Abstract
While it is well established that the KEAP1-NRF2 pathway regulates the main inducible cellular response to oxidative stress, this cytoprotective function of NRF2 could become deleterious to the host if it confers survival onto irreparably damaged cells. In this regard, we have found that in diseased states, NRF2 promotes the transcriptional activation of a specific subset of the senescence-associated secretory phenotype (SASP) gene program, which we have named the NRF2-induced secretory phenotype (NISP). In two models of hepatic disease using Pten::Keap1 and Keap1::Atg7 double knockout mice, we found that the NISP functions in the liver to recruit CCR2 expressing monocytes, which function as immune system effector cells to directly remove the damaged cells. Through activation of this immune surveillance pathway, in non-transformed cells, NRF2 functions as a tumour suppressor to mitigate the long-term survival of damaged cells which otherwise would be detrimental for host survival. This pathway represents the final stage of the oxidative stress response, as it allows cells to be safely removed if the macromolecular damage caused by the original stressor is so extensive that it is beyond the repair capacity of the cell.
Keywords: KEAP1, NRF2, NFE2L2, Stress response, Oxidative stress, Senescence, SASP, NISP, Immune surveillance, Efferocytosis
2. Introduction
Through the regulation of cytoprotective gene expression, the transcription factor NRF2 controls the main inducible oxidative stress response in humans [[1], [2], [3]]. The inducible nature of this response is mediated by the E3 ubiquitin ligase adaptor KEAP1, which, under homeostatic conditions, maintains basal cytoprotective gene expression due to the constitutive ubiquitination and proteasome-dependent degradation of NRF2 [4,5]. The function of KEAP1 is directly modulated by the action of stressor molecules, which can bind to reactive cysteine residues within KEAP1, resulting in a conformation change in the protein, the loss of its ubiquitination activity, and thus the stabilization of NRF2 [6].
Due to the central positioning of the KEAP1-NRF2 pathway in redox homeostasis and the cellular response to oxidative stress, it plays an important role in the development and progression of many pathologies, including autoimmune, respiratory and metabolic diseases [7]. However, some important unexplained observations based on data from mouse models suggest that NRF2 activation has additional physiological outputs which have yet to be described. For example, despite the fact that oxidative stress plays an important role in the etiology of many aging-related diseases, genetic activation of Nrf2 in mice shows that its function in longevity is more nuanced [8,9]. Thus, while correlative analysis of Nrf2 activity suggests that it has a pro-longevity function, lifelong genetic activation of Nrf2 in mice is associated with decreased lifespan, while heterozygous Nrf2 mice do not exhibit a decrease in lifespan [[10], [11], [12], [13], [14]]. Given the well-established cytoprotective functions of Nrf2, this decrease in longevity upon lifelong Nrf2 activation suggests that it may function, through a currently unknown mechanism, as an antagonistic pleiotropy factor, such that it is beneficial early in life, and costly later.
One well described example of antagonistic pleiotropy is cellular senescence, which benefits organismal survival through its tumor suppressor function, but can also contribute to the pathology of diseases later in life [15]. Specifically, senescence functions as an important endogenous anti-tumour mechanism by inhibiting the proliferation of damaged, precancerous cells [16,17]. These senescent cells can either be removed by immune effectors through the immune surveillance pathway or maintained in a non-proliferative state in situ [18]. Through the development of the senescence-associated secretory phenotype (SASP), which consists of various cytokines, chemokines and proteases, the senescent cells that continue to reside in tissues can contribute to the etiology of many aging-associated diseases [19]. In this way, senescence benefits organismal survival early in life, but may deleteriously impact longevity through the multiple functions of the SASP.
The best characterized function of NRF2 in human disease relates to its function in cancer, where its cytoprotective, metabolic and stress resistant attributes are frequently hijacked by the tumour cells [20]. This leads to aggressive tumour growth, anticancer therapy resistance, and a poor prognosis for patients, particularly those with non-small cell lung carcinoma [21,22]. Despite these facts, constitutive activation of NRF2 signaling alone is not sufficient to induce tumor formation [23,24], which suggests that it may also have a hitherto undescribed parallel tumour suppressor role which can counteract its facilitation of oncogenic processes.
Thus, taken together, the enigmatic cancer and longevity phenotypes suggest that NRF2 has additional important physiological functions which are yet to be uncovered. In this study, we found that NRF2 functions as a senescence promoting factor, such that cells with high levels of NRF2 activity acquired through genetic and epigenetic events can easily enter a senescent state. Furthermore, NRF2 regulates the transcription of a unique subset of SASP genes, which we have named the NRF2-induced secretory phenotype (NISP). The physiological function of the NISP is to recruit immune effector cells to remove these damaged NRF2-activated cells through the immune surveillance pathway. Thus, the NRF2-NISP-Immune surveillance axis functions as a tumour suppressor pathway in order to mitigate the survival of damaged and pre-cancerous NRF2-activated cells. This pathway represents the final stage of the oxidative stress response, as it allows cells to be safely removed if the macromolecular damage caused by the original stressor is so extensive that it is beyond the repair capacity of the cell.
3. Results
3.1. KEAP1 mutant cells cannot proliferate after mitomycin C removal
We recently identified two classes of compounds, the DNA damaging agent mitomycin C (MMC) and the quinone-containing geldanamycin family of HSP90 inhibitors, to be synthetic lethal with NRF2 [25,26]. As under most conditions, NRF2 promotes enhanced survival of cancer cells in response to chemotherapeutic drug treatment [27,28], we wanted to determine whether cells with augmented NRF2 signaling could become resistant to the aforementioned synthetic lethal compounds, and if so, through what mechanism this resistance is achieved. To explore this problem, we utilized a previously established co-culture system using fluorescently-labelled isogenic WT and Keap1 KO Hepa1 cells (Figs. S1A and S1B) [25]. Consistent with our previous results, MMC displayed enhanced toxicity in Keap1 KO cells (Fig. 1A), and therefore we decided to focus on MMC as it is already approved for clinical use.
Fig. 1.
NRF2 primes cells to become senescent
A. Viabilities of co-cultured isogenic WT-GFP and Keap1 KO-mCherry cells, determined by increase in fluorescence intensity relative day 0, treated with either DMSO or 50 nM mitomycin C, for 7 days. B. Overview of the mitomycin C (MMC) washout experimental protocol using isogenic Hepa1 cells. C. Viabilities of co-cultured isogenic WT-GFP and Keap1 KO-mCherry cells, determined by increase in fluorescence intensity relative to day 0, treated with either DMSO, 50 nM mitomycin C for the full duration of the experiment (Full MMC), or 72 h MMC treatment followed by recovery in fresh media. Each experiment was performed at least 3 times. Data represent mean ± SD, N = 6, *p < 0.001 when comparing growth between day 5 and day 7. ns = not significant. D. ATAC-Seq-derived chromatin accessibility data for 4 different transcription factors showing the time-dependent changes in chromatin accessibility during RAS-induced senescence. E. The relative expression of representative senescence-associated genes in isogenic WT and Keap1-Nrf2 DKO Hepa1 cells after treatment with 33 nM doxorubicin for 24 h, followed by 48-h recovery in fresh media, determined using RT-qPCR. F. Viabilities of co-cultured isogenic WT-GFP and Keap1 KO-mCherry Hepa1 cells, determined by fluorescence intensity relative to the DMSO control, exposed to the indicated concentrations of mitomycin C alone, or the co-treatment with 100 nM ABT-263, for 8 days. Note that in Keap1 KO cells, the co-treatments give a significantly reduced survival relative to the single treatment with mitomycin C alone. Each experiment was performed at least 3 times. Data represent mean ± SD, N = 6, *p < 0.05, **p < 0.005.
To determine whether Keap1 KO cells, which exhibit enhanced NRF2 signaling, could develop resistance to MMC, we designed and conducted a series of washout experiments, as shown in Fig. 1B. We treated the co-cultures with a low concentration of mitomycin C (50 nM) for 72 h, after which fresh media was added, and the recovery of the cells was monitored for a further 4 days. Interestingly, while the WT cells were able to grow in either the presence of MMC, or after MMC washout, the Keap1 KO cells were unable to do so under either condition (Fig. 1C, S1C). This failure of the Keap1 KO cells to begin proliferating again, even up to 4 days after MMC removal, suggested that, contrary to our expectations, resistance to MMC could not be acquired.
As MMC is a cytotoxic compound which induces DNA damage, under these NRF2-dependent synthetic lethal conditions, the cellular damage caused by MMC may be irreparable. Because NRF2 functions as a stress response factor, we hypothesized that NRF2 activation in the context of irreparable cellular damage may activate a stress response which inhibits the proliferation and survival of these damaged cells. One candidate cellular state which is induced by cellular damage, and is consistent with the phenotype that we observed, is cellular senescence. We thus hypothesized that, in response to irreparable cellular damage, NRF2 may prime cells to become senescent by upregulating genes required for senescence induction. In support of this idea, after MMC washout, Keap1 KO cells, but not WT cells, stained positively for the senescence marker SA-β-gal (Fig. S1D). Similarly, relative to the isogenic WT cells, the Keap1 KO cells exhibited enhanced induction of representative senescence-associated gene expression in response to MMC treatment (Fig. S1E).
To test the generality of this NRF2-dependent effect, we conducted MMC washout experiments in human cancer cells lines with mutation-driven activation of NRF2 signaling. In agreement with our data from the isogenic Hepa1 cells, in all cases, the NRF2 activated cancer cells were unable to proliferate after removal of MMC from the culture media (Figs. S1F and G). Furthermore, after MMC washout, all of the NRF2-activated human cancer cell lines stained positively for the senescence marker SA-β-gal (Fig. S1H).
Taken together, these data suggest that NRF2-activated cell lines may be predisposed to enter cellular senescence more easily than their non-activated equivalents. As this represents a hitherto unexplored mechanism of NRF2 activity, we wished to explore the mechanism through which NRF2 may contribute to senescence induction.
3.2. NRF2 functions as a senescence promoting factor
Through the time-course-dependent evaluation of gene enhancer activity, senescence priming has recently been ascribed to the AP-1 transcription factor, which has been labelled a “senescence pioneer” due to its ability to imprint a reversible transcriptional program onto senescent cells [29]. To determine whether NRF2 is also able to function to promote cellular senescence, we analyzed the senescence time-course ATAC-Seq dataset, focusing on the accessibility of the antioxidant response element of NRF2 during senescence induction [29]. Evaluation of the “chromatin opening index” metric revealed that, like the FOS:JUN AP-1 complex, NRF2 is also able to promote cellular senescence, while other factors like p53 and FOXD3 function as senescence settlers and migrants, respectively (Fig. 1D). Therefore, these ATAC-Seq data provide compelling support for our contention that activation of NRF2 signaling primes cells to become senescent.
While senescent cells are characterized by withdrawal from the cell cycle, they are still metabolically active, and are able to interact with their environment through the secretion of a multitude of cytokines, chemokines, growth factors and proteases, which are collectively referred to as the senescence-associated secretory phenotype (SASP) [19,30]. The core SASP program consists of IL-1α, IL-6 and IL-8, which function in concert in both a paracrine and autocrine manner to establish and maintain the senescence state [17,31]. Transcriptional regulation of the SASP program is mediated by a select group of transcription factors, which includes C/EBPβ, NF-κB and GATA4 [17,31,32]. If NRF2 is an integral part of the senescence program, we hypothesized that it should directly control the transcription of the central SASP factors IL-1α, IL-6 and IL-8. Importantly, analysis of ChIP-Seq data from the UCSC genome browser revealed that NRF2 binds to enhancer elements in the Il1α, Il6 and Il8 loci, which suggests it could play an important role in their regulation (Fig. S2A). As these ChIP-Seq data were generated in the absence of a senescence inducing stimuli, they are consistent with our contention that NRF2 functions to promote cellular senescence. Furthermore, the NRF2 binding sites are adjacent to those of the well-characterized senescence master regulator C/EBPβ, which suggests that these enhancers are functionally important for induction and maintenance of senescence (Fig. S2A). Of note, NRF2 has recently been shown to directly bind to, and activate transcription in a complex with, C/EBPβ, which may facilitate its role in senescence-associated gene induction [33].
