Abstract
Shiga toxigenic Escherichia coli (STEC) comprises a diverse group of organisms capable of causing severe gastrointestinal disease in humans. Within the STEC family, certain strains appear to be of greater virulence for humans, for example, those belonging to serogroups O111 and O157 and those with particular combinations of other putative virulence traits. We have developed two multiplex PCR assays for the detection and genetic characterization of STEC in cultures of feces or foodstuffs. Assay 1 utilizes four PCR primer pairs and detects the presence of stx1, stx2 (including variants of stx2), eaeA, and enterohemorrhagic E. coli hlyA, generating amplification products of 180, 255, 384, and 534 bp, respectively. Assay 2 uses two primer pairs specific for portions of the rfb (O-antigen-encoding) regions of E. coli serotypes O157 and O111, generating PCR products of 259 and 406 bp, respectively. The two assays were validated by testing 52 previously characterized STEC strains and observing 100% agreement with previous results. Moreover, assay 2 did not give a false-positive O157 reaction with enteropathogenic E. coli strains belonging to clonally related serogroup O55. Assays 1 and 2 detected STEC of the appropriate genotype in primary fecal cultures from five patients with hemolytic-uremic syndrome and three with bloody diarrhea. Thirty-one other primary fecal cultures from patients without evidence of STEC infection were negative.
Shiga toxigenic Escherichia coli (STEC) is an important cause of gastrointestinal disease in humans, particularly since such infections may result in life-threatening sequelae such as hemolytic-uremic syndrome (HUS) and thrombotic thrombocytopenic purpura (22). The morbidity and mortality associated with several recent large outbreaks of STEC disease have highlighted the threat these organisms pose to public health (17, 25, 28, 32). For this reason, there is an increasing demand for improved diagnostic procedures for the detection of STEC in fecal samples and, in particular, in foods such as meat and dairy products. It has been recognized for a number of years that STEC strains causing human disease may belong to a very broad range of O serogroups (22). However, many of the STEC strains found in the gastrointestinal tracts of domestic animals (the principal source of human infections) may have a low degree of virulence in humans. These strains are less likely to produce putative accessory virulence factors such as intimin (encoded by eaeA) and the plasmid-encoded enterohemolysin (encoded by enterohemorrhagic E. coli (EHEC) hlyA) (1, 5, 6, 33, 34). Within the human disease-associated strains, those producing Shiga toxin type 2 (Stx2, encoded by stx2) appear to be more commonly responsible for serious complications such as HUS than those producing only Shiga toxin type 1 (Stx1, encoded by stx1) (23, 26). Furthermore, STEC belonging to serogroup O157 and, to a lesser extent, serogroup O111 are responsible for the vast majority of HUS outbreaks (9, 16, 25, 28, 32). Accordingly, the capacity to rapidly determine whether a patient is infected, or food is contaminated, with STEC belonging to serogroup O111 or O157 or whether the STEC produces virulence factors associated with more serious disease would be highly advantageous. For this reason, we have developed multiplex PCR assays for the simultaneous detection of (i) stx1, stx2, eaeA, and EHEC hlyA and (ii) portions of the rfb (O-antigen-encoding) regions of E. coli O111 and O157.
Examination of STEC isolates by multiplex PCR.
PCR primer pairs were designed with reference to published sequence data for stx1 (19), stx2 (18), eaeA (35), EHEC hlyA (33), and portions of the rfb regions of E. coli O111 (2) and O157 (7). Details of the nucleotide sequence, the specific gene region amplified, and the size of the PCR product for each primer pair are listed in Table 1. Crude DNA extracts were prepared from characterized STEC strains, as described previously (27). A total of 52 STEC strains were examined, and these were from our own collection or were kindly provided by Roy Robins-Browne, Royal Children’s Hospital, Melbourne, Australia, or Lothar Beutin, Robert Koch Institute, Berlin, Germany. Twenty eight strains were isolated from human feces (patients with diarrhea or HUS), 7 were from domestic animals, and 17 were from foods. The STEC serogroups included O157 (19 isolates), O111 (10 isolates), O26 and OX3 (2 isolates each), and O48, O91, O98, O113, O128, O141, and O159 (1 isolate each); a further 12 STEC isolates were not typeable by the O serogroup. Four stx-negative enteropathogenic E. coli (EPEC) strains belonging to serogroups O55 (three isolates provided by R. Robins-Browne) and O111 (one isolate) and a sorbitol-fermenting, nontoxigenic O157:H20 isolate (also provided by R. Robins-Browne) were also tested.
