Highlights
-
•
PIK3CA mutation was significantly associated with lower pCR rate in HER2-positive breast cancer patients treated with neoadjuvant anti-HER2 therapy.
-
•
The adverse effect of PIK3CA mutation on pCR remained significant regardless of single- or dual-agent anti-HER2 therapy, and hormone receptor status.
-
•
There was no significant association between mutated and wild-type PIK3CA neither in the neoadjuvant setting nor in the adjuvant setting.
-
•
In the metastatic setting, PIK3CA mutation was predictive of worse overall response rate, progression-free survival, but not for overall survival.
Keywords: Breast cancer, HER2-positive, Anti-HER2 therapy, PIK3CA mutation, Predictive
Abstract
Background
This study aimed to comprehensively explore the clinical significance of PIK3CA mutation in human epidermal growth factor receptor 2 (HER2)-positive breast cancer treated with anti-HER2 therapy.
Methods
We systematically searched PubMed, Embase, and the Cochrane databases for eligible studies assessing the association between PIK3CA mutation and outcomes in patients with HER2-positive breast cancer receiving anti-HER2 therapy. The main outcomes included: (1) pathological complete response (pCR) or disease-free survival (DFS) for the neoadjuvant setting; (2) DFS or invasive DFS for the adjuvant setting; (3) objective response rate (ORR), progression-free survival (PFS), time-to-progression (TTP), or overall survival (OS) for the metastatic setting. The mutational landscape of HER2-positive breast cancer according to PIK3CA mutation status was examined based on TCGA breast cancer dataset.
Results
Totally, 43 eligible studies, covering 11,099 patients with available data on PIK3CA mutation status, were identified. In the neoadjuvant setting, PIK3CA mutation was significantly associated with a lower pCR rate (OR=0.23, 95% CI 0.19–0.27, p<0.001). This association remained significant irrespective of the type of anti-HER2 therapy (single-agent or dual-agent) and hormone receptor status. There were no significant differences in DFS between PIK3CA mutated and wild-type patients in either the neoadjuvant or adjuvant settings. In the metastatic setting, PIK3CA mutation predicted worse ORR (OR=0.26, 95%CI 0.17–0.40, p<0.001), PFS (HR=1.28, 95%CI 1.03–1.59, p = 0.024) and TTP (HR=2.27, 95%CI 1.54–3.34, p<0.001). However, no significant association was observed between PIK3CA mutation status and OS. Distinct mutational landscapes were observed in HER2-positive breast cancer between individuals with PIK3CA mutations and those with wild-type PIK3CA.
Conclusions
PIK3CA mutation was significantly associated with a lower pCR rate in HER2-positive breast cancer treated with neoadjuvant anti-HER2 therapy. In the metastatic setting, PIK3CA mutation was predictive of worse ORR, PFS and TTP. These results suggest the potential for developing PI3K inhibitors as a therapeutic option for these patients.
Introduction
Human epidermal growth factor receptor 2 (HER2) amplification is identified in about 20–25% of breast cancers, contributing to a more aggressive phenotype and poor outcomes [1]. Despite the remarkable progress with the availability of anti-HER2 agents for HER2-positive breast cancer across all disease stages, resistance often develops, resulting in treatment failure [2]. Currently, apart from HER2, there is a lack of robust predictive markers for the selection of anti-HER2 therapy. Hence, it is crucial to identify additional promising biomarkers for further selection of patients who may benefit from or be non-responsive to anti-HER2 therapy.
Phosphoinositide 3-kinase (PI3K) p110α, encoded by the PIK3CA gene, is a subunit of PI3K, one of the crucial kinases in the PI3K/AKT1/MTOR pathway [3]. PIK3CA mutation is common in breast cancer, with the highest prevalence observed within hormone receptor-positive /HER2-negative tumors [4,5]. Based on the SOLAR-1 study, PIK3CA mutation is recommended to select candidates for alpelisib in patients with hormone receptor-positive-positive/HER2-negative metastatic breast cancer [6]. In HER2-positive breast cancer, PIK3CA mutation was reported in approximately 20% to 25% of all cases [5]. PI3K pathway activation, as judged by PIK3CA mutation or loss of PTEN expression, has been considered as a key effector of HER2 signaling, and can drive intrinsic resistance to trastuzumab [7].
