M. genavense (3) | 5.6 (5.2–6.1) | 0.7 (0.4–1.3) | 1.3 (1.1–1.4) | 7.9 (7.5–8.3) | 32.1 (30.5–33.0) | 0.9 (0.8–1.0) | 3.6 (3.2–3.8) | 28.0 (24.5–32.5) | 3.9 (3.6–4.2) | 10.8 (8.4–15.6) | 0.4 (0.3–0.5) | 2.2 (2.0–2.4) | 4.8 (4.2–5.0) | |||
| M. simiae (3) | 7.8 (6.3–8.5) | 1.1 (1.0–1.2) | 1.9 (1.8–2.0) | 6.3 (5.9–6.7) | 31.0 (30.2–31.8) | 1.4 (1.2–1.6) | 1.3 (1.2–1.4) | 25.2 (23.5–27.2) | 3.1 (3.0–3.3) | 13.9 (13.0–14.8) | 0.8 (0.6–1.0) | 2.3 (2.0–2.5) | 4.6 (4.2–5.1) | |||
| M. tuberculosis (3) | 1.1 (0.8–1.4) | 0.5 (0.3–0.7) | 5.6 (5.3–5.8) | 35.8 (33.5–37.0) | 1.5 (1.3–1.7) | 0.2 (0.2–0.2) | 18.3 (17.5–19.5) | 7.2 (7.0–7.3) | 19.3 (17.5–21.5) | 0.3 (0.2–0.3) | 3.1 (2.9–3.3) | 5.2 (4.7–5.5) | ||||
Values are means and ranges. For each fatty acid designation, the number to the left of the colon is the number of carbon atoms, and the number to the right of the colon is the number of double bonds. cis-6, cis-10, and cis-11 indicate the double bond position from the hydrocarbon end of the cis isomer. TBSA, tuberculosteric acid (10-methyloctadecanoic acid).
FIG. 1.
Gas chromatograms of M. fortuitum, M. genavense, M. simiae, and M. tuberculosis. See text for derivations of fatty acid designations. TBSA, tuberculosteric acid (10-methyloctadecanoic acid).
The mistaken identification of M. genavense as M. fortuitum by GLC as reported by Coyle et al. (6) has been referred to as an example of the severe limitations of GLC fatty acid applications (2). Our study reveals that this misidentification is based on the limitations of the database of the integrator and microcomputer of the Microbial Identification System, which extends only up to the detection of carbon chain 20:0. The detection of fatty acids, MACP, secondary alcohols, and certain markers with carbon chains between 20:0 and 26:0 are essential for the correct identification of mycobacterial species.
In order to provide reliable identification by GLC, the chromatographer should also be aware that the peak separations and shapes, as well as the retention indices, change with the age of the column and the occurrence of contamination. Each reinstallation after column rejuvenation reduces the length of the column to a certain extent and changes the retention indices. Therefore, frequent calibration of the database, to respond to variations in the column condition, is critical for peak identification. Furthermore, the retention times for certain markers may overlap. For example, the retention times of nonadecenoic acid (19:1) and 2-eicosanol (2-OH-20:0) overlap. These are differential markers for Mycobacterium flavescens and Mycobacterium avium-M. intracellulare, respectively. In order to avoid misidentification, confirmation of the peaks should be established by mass spectrometry or by a combination of techniques including comparing the retention times with known standards obtained from two columns with considerably different polarity (4), hydrogenation of unsaturated acids, and trifluoroacetylation of hydroxy acids and secondary alcohols (12).
Acknowledgments
We thank A. Lazlo, Laboratory Centre for Disease Control, Ottawa, Canada, and L. Thibert, Laboratoire de Santé Publique du Québec, Quebec, Canada, for provision of reference strains. We also thank M. Ali for preparing media and M. Antoine for clerical assistance.