TABLE 1.
PCR primers
| Primer | Sequence (5′–3′) | Specificitya | Amplicon size (bp) |
|---|---|---|---|
| Assay 1 | |||
| stx1F | ATAAATCGCCATTCGTTGACTAC | nt 454–633 of A subunit coding region of stx1 | 180 |
| stx1R | AGAACGCCCACTGAGATCATC | ||
| stx2F | GGCACTGTCTGAAACTGCTCC | nt 603–857 of A subunit coding region of stx2 (including stx2 variants) | 255 |
| stx2R | TCGCCAGTTATCTGACATTCTG | ||
| eaeAF | GACCCGGCACAAGCATAAGC | nt 27–410 of eaeA (this region is conserved between EPEC and STEC) | 384 |
| eaeAR | CCACCTGCAGCAACAAGAGG | ||
| hlyAF | GCATCATCAAGCGTACGTTCC | nt 70–603 of EHEC hlyA | 534 |
| hlyAR | AATGAGCCAAGCTGGTTAAGCT | ||
| Assay 2 | |||
| O157F | CGGACATCCATGTGATATGG | nt 393–651 of rfbEO157:H7 | 259 |
| O157R | TTGCCTATGTACAGCTAATCC | ||
| O111F | TAGAGAAATTATCAAGTTAGTTCC | nt 24–429 of ORF 3.4 of E. coli O111 rfb region | 406 |
| O111R | ATAGTTATGAACATCTTGTTTAGC |
nt, nucleotide; ORF, open reading frame.
Samples (2 μl) of each extract were amplified in 50-μl reaction mixtures containing 200 μM concentrations of deoxynucleoside triphosphates, an approximately 250 nM concentration of each primer, and 1 U of Taq polymerase (Boehringer GmbH, Mannheim, Germany) in 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 2 mM MgCl2, 0.1% gelatin, 0.1% Tween 20, and 0.1% Nonidet P-40. Samples were subjected to 35 PCR cycles, each consisting of 1 min of denaturation at 95°C; 2 min of annealing at 65°C for the first 10 cycles, decrementing to 60°C by cycle 15; and 1.5 min of elongation at 72°C, incrementing to 2.5 min from cycles 25 to 35. PCR reaction mixtures were electrophoresed on 2% agarose gels and stained with ethidium bromide. Figure 1 shows a representative gel for eight previously characterized STEC strains subjected to PCR assay 1. Clear PCR products of the expected sizes were observed, consistent with the presence of stx1, stx2, eaeA, and EHEC hlyA. Assay 1 PCR results for all 52 STEC strains tested are shown in Table 2. Again, there was 100% agreement with previously determined genotype data (28, 29).
FIG. 1.
Characterization of reference STEC strains by multiplex PCR assay 1. Lanes: M, DNA size markers (pUC19 DNA digested with HpaII; fragment sizes visible are 501/489, 404, 331, 242, 190, 147, and 111 bp); 1, negative control; 2, O157:H− strain 96/2998 (stx1+ stx2+ eaeA+ EHEC hlyA+); 3, O157 strain 94-8628 (stx2+ eaeA+ EHEC hlyA+); 4, O157 strain 96/0052 (stx1+ stx2+ eaeA+); 5, O48:H21 strain 94CR (stx1+ stx2+ EHEC hlyA+); 6, O128 strain 95AS1 (stx1+ stx2+); 7, O91 strain 95HE4 (stx1+ EHEC hlyA+); 8, O113 strain MW10 (stx2+ EHEC hlyA+); 9, OX3:H21 strain O31 (stx2+). The expected mobilities for the various specific PCR products are also indicated.
TABLE 2.