The evidence from previous studies on the role of PIK3CA mutation in HER2-positive breast cancer is heterogeneous. In the neoadjuvant setting, a pooled analysis of five prospective trials confirmed that PIK3CA mutation was significantly linked to lower pathological complete response (pCR) rates in HER2-positive breast cancer patients after anti-HER2 therapy and chemotherapy [8]. However, this association was only confined to dual anti-HER2 therapy with trastuzumab and lapatinib, but not for single anti-HER2 therapy with trastuzumab or lapatinib [8]. In contrast, the TRYPHAENA study did not find a predictive effect of PIK3CA mutation on pCR in patients receiving dual anti-HER2 therapy with pertuzumab plus trastuzumab as neoadjuvant treatment [9]. In the adjuvant setting, several clinical trials failed to demonstrate a predictive effect of PIK3CA mutation on anti-HER2 therapy or its prognostic significance [10], [11], [12], [13]. Additionally, although several studies in the metastatic setting reported an adverse prognostic effect of PIK3CA mutation in HER2-positive patients treated with anti-HER2 therapy [14,15], others failed to confirm this effect [16], [17], [18]. Therefore, the predictive and prognostic relevance of PIK3CA mutation within HER2-positive breast cancer treated with anti-HER2 therapy has not been clearly defined.
This study aimed to comprehensively explore the predictive and prognostic impact of PIK3CA mutation in HER2-positive breast cancer treated with anti-HER2 therapy. Additionally, we examined the mutational profile of HER2-positive breast cancer according to the status of PIK3CA mutation.
Materials and methods
This systematic review and meta-analysis is reported in accordance with the guidelines of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses [19]. The protocol was registered in the Prospective Register of Systematic Reviews (PROSPERO) (CRD42022373328).
Search strategy
A comprehensive literature search of PubMed, Embase, and the Cochrane Library Central Register of Controlled Trials databases was conducted to identify relevant studies from the inception of each database to Sep 30, 2022. No language or study type restrictions were applied for the search. The combinations of following Medical Subject Heading terms or keywords were used: ‘Breast Neoplasms’ AND ‘HER2-positive’ OR ‘ErbB2-positive’ AND ‘PI3K’ OR ‘PIK3CA’ AND ‘mutation’ OR ‘mutated’ AND ‘prognostic’ OR ‘prognosis’ OR ‘prediction’ OR ‘outcome’ OR ‘response’ OR ‘survival’ OR ‘death’. The detailed search strategy is provided in Supplementary Table S1. Conference abstracts from the American Society of Clinical Oncology and the San Antonio Breast Cancer Symposium were also carefully reviewed to identify unpublished studies. Additionally, the references of the selected studies, relevant meta-analyses or reviews were further manually scrutinized to ensure completeness.
Selection criteria
Two investigators (HZC and XBH) independently performed the search and reviewed the list of retrieved records to select potentially eligible articles. In case of disagreements, the study was discussed, and a consensus was reached with all investigators. Full-text publications or unpublished abstracts of original prospective or retrospective studies were included. To be eligible, studies had to have data available for assessing the association of PIK3CA mutation with outcome measures in patients with HER2-positive breast cancer treated with anti-HER2 therapy. There were no restrictions on specific disease stages or treatment settings.
Studies evaluating the effect of PIK3CA mutation on other molecular subtypes of breast cancer rather than HER2-positive tumors were excluded. Studies that enrolled patients with HER2-positive breast cancer but not involved anti-HER2 therapy were also excluded. Reviews, letters, comments, case reports, study protocols, preclinical or animal studies, and articles not written in English were excluded. In case of studies with overlapping patient populations, only the most recent and complete study was included.