To demonstrate the requirement for NRF2 for the induction of the senescence gene program, we treated WT and isogenic Keap1-Nrf2 double knockout (DKO) Hepa1 cells with the DNA damaging agent, and senescence inducer, doxorubicin for 24 h (Fig. 1E). Senescence associated gene expression was then measured after 48 h of recovery in regular cell culture media. Doxorubicin was used in place of MMC to eliminate the impact of the synthetic lethal relationship between Nrf2 and MMC [26]. While WT cells showed clear induction of the senescence master regulators Il1α and Il1β, in addition to a number of SASP factors [19,34], this gene induction was not observed in the absence of Nrf2 in Keap1-Nrf2 DKO cells (Fig. 1E, orange bars). Of note, in WT cells, the senescence gene induction was accompanied by induction of the prototypical Nrf2 target genes Nqo1, Gclc, Gclm and Txnrd1, which supports the idea that activation of Nrf2 signaling plays an important role in senescence induction (Fig. S2B).
Furthermore, as senescent cells acquire a characteristic flattened morphology upon cell-cycle withdrawal [35], we analyzed the morphology of the isogenic WT and Keap1 KO cells in response to the senescence inducers MMC, doxorubicin, cisplatin, and paclitaxel. In all cases, the surviving Keap1 KO-mCherry cells assumed a flattened morphology when compared to the co-cultured WT cells (Fig. S2C).
Finally, to determine whether NRF2 activation alone is sufficient to induce SASP gene expression, we analyzed senescence-associated gene expression under basal conditions in the isogenic WT and Keap1 KO Hepa1 cells. This experiment revealed that in cancer cells, under basal conditions and in the absence of any stressors, Nrf2 activation alone is sufficient to induce SASP gene expression (Fig. S2D), which supports our contention that the SASP represents an important component of the NRF2 response. Taken together, these data demonstrate that NRF2 functions to promote cellular senescence, such that NRF2 activity primes cells to become senescent, while conversely, the absence of NRF2 delays induction of senescence-associated gene expression.
3.3. NRF2 activation sensitizes cells to senolytic compounds
As senescent cells play an important role in the progression of many human diseases, a diverse range of compounds have been developed which display toxicity specifically in senescent cells. These compounds, collectively referred to as senolytics, include inhibitors of anti-apoptotic factors, pro-survival pathways, and proteostasis chaperones [16,17].
In order to directly test our hypothesis that NRF2 primes cells to become senescent, we treated the co-cultured isogenic WT and Keap1 KO Hepa1 cells with the senolytic ABT-263, which targets BCL2, using a co-treatment protocol utilizing MMC to induce senescence [36]. If Nrf2 functions as to promote cellular senescence, cells with activated Nrf2 should be uniquely sensitive to the co-treatment of MMC together with the senolytics, resulting in enhanced toxicity only in the Keap1 KO cells (Fig. S3A). Consistent with this idea, while in the absence of MMC, ABT-263 had no impact on Keap1 KO cell viability (Fig. S3B), the co-treatment with MMC resulted in reduced cell survival specifically in Keap1 KO cells (Fig. 1F, red bars). To expand these findings beyond the isogenic Hepa1 cell system, we assayed the synergistic effect of MMC plus ABT-263 in the human liver cancer cell lines, JHH2 and JHH5. Importantly, activation of NRF2 in the KEAP1 mutated JHH5 cells also conferred specific sensitivity to ABT-263 (Fig. S3C). This sensitivity to senolytics is a hallmark characteristic of cellular senescence, and strongly supports our assertion that NRF2 functions as a senescence priming factor.
To test the generality of this NRF2-dependent senolytic sensitivity, we treated a range of human lung cancer cell lines harboring either mutation-driven hyperactivation of NRF2 (H460, H2023, A549) or wild-type KEAP1-NRF2 signaling (HCC827, COR-L105, HCC4006) with MMC plus ABT-263. While ABT-263 treatment alone had no impact on cell viability (Fig. S3B), in all cases, the cell lines with augmented NRF2 signaling exhibited enhanced sensitivity to treatment with MMC plus the senolytic ABT-263 (Fig. S3D), while in stark contrast, no enhanced sensitivity was observed in the cells with normal NRF2 function (Fig. S3E). This result is consistent with a previous PDX model-based study, which showed that NRF2 activity conferred sensitivity to the co-treatment of Trametinib with ABT-263 [37,38].
3.4. NRF2 regulates the expression of a subset of SASP genes
As NRF2 regulates an inducible stress response, we posited that the induction of cellular senescence, and the accompanying secretory phenotype of senescent cells, represents an important and uncharacterized component of the overall NRF2 stress response which becomes physiologically important when NRF2 is activated in the context of irreparable cellular damage. In this regard, as liver-specific models of senescence are associated with a diverse range of interactions between the senescent cells and the healthy host [18,39,40], we surmised that a hepatocyte model may be particularly informative when trying to determine the physiological function of NRF2 in the context of irreparable damage. Therefore, in order to test our hypothesis, we used the hepatocyte-specific Pten::Keap1 conditional knockout (DKO) mouse model, in which Nrf2 is activated in the context of significant liver damage. Specifically, these mice are characterized by a striking elevation in the liver damage markers ALT, AST and LDH, which ultimately results in the death of the mice within one month of birth [41]. Thus, we postulated that the severe disease phenotype in the DKO mouse would allow us to examine the role of Nrf2 when activated in the context of irreparable damage.
As the senescence state is maintained by the activity of cyclin-dependent kinase inhibitors (CDKi), such as p16INK4A, we first analyzed the CDKi expression levels in the DKO mice using a microarray-based analysis. Within the CDKi family, we found that Cdkn2b, which encodes p15INK4B, is specifically upregulated in an Nrf2-dependent manner (Fig. 2A). This provided the first evidence that senescence may play an important role in the Nrf2 response in vivo.
Fig. 2.
Nrf2 regulates the expression of a specific subset of SASP genes
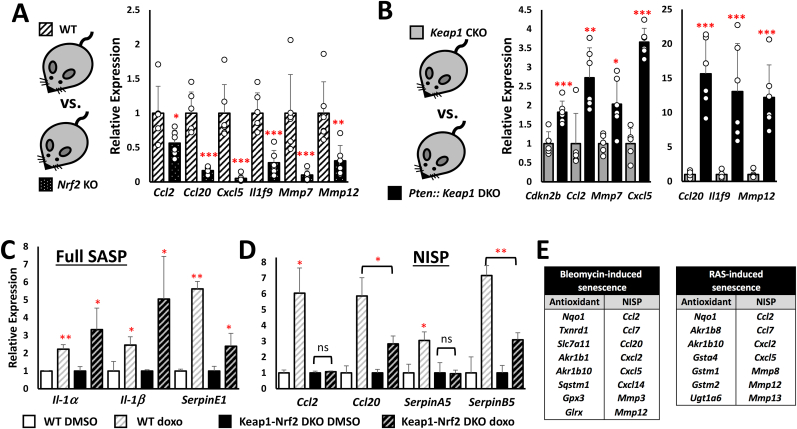
A. Relative expression of differentially regulated senescence-associated genes (SASP) in the livers of Pten::Keap1 DKO mice relative to Pten CKO mice as determined by microarray. CDKi = Cyclin-dependent kinase inhibitor, MMP = matrix metalloproteinase. B. Relative expression of well-described classical SASP genes, as defined by Ref. [19]; in the livers of Pten::Keap1 DKO mice relative to Pten CKO mice as determined by microarray. C. Relative expression of a liver-specific SASP gene signature, as defined by Ref. [40]; in the livers of Pten::Keap1 DKO mice relative to Pten CKO mice as determined by microarray.
To expand on this finding, we then extended our microarray analysis to SASP gene expression in the DKO mice, as this represents a second fundamental characteristic of senescent cells [19,30], by focusing primarily on the chemokine, cytokine, matrix metalloproteinase (MMP) and serine protease (Serpin) gene families (Fig. 2A). Consistent with our expectations, we observed increased Nrf2-dependent expression of a number of well characterized SASP genes, including Ccl2, Cxcl5 and Mmp12 [19]. In contrast, and to our surprise, we did not observe a global induction of SASP gene expression, as the expression of important and classical SASP factors such as Ccl1, Cxcl1 and Il1α was unchanged in the DKO mouse liver (Fig. 2B). From these data, we conclude that in vivo, Nrf2 regulates the expression of a specific subset of SASP genes.
Of note, there is very little overlap between the Nrf2-dependent SASP (this study) and NRas-induced liver-specific SASP [40] (Fig. 2C), suggesting that the Nrf2 phenotype is unique. Interestingly, while NF-κB is known to be a major regulator of SASP gene expression [17], we observed increased expression of a number of NF-κB repressors, including FoxJ1, in the DKO mice, which suggests that competition between Nrf2 and NF-κB may be in part responsible for the specific SASP found in Nrf2-activated cells [42].
3.5. The NRF2-induced secretory phenotype
To confirm the microarray results, we validated the in vivo Nrf2-specific senescence-associated gene expression pattern of a representative set of genes by RT-qPCR using liver-derived cDNA from Pten CKO and Pten::Keap1 DKO mice (Fig. 3A). These results confirmed that Cdkn2b and Il1f9 are significantly induced 2-3-fold, while Ccl2, Ccl20 and Mmp12 are significantly upregulated by at least 19-fold in the DKO mice. Furthermore, Pten CKO alone is insufficient to induce Nrf2-dependent gene expression, which highlights the fact that the specific combination of Nrf2 activation in the context of the diseased liver is required for SASP gene induction (Fig. S4A). Importantly, however, RT-qPCR revealed that the well-characterized senescence factors Il1α, Il1β and Il6 are not upregulated in the DKO mice (Fig. 3B), which supports our view that Nrf2 regulates the expression of a specific subset of SASP genes.
Fig. 3.
Nrf2 directly regulates NISP gene expression
A. RT-qPCR validation of the Nrf2-dependent senescence-associated gene expression changes observed in the livers of 15-day old Pten::Keap1 DKO mice. Each experiment was performed at least 3 times. Data represent mean ± SD, N = 6 mice per genotype, *p < 0.05, **p < 0.005. B. Confirmation by RT-qPCR that a global increase in senescence-associated gene expression is not observed in 15- day old Pten::Keap1 DKO mouse livers. Each experiment was performed at least 3 times. Data represent mean ± SD, N = 6 mice per genotype, with no significant change between genotypes for any of the genes. C. ChIP-Seq data of the human CCL2 locus on chromosome 17, taken from the UCSC genome browser. The ChIP-Seq track for NRF2 (also named NFE2L2) from IMR-90 cells shows a binding site in the histone H3K27Ac marked enhancer region adjacent to the CCL2 promoter. Note that this ChIP-Seq data was generate in the basal state, without any additional senescence-inducing stimuli. D. A summary of the NRF2 binding sites adjacent to the NISP (Nrf2-induced secretory phenotype) gene loci, identified using the IMR-90 ChIP-Seq data set from the UCSC genome browser. E. ChIP-qPCR in A549 cells showing enrichment of NRF2 upon MMC treatment at the enhancer elements of NISP genes relative to the negative control, p21. Each experiment was performed at least 3 times. Data represent mean ± SD, **p < 0.005. F. ELISA for secreted CCL2 protein, using conditioned media from WT and Keap1 KO Hepa1 cells, in both the basal state, and in response to MMC treatment. Each experiment was performed at least 3 times. Data represent mean ± SD, **p < 0.005. G. Visual representation of the relationship between the newly characterized Nrf2-induced secretory phenotype (NISP) and the classical senescence-associated secretory phenotype (Full SASP), in which the NISP represents a specific sub-set of SASP genes.