Characterization of STEC by multiplex PCR
| STEC genotypea
|
No. of isolates by source
|
||||||
|---|---|---|---|---|---|---|---|
| stx1 | stx2 | eaeA | hlyA | Human | Animal | Food | Total |
| + | − | − | − | 0 | 0 | 2 | 2 |
| − | + | − | − | 1 | 0 | 2 | 3 |
| + | + | − | − | 3 | 1 | 1 | 5 |
| + | − | − | + | 1 | 0 | 2 | 3 |
| − | + | − | + | 0 | 0 | 2 | 2 |
| + | + | − | + | 1 | 0 | 6 | 7 |
| + | − | + | − | 0 | 0 | 0 | 0 |
| − | + | + | − | 0 | 0 | 0 | 0 |
| + | + | + | − | 1 | 0 | 0 | 1 |
| + | − | + | + | 6 | 0 | 0 | 6 |
| − | + | + | + | 6 | 1 | 2 | 9 |
| + | + | + | + | 9 | 5 | 0 | 14 |
+, gene present; −, gene absent.
Figure 2 shows a representative gel for eight reference STEC isolates (four O111 strains and four O157 strains), as well as for an O55 EPEC isolate (which is genetically related to O157:H7 STEC strains), analyzed by PCR assay 2. Clear PCR products of the expected sizes were seen for each of the O111 and O157 strains (406 bp and 259 bp, respectively), but the O55 strain did not yield a PCR product. An analysis of the entire STEC collection showed that all 10 O111 strains tested as well as O111 EPEC strain 87A yielded a 406-bp PCR product. The 19 O157 STEC strains tested (including O157:H− and O157:H7 strains) all yielded a 259-bp PCR product, as did the sorbitol-fermenting, nontoxigenic O157:H20 isolate. The 23 STEC isolates tested that did not belong to either serogroup O111 or O157 yielded no PCR products (result not presented). All three O55 EPEC isolates tested were negative by assay 2, although they did give a positive PCR result for eaeA by assay 1 (as did O111 EPEC strain 87A) (result not presented).
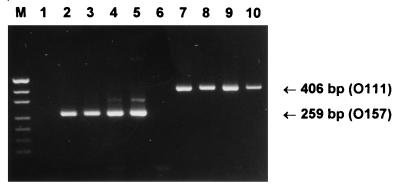
FIG. 2.
Characterization of reference STEC strains by multiplex PCR assay 2. Lanes: M, DNA size markers (pUC19 DNA digested with HpaII; fragment sizes visible are 501/489, 404, 331, 242, 190, 147, and 111 bp); 1, negative control; 2, O157:H− strain 95SF2; 3, O157:H− strain 96GR1; 4, O157 strain 96/0052; 5, O157:H7 strain 90/103; 6, O55 EPEC strain 93/282; 7, O111:H− strain 95NR1; 8, O111:H− strain 96RO1; 9, O111:H− strain PH; 10, O111:H− strain CB168. The expected mobilities for the O111- and O157-specific PCR products are also indicated.
Analysis of primary fecal cultures by multiplex PCR.
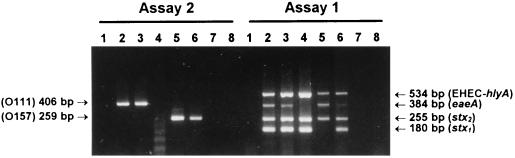
To demonstrate the diagnostic utility of multiplex PCR, crude DNA extracts of primary fecal cultures from four sporadic HUS cases and one patient with bloody diarrhea were analyzed by assays 1 and 2 (Fig. 3). PCR analysis indicated that fecal cultures from HUS patients 1 and 2 contained O111 STEC strains which were positive for stx1, stx2, eaeA, and EHEC hlyA. The fecal culture from HUS patient 3 contained a STEC isolate not belonging to serogroup O111 or O157 which was positive for stx1, stx2, and EHEC hlyA but which was negative for eaeA, while HUS patient 4 had an O157 STEC isolate which was positive for stx2, eaeA, and EHEC hlyA. The fecal culture from patient 5, who had bloody diarrhea, contained an O157 STEC isolate which was positive for stx1, stx2, eaeA, and EHEC hlyA. Fecal cultures from a further three cases of STEC infection were also tested (results not presented). These included one HUS case caused by an O157 STEC isolate positive for stx2, eaeA, and EHEC hlyA, one bloody diarrhea case caused by an STEC isolate not of serogroup O111 or O157 that was positive for stx2, eaeA, and EHEC hlyA, and one bloody diarrhea case complicated with microangiopathic hemolytic anemia and thrombocytopenia caused by an O113 STEC isolate positive for stx2 and EHEC hlyA. In all eight cases, the direct PCR results were 100% compatible with the genotype of the STEC which was isolated from the cultures. A total of 32 other primary cultures of fecal samples submitted for bacteriological investigation were also tested (results for two of which [patients 6 and 7] are also shown in Fig. 3). Three of these samples had yielded Salmonella sp., one grew Shigella flexneri, and one was positive for rotavirus; the remainder did not yield a pathogen. Thirty one of these samples were negative by both assays 1 and 2 and were also negative when tested by an independent stx-specific PCR assay (27). The remaining sample (from a patient who had diarrhea but was negative for other pathogens) yielded a very weak positive stx2 reaction by assay 1, which was subsequently confirmed by the independent stx-specific PCR assay (27), but no PCR products were seen by assay 2. Isolation of STEC from this culture was not attempted, as the weak PCR signal suggested that only very small numbers of STEC would have been present.