Study endpoints and definitions
Studies should have one of the following outcome measures: (1) pCR, or disease-free survival (DFS) for the neoadjuvant setting; (2) DFS, or invasive DFS (iDFS) for the adjuvant setting; (3) objective response rate (ORR), progression-free survival (PFS), time-to-progression (TTP), or overall survival (OS) for the metastatic setting. The definitions of study endpoints depended on each study. Generally, pCR was defined as no invasive and no non-invasive residuals in breast and lymph nodes (ypT0 ypN0). DFS was defined as the time from randomization of neoadjuvant therapy [8] or the date of diagnosis [20] (for the neoadjuvant setting), or the time from randomization of adjuvant therapy (for the adjuvant setting) [11,13] to disease recurrence (local or distant), contralateral breast cancer, secondary malignancy or death due to any cause. iDFS was defined as the time from surgery until invasive breast cancer recurrence, secondary malignancy or death due to any cause. PFS was defined as the time from randomization to the first documented progressive disease or death resulting from any cause. TTP was defined as the time from randomization to the first documented progressive disease. OS was defined as the time from randomization to death due to any cause.
Data extraction and quality assessment
Two investigators (HZC and XBH) independently extracted the data. For each eligible study, the following data were extracted: the first author and year of publication, country where the study was conducted, study design, treatment setting, treatment, number of patients with PIK3CA mutation and those with wild-type PIK3CA, and outcome measures stratified by the status of PIK3CA mutation. The pCR rate and ORR were derived from studies, separately for PIK3CA mutated and PIK3CA wild-type patients. For time-to-event outcomes, including DFS, iDFS, PFS, TTP and OS, hazard ratios (HRs) with the corresponding 95% confidence intervals (CIs) for patients with PIK3CA mutation versus those with wild-type PIK3CA should be extracted. When HRs from both univariate and multivariate analyses were available, results from the multivariate analysis were preferred. When the HR for time-to-event outcomes was not provided, it was estimated from the Kaplan-Meier curves based on the approach by Tierney [21]. The quality of included studies was assessed by two investigators (HZC and DQW) using the Newcastle-Ottawa scale (NOS) [22].
Bioinformatic analysis of TCGA‑BRCA data
Somatic mutation data for breast cancer were downloading from The Cancer Genome Atlas (TCGA) Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/), and related clinical data were extracted from cBioPortal (https://www.cbioportal.org/). A total of 160 HER2-positive breast cancer patients with PIK3CA mutation data were extracted for further analysis, comprising 48 PIK3CA mutated patients and 112 wild-type patients. The somatic mutations were visualized using the R maftools package. Somatic alterations in ten canonical signaling pathways, including Notch, Hippo, cell cycle, MYC, WNT, TP53, PI3K, TGFβ, NRF2 and RTK/RAS [23], were compared between PIK3CA mutated patients and wild-type patients.
Statistical analysis
The associations of PIK3CA with pCR rate and ORR were assessed by pooled estimates of the odds ratio (OR) and associated 95% CIs using the Mantel-Haenszel fixed-effects model [24] or the DerSimonian-Laird random-effects model [25]. For time-to-event outcomes, the pooled HRs with 95%CIs were calculated. Heterogeneity between studies was estimated by the Cochran's Q-test and I2 statistic. Heterogeneity was classified as low (I2<25%), moderate (25%≤I2<50%), and high (I2≥50) [26]. When the heterogeneity was less than 50%, the fixed-effects model was applied to pool the results; otherwise, the random-effects model was used. To assess the stability and consistency of the pooled results, sensitivity analyses were conducted using a leave-one-out approach. The publication bias was evaluated using visual inspection of funnel plots, as well as the Begg's and Egger's tests [27,28]. All reported p-values are two-sized, with a p-value of <0.05 indicating statistically significant. All statistical analyses were done with Stata version 15.0 (Stata Corporation, College Station, TX, USA).