In order to support our contention that Nrf2 directly regulates the expression of these SASP factors, we analyzed the binding status of NRF2 at the enhancer regions of the SASP genes identified in the microarray, using NRF2 ChIP-Seq data from the UCSC genome browser database, an example of which is shown in Fig. 3C. Importantly, these ChIP-Seq data revealed that NRF2 directly binds to the enhancers of a broad range of the SASP factors which we identified in the Pten::Keap1 DKO mouse livers (Fig. 3D). Furthermore, and consistent with our model, ChIP-qPCR revealed enhanced binding of NRF2 to the enhancers of these SASP genes in response to MMC treatment (Fig. 3E). Specifically, NRF2 was significantly enriched at the enhancer elements of the SASP factors Ccl2, Cxcl2, Cxcl5 and Mmp7, which supports our contention that NRF2-dependent regulation of these SASP factors plays an important role in the damage response to MMC.
To confirm that NRF2-dependent transcription results in SASP factor secretion, we focused on the chemokine CCL2, as our data have revealed it to be positively regulated by NRF2 across all of our experimental conditions. Using the isogenic WT and Keap1 KO Hepa1 cells, ELISA assays revealed that both in the basal state, and in response to MMC-mediated senescence induction, significantly more CCL2 protein is secreted by Keap1 KO cells when compared to their isogenic WT counterparts (Fig. 3F). This strongly supports our thesis that NRF2-dependent regulation of SASP gene expression is functionally important.
Taken together, these results clearly demonstrate that within the senescence program, NRF2 directly regulates the expression of a specific subset of SASP genes. Due to the specificity of this response, we have named this gene program the NRF2-induced secretory phenotype (NISP) (Fig. 3G).
3.6. NRF2 regulates NISP expression across a range of conditions
In order to determine the extent of Nrf2 regulation on NISP gene expression, we first analyzed the expression of a representative set of NISP genes in Nrf2 KO mice [1,41] (Fig. 4A). This RT-qPCR analysis revealed that in the basal state, and thus absence of any substantial senescence-inducing stimuli, NISP gene expression is significantly downregulated in the absence of Nrf2 (Fig. 4A). This result is consistent with the Nrf2-dependent expression pattern of cytoprotective genes, where Nrf2 regulates both their basal and inducible expression, which in turn indicates that the NISP genes represent a core component of the physiological Nrf2 response [1,45].
Fig. 4.
Full NISP induction requires NRF2 activation in the context of cellular damage
A. Relative expression of representative NISP genes in the livers of 10-week-old Nrf2 KO mice relative to WT mice as determined by RT-qPCR. Each experiment was performed at least 3 times. Data represent mean ± SD, N = 6 mice per genotype, *p < 0.05, **p < 0.01, ***p < 0.005. B. Relative expression of representative NISP genes in the livers of 15-day old Pten::Keap1 DKO mice relative to Keap1 CKO mice as determined by RT-qPCR. Each experiment was performed at least 3 times. Data represent mean ± SD, N = 6 mice per genotype, *p < 0.05, **p < 0.01, ***p < 0.005. C, D. The relative expression of representative (C) Full SASP and (D) NISP genes in isogenic WT and Keap1-Nrf2 DKO Hepa1 cells after treatment with 33 nM doxorubicin for 24 h, followed by 96-h recovery in fresh media, determined using RT-qPCR. Each experiment was performed at least 3 times. Data represent mean ± SD, N = 6, *p < 0.005 ns = not significant. E. Significant co-induction of the NRF2-dependent antioxidant and NISP gene expression signatures in bleomycin and RAS-induced senescence models. The data were sourced from Refs. [43,44]; and [29].
To elucidate the role of Nrf2 in the inducible activation of the expression of NISP genes, we compared Keap1 conditional knockout (Keap1 CKO) (Keap1flox/flox:Albumin-Cre) mice with Pten::Keap1::Albumin-Cre DKO mice (Fig. 4B). The DKO mice are characterized by hyperactivation of hepatic Nrf2 levels and signaling relative to Keap1-CKO mice, coupled with a disease phenotype in which the mice die within 28 days of birth [41]. In contrast, the deletion of Keap1 alone, while substantially activating Nrf2 above wild-type levels, does not result in any obvious liver damage or deleterious phenotype in the mice [41]. At 15 days post-birth, the DKO mice exhibit increased expression of the CDK inhibitor, and senescence marker, Cdkn2b, in addition to a range of representative NISP genes (Fig. 4B). Taken together, these data indicate that full activation of the NISP is only realized in response to the irreparable damage observed in the diseased state.
3.7. Nrf2 regulates NISP gene expression in response to DNA-damage-induced senescence
As under normal conditions, the function of Nrf2 within the senescence program will be integrated into a cellular response consisting of multiple pathways, we wanted to investigate the role of Nrf2 in NISP induction in response to DNA-damage-mediated senescence induction. To this end, we treated WT and isogenic Keap1-Nrf2 DKO Hepa1 cells with doxorubicin for 24 h, and then allowed the cells to recover in fresh media for 4 days, before collecting the RNA for analysis by RT-qPCR. Unlike at the 48-hr time point (Fig. 1E), after 4 days of recovery in fresh media, the Keap1-Nrf2 DKO cells were able to induce the expression of the SASP factors Il1α, Il1β and SerpinE1 at similar levels to WT cells (Fig. 4C). This shows that, while Nrf2 functions as an important senescence promoter, in the absence of Nrf2, SASP induction is delayed, but not abolished. In contrast to the expression dynamics of the classical or “Full SASP” genes, NISP gene expression was significantly impacted by the loss of Nrf2. Specifically, Ccl2 and SerpinA5 were not induced by doxorubicin treatment in the absence of Nrf2 (Fig. 4D). In addition, while Ccl20 and SerpinB5 were induced by doxorubicin in the Keap1-Nrf2 DKO cells, this was at a significantly lower level than in the WT cells. Treatment of Keap1 KO cells with a C/EBPβ inhibitor produced a variable effect on NISP gene expression, which suggests that, while C/EBPβ may be an important co-regulator of some SASP genes, including CCL2 and CCL20, it is not required for the induction of all NISP genes (Fig. S4B). Together, these data strongly support the in vivo data from the Pten::Keap1 DKO mice, namely that Nrf2 plays a critical role in the induction of NISP gene expression in response to senescence-inducing stimuli.
To expand upon these findings, we analyzed the role of Nrf2 in NISP gene induction in other models of senescence. As our data suggested that Nrf2 activity may be induced as part of the senescence response (Fig. S2B), we hypothesized that we could use increased expression of Nrf2-target genes as a marker for Nrf2 activity, and then determine whether the expression of these classical antioxidant genes correlates with NISP gene expression in response to different senescence inducing agents. Interestingly, in both bleomycin and RAS-induced senescence, enhanced antioxidant and NISP gene expression were significantly enriched together (Fig. 4E) [29,43,44]. Taken together, these data clearly demonstrate that NRF2 regulates NISP gene expression under a diverse range of cell damaging conditions.
3.8. The NISP promotes the immune-surveillance-mediated removal of damaged cells
Due to the fact that NRF2 regulates an inducible cytoprotective pathway, while cellular senescence functions to limit the proliferation of damaged cells, we hypothesized that senescence induction may represent a previously unrecognized component of the NRF2 stress response program. To test this idea, and to determine the physiological significance of NRF2-dependent senescence induction, we analyzed the liver-specific phenotype of the Pten:Keap1 DKO mice, focusing on how the unique features of this disease model may reveal important aspects of the role of NRF2 in the cellular damage response.
There are two particularly interesting features of the DKO phenotype which suggested that the senescence-mediated immune surveillance pathway may be activated in these mice. First, loss of hepatocyte marker expression, which was revealed to be widespread based on the well characterized markers from the Human Protein Atlas database [46], suggested that there was significant hepatocyte depletion in the DKO mice (Fig. S5A). Second, we found that the DKO mouse livers were characterized by substantial infiltration of cells from the immune system (Fig. S5B). Taken together, these phenotypic features encouraged us to explore whether the immune surveillance pathway is activated in the diseased livers of the Pten:Keap1 CKO mice.
Senescence-mediated immune surveillance is the process through which damaged and pre-malignant cells are actively removed by the immune system [18]. In the liver, this important tumour suppressor response is mediated by the secretion of chemokines and cytokines, which together function to attract and activate monocytes/macrophages to remove the damaged cells [18]. As the DKO mice are characterized by NISP expression, which includes various chemokines and cytokines, activation of the immune surveillance pathway would explain all of the phenotypic features that we observe in these mice.
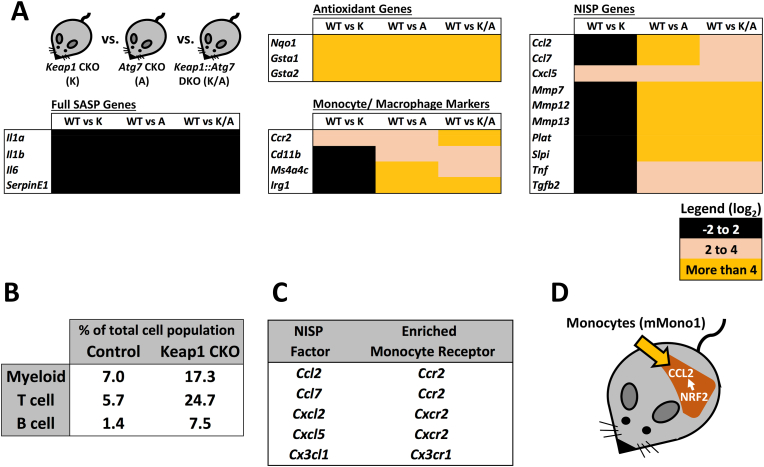
To determine which immune cells accumulate in the DKO livers, we first analyzed lineage-specific gene expression markers for monocytes/macrophages, T cells and B cells (Fig. 5A–C). This analysis revealed that the DKO liver phenotype is not characterized by a broad-spectrum immune response, but instead consists of a specific monocyte/macrophage infiltration profile.
Fig. 5.
The NISP functions to recruit CCR2-expressing monocyte-derived macrophages
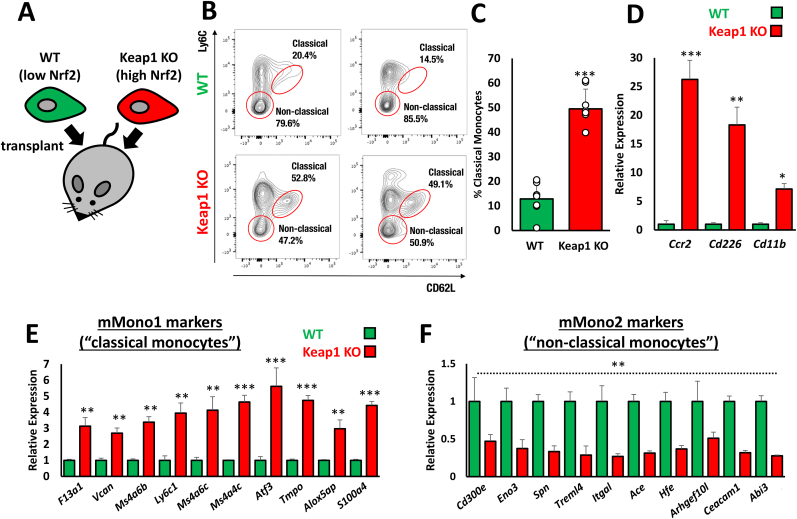
A. Enrichment of monocyte/macrophage markers, and not those of liver-resident Kupffer cells, specifically in the livers of Pten::Keap1 DKO mice, as determined by microarray. B. Relative expression of T cell and B cell markers in the livers of Pten::Keap1 DKO mice relative to Pten CKO mice, as determined by microarray. Note that, the immune infiltration into the livers of Pten::Keap1 DKO mice is specific to monocytes/macrophages, and does not represent a general immune cell infiltration phenotype. C. Confirmation by RT-qPCR that 15-day old Pten::Keap1 DKO mice are characterized by monocyte/macrophage infiltration. Each experiment was performed at least 3 times. Data represent mean ± SD, N = 6 mice per genotype, *p < 0.01, **p < 0.001. D, E. RT-qPCR analysis of markers of mMono1 and mMono2 cells, which correspond to “classical” and “non-classical” monocytes, based on specific and comprehensive gene expression patterns identified by Zilionis et al. using scRNA-Seq. These data show that 15-day old Pten::Keap1 DKO mice are characterized specifically by infiltration of mMono1 (“classical”) monocytes. Each experiment was performed more than 3 times. Data represent mean ± SD, N = 6 mice per genotype, **p < 0.001. F. Markers specific to CCR2+ monocyte-derived macrophages are specifically enriched in 15-day old Pten::Keap1 DKO mouse livers, as determined by RT-qPCR. Each experiment was performed at least 3 times. Data represent mean ± SD, N = 6 mice per genotype, *p < 0.01, **p < 0.001.