FIG. 3.
Multiplex PCR analysis of primary fecal cultures. Crude DNA extracts of primary fecal cultures were analyzed by multiplex PCR assay 1 or assay 2, as indicated. Lanes: 1, negative control; 2, patient 1 (HUS); 3, patient 2 (HUS); 4, patient 3 (HUS); 5, patient 4 (HUS); 6, patient 5 (bloody diarrhea); 7, patient 6 (control); 8, patient 7 (control). The expected mobilities for the various specific PCR products are also indicated.
Sensitivity of multiplex PCR.
To assess sensitivity, one of the STEC-negative fecal cultures tested above was spiked with serial dilutions of cultures of STEC strains 95NR1 (O111:H−) or 95SF2 (O157:H−). Extracts of these samples were then subjected to assay 2 (Fig. 4). For both the O111 and the O157 serogroup strains, a PCR product of the appropriate size could still be seen in the sample that contained the 104-fold-diluted STEC culture (equivalent to approximately 103 STEC CFU per assay), but not in the sample containing 105-fold dilutions. Also, the presence of both O111 and O157 STEC (each diluted 10-fold) in a sample did not appear to interfere with the strength of the PCR signal (Fig. 4).
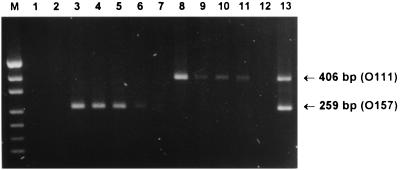
FIG. 4.
Sensitivity of multiplex PCR assay 2. An STEC-negative fecal culture was spiked with serial 10-fold dilutions of cultures of STEC strains 95NR1 (O111:H−) or 95SF2 (O157:H−). Extracts of these samples were then subjected to assay 2. Lanes: M, DNA size markers (pUC19 DNA digested with HpaII; fragment sizes visible are 501/489, 404, 331, 242, 190, 147, and 111 bp); 1, negative control; 2, STEC-negative fecal culture (FC); 3, FC plus a 10-fold dilution of a strain 95SF2 culture (10−1 95SF2); 4, FC plus 10−2 95SF2; 5, FC plus 10−3 95SF2; 6, FC plus 10−4 95SF2; 7, FC plus 10−5 95SF2; 8, FC plus 10−1 95NR1; 9, FC plus 10−2 95NR1; 10, FC plus 10−3 95NR1; 11, FC plus 10−4 95NR1; 12, FC plus 10−5 95NR1; 13, FC plus 10−1 95SF2 plus 10−1 95NR1. The expected mobilities for the O111- and O157-specific PCR products are also indicated.
Discussion and conclusions.
PCR is generally considered to be the most sensitive means of determining whether a fecal specimen or a food sample contains STEC. Although direct extracts of feces or foods can be used as templates for PCR, the best results are usually obtained by testing extracts of primary broth cultures (3, 14, 27). Broth enrichment serves two purposes: inhibitors in the sample are diluted, and bacterial growth increases the number of copies of the target sequence. Sensitivity is important when testing fecal samples, because although STEC numbers may be very high in the early stages of infection, they may drop dramatically as disease progresses. Sensitivity is also particularly important when testing suspected foods because, at least for certain O111 and O157 STEC strains, the infectious dose for humans may be of the order of 1 to 10 CFU (17, 28). Thus, foods destined to be consumed without further cooking or processing need to be totally free of such STEC strains. Some of the PCR assays for detection of STEC described to date use single pairs of primers based on consensus sequences, which are capable of amplifying all stx-related genes, with subsequent identification of the stx type requiring Southern or dot blot hybridization with labelled oligonucleotides directed against type-specific sequences within the amplified fragment (21, 27, 31). Others combine different primer pairs for stx1 and stx2, and in some cases stx2 variants, in the same reaction and direct the amplification of fragments which differ in size for each toxin type (4, 8, 14, 20, 30).