Results
Literature search results
A total of 961 publications were retrieved through database searching, while an additional 20 records were identified from other sources. After removing duplicated records, 772 studies were retained. Following the review of titles and abstracts, 664 publications were excluded. Upon assessing the full text of articles, 42 studies fulfilled the inclusion criteria. Additionally, one recently published study that met the inclusion criteria was included after the initial database searching [29]. Consequently, 43 studies were included in this meta-analysis, consisting of 41 publications [8,9,11,[13], [14], [15], [16], [17], [18],20,[29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46], [47], [48], [49], [50], [51], [52], [53], [54], [55], [56], [57], [58], [59]]and two abstracts [60,61] (Fig. 1).
Fig. 1.
Flow diagram of study selection. HER2, human epidermal growth factor receptor 2.
Characteristics of identified studies and quality assessment
The baseline characteristics of the included studies are presented in Table 1. Of the 43 studies, 20 studies enrolled patients treated with anti-HER2 therapies in the neoadjuvant setting [8,9,20,29,30,[32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42],[45], [46], [47],60], with five studies in the adjuvant setting [11,13,48,49,61], 14 in the metastatic setting [[14], [15], [16], [17], [18],50,[52], [53], [54], [55], [56], [57], [58], [59]], one study in both the neoadjuvant and adjuvant settings [31], and three in neoadjuvant, adjuvant and metastatic settings [43,44,51]. Most of these studies were exploratory or post-hoc analyses of prospective clinical trials. Notably, the study by Loibl et al. published in 2016 was a pooled analysis using individual patient data from five prospectively randomized neoadjuvant trials that assessed the effect of PIK3CA status on pCR [8], including the GeparQuattro [62], GeparQuinto (NCT00288002), GeparSixto [63], NeoALTTO [64], and CHERLOB [65] trials. Due to overlapping patient populations, these five studies were not included in this meta-analysis. All included studies were published between 2011 and 2022, covering 11,099 patients with available PIK3CA mutation data. Nine studies were conducted at a single center [20,29,31,32,37,40,[43], [44], [45]], and the remaining 34 were muti-center studies. The majority of studies employed anti-HER2 therapy in combination with chemotherapy or endocrine therapy, except for two studies [57,58], and one treatment arm within other five studies [30,34,54,59,60]. Regarding anti-HER2 agents, 24 studies utilized single-agent anti-HER2 therapy with trastuzumab, lapatinib, neratinib or trastuzumab emtansine (T-DM1) [11,13,[15], [16], [17], [18],20,31,32,34,35,40,43,46,[48], [49], [50], [51], [52], [53],[55], [56], [57],59]. Conversely, the remaining 19 studies employed single-agent, or dual-agent anti-HER2 therapy involving the combination of trastuzumab and pertuzumab, trastuzumab plus lapatinib, afatinib plus trastuzumab, T-DM1 plus pertuzumab, or pyrotinib plus trastuzumab [8,9,14,29,30,33,[36], [37], [38], [39],41,42,44,45,47,54,58,60,61]. The overall rate of PIK3CA mutation was 36.8%, with the frequency for each study ranging from 12.3% to 55%.
Table 1.
Main characteristics of the studies in primary HER2-positive breast cancer treated with anti-HER2 therapy.