As we hypothesized that the NISP is responsible for the infiltration of monocytes and macrophages, we wanted to determine which NISP factor(s) are responsible for this specific immune response. In the liver, immune surveillance is mediated by the secretion of the chemokine CCL2, which recruits CCR2 expressing classical monocytes to the damaged or pre-malignant hepatocytes [18,39,40]. Since CCL2 is a NISP factor (Fig. 2, Fig. 3), it represented the perfect candidate for our Nrf2-dependent phenotype. Importantly, the gene expression analysis provided no evidence to suggest that the increase in monocytes/macrophages was due to an expansion of liver-resident macrophages (Kupffer cells) (Fig. 5A), thus providing further support to the idea that the increase in monocyte/macrophage infiltration observed in the DKO mice was due to the recruitment of cells from outside of the liver.
Monocytes, which can differentiate into macrophages upon tissue infiltration, are traditionally classified into two populations, “classical” (CCR2+ Ly6Chi) and “non-classical” (CCR2- Ly6Clo), both of which originate from the bone marrow [47]. In order to more accurately determine which monocyte population is enriched in Pten:Keap1 DKO livers, we utilized additional specific markers of the “classical” and “non-classical” populations derived from scRNA-Seq [48]. Analysis of the gene expression data from this study revealed that a panel of 20 genes could be used to thoroughly distinguish between the two monocyte populations, named mMono1 (classical) and mMono2 (non-classical) based on the scRNA-Seq gene expression profiles (Figs. S5C and D). RT-qPCR analysis of these markers revealed that mMono1, which corresponds to “classical” monocytes, were specifically enriched in Pten:Keap1 DKO livers (Fig. 5D and E). Furthermore, in the liver, monocyte-derived macrophages can be distinguished from the tissue resident Kupffer cells based on the expression of Clec4d, Clec4e and Clec5a, which are specific for classical monocyte-derived cells [47]. Consistent with our model, Clec4d, Clec4e and Clec5a, were all upregulated in the Pten:Keap1 DKO livers, which supports our contention that classical monocytes are specifically recruited into the Nrf2-activated DKO livers (Fig. 5F).
Taken together, these data support a model in which, in vivo, the NISP functions to recruit CCR2+ classical monocytes to the liver to remove the damaged hepatocytes through the senescence-mediated immune surveillance pathway. This results in a net loss in hepatocyte cell number, which explains the relative reduction in hepatocyte gene expression which is a characteristic feature of this disease model [41].
3.9. The NISP is also expressed in Keap1::Atg7 DKO mice
In order to determine whether the NISP represents a general component of the Nrf2 response, and is not in some way restricted to the co-deletion of Pten, we carried out an additional microarray using liver samples from Keap1::Atg7-Alb mice (Keap1::Atg7 DKO) (Fig. 6A). In this model, loss of Atg7 disrupts cellular autophagy, which results in significant liver damage, and thus, like the Pten::Keap1 DKO mice, the Keap1::Atg7 DKO mice also display a disease phenotype, in which 20% of mice die within 20 weeks of birth [49]. Therefore, this Keap1::Atg7 DKO model represents a second phenotype in which Nrf2 is activated in the diseased state [49]. Furthermore, Atg7 knockout alone is sufficient to activate Nrf2 through the non-canonical p62 pathway, and also causes significant liver damage [50], and therefore this model also allowed us to explore NISP expression in the context of a functional Keap1 protein.
Fig. 6.
The NRF2-NISP-Immune surveillance axis is also functional in Atg7 conditional knockout mice.
A. The gene expression profiles of liver-specific Keap1 conditional knockout (K), Atg7 conditional knockout (A) and Keap1::Atg7 double conditional knockout mice (K/A) were compared to wild-type mice in order to determine the contribution of Nrf2 activation in the diseased state (A and K/A) to NISP induction and monocyte infiltration. Note that while antioxidant genes were upregulated to a similar extent across all three mouse lines, NISP gene induction and monocyte infiltration was restricted to the (A) and (K/A) mouse, which exhibit significant liver damage. B. scRNA-Seq analysis of oesophagus-specific Keap1 CKO mice reveals significant immune cell infiltration into the oesophagi upon Nrf2 activation. The data were sourced from Hirose et al., 2023.
C. Five upregulated NISP chemokines function as ligands for monocyte receptors that are enriched in the livers of Pten::Keap1 DKO mice, which argues that a key function of the NISP is to attract monocytes to the damaged cells. CXCR2 is encoded by the Il8rb gene from Fig. 7. D. Summary of the NRF2-dependent immune infiltration model. In Pten::Keap1 DKO livers, activated Nrf2 promotes transcription of the NISP factor CCL2, which functions to recruit CCR2+ monocyte-derived macrophages into the damaged liver.
We compared NISP gene expression between Atg7 CKO mice, Keap1 CKO mice and Keap1::Atg7 DKO mice in order to fully elucidate the requirement for the diseased state in Nrf2-dependent NISP-induction (Fig. 6A). While induction of Nrf2's typical antioxidant target genes was comparable between all three genotypes, NISP gene induction, and a concomitant increase in monocyte infiltration, was only observed in the livers of diseased Atg7 CKO and Keap1::Atg7 DKO mice, which strongly supports our contention that NISP induction only occurs as part of a damage response, and functions to recruit monocytes directly to the site of tissue damage (Fig. 6A).
Thus, the Atg7 CKO and Keap1::Atg7 DKO mice recapitulate all of the central components of the Nrf2-NISP-monocyte infiltration response observed in the Pten::Keap1 DKO mice. While the Pten::Keap1 DKO, Atg7 CKO and Keap1::Atg7 DKO mice represent genetic model systems which would not normally be observed in healthy animals, the tissue damage which these models induce is representative of that which can be observed under physiological conditions. Taken together, these results strongly suggest that this NISP induction represents an important, and previously unrecognized component of the KEAP1-NRF2 stress response pathway.
To determine the broader relevance of Nrf2-mediated immune infiltration, we re-analyzed our recent scRNA-Seq dataset in which Keap1 was conditionally knocked out specifically in the mouse oesophagus (Hirose et al., 2022) [79]. Consistent with our new model, these data also showed the specific recruitment of immune cells, including both myeloid and T cells, into the oesophagi of the Keap1 knockout, and thus Nrf2-activated, tissue (Fig. 6B).
As the NISP includes a number of additional chemokines, we explored whether any other NISP factors may function in concert with CCL2 to recruit monocytes to the liver (Fig. 6C). This analysis revealed that monocyte-expressed receptors for the additional NISP factors Ccl7, Cxcl2, Cxcl5 and Cx3cl1 were all enriched in the Pten:Keap1 DKO livers. This enrichment of five monocyte-targeting chemokine-receptor pairs strongly supports our contention that the NISP functions to recruit monocytes into the damaged liver [51,52].
Furthermore, as Nrf2 also promotes monocyte and macrophage invasion in mouse disease models of steatohepatitis, colitis, pancreatitis and autoimmune nephritis, we would posit that it represents a central component of the Nrf2 response in damaged epithelial tissues, and that the NRF2-NISP-Immune recruitment model represents a framework through which these disease phenotypes can be understood (Table 1) (Fig. 6D).
Table 1.
Nrf2 promotes monocyte and macrophage tissue invasion in a broad range of disease states
Although the function of Nrf2 is most commonly described as anti-inflammatory, Nrf2 actively promotes inflammation and monocyte and macrophage invasion into damaged epithelial tissues across a range of mouse disease models, and in multiple tissue types.
| Disease Model | Experimental System | NRF2 phenotype | Reference |
|---|---|---|---|
| Steatohepatitis | Hepatocyte-specific NEMO CKO mouse | Keap1 DKO mice exhibit increased monocyte-derived | [53] |
| macrophages in the diseased livers | |||
| DSS-induced colitis | Adipocyte-specific Atg7 CKO mouse | Nrf2 activation by the non-canonical p62 pathway resulted | [54] |
| in increased monocyte invasion into the damage colons | |||
| Pancreatitis | Pancreatic acinar-specific VMP1 CKO mouse | Nrf2 DKO mice exhibit decreased inflammation and | [55] |
| macrophage infiltration into the damaged pancreases | |||
| Autoimmune nephritis | Fas mutant lpr/lpr mice | Nrf2 DKO mice exhibit decreased macrophage, CD4 | [56] |
| and CD8 T cell infiltration into the damaged kidneys | |||
| Lung cancer | scRNA-Seq analysis of human lung tumours | KEAP1 mutant tumours are enriched with proinflammatory | [57] |
| monocyte-derived macrophages (CP2E pattern) |
3.10. The NISP promotes the recruitment of classical monocytes to NRF2-dependent tumours
In addition to regulating a cytoprotective stress response pathway, NRF2 also plays an important role during tumorigenesis, where the hijacking of this protective function of NRF2 confers a selective survival advantage upon the tumour cells [20,[58], [59], [60]]. In this context, spheroid cultures of non-small cell lung carcinoma-derived KEAP1 mutant A549 cells have been shown to secrete large quantities of the chemokines CCL2 and CXCL5; both of which function as monocyte attractants [61]. As we have defined CCL2 and CXCL5 to be NISP factors, these data encouraged us to investigate NISP induction in NRF2-activated cancers.
In order to determine whether the NISP plays a role in Nrf2-dependent tumours, we utilized the isogenic WT and Keap1 KO Hepa1 cells in an allograft tumour model (Fig. 7A, Fig. S6A). Upon transplantation into nude mice, relative to the isogenic WT cells, Keap1 KO tumours displayed enhanced Cdkn2b and NISP expression, including Ccl20 and Cxcl5, which is consistent with the expression profile of the Pten::Keap1 DKO mice (Fig. S6B).
Fig. 7.
Activation of Nrf2 recruits proinflammatory classical monocytes into tumours
A. Overview of the isogenic WT and Keap1 KO Hepa1 cell allograft mouse tumour model. B, C. FACS analysis of the relative proportion of classical and non-classical monocytes that infiltrated the WT and Keap1 KO Hepa1 allograft tumours. Data represent mean ± SD, N = 7–8 tumours per cell type, ***p < 0.0001. D. Relative expression of monocyte/macrophage markers in Keap1 KO tumours relative to the isogenic WT tumours, as determined by RT-qPCR. Each experiment was performed at least 3 times. Data represent mean ± SD, N = 6 tumours per cell type, *p < 0.05, **p < 0.01, ***p < 0.0001. E, F. RT-qPCR analysis of markers of mMono1 and mMono2 cells, which correspond to “classical” and “non-classical” monocytes, based on specific and comprehensive gene expression patterns identified by Zilionis et al. using scRNA-Seq. These data show that Keap1 KO tumours are specifically enriched with mMono1 (“classical”) monocytes. Each experiment was performed at least 3 times. Data represent mean ± SD, N = 6 mice per genotype, **p < 0.001, ***p < 0.0001..