PCR has also been used for the detection of genes encoding accessory STEC virulence factors, such as eaeA and EHEC hlyA (15, 33). Fratamico et al. (12) combined previously described stx- (21) and eaeA-specific (15) PCR primer pairs with those specific for a portion of the 60-MDa virulence plasmid from an O157:H7 STEC in a multiplex assay. They concluded that this assay was suitable for the identification of STEC belonging to serogroup O157. However, the O157 virulence plasmid primers used actually recognize a portion of the EHEC hlyA gene, which of course is not confined to serogroup O157. Gannon et al. (13) have recently described two other multiplex PCR formats based on a stx-specific primer pair and two distinct eaeA-specific primer pairs, as well as a primer pair specific for a portion of the fliCh7 gene, which encodes the H7 antigen. The two eaeA-specific primer sets recognized either the highly conserved 5′ portion of eaeA or a region at the (variable) 3′ end of the gene which was specific for O157 strains and a small number of other serogroups, including O55 (15). A further multiplex PCR has been used to detect O157:H7 STEC; the PCR uses stx1- and stx2-specific primer pairs in combination with a primer pair that recognizes a single base mutation in the β-glucoronidase-encoding uidA gene (10). This mutation in uidA results in a β-glucoronidase-negative phenotype, a feature strongly (although not absolutely) associated with O157:H7 strains. Louie et al. (24) also exploited the heterogeneity of the 3′ end of eaeA to design two PCR assays, one of which was specific for eaeA from O157 STEC strains and O55 EPEC strains. The other primer pair gave a positive reaction with 16 of 22 O111 STEC strains but not with O111 EPEC strains. The sequence heterogeneity of eaeA within O111 STEC is presumably responsible for the fact that six of the strains (all of which were O111:H11) were found to be negative by PCR even though they hybridized to a probe specific for the conserved region of eaeA.
In the present study, we have designed two multiplex PCR assays for the detection and characterization of STEC. Assay 1 detects the presence of stx1, stx2, eaeA, and EHEC hlyA, generating PCR products of distinct sizes which are easily distinguished after agarose gel electrophoresis. Mismatch between the primer and target sequences due to the known heterogeneity of stx2 genes has the potential to interfere with PCR assays, but the primers used in the present study were designed to amplify all stx2 variants currently deposited with GenBank. Similarly, the eaeA primers are specific for a region at the 5′ end of the gene that appears to be conserved among all STEC and EPEC strains examined to date. Moreover, the PCR primers for the amplification of EHEC hlyA were designed to eliminate the possibility of cross-reaction with the gene encoding E. coli alpha-hemolysin, with which it shares about 70% DNA sequence homology. Multiplex PCR assay 2 detects the presence of genes involved in the biosynthesis of serogroup O111 and O157 O antigen. The actual region of the O111 rfb gene cluster (open reading frame 3.4) that is amplified is similar to that described previously (28), but the primer sequences have been modified such that their melting temperatures are similar to those for the primers designed to amplify the O157 rfbE gene. These primers are the only ones described to date which directly target O-antigen-encoding genes, and the assay is not dependent on clonal association between particular variants of unrelated genes (e.g., eaeA or uidA) and STEC strains of a given serotype. Thus, all O111 and O157 strains tested positive, and for the O157-specific PCR, there was no reaction with clonally related EPEC strains belonging to serogroup O55.