| First Author | Year | Country | Study center | Original study design | Treatment setting | Treatment | Anti-HER2 therapy | No. of PIK3CA MT/WT* | Rate of PIK3CA MT* | End points |
|---|---|---|---|---|---|---|---|---|---|---|
| Bianchini [30] | 2017 | Global | Multicenter | Prospective cohort | Neoadjuvant | T + H vs. T + H + P vs. H + P vs. T + P | H or H + P | 88/185 | 32.2% | pCR |
| Cizkova [31] | 2013 | France | Single center | Prospective phase II, randomized, two arms | Neoadjuvant and adjuvant | A-based Chemo→T+ H (neoadjuvant and adjuvant or only adjuvant) | H | 17/63 | 21.3% | DFS |
| Cocciolone [32] | 2018 | Italy | Single center | Prospective phase I/II, single arm | Neoadjuvant | ddAC→ddT + H | H | 4/17 | 19.0% | pCR, DFS |
| Hanusch [33] | 2015 | Germany | Multicenter | Prospective phase II, single arm | Neoadjuvant | Afatinib + H→Afatinib + T +H→AC + H | T-DM1 or H | 13/48 | 21.3% | pCR |
| Harbeck [34] | 2021 | Germany | Multicenter | Prospective phase II, randomized, three arms | Neoadjuvant | T-DM1 vs. T-DM1 + ET vs. H + ET | H | 31/159 | 16.3% | pCR |
| Huang [35] | 2015 | China | Multicenter | Prospective phase II, randomized, two arms | Neoadjuvant | TCb + H vs. H + AT | H | 30/47 | 39.0% | pCR |
| Irelli [36] | 2022 | Italy | Multicenter | Retrospective | Neoadjuvant | AT + H or AT + H + P | H or H + L | 8/30 | 21.1% | pCR |
| Li [37] | 2021 | China | Single center | Retrospective | Neoadjuvant | TCb + H or TCb + H + L | H, L, or H + L | 22/18 | 55.0% | pCR |
| Loibl [8] | 2016 | Global | Multicenter | Prospective | Neoadjuvant | T-based chemo + H vs. T-based chemo + L vs. T-based chemo + H + L | H + P | 210/757 | 21.7% | pCR, DFS, OS |
| Loibl [38] | 2019 | Germany | Multicenter | Prospective, randomized, two arms | Neoadjuvant | Nab-paclitaxe + H+ P→AC vs. T+ H+ P→AC | H + L | 63/232 | 21.4% | pCR |
| Rimawi [39] | 2018 | USA | Multicenter | Prospective phase II, single arm | Neoadjuvant | H+ L + ET | H | 14/32 | 30.4% | pCR |
| Sueta [40] | 2014 | Japan | Single center | Retrospective | Neoadjuvant | H-based combinations | H or H + L | 7/36 | 16.3% | pCR |
| Toomey [41] | 2017 | Ireland | Multicenter | Prospective phase II, randomized, three arms | Neoadjuvant | TCb + H vs. TCb + L vs. TCb + H + L | H or H + L | 17/52 | 24.6% | pCR |
| Dave [42] | 2011 | USA | Multicenter | Two prospective phase II, single arm | Neoadjuvant | T + H, T + H + L | H + P | 15/47 | 24.6% | pCR |
| Schneeweiss [9] | 2014 | Global | Multicenter | Prospective phase II, randomized, three arms | Neoadjuvant | FAC + H + P vs. FAC→T + H + P vs. TCb + H + P | H | 39/126 | 23.6% | pCR |
| Barbareschi [43] | 2012 | Italy | Single center | Retrospective | Neoadjuvant, adjuvant, and metastatic | H-based combinations | H or H + P | 25/104 | 19.4% | pCR, ORR |
| Kim [44] | 2022 | Korea | Single center | Retrospective | Neoadjuvant, adjuvant, and metastatic | H or H+ P based combinations | Pyrotinib + H | 34/56 | 37.8% | pCR, PFS |
| Yin [45] | 2022 | China | Single center | Prospective phase II, single arm | Neoadjuvant | Pyrotinib + H+ T+ cisplatin | H | 13/40 | 24.5% | pCR |
| Yuan [20] | 2015 | China | Single center | Retrospective | Neoadjuvant | H-based combinations | T-DM1+P, or H + P | 16/25 | 39.0% | pCR, DFS, DDFS |
| Haas [60] | 2017 | Global | Multicenter | Prospective phase III, randomized, two arms | Neoadjuvant | T-DM1 + P vs. TCb + H + P | H | 114/311 | 26.8% | pCR |
| Loibl [46] | 2017 | Global | Multicenter | Prospective phase II, randomized, two arms | Neoadjuvant | Buparlisib + H + T vs. T + H | H, L, or H + L | 8/42 | 16.0% | pCR |