As our working model is that the NISP functions to recruit monocytes to remove damaged cells, we specifically focused on the infiltration of monocytes into the Keap1 KO tumours. Monocytes can be broadly divided into two types, “classical” which can enter tissues in response to chemokine signals, and “non-classical”, which mostly patrol the vasculature and support endothelial cell functions [62]. In perfect agreement with our model, FACS analysis revealed significantly enhanced classical monocyte infiltration into the Keap1 KO tumours (Fig. 7B, C, S7). Specifically, classical monocytes were enriched almost 4-fold in the Keap1 KO tumours relative to the isogenic WT equivalents, which strongly suggests that the Nrf2-NISP axis is a major determinant of classical monocyte infiltration. In accordance with this model, gene expression analysis confirmed a significant increase in the expression of classical monocyte markers, including Ccr2, which we had previously shown to be important for the liver damage response phenotype (Fig. 7D–F, Fig. 5).
3.11. The NISP promotes macrophage efferocytosis to remove damaged cells
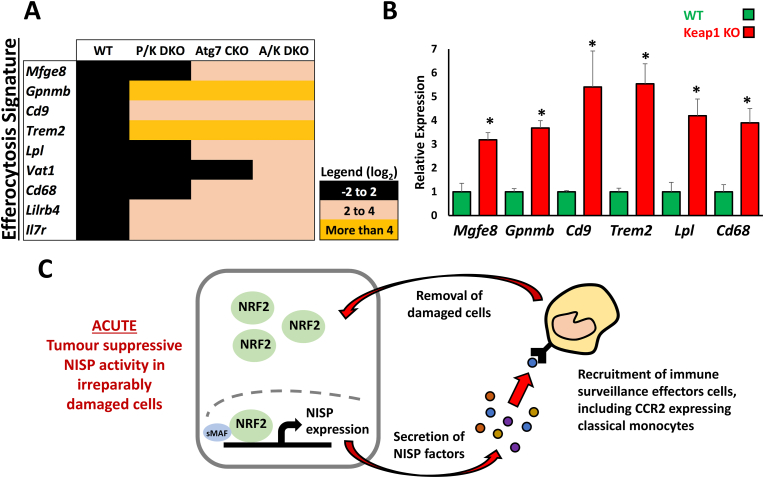
Damaged and apoptotic cells are removed from tissues by phagocytes through the process of efferocytosis [63]. This phagocytic removal of apoptotic cells is mediated by the secretion of CX3CL1 by dying cells, which functions as a potent “find-me” signal to the immune system [63]. As we identified CX3CL1 to be a NISP factor (Fig. 2), we hypothesized that the NISP may function to remove damaged cells through efferocytosis. To determine whether the monocytes and monocyte-derived macrophages that are recruited by the NISP actually function to remove damaged cells, we analyzed the expression of a recently described macrophage-specific efferocytosis gene signature [64]. Consistent with our model, in all three genetic models of Nrf2 activation in the context of liver damage, we found significant upregulation of efferocytosis gene expression (Fig. 8A). Furthermore, RT-qPCR revealed the same efferocytosis genes were also significantly upregulated in the Keap1 KO allograft tumours relative to WT tumours (Fig. 8B), which supports our model in which monocyte-derived macrophages are recruited by the NRF2-dependent NISP in order to remove damaged or oncogenic cells.
Fig. 8.
The NISP mediates the removal of damaged cells when the NRF2-dependent oxidative stress response is insufficient to restore homeostasis
A. Gene expression profiles of a macrophage-specific efferocytosis gene signature in liver-specific Pten:Keap1 double conditional knockout (P/K DKO), Atg7 conditional knockout (Atg7 CKO) and Keap1::Atg7 double conditional knockout mice (A/K DKO). Note that efferocytosis markers were specifically upregulated in the mouse models exhibiting genetic activation of Nrf2 in the context of cellular damage. B. RT-qPCR analysis of a macrophage-specific efferocytosis gene signature in allograft WT and Keap1 KO tumours. Each experiment was performed at least 3 times. Data represent mean ± SD, N = 6 mice per genotype, *p < 0.001. C. In irreparably damaged cells, the acute NRF2-dependent stress response is terminated by the induction of NISP gene expression. Their protein products function to recruit immune cells, which remove the damaged cells through the immune surveillance pathway. This mechanism maintains organismal homeostasis by limiting the accumulation of damaged cells with high NRF2 activity, as they may otherwise become deleterious to the host due to the pro-survival characteristics which NRF2 confers upon cells..
Taken together, these data indicate that in irreparably damaged cells, or during early stages of tumourigenesis, high levels of NRF2 promote NISP expression to enable the acute recruitment of CCR2+ classical monocytes, which in turn function as important components of the immune surveillance pathway (Fig. 8C). This pathway represents a previously unrecognized tumour suppressor component of the NRF2 response, and functions to eliminate irreparably damaged cells, which may otherwise become tumorigenic if maintained within the tissue in situ.
4. Discussion
It is currently unknown how the cytoprotective functions of NRF2 are leveraged when NRF2 is activated in irreparably damaged cells. In this study, we found that NRF2 functions to promote cellular senescence, such that cells with high levels of NRF2 activity can easily enter a senescent state. In this regard, we found that NRF2 regulates the transcription of a unique subset of senescence-associated secretory phenotype genes, which we have named the NRF2-induced secretory phenotype (NISP). Analysis of genetic activation of Nrf2 in both Pten::Keap1 and Keap1::Atg7 double knockout mouse liver models revealed that the physiological function of the NISP is to recruit immune effector cells to remove these damaged NRF2-activated cells through the immune surveillance pathway. Thus, the NRF2-NISP-Immune surveillance axis functions as a tumour suppressor pathway in order to mitigate the survival of pro-oncogenic NRF2-activated cells.
The KEAP1-NRF2 pathway is a stress response pathway which has been maintained by natural selection due to its ability to benefit the survival of the host organism. One important distinction between this pathway and other stress response pathways, such as p53, is that the chronic activation of NRF2 has not been associated directly with a mechanism to promote cell death if the survival of the cell becomes deleterious to the host. In the case of p53, a dichotomy of pathway outputs, represented by the pro-survival induction of cell cycle arrest coupled with activation of DNA repair pathways, or alternatively, the promotion of apoptosis, benefits organismal survival if the cellular damage cannot be repaired [65]. From the perspective of the host organism, this dichotomy of pathway outputs is advantageous, as chronic activation of a stress response pathway signifies that the cellular damage may be insurmountable, and therefore switching from a cytoprotective function to a cell death-inducing output is beneficial for host survival. In this study, we found that NRF2 activation can also lead to a dichotomy of pathway outputs, which together function to benefit the survival of the host under a broad range of conditions. In addition to its well characterized role in the adaptive response to oxidative stress, we found that, in the diseased state, NRF2 functions to promote cellular senescence by activating a unique subset of senescence-associated secretory phenotype (SASP) genes that we have named the NRF2-induced secretory phenotype (NISP). As summarized in Fig. 8C, the physiological function of the NISP is to recruit immune effector cells, including CCR2+ classical monocytes, to facilitate the removal of the irreparably damaged, chronically NRF2-activated cells, though the immune surveillance pathway. Through this mechanism, the KEAP1-NRF2 pathway functions in a tumor suppressor capacity in order to mitigate the survival of damaged, pre-cancerous cells, which may otherwise benefit from the cytoprotective properties conferred through NRF2 activation.
Some important features of genetically modified mouse models of Nrf2 activation suggest that, in addition to its effects facilitating tumor growth and therapeutic resistance, NRF2 also has a currently undescribed tumour suppressor function. For example, the expression of an inactivated mutant form of Keap1 found in human tumours does not give rise to tumours in mice [23]. Similarly, the expression of constitutive active Nrf2, again derived from a mutation found in human tumours, also does not result in cancer formation [24]. These findings show clearly that NRF2 is not a classical oncogene, but is only able to promote cancer formation in the context of additional mutations, such as p53, KRAS or PTEN [[58], [59], [60]]. Taken together, these findings are somewhat perplexing considering the proliferative, metabolic and stress resistant phenotypes that constitutive NRF2 activation confers upon cells [66]. Therefore, the existing data suggest that the functions of NRF2 facilitating cancer development are potentially counter-balanced by a parallel, but undescribed, tumour suppressive function.
In contrast, in malignant tumours the function of NRF2 is currently most commonly characterized as tumour promoting, as the cytoprotective and proliferative properties of NRF2 are hijacked by the tumour cells to generate aggressive, therapy-resistant tumours [20]. If the NRF2-NISP-immune surveillance pathway is functional in vivo, how can we reconcile this tumor suppressor function with the well-demonstrated oncogenic-enhancing activities of NRF2 in patients?
While activating mutations in the KEAP1-NRF2 pathway are associated with a poor prognosis for patients, importantly, this is only true in late stage cancer patients, as in early stage tumours of both the lung and oesophagus, activation of NRF2 is not associated with a worse prognosis [[67], [68], [69]]. These clinical data clearly show that during early stages of tumorigenesis, NRF2 activity is not tumour promoting. Of note, in early stage lung adenocarcinoma patients, KEAP1 mutant tumours are associated with increased infiltration of NK cells, which form an important component of the immune surveillance pathway and can also be recruited by the NISP factor CCL2 [70,71]. Taken together, these data support our contention that in early stage tumours, NRF2 activation functions as a tumour suppressor by promoting the immune surveillance-mediated removal of damaged cancer cells, and that this negative selective pressure must be overcome during later stages of tumourigenesis in order for NRF2-activated tumours to become malignant. Consistent with this model, the analysis of NRF2 activation during hepatocarcinogenesis in a rodent model revealed that, while activating mutations in Nrf2 are found in 90% of early-stage tumours, this mutation incidence is reduced to only 25% at later, malignant stages [72]. This time-course-dependent analysis means that 72% of the Nrf2-mutated early-stage tumours are resolved in situ by a mechanism which is currently unknown. We would posit that NRF2-induced immune surveillance may be responsible for the removal of these early-stage liver tumours.
Thus, in the context of NRF2's myriad of properties in tumours, we believe that activation of the NRF2-NISP-immune surveillance pathway explains many of the enigmatic tumorigenesis results from the literature, and therefore constitutes the hidden tumour suppressor function that NRF2 displays in vivo.
In addition to its role in tumorigenesis, an additional clinically important function of NRF2 relates to its regulation of the response to oxidative stress [1]. As oxidative stress plays a critical role in many aging-related maladies, including neurodegenerative and chronic kidney diseases, it has been hypothesized that antioxidant therapy, of which activation of NRF2 would represents a prototypic example, would benefit organismal aging and longevity [8,9]. Indeed, in support of this idea, correlative analysis in mice has suggested that activation of Nrf2 has a pro-longevity effect [10,11]. In this context, it is therefore surprising that the global constitutive activation of Nrf2 signaling does not extend longevity in mice, and instead, is associated with reduced overall lifespan [12,13]. These findings are enigmatic given the well-established cytoprotective properties of NRF2. In the context of these results, our contention that NRF2 promotes cellular senescence provides a simple framework through which these anomalous longevity data can be understood. When viewed through a senescence lens, constitutively active NRF2 functions as an antagonistic pleiotropy factor, which promotes survival early in life through its cytoprotective activities, but as this is coupled to a senescence-inducing function, is deleterious during the post-reproductive stages of life. This simple framework also explains why, under physiological conditions, the cytoprotective functions of NRF2 are tightly regulated by KEAP1, as the antagonistic pleiotropy characteristics are only evident in constitutive active mouse models. Thus, KEAP1 functions to limit the deleterious impact of constitutive NRF2 activation. In support of this model, a recent study showed that in glioblastoma, NRF2 activation promotes a cellular senescence phenotype in human patients [73].