The two multiplex PCR assays were also found to be effective for the direct detection and characterization of STEC in primary fecal cultures from eight patients with HUS or bloody diarrhea. The absence of reactivity of fecal cultures from persons without evidence of STEC infection (including those known to be infected with other pathogens) is further evidence for the specificity of both assay 1 and assay 2. Information on the genotype of a STEC isolate may be of considerable microbiological and clinical significance because, as discussed previously, there appears to be a link between certain combinations of traits and the capacity of a STEC isolate to cause serious gastrointestinal disease in humans and complications such as HUS. The presence of these particular STEC genotypes in a food source (e.g., meat) is also extremely significant from a food safety or public health viewpoint. Previous studies have shown that PCR assays based on stx sequences can detect the presence of very low numbers of STEC organisms in microbiologically complex samples (8, 27, 28). The analysis of spiked fecal cultures in the present study demonstrated that serogroup-specific PCR assay 2 was also very sensitive. Fecal cultures spiked with 104-fold dilutions of O111 or O157 STEC cultures (i.e., the STEC constituted 0.01% of the total flora) generated a PCR product which was visible on an ethidium bromide-stained agarose gel. Other direct methods (e.g., plating on cefixime-tellurite sorbitol MacConkey agar or latex agglutination for O157 STEC, enzyme-linked immunosorbent assay for either lipopolysaccharide or Stx, etc.) are unlikely to detect such low levels of STEC. The sensitivity of the PCR could undoubtedly be improved even further by increasing the volume of sample assayed or by secondary hybridization of the PCR products with labelled rfb-specific probes.
Although much useful information can be gained from multiplex PCR analysis of fecal or food samples, the interpretation is complicated by the possibility that the composite genotypic profile may represent the sum of genotypes of more than one STEC organism. For this reason, isolation of STEC from plate cultures, for example by colony hybridization with stx-specific probes, should always be attempted. However, given the sensitivity of PCR screens, there is a likelihood that a proportion of genuine STEC PCR-positive specimens will not yield an isolate even after heroic efforts. The probability of isolating STEC is considerably greater, however, if the infecting serogroup is known, as this permits the deployment of immunomagnetic bead enrichment techniques, which have already been described for serogroups O157 (11) and O111 (28). The capacity to target these procedures at specimens that have been proven by multiplex PCR to contain STEC belonging to these serogroups should result in significant savings in terms of labor.
Acknowledgments
We are grateful to Roy Robins-Browne and Lothar Beutin for providing STEC strains, to Jan Lanser for referring fecal samples, and to Matthew Woodrow for technical assistance.
This work was supported by a grant from the National Health and Medical Research Council of Australia. A.W.P. holds an NHMRC Australian Postdoctoral Fellowship.
REFERENCES
- 1.Barrett T J, Kaper J B, Jerse A E, Wachsmuth I K. Virulence factors in Shiga-like toxin-producing Escherichia coli isolated from humans and cattle. J Infect Dis. 1992;165:979–980. doi: 10.1093/infdis/165.5.979. [DOI] [PubMed] [Google Scholar]
- 2.Bastin D A, Reeves P R. Sequence analysis of the O antigen gene (rfb) cluster of Escherichia coli O111. Gene. 1995;164:17–23. doi: 10.1016/0378-1119(95)00459-j. [DOI] [PubMed] [Google Scholar]
- 3.Begum D, Jackson M P. Direct detection of Shiga-like toxin-producing Escherichia coli in ground beef using the polymerase chain reaction. Mol Cell Probes. 1995;9:259–264. doi: 10.1016/s0890-8508(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 4.Begum D, Strockbine N A, Sowers E G, Jackson M P. Evaluation of a technique for identification of Shiga-like toxin-producing Escherichia coli by using polymerase chain reaction and digoxigenin-labeled probes. J Clin Microbiol. 1993;31:3153–3156. doi: 10.1128/jcm.31.12.3153-3156.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beutin L, Geier D, Steinruck H, Zimmermann S, Scheutz F. Prevalence and some properties of verotoxin (Shiga-like toxin)-producing Escherichia coli in seven different species of healthy domestic animals. J Clin Microbiol. 