REFERENCES
- 1.Böttger E C, Teske A, Kirschner P, Boost S, Chang H R, Beer V, Hirschel B. Disseminated “Mycobacterium genavense” infection in patients with AIDS. Lancet. 1992;340:76–80. doi: 10.1016/0140-6736(92)90397-l. [DOI] [PubMed] [Google Scholar]
- 2.Böttger E C. Approaches for identification of microorganisms: despite longer experience with fatty acid profiles, DNA-based analysis offers several advantages. ASM News. 1996;62:247–250. [Google Scholar]
- 3.Chedore P. Case study: “Two 1996, isolation of Mycobacterium genavense, from a case of disseminated tuberculosis.”. Ont Soc Med Tech Advocate. 1996;1996(Spring):21. [Google Scholar]
- 4.Chou S, Aldova E, Kasatiya S. Cellular fatty acid composition of Plesiomonas shigelloides. J Clin Microbiol. 1991;29:1072–1074. doi: 10.1128/jcm.29.5.1072-1074.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chou S, Chedore P, Haddad A, Paul N R, Kasatiya S. Direct identification of Mycobacterium species in Bactec 7H12B medium by gas-liquid chromatography. J Clin Microbiol. 1996;34:1317–1320. doi: 10.1128/jcm.34.5.1317-1320.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coyle M B, Carlson L C, Wallis C K, Leonard R B, Raieys V A, Kilburn J O, Samadpour M, Böttger E C. Laboratory aspects of “Mycobacterium genavense,” a proposed species isolated from AIDS patients. J Clin Microbiol. 1992;30:3206–3212. doi: 10.1128/jcm.30.12.3206-3212.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fourche J, Capdepuy M, Texier-Maugein J. Gas chromatographic fatty acid detection to differentiate Nocardia asteroides, Mycobacterium fortuitum and Mycobacterium chelonei. J Chromatogr. 1989;487:142–146. doi: 10.1016/s0378-4347(00)83016-9. [DOI] [PubMed] [Google Scholar]
- 8.Hirschel B, Chang H R, Mach N, Piguet P F, Cox J, Piguet J D, Silva M T, Larsson L, Klatser P R, Thole J E R, Rigorts L, Portaets F. Fatal infection with novel, unidentified mycobacterium in a man with the acquired immunodeficiency syndrome. N Engl J Med. 1990;323:109–113. doi: 10.1056/NEJM199007123230207. [DOI] [PubMed] [Google Scholar]
- 9.Kiehn T E, Hoefer H, Bottger E C, Ross R, Wong M, Edwards F, Antinoff N, Armstrong D. Mycobacterium genavense infection in pet animals. J Clin Microbiol. 1996;34:1840–1842. doi: 10.1128/jcm.34.7.1840-1842.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luquin M, Ausina V, Lopez Calahorra F, Belda F, Garcia Barcelo M, Celma C, Prats G. Evaluation of practical chromatographic procedures for identification of clinical isolates of mycobacteria. J Clin Microbiol. 1991;29:120–130. doi: 10.1128/jcm.29.1.120-130.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maschek H, Georgü A, Schmidt R E, Kirschner P, Böttger E C. Mycobacterium genavense: autopsy findings in three patients. Am J Clin Pathol. 1994;101:95–99. doi: 10.1093/ajcp/101.1.95. [DOI] [PubMed] [Google Scholar]
- 12.Moss C W, Dees S B. Identification of microorganisms by gas-chromatographic mass spectrometric analysis of cellular fatty acid. J Chromatogr. 1975;112:595–604. doi: 10.1016/s0021-9673(00)99988-6. [DOI] [PubMed] [Google Scholar]
- 13.Nadal D, Caduff R, Kraft R, Salfinger M, Bodmer T, Krischner P, Böttger E C, Schaad U B. Invasive infection with “Mycobacterium genavense” in children with the acquired immunodeficiency syndrome. Eur J Clin Microbiol Infect Dis. 1993;12:5–11. doi: 10.1007/BF01997055. [DOI] [PubMed] [Google Scholar]
- 14.Portaets F, Realini L, Bauwens L, Hirschel B, Meyers W M, De Meurichy W. Mycobacteriosis caused by Mycobacterium genavense in birds kept in a zoo: 11-year survey. J Clin Microbiol. 1996;34:319–323. doi: 10.1128/jcm.34.2.319-323.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shafran S D, Taylor G D, Talbot J S. Disseminated Mycobacterium genavense infection in Canadian AIDS patients. Tubercle Lung Dis. 1995;76:168–170. doi: 10.1016/0962-8479(95)90562-6. [DOI] [PubMed] [Google Scholar]
- 16.Springer B, Wu W-K, Bodmer T, Haase G, Pfyffer G E, Kroppenstedt R M, Schröder K-H, Emler S, Kilburn J O, Kirschner P, Telenti A, Coyle M B, Böttger E C. Isolation and characterization of a unique group of slowly growing mycobacteria: description of Mycobacterium lentiflavum sp. nov. J Clin Microbiol. 1996;34:1100–1107. doi: 10.1128/jcm.34.5.1100-1107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tortoli E, Bartoloni A, Burrini C, Mantella A, Simonetti M T. Utility of high-performance liquid chromatography for identification of mycobacterial species rarely encountered in clinical laboratories. Eur J Clin Microbiol Infect Dis. 1995;14:240–243. doi: 10.1007/BF02310365. [DOI] [PubMed] [Google Scholar]