In contrast to the senescence and NISP phenotypes observed in Pten::Keap1 DKO mouse livers, Nrf2 has previously been shown to promote cellular proliferation in other liver experimental systems, including partial hepatectomy and portal vein branch ligation (PVBL) models [[74], [75], [76]]. One significant difference between these models and the Pten::Keap1 DKO model is the role of Nrf2 activation in the generation of liver damage, as in Pten::Keap1 DKO mice, Nrf2 activation results in significantly increased liver damage relative to the control mice [41]. This requirement for tissue damage to elicit the senescence and NISP response suggests that this phenotype is elicited specifically in the context of significant cellular damage, and that in the absence of such damage, Nrf2 activation can promote other distinct cellular responses, including cell proliferation.
Consistent with this idea, other transcription factors which play an important role in the regulation of senescence can also promote a diverse range of context-dependent cellular outcomes. For example, C/EBPβ promotes cellular proliferation in response to partial hepatectomy, despite the fact that in other cellular contexts, C/EBPβ is required for the establishment and maintenance of the senescence state [31,77]. Thus, due to the unique Nrf2-dependent tissue damage phenotype observed in the Pten::Keap1 DKO mice, there was no change in gene expression of Nrf2 or C/EBPβ target genes that have previously been shown to be upregulated in partial hepatectomy or portal vein branch ligation studies (Fig. S8). One limitation of this study is the reliance on gene expression data as the major source of evidence for monocyte invasion into damaged tissues in the Pten and Atg7-based mouse models. In the future, additional studies will be carried out to further characterize the immune cell recruitment and infiltration phenotypes caused by Nrf2 activation in damaged tissues.
In summary, we have found that NRF2 functions to prime cells to become senescent in response to irreparable damage. In the senescent state, NRF2 drives expression of a unique secretory phenotype (NISP), which functions in a tumour suppressor capacity by recruiting immune effector cells in order to remove the damaged, senescent cells through the immune surveillance pathway. This pathway represents the final stage of the oxidative stress response, as it allows cells to be safely removed if the macromolecular damage caused by the original stressor is so extensive that it is beyond the repair capacity of the cell.
Author contributions
L.B. conceived the project and designed all of the experiments. L.B., K.T., A.Z., Y.T and T.S. carried out the experiments. M.Y. supervised the project. L.B., M.Y. and T.W.K. analyzed the results and wrote the manuscript.
Materials and methods
Mitomycin C washout and recovery
For the co-cultured isogenic WT and Keap1 KO cells, on “day −1” 2000 cells from each cell type were seeded into individual wells of a black flat-bottom 96-well plate (Corning #3904). On the following day (day 0), 50 nM mitomycin C, or 0.1% DMSO was added to each well. Immediately after the addition of the compounds, the fluorescence intensities of GFP and mCherry were measured using a PHERAstar FS microplate reader (BMG Labtech, Ortenberg, Germany). The plates were then returned to the 37 °C incubator until day 3, when the mitomycin C media was replaced with fresh media in the “washout” samples. The fluorescence intensity was measured again on days 5, 6 and 7 to determine cell growth relative to the initial fluorophore intensities measured on day 0. For the human cancer cell lines (H460, A549 and H2023), on “day −1” 2000 cells from each cell type were seeded into individual wells of a clear flat-bottom 96-well plate. On the following day (day 0), 50 nM mitomycin C, or 0.1% DMSO was added to each well. The 96-well plates were then incubated at 37 °C for 48 h, when the mitomycin C media was replaced with fresh media in the “washout” samples. The plates were then returned to the incubator and allowed to recover for a further 72 h. After this, they were washed with PBS, lysed with 25 μl RIPA buffer (50 mM Tris, 150 mM NaCl, 1% (v/v) NP-40, 0.5% (w/v) deoxycholic acid, 0.1% (w/v) SDS, pH7.4), and then frozen at −30 °C. Total protein concentrations for each well were determined using the BCA protein assay (Pierce) following the manufacturer's instructions, and were normalized to the DMSO controls.
Mice
Ptenflox/flox (WT), Ptenflox/flox:Albumin-Cre (Pten CKO), Keap1flox/flox:Albumin-Cre (Keap1 CKO), Ptenflox/flox: Keap1flox/flox: Albumin-Cre (PtenKeap1 DKO) and Nrf2−/− (Nrf2 KO) mice were all previously described [41]. For the Atg7 mouse experiments, Atg7flox/flox (control), Atg7flox/flox:Albumin-Cre (Atg7 CKO), Keap1flox/flox:Albumin- Cre (Keap1 CKO) and Atg7flox/flox:Keap1flox/flox:Albumin-Cre (Keap1Atg7 DKO) mice were generated and maintained as described previously [49]. The mice were provided with water and rodent chow ad libitum. All mice were maintained under specific-pathogen-free conditions and treated according to the regulations of The Standards for Human Care and Use of Laboratory Animals of Tohoku University and Guidelines for Proper Conduct of Animal Experiments of the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Allograft tumor implantation
Suspensions of WT or Keap1 KO Hepa1 cells (2 x106 cells in 100 μl PBS) were injected subcutaneously into the left or right trunk of 5-week-old Balb/c-nu/nu female mice. The allografts were allowed to grow for 5 weeks, after which the tumours were removed and frozen in liquid nitrogen. All animals were housed in specific pathogen-free conditions, and monitored according to the regulations of The Standards for Human Care and Use of Laboratory Animals of Tohoku University and the Guidelines for Proper Conduct of Animal Experiments by the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Flow cytometry and cell sorting
Tumours were harvested from nude mice, which were transplanted with wild-type Hepa1 cells or Keap1 knockout Hepa1 cells. The tumour cells were isolated using Tumor Dissociation Kit, mouse (Miltenyi Biotec), following the protocol provided by the manufacturer. Red blood cells were removed using RBC Lysis buffer (0.015 M NH4Cl, 1 mM KHCO3 and 10 μM EDTA-2Na). Debris was removed using Debris removal solution (Miltenyi Biotec). Propidium iodide (1 μg/mL) was used to remove dead cells. Analyses and cell sorting were performed using FACSVerse (BD Biosciences). Data were analyzed using FlowJo software (BD Biosciences). The following antibodies were used for the flow cytometric analyses: Brilliant Violet 421™ anti-mouse/human CD11b Antibody (Biolegend, Cat: 101236), FITC anti-mouse Ly-6C (BD Pharmingen™, Cat: 553104), and Anti-Mo CD62L (L-Selectin) APC (Invitrogen, REF: 17-0621-82).
Cell culture
All cells were maintained in high glucose Dulbecco's modified Eagle's medium (DMEM), except for the JHH2 and JHH5 cells, which were maintained William's E media, supplemented with 10% fetal bovine serum (FBS), and antibiotics. All cells were cultured in a humidified atmosphere with 5% CO2 at 37 °C.
Gene expression analysis and ChIP-qPCR
For RT-qPCR, total RNA was prepared from cell lysates using TRIzol reagent (Life Technologies, Carlsbad, CA) in accordance with the manufacturer's instructions. A 1 μg aliquot of total RNA was reverse transcribed with ReverTra Ace (Toyobo, Osaka, Japan). The resultant cDNA was used as a template for quantitative reverse transcription-PCR (qRT-PCR) on a SYBR green 7300 real time PCR analyzer (Life Technologies). Chromatin precipitation for ChIP-qPCR was carried out as previously described [78]. The ChIP-qPCR primers were designed based on the NRF2 binding peaks in the genes of interest in the UCSC genome browser. The primers used during the qPCR analysis are available upon request. Microarray analysis of PtenKeap1 DKO mice was carried out using the previously published GSE50575 dataset [41].
Microarray analysis
Total RNA from the livers of Atg7flox/flox (control), Atg7flox/flox:Albumin-Cre (Atg7 CKO), Keap1flox/flox:Albumin- Cre (Keap1 CKO) and Atg7flox/flox:Keap1flox/flox: Albumin-Cre (Keap1Atg7 DKO) mice were labelled with Cy3. The samples were hybridized to whole-mouse-genome Oligo DNA Microarray kit ver2.0 (Agilent Technologies, Inc., Santa Clara, CA) according to the manufacturer's protocol. Arrays were scanned using a G2539A microarray scanner system (Agilent), and the resulting data were analyzed using GeneSpring GX software (Agilent). The microarray data produced in this study have been submitted to the Gene Expression Omnibus (GEO) database, and assigned the GEO accession number (under application).
Histological analysis
The livers were fixed in Mildform 10 N (Wako Pure Chemical Corporation, Osaka, Japan) and embedded in paraffin for staining with hematoxylin and eosin.
ELISA
The ELISA for CCL2 was carried out using the Mouse MCP1 ELISA Kit (ab208979, Abcam) following the protocol provided by the manufacturer. The samples were generated from conditioned media taken from WT and Keap1 KO Hepa1 cells, which were treated with either DMSO or 200 nM MMC for 5 days.
Senescence associated-β-gal staining
SA-β-gal staining was carried out using the Senescence Detection Kit (ab65351, Abcam) following the protocol provided by the manufacturer. The samples used in this assay were generated using MMC washout conditions, as described above.
Reagents
Mitomycin C and doxorubicin were purchased from Sigma-Aldrich (Missouri, USA). ABT-263 was purchased from Cayman Chemical (Ann Arbor, Michigan, USA). Celastrol was purchased from Selleck Chemicals (Texas, USA).
Statistical analysis
The data are presented as mean ± SD. Student's T-test (two-tailed) was used to determine the statistical significance of the results.
Declaration of competing interest
None.
Acknowledgements
We would like to thank all of the members of the Yamamoto lab for their thoughtful and stimulating discussions. This research was partially supported by JSPS KAKENHI Grants-in-Aid for Early Career Scientists 19K16512 (to L.B.), the Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)) from AMED under Grant Number JP17am0101001 (support number 1234), AMED-P-CREATE (JP19cm0106101 to M.Y.) and JSPS KAKENHI 19H05649 (to M.Y.).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102845.
Contributor Information
Liam Baird, Email: liambaird@med.tohoku.ac.jp.