1993;31:2483–2488. doi: 10.1128/jcm.31.9.2483-2488.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beutin L, Geier D, Zimmermann S, Karch H. Virulence markers of Shiga-like toxin-producing Escherichia coli strains originating from healthy domestic animals of different species. J Clin Microbiol. 1995;33:631–635. doi: 10.1128/jcm.33.3.631-635.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bilge S S, Vary J C, Jr, Dowell S F, Tarr P I. Role of the Escherichia coli O157:H7 O side chain in adherence and analysis of an rfb locus. Infect Immun. 1996;64:4795–4801. doi: 10.1128/iai.64.11.4795-4801.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brian M J, Frosolono M, Murray B E, Miranda A, Lopez E L, Gomez H F, Cleary T G. Polymerase chain reaction for diagnosis of enterohemorrhagic Escherichia coli infection and hemolytic-uremic syndrome. J Clin Microbiol. 1992;30:1801–1806. doi: 10.1128/jcm.30.7.1801-1806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caprioli A, Luzzi I, Rosmini F, Resti C, Edefonti A, Perfumo F, Farina C, Goglio A, Gianviti A, Rizzone G. Communitywide outbreak of hemolytic uremic syndrome associated with non-O157 verocytotoxin-producing Escherichia coli. J Infect Dis. 1994;169:208–211. doi: 10.1093/infdis/169.1.208. [DOI] [PubMed] [Google Scholar]
- 10.Cebula T A, Payne W L, Feng P. Simultaneous identification of strains of Escherichia coli serotype O157:H7 and their Shiga-like toxin type by mismatch amplification mutation assay-multiplex PCR. J Clin Microbiol. 1995;33:248–250. doi: 10.1128/jcm.33.1.248-250.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chapman P A, Siddons C A. A comparison of immunomagnetic separation and direct culture for the isolation of verocytotoxin-producing Escherichia coli O157 from cases of bloody diarrhoea, non-bloody diarrhoea and asymptomatic contacts. J Med Microbiol. 1996;44:267–271. doi: 10.1099/00222615-44-4-267. [DOI] [PubMed] [Google Scholar]
- 12.Fratamico P M, Sackitey S K, Wiedmann M, Deng M Y. Detection of Escherichia coli O157:H7 by multiplex PCR. J Clin Microbiol. 1995;33:2188–2191. doi: 10.1128/jcm.33.8.2188-2191.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gannon V P J, D’Souza S, Graham T, King R K, Rahn K, Read S. Use of the flagellar H7 gene as a target in multiplex PCR assays and improved specificity in identification of enterohemorrhagic Escherichia coli strains. J Clin Microbiol. 1997;35:656–662. doi: 10.1128/jcm.35.3.656-662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gannon V P J, King R K, Kim J Y, Thomas E J. Rapid and sensitive method for detection of Shiga-like toxin-producing Escherichia coli in ground beef using the polymerase chain reaction. Appl Environ Microbiol. 1992;58:3809–3815. doi: 10.1128/aem.58.12.3809-3815.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gannon V P J, Rashed M, King R K, Golsteyn Thomas E J. Detection and characterization of the eae gene of Shiga-like toxin-producing Escherichia coli using polymerase chain reaction. J Clin Microbiol. 1993;31:1268–1274. doi: 10.1128/jcm.31.5.1268-1274.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffin P M. Escherichia coli O157:H7 and other enterohemorrhagic Escherichia coli. In: Blaser M J, Smith P D, Ravdin J I, Greenberg H B, Guerrant R I, editors. Infections of the gastrointestinal tract. New York, N.Y: Raven Press; 1995. pp. 739–761. [Google Scholar]
- 17.Griffin P M, Bell B P, Cieslak P R, Tuttle J, Barrett T J, Doyle M P, McNamara A M, Shefer A M, Wells J G. Large outbreak of Escherichia coli O157:H7 infections in the western United States: the big picture. In: Karmali M A, Goglio A G, editors. Recent advances in verocytotoxin-producing Escherichia coli infections. Amsterdam, The Netherlands: Elsevier Science B.V.; 1994. pp. 7–12. [Google Scholar]
- 18.Jackson M P, Neill R J, O’Brien A D, Holmes R K, Newland J W. Nucleotide sequence analysis and comparison of the structural genes for Shiga-like toxin I and Shiga-like toxin II encoded by bacteriophages from Escherichia coli. FEMS Microbiol Lett. 1987;44:109–114. doi: 10.1016/0882-4010(87)90106-9. [DOI] [PubMed] [Google Scholar]
- 19.Jackson M P, Newland J W, Holmes R K, O’Brien A D. Nucleotide sequence analysis of the structural genes for Shiga-like toxin I encoded by bacteriophage 933J from Escherichia coli. Microb Pathog. 1987;2:147–153. doi: 10.1016/0882-4010(87)90106-9. [DOI] [PubMed] [Google Scholar]