| Carey [47] | 2016 | Global | Multicenter | Prospective phase III, randomized, three arms | Neoadjuvant | T + H vs. T + L vs. T + H + L | Pyrotinib + H | 36/145 | 19.9% | pCR |
| Shi [29] | 2022 | China | Single center | Prospective phase II, single arm | Neoadjuvant | AC + pyrotinib→T+ H + pyrotinib | H | 19/26 | 42.2% | pCR |
| Fountzilas [48] | 2016 | Greece | Multicenter | Retrospective | Adjuvant | AT→CMF, or A→CMF→T, both followed by H | H | 63/214 | 22.7% | DFS |
| Guarneri [13] | 2020 | Italy, Spain | Multicenter | Prospective phase III, randomized, two arms | Adjuvant | AC→T+ H vs. T+ H→FEC | H | 174/629 | 21.7% | DFS |
| Jensen [49] | 2012 | Denmark | Multicenter | Prospective, single arm | Adjuvant | EFC, followed by H | H + P, or T-DM1 + P | 61/176 | 25.7% | iDFS, OS |
| Metzger [61] | 2021 | Global | Multicenter | Prospective phase III, randomized, two arms | Adjuvant | AC→T + H + P vs. AC→T-DM1 + P | H | 525/1251 | 29.6% | iDFS |
| Pogue-Geile [11] | 2015 | Global | Multicenter | Prospective phase III, randomized, two arms | Adjuvant | AC→T+ H vs. AC→T | H or H + P | 166/505 | 24.7% | DFS |
| Baselga [14] | 2014 | Global | Multicenter | Prospective phase III, randomized, two arms | Metastatic, first-line | T + H + P vs. T + H | T-DM1 or L | 176/381 | 31.6% | PFS |
| Baselga [15] | 2016 | Germany | Multicenter | Prospective phase III, randomized, two arms | Metastatic, previously treated | T-DM1 vs. X + L | H | 79/180 | 30.5% | PFS, ORR, OS |
| Gogas [50] | 2016 | Greece | Multicenter | Retrospective | Metastatic, any line | H-based combinations | H | 17/88 | 16.2% | TTP, OS |
| Guo [51] | 2021 | China | Multicenter | Prospective | Neoadjuvant, adjuvant, and metastatic | H-based combinations | H | 174/439 | 28.4% | ORR |
| Kotoula [52] | 2019 | Greece, Australia | Multicenter | Retrospective | Metastatic, first-line | H-based combinations | L | 26/80 | 24.5% | TTP, OS |
| Nishimura [53] | 2017 | Japan | Multicenter | Retrospective | Metastatic, previously treated | X + L | H, T-DM1, or T-DM1 + P |
20/49 | 29.0% | ORR, PFS, OS |
| Perez [54] | 2019 | Global | Multicenter | Prospective phase III, randomized, three arms | Metastatic, first-line | T + H vs. T-DM1 vs. T-DM1 + P | N, L | 263/723 | 26.7% | PFS |
| Saura [16] | 2021 | Spain | Multicenter | Prospective phase III, randomized, two arms | Metastatic, previously treated | X + N vs. L + X | L | 143/277 | 34.0% | PFS |
| Wang [55] | 2011 | China | Multicenter | Prospective, single arm | Metastatic, previously treated | L + X | L | 7/50 | 12.3% | ORR |
| Xu [56] | 2014 | Global | Multicenter | Prospective phase III, randomized, two arms | Metastatic, first-line | T+ L vs. T | L | 65/106 | 38.0% | ORR |
| Xu [18] | 2011 | China | Multicenter | Prospective, single arm | Metastatic, any line | L + X | L or H | 11/27 | 28.9% | ORR, PFS |
| Kim [17] | 2019 | Five Asian countries | Multicenter | Retrospective | Metastatic, any line | L or H-based combinations | T-DM1 + P | 48/106 | 31.2% | PFS |
| Krop [57] | 2012 | USA | Multicenter | Prospective phase II, single arm | Metastatic, previously treated | T-DM1 | T-DM1 + P | 11/49 | 18.3% | ORR |
| Miller [58] | 2014 | Global | Multicenter | Prospective phase IIa, single arm | Metastatic, any line | T-DM1 + P | T-DM1 or H | 12/35 | 25.5% | ORR |
| Kim [59] | 2016 | Global | Multicenter | Prospective phase III, randomized, two arms | Metastatic, previously treated | T-DM1 vs. TPC | H or H + P | 65/187 | 25.8% | PFS |
The number of patients shown here was based on cases with HER2-positive breast cancer receiving anti-HER2 therapy in each study. The number of patients who had breast cancer with other subtypes or did not receive anti-HER2 therapy was not presented here.