Masayuki Yamamoto, Email: masiyamamoto@med.tohoku.ac.jp.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Itoh K., Chiba T., Takahashi S., Ishii T., Igarashi K., Katoh Y., Oyake T., Hayashi N., Satoh K., Hatayama I., et al. An nrf2/small maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 2.Yamamoto M., Kensler T.W., Motohashi H. The KEAP1-NRF2 system: a thiol-based sensor-effector apparatus for maintaining redox homeostasis. Physiol. Rev. 2018;98:1169–1203. doi: 10.1152/physrev.00023.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baird L., Yamamoto M. The molecular mechanisms regulating the KEAP1-NRF2 pathway. Mol. Cell Biol. 2020;40 doi: 10.1128/MCB.00099-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Itoh K., Wakabayashi N., Katoh Y., Ishii T., Igarashi K., Engel J.D., Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kobayashi A., Kang M.-I., Okawa H., Ohtsuji M., Zenke Y., Chiba T., Igarashi K., Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baird L., Llères D., Swift S., Dinkova-Kostova A.T. Regulatory flexibility in the Nrf2-mediated stress response is conferred by conformational cycling of the Keap1-Nrf2 protein complex. Proc. Natl. Acad. Sci. U.S.A. 2013;110(38):15259–15264. doi: 10.1073/pnas.1305687110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cuadrado A., Rojo A.I., Wells G., Hayes J.D., Cousin S.P., Rumsey W.L., Attucks O.C., Franklin S., Levonen A.-L., Kensler T.W., et al. Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat. Rev. Drug Discov. 2019;18:295–317. doi: 10.1038/s41573-018-0008-x. [DOI] [PubMed] [Google Scholar]
- 8.Finkel T., Holbrook N.J. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 9.Liguori I., Russo G., Curcio F., Bulli G., Aran L., Della-Morte D., et al. Oxidative stress, aging, and diseases. Clin. Interv. Aging. 2018;13:757–772. doi: 10.2147/CIA.S158513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis K.N., Wason E., Edrey Y.H., Kristan D.M., Nevo E., Buffenstein R. Regulation of Nrf2 signaling and longevity in naturally long-lived rodents. Proc. Natl. Acad. Sci. U.S.A. 2015;112:3722–3727. doi: 10.1073/pnas.1417566112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tyshkovskiy A., Bozaykut P., Borodinova A.A., Gerashchenko M.V., Ables G.P., Garratt M., Khaitovich P., Clish C.B., Miller R.A., Gladyshev V.N. Identification and application of gene expression signatures associated with lifespan extension. Cell Metabol. 2019;30:573–593.e8. doi: 10.1016/j.cmet.2019.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taguchi K., Maher J.M., Suzuki T., Kawatani Y., Motohashi H., Yamamoto M. Genetic analysis of cytoprotective functions supported by graded expression of Keap1. Mol. Cell Biol. 2010;30:3016–3026. doi: 10.1128/MCB.01591-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki T., Seki S., Hiramoto K., Naganuma E., Kobayashi E.H., Yamaoka A., Baird L., Takahashi N., Sato H., Yamamoto M. Hyperactivation of Nrf2 in early tubular development induces nephrogenic diabetes insipidus. Nat. Commun. 2017;8 doi: 10.1038/ncomms14577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoh K., Itoh K., Enomoto A., Hirayama A., Yamaguchi N., Kobayashi M., Morito N., Koyama A., Yamamoto M., Takahashi S. Nrf2-deficient female mice develop lupus-like autoimmune nephritis. Kidney Int. 2001;60:1343–1353. doi: 10.1046/j.1523-1755.2001.00939.x. [DOI] [PubMed] [Google Scholar]
- 15.Rodríguez J.A., Marigorta U.M., Hughes D.A., Spataro N., Bosch E., Navarro A. Antagonistic pleiotropy and mutation accumulation influence human senescence and disease. Nat Ecol Evol. 2017;1 doi: 10.1038/s41559-016-0055. [DOI] [PubMed] [Google Scholar]
- 16.Childs B.G., Gluscevic M., Baker D.J., Laberge R.-M., Marquess D., Dananberg J., van Deursen J.M. Senescent cells: an emerging target for diseases of ageing. Nat. Rev. Drug Discov. 2017;16:718–735. doi: 10.1038/nrd.2017.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Micco R., Krizhanovsky V., Baker D., d'Adda di Fagagna F. Cellular senescence in ageing: from mechanisms to therapeutic opportunities. Nat. Rev. Mol. Cell Biol. 2021;22:75–95. doi: 10.1038/s41580-020-00314-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang T.-W., Yevsa T., Woller N., Hoenicke L., Wuestefeld T., Dauch D., Hohmeyer A., Gereke M., Rudalska R., Potapova A., et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature. 2011;479:547–551. doi: 10.1038/nature10599. [DOI] [PubMed] [Google Scholar]
- 19.Coppé J.-P., Desprez P.-Y., Krtolica A., Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu. Rev. Pathol. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baird L., Kensler T.W., Yamamoto M. Novel NRF2-activated cancer treatments utilizing synthetic lethality. IUBMB Life. 2022;74:1209–1231. doi: 10.1002/iub.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shibata T., Ohta T., Tong K.I., Kokubu A., Odogawa R., Tsuta K., Asamura H., Yamamoto M., Hirohashi S. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc. Natl. Acad. Sci. USA. 2008;105:13568–13573. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Solis L.M., Behrens C., Dong W., Suraokar M., Ozburn N.C., Moran C.A., Corvalan A.H., Biswal S., Swisher S.G., Bekele B.N., et al. Nrf2 and Keap1 abnormalities in non–small cell lung carcinoma and association with clinicopathologic features. Clin. Cancer Res. 2010;16:3743–3753. doi: 10.1158/1078-0432.CCR-09-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang Y.P., Torrente L., Falzone A., Elkins C.M., Liu M., Asara J.M., Dibble C.C., DeNicola G.M. Cysteine dioxygenase 1 is a metabolic liability for non-small cell lung cancer. Elife. 2019;8 doi: 10.7554/eLife.45572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bowman B.M., Montgomery S.A., Schrank T.P., Simon J.M., Ptacek T.S., Tamir T.Y., Mulvaney K.M., Weir S.J., Nguyen T.T., Murphy R.M., et al. A conditional mouse expressing an activating mutation in NRF2 displays hyperplasia of the upper gastrointestinal tract and decreased white adipose tissue. J. Pathol. 2020;252:125–137. doi: 10.1002/path.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baird L., Suzuki T., Takahashi Y., Hishinuma E., Saigusa D., Yamamoto M. Geldanamycin-derived HSP90 inhibitors are synthetic lethal with NRF2. Mol. Cell Biol. 2020;40 doi: 10.1128/MCB.00377-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baird L., Yamamoto M. NRF2-Dependent bioactivation of mitomycin C as a novel strategy to target KEAP1-NRF2 pathway activation in human cancer. Mol. Cell Biol. 2021;41 doi: 10.1128/MCB.00473-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh A., Misra V., Thimmulappa R.K., Lee H., Ames S., Hoque M.O., Herman J.G., Baylin S.B., Sidransky D., Gabrielson E., et al. Dysfunctional KEAP1–NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3 doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X.-J., Sun Z., Villeneuve N.F., Zhang S., Zhao F., Li Y., Chen W., Yi X., Zheng W., Wondrak G.T., et al. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis. 2008;29:1235–1243. doi: 10.1093/carcin/bgn095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martínez-Zamudio R.I., Roux P.-F., de Freitas J.A.N.L.F., Robinson L., Doré G., Sun B., Belenki D., Milanovic M., Herbig U., Schmitt C.A., et al. AP-1 imprints a reversible transcriptional programme of senescent cells. Nat. Cell Biol. 2020;22:842–855. doi: 10.1038/s41556-020-0529-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coppé J.-P., Patil C.K., Rodier F., Sun Y., Muñoz D.P., Goldstein J., Nelson P.S., Desprez P.-Y., Campisi J. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6 doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuilman T., Michaloglou C., Vredeveld L.C.W., Douma S., van Doorn R., Desmet C.J., Aarden L.A., Mooi W.J., Peeper D.S. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133:1019–1031. doi: 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- 32.Gomis R.R., Alarcón C., Nadal C., Van Poznak C., Massagué J. C/EBPβ at the core of the TGFβ cytostatic response and its evasion in metastatic breast cancer cells. Cancer Cell. 2006;10:203–214. doi: 10.1016/j.ccr.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 33.Okazaki K., Anzawa H., Liu Z., Ota N., Kitamura H., Onodera Y., Alam M.M., Matsumaru D., Suzuki T., Katsuoka F., Tadaka S., Motoike I., Watanabe M., Hayasaka K., Sakurada A., Okada Y., Yamamoto M., Suzuki T., Kinoshita K., Sekine H., Motohashi H. Enhancer remodeling promotes tumor-initiating activity in NRF2-activated non-small cell lung cancers. Nat. Commun. 2020;11:5911. doi: 10.1038/s41467-020-19593-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Acosta J.C., Banito A., Wuestefeld T., Georgilis A., Janich P., Morton J.P., Athineos D., Kang T.-W., Lasitschka F., Andrulis M., et al. A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nat. Cell Biol. 2013;15:978–990. doi: 10.1038/ncb2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cho K.A., Ryu S.J., Oh Y.S., Park J.H., Lee J.W., Kim H.-P., Kim K.T., Jang I.S., Park S.C. Morphological adjustment of senescent cells by modulating caveolin-1 status. J. Biol. Chem. 2004;279:42270–42278. doi: 10.1074/jbc.M402352200. [DOI] [PubMed] [Google Scholar]
- 36.Chang J., Wang Y., Shao L., Laberge R.-M., Demaria M., Campisi J., Janakiraman K., Sharpless N.E., Ding S., Feng W., et al. Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat. Med. 2016;22:78–83. doi: 10.1038/nm.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X., Zhang R., Chen H., Wang L., Ren C., Pataer A., Wu S., Meng Q.H., Ha M.J., Morris J., Xi Y., Wang J., Zhang J., Gibbons D.L., Heymach J.V., Meric-Bernstam F., Minna J., Swisher S.G., Roth J.A., Fang B. KRT-232 and navitoclax enhance trametinib's anti-Cancer activity in non-small cell lung cancer patient-derived xenografts with KRAS mutations. Am. J. Cancer Res. 2020;10:4464–4475. [PMC free article] [PubMed] [Google Scholar]
- 38.Krall E.B., Wang B., Munoz D.M., Ilic N., Raghavan S., Niederst M.J., Yu K., Ruddy D.A., Aguirre A.J., Kim J.W., Redig A.J., Gainor J.F., Williams J.A., Asara J.M., Doench J.G., Janne P.A., Shaw A.T., McDonald R.E., Iii, Engelman J.A., Stegmeier F., Schlabach M.R., Hahn W.C. KEAP1 loss modulates sensitivity to kinase targeted therapy in lung cancer. Elife. 2017;6 doi: 10.7554/eLife.18970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xue W., Zender L., Miething C., Dickins R.A., Hernando E., Krizhanovsky V., Cordon-Cardo C., Lowe S.W. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eggert T., Wolter K., Ji J., Ma C., Yevsa T., Klotz S., Medina-Echeverz J., Longerich T., Forgues M., Reisinger F., et al. Distinct functions of senescence-associated immune responses in liver tumor surveillance and tumor progression. Cancer Cell. 2016;30:533–547. doi: 10.1016/j.ccell.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taguchi K., Hirano I., Itoh T., Tanaka M., Miyajima A., Suzuki A., Motohashi H., Yamamoto M. Nrf2 enhances cholangiocyte expansion in pten-deficient livers. Mol. Cell Biol. 2014;34:900–913. doi: 10.1128/MCB.01384-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin L., Spoor M.S., Gerth A.J., Brody S.L., Peng S.L. Modulation of Th1 activation and inflammation by the NF-κB repressor Foxj1. Science. 2004;303:1017–1020. doi: 10.1126/science.1093889. [DOI] [PubMed] [Google Scholar]