- 20.Johnson W M, Pollard D R, Lior H, Tyler S D, Rozee K R. Differentiation of genes coding for Escherichia coli verotoxin 2 and the verotoxin associated with porcine edema disease (VTe) by the polymerase chain reaction. J Clin Microbiol. 1990;28:2351–2353. doi: 10.1128/jcm.28.10.2351-2353.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karch H, Meyer T. Single primer pair for amplifying segments of distinct Shiga-like toxin genes by polymerase chain reaction. J Clin Microbiol. 1989;27:2751–2757. doi: 10.1128/jcm.27.12.2751-2757.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karmali M A. Infection by verotoxin-producing Escherichia coli. Clin Microbiol Rev. 1989;2:15–38. doi: 10.1128/cmr.2.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kleanthous H, Smith H R, Scotland S M, Gross R J, Rowe B, Taylor C M, Milford D V. Haemolytic uraemic syndromes in the British Isles, 1985-8: association with verocytotoxin producing Escherichia coli. Part 2: microbiological aspects. Arch Dis Child. 1990;65:722–727. doi: 10.1136/adc.65.7.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Louie M, De-Azavedo J, Clarke R, Borczyk A, Lior H, Richter M, Brunton J. Sequence heterogeneity of the eae gene and detection of verotoxin-producing Escherichia coli using serotype-specific primers. Epidemiol Infect. 1994;112:449–461. doi: 10.1017/s0950268800051153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minami S. On measures for the control of enterohaemorrhagic E. coli O157 in Japan, background paper number 9. WHO Consultation on the Prevention and Control of Enterohaemorrhagic Escherichia coli (EHEC) Infections. Geneva, Switzerland: World Health Organization; 1997. [Google Scholar]
- 26.Ostroff S M, Tarr P I, Neill M A, Lewis J H, Hargrett-Bean N, Kobayashi J M. Toxin genotypes and plasmid profiles as determinants of systemic sequelae in Escherichia coli O157:H7 infections. J Infect Dis. 1989;160:994–999. doi: 10.1093/infdis/160.6.994. [DOI] [PubMed] [Google Scholar]
- 27.Paton A W, Paton J C, Goldwater P N, Manning P A. Direct detection of Escherichia coli shiga-like toxin genes in primary fecal cultures using the polymerase chain reaction. J Clin Microbiol. 1993;31:3063–3067. doi: 10.1128/jcm.31.11.3063-3067.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paton A W, Ratcliff R, Doyle R M, Seymour-Murray J, Davos D, Lanser J A, Paton J C. Molecular microbiological investigation of an outbreak of hemolytic uremic syndrome caused by dry fermented sausage contaminated with Shiga-like toxin-producing Escherichia coli. J Clin Microbiol. 1996;34:1622–1627. doi: 10.1128/jcm.34.7.1622-1627.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paton A W, Voss E, Manning P A, Paton J C. Shiga toxin-producing Escherichia coli isolates from cases of human disease show enhanced adherence to intestinal epithelial (Henle 407) cells. Infect Immun. 1997;65:3799–3805. doi: 10.1128/iai.65.9.3799-3805.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pollard D R, Johnson W M, Lior H, Tyler S D, Rozee K R. Rapid and specific detection of verotoxin genes in Escherichia coli by the polymerase chain reaction. J Clin Microbiol. 1990;28:540–545. doi: 10.1128/jcm.28.3.540-545.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Read S C, Clarke R C, Martin A, De-Grandis S A, Hii J, McEwen S, Gyles C L. Polymerase chain reaction for detection of verocytotoxigenic Escherichia coli isolated from animal and food sources. Mol Cell Probes. 1992;6:153–161. doi: 10.1016/0890-8508(92)90060-b. [DOI] [PubMed] [Google Scholar]
- 32.Reilly W J. Verotoxigenic E. coli O157 in Scotland, background paper number 4. WHO Consultation on the Prevention and Control of Enterohaemorrhagic Escherichia coli (EHEC) Infections. Geneva, Switzerland: World Health Organization; 1997. [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidt H, Beutin L, Karch H. Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL933. Infect Immun. 1995;63:1055–1061. doi: 10.1128/iai.63.3.1055-1061.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt H, Karch H. Enterohemolytic phenotypes and genotypes of Shiga toxin-producing Escherichia coli O111 strains from patients with diarrhea and hemolytic-uremic syndrome. J Clin Microbiol. 1996;34:2364–2367. doi: 10.1128/jcm.34.10.2364-2367.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu J, Kaper J B. Cloning and characterization of the eae gene of enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol. 1992;6:411–417. doi: 10.1111/j.1365-2958.1992.tb01484.x. [DOI] [PubMed] [Google Scholar]