- 43.Pazolli E., Luo X., Brehm S., Carbery K., Chung J.-J., Prior J.L., Doherty J., Demehri S., Salavaggione L., Piwnica-Worms D., et al. Senescent stromal-derived osteopontin promotes preneoplastic cell growth. Cancer Res. 2009;69:1230–1239. doi: 10.1158/0008-5472.CAN-08-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hiebert P., Wietecha M.S., Cangkrama M., Haertel E., Mavrogonatou E., Stumpe M., Steenbock H., Grossi S., Beer H.-D., Angel P., et al. Nrf2-Mediated fibroblast reprogramming drives cellular senescence by targeting the matrisome. Dev. Cell. 2018;46:145–161.e10. doi: 10.1016/j.devcel.2018.06.012. [DOI] [PubMed] [Google Scholar]
- 45.McMahon M., Itoh K., Yamamoto M., Chanas S.A., Henderson C.J., McLellan L.I., Wolf C.R., Cavin C., Hayes J.D. The Cap'n'Collar basic leucine zipper transcription factor Nrf2 (NF-E2 p45-related factor 2) controls both constitutive and inducible expression of intestinal detoxification and glutathione biosynthetic enzymes. Cancer Res. 2001;61:3299–3307. [PubMed] [Google Scholar]
- 46.Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A., et al. Tissue-based map of the human proteome. Science. 2015;347 doi: 10.1126/science.1260419. 1260419–1260419. [DOI] [PubMed] [Google Scholar]
- 47.Guillot A., Tacke F. Liver macrophages: old dogmas and new insights. Hepatol Commun. 2019;3:730–743. doi: 10.1002/hep4.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zilionis R., Engblom C., Pfirschke C., Savova V., Zemmour D., Saatcioglu H.D., Krishnan I., Maroni G., Meyerovitz C.V., Kerwin C.M., et al. Single-cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity. 2019;50:1317–1334.e10. doi: 10.1016/j.immuni.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taguchi K., Fujikawa N., Komatsu M., Ishii T., Unno M., Akaike T., Motohashi H., Yamamoto M. Keap1 degradation by autophagy for the maintenance of redox homeostasis. Proc. Natl. Acad. Sci. USA. 2012;109:13561–13566. doi: 10.1073/pnas.1121572109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Komatsu M., Kurokawa H., Waguri S., Taguchi K., Kobayashi A., Ichimura Y., Sou Y.-S., Ueno I., Sakamoto A., Tong K.I., et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 51.Vilgelm A.E., Richmond A. Chemokines modulate immune surveillance in tumorigenesis, metastasis, and response to immunotherapy. Front. Immunol. 2019;10:333. doi: 10.3389/fimmu.2019.00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Metzemaekers M., Mortier A., Vacchini A., Boff D., Yu K., Janssens R., Farina F.M., Milanesi S., Berghmans N., Pörtner N., et al. Endogenous modification of the chemoattractant CXCL5 alters receptor usage and enhances its activity toward neutrophils and monocytes. Sci. Signal. 2021;14 doi: 10.1126/scisignal.aax3053. [DOI] [PubMed] [Google Scholar]
- 53.Mohs A., Otto T., Schneider K.M., Peltzer M., Boekschoten M., Holland C.H., Hudert C.A., Kalveram L., Wiegand S., Saez-Rodriguez J., Longerich T., Hengstler J.G., Trautwein C. Hepatocyte-specific NRF2 activation controls fibrogenesis and carcinogenesis in steatohepatitis. J. Hepatol. 2021;74:638–648. doi: 10.1016/j.jhep.2020.09.037. [DOI] [PubMed] [Google Scholar]
- 54.Richter F.C., Friedrich M., Kampschulte N., Piletic K., Alsaleh G., Zummach R., Hecker J., Pohin M., Ilott N., Guschina I., Wideman S.K., Johnson E., Borsa M., Hahn P., Morriseau C., Hammock B.D., Schipper H.S., Edwards C.M., Zechner R., Siegmund B., Weidinger C., Schebb N.H., Powrie F., Simon A.K. Adipocyte autophagy limits gut inflammation by controlling oxylipin and IL-10. EMBO J. 2023 doi: 10.15252/embj.2022112202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang S., Chao X., Jiang X., Wang T., Rodriguez Y., Yang L., Pacher P., Ni H.-M., Ding W.-X. Loss of acinar cell VMP1 triggers spontaneous pancreatitis in mice. Autophagy. 2022;18:1572–1582. doi: 10.1080/15548627.2021.1990672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morito N., Yoh K., Hirayama A., Itoh K., Nose M., Koyama A., Yamamoto M., Takahashi S. Nrf2 deficiency improves autoimmune nephritis caused by the fas mutation lpr. Kidney Int. 2004;65:1703–1713. doi: 10.1111/j.1523-1755.2004.00565.x. [DOI] [PubMed] [Google Scholar]
- 57.Bischoff P., Trinks A., Obermayer B., Pett J.P., Wiederspahn J., Uhlitz F., Liang X., Lehmann A., Jurmeister P., Elsner A., Dziodzio T., Rückert J.-C., Neudecker J., Falk C., Beule D., Sers C., Morkel M., Horst D., Blüthgen N., Klauschen F. Single-cell RNA sequencing reveals distinct tumor microenvironmental patterns in lung adenocarcinoma. Oncogene. 2021;40:6748–6758. doi: 10.1038/s41388-021-02054-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Romero R., Sayin V.I., Davidson S.M., Bauer M.R., Singh S.X., LeBoeuf S.E., Karakousi T.R., Ellis D.C., Bhutkar A., Sánchez-Rivera F.J., et al. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat. Med. 2017;23:1362–1368. doi: 10.1038/nm.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jeong Y., Hoang N.T., Lovejoy A., Stehr H., Newman A.M., Gentles A.J., Kong W., Truong D., Martin S., Chaudhuri A., et al. Role of KEAP1/NRF2 and TP53 mutations in lung squamous cell carcinoma development and radiation resistance. Cancer Discov. 2017;7:86–101. doi: 10.1158/2159-8290.CD-16-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Best S.A., De Souza D.P., Kersbergen A., Policheni A.N., Dayalan S., Tull D., Rathi V., Gray D.H., Ritchie M.E., McConville M.J., et al. Synergy between the KEAP1/NRF2 and PI3K pathways drives non-small-cell lung cancer with an altered immune microenvironment. Cell Metabol. 2018;27:935–943.e4. doi: 10.1016/j.cmet.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 61.Tazzyman S., Barry S.T., Ashton S., Wood P., Blakey D., Lewis C.E., Murdoch C. Inhibition of neutrophil infiltration into A549 lung tumors in vitro and in vivo using a CXCR2-specific antagonist is associated with reduced tumor growth. Int. J. Cancer. 2011;129:847–858. doi: 10.1002/ijc.25987. [DOI] [PubMed] [Google Scholar]
- 62.Jakubzick C.V., Randolph G.J., Henson P.M. Monocyte differentiation and antigen-presenting functions. Nat. Rev. Immunol. 2017;17:349–362. doi: 10.1038/nri.2017.28. [DOI] [PubMed] [Google Scholar]
- 63.Doran A.C., Yurdagul A., Tabas I. Efferocytosis in health and disease. Nat. Rev. Immunol. 2020;20:254–267. doi: 10.1038/s41577-019-0240-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Park M.D., Reyes-Torres I., LeBerichel J., Hamon P., LaMarche N.M., Hegde S., Belabed M., Troncoso L., Grout J.A., Magen A., Humblin E., Nair A., Molgora M., Hou J., Newman J.H., Farkas A.M., Leader A.M., Dawson T., D'Souza D., Hamel S., Sanchez-Paulete A.R., Maier B., Bhardwaj N., Martin J.C., Kamphorst A.O., Kenigsberg E., Casanova-Acebes M., Horowitz A., Brown B.D., De Andrade L.F., Colonna M., Marron T.U., Merad M. TREM2 macrophages drive NK cell paucity and dysfunction in lung cancer. Nat. Immunol. 2023;24:792–801. doi: 10.1038/s41590-023-01475-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bieging K.T., Mello S.S., Attardi L.D. Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Cancer. 2014;14:359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kansanen E., Kuosmanen S.M., Leinonen H., Levonen A.-L. The Keap1-Nrf2 pathway: mechanisms of activation and dysregulation in cancer. Redox Biol. 2013;1:45–49. doi: 10.1016/j.redox.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim J.-H., Bogner P.N., Ramnath N., Park Y., Yu J., Park Y.-M. Elevated peroxiredoxin 1, but not NF-E2-related factor 2, is an independent prognostic factor for disease recurrence and reduced survival in stage I non-small cell lung cancer. Clin. Cancer Res. 2007;13:3875–3882. doi: 10.1158/1078-0432.CCR-06-2893. [DOI] [PubMed] [Google Scholar]
- 68.Tong Y.-H., Zhang B., Yan Y.-Y., Fan Y., Yu J.-W., Kong S.-S., Zhang D., Fang L., Su D., Lin N.-M. Dual-negative expression of Nrf2 and NQO1 predicts superior outcomes in patients with non-small cell lung cancer. Oncotarget. 2017;8:45750–45758. doi: 10.18632/oncotarget.17403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li M., Zhang Z., Wang Q., Yi Y., Li B. Integrated cohort of esophageal squamous cell cancer reveals genomic features underlying clinical characteristics. Nat. Commun. 2022;13:5268. doi: 10.1038/s41467-022-32962-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kadara H., Choi M., Zhang J., Parra E.R., Rodriguez-Canales J., Gaffney S.G., Zhao Z., Behrens C., Fujimoto J., Chow C., Yoo Y., Kalhor N., Moran C., Rimm D., Swisher S., Gibbons D.L., Heymach J., Kaftan E., Townsend J.P., Lynch T.J., Schlessinger J., Lee J., Lifton R.P., Wistuba, Herbst R.S. Whole-exome sequencing and immune profiling of early-stage lung adenocarcinoma with fully annotated clinical follow-up. Ann. Oncol. 2017;28:75–82. doi: 10.1093/annonc/mdw436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morrison B.E., Park S.J., Mooney J.M., Mehrad B. Chemokine-mediated recruitment of NK cells is a critical host defense mechanism in invasive aspergillosis. J. Clin. Invest. 2003;112:1862–1870. doi: 10.1172/JCI18125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Orrù C., Perra A., Kowalik M.A., Rizzolio S., Puliga E., Cabras L., Giordano S., Columbano A. Distinct mechanisms are responsible for nrf2-keap1 pathway activation at different stages of rat hepatocarcinogenesis. Cancers. 2020;12:2305. doi: 10.3390/cancers12082305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Salam R., Saliou A., Bielle F., Bertrand M., Antoniewski C., Carpentier C., Alentorn A., Capelle L., Sanson M., Huillard E., Bellenger L., Guégan J., Le Roux I. Cellular senescence in malignant cells promotes tumor progression in mouse and patient Glioblastoma. Nat. Commun. 2023;14:441. doi: 10.1038/s41467-023-36124-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Beyer T.A., Xu W., Teupser D., auf dem Keller U., Bugnon P., Hildt E., Thiery J., Kan Y.W., Werner S. Impaired liver regeneration in Nrf2 knockout mice: role of ROS-mediated insulin/IGF-1 resistance. EMBO J. 2008;27:212–223. doi: 10.1038/sj.emboj.7601950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shirasaki K., Taguchi K., Unno M., Motohashi H., Yamamoto M. NF-E2-related factor 2 promotes compensatory liver hypertrophy after portal vein branch ligation in mice. Hepatology. 2014;59:2371–2382. doi: 10.1002/hep.27020. [DOI] [PubMed] [Google Scholar]
- 76.Chan B.K.Y., Elmasry M., Forootan S.S., Russomanno G., Bunday T.M., Zhang F., Brillant N., Starkey Lewis P.J., Aird R., Ricci E., Andrews T.D., Sison-Young R.L., Schofield A.L., Fang Y., Lister A., Sharkey J.W., Poptani H., Kitteringham N.R., Forbes S.J., Malik H.Z., Fenwick S.W., Park B.K., Goldring C.E., Copple I.M. Pharmacological activation of Nrf2 enhances functional liver regeneration. Hepatology. 2021;74:973–986. doi: 10.1002/hep.31859. [DOI] [PubMed] [Google Scholar]
- 77.Greenbaum L.E., Li W., Cressman D.E., Peng Y., Ciliberto G., Poli V., Taub R. CCAAT enhancer- binding protein beta is required for normal hepatocyte proliferation in mice after partial hepatectomy. J. Clin. Invest. 1998;102:996–1007. doi: 10.1172/JCI3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Baird L., Tsujita T., Kobayashi E.H., Funayama R., Nagashima T., Nakayama K., Yamamoto M. A homeostatic shift facilitates endoplasmic reticulum proteostasis through transcriptional integration of proteostatic stress response pathways. Mol. Cell Biol. 2017;37(4) doi: 10.1128/MCB.00439-16. 2017 Feb 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hirose W., Horiuchi M., Li D., Motoike I.N., Zhang L., Nishi H., Taniyama Y., Kamei T., Suzuki M., Kinoshita K., Katsuoka F., Taguchi K., Yamamoto M. Selective elimination of NRF2-activated cells by competition with neighboring cells in the esophageal epithelium. Cell Mol Gastroenterol Hepatol. 2023;15:153–178. doi: 10.1016/j.jcmgh.2022.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.