Abbreviations: HER2, human epidermal growth factor receptor-2; MT, mutation; WT, wild type; pCR, pathological complete response; DFS, disease-free survival; OS, overall survival; PFS, progression-free survival; ORR, overall response rate; DDFS, distant disease-free survival; iDFS, invasive disease-free survival; TTP, time to progression; H, trastuzumab; L, lapatinib; P, pertuzumab; T, taxanes; A, anthracyclines; C, cyclophosphamide; Cb, carboplatin; F, fluorouracil; M, methotrexate; X, capecitabine; N, neratinib; Chemo, chemotherapy; ET, endocrine therapy; TPC, treatment of physician's choice; dd, dose-dense; T-DM1, trastuzumab emtansine.
The quality scores of included studies according to the NOS were listed in Supplementary Table S2. Two studies had a score of six points, and 18 studies had a score of seven points, indicating a moderate quality. The remaining studies were considered as having high quality, with 13 and 10 studies achieving eight and nine points, respectively.
Effect of PIK3CA mutation on pCR and DFS in the neoadjuvant setting
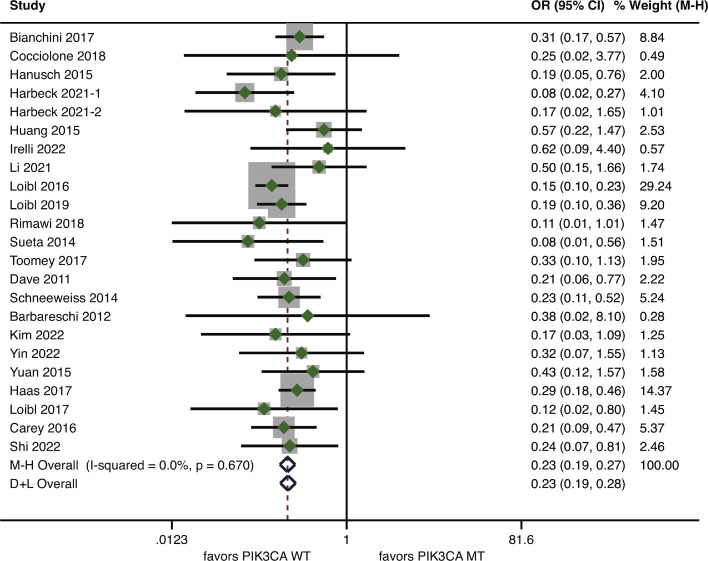
In the neoadjuvant setting, 22 studies covering 3361 patients reported data on pCR rate according to PIK3CA status [8,9,20,[29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46], [47]]. The reported pCR rates varied from 0% to 80% for patients with PIK3CA mutation and from 12.8% to 80.8% for PIK3CA wild-type patients (Supplementary Table S3). According to the fixed-effects model, PIK3CA mutation was significantly associated with a reduced pCR rate, with a pooled OR of 0.23 (95%CI 0.19–0.27, p<0.001) (Fig. 2). There was no evidence of inter-study heterogeneity (I2=0%, p = 0.670).
Fig. 2.
Forest plot of association between PIK3CA mutation and pathological complete response for HER2-positve breast cancer patients treated with anti-HER2 therapy in the neoadjuvant setting. HER2, human epidermal growth factor receptor 2.
Only four studies in the neoadjuvant setting provided DFS data according to PIK3CA status [8,20,31,32]. None of these four studies found a significant association between PIK3CA status and DFS. The pooled analysis found no significant difference in DFS between PIK3CA mutated patients and wild-type patients (HR=1.18, 95%CI 0.85–1.63, p = 0.329; I2=29.0%, p = 0.238) (Supplementary Figure S1).
Effect of PIK3CA mutation on pCR according to anti-HER2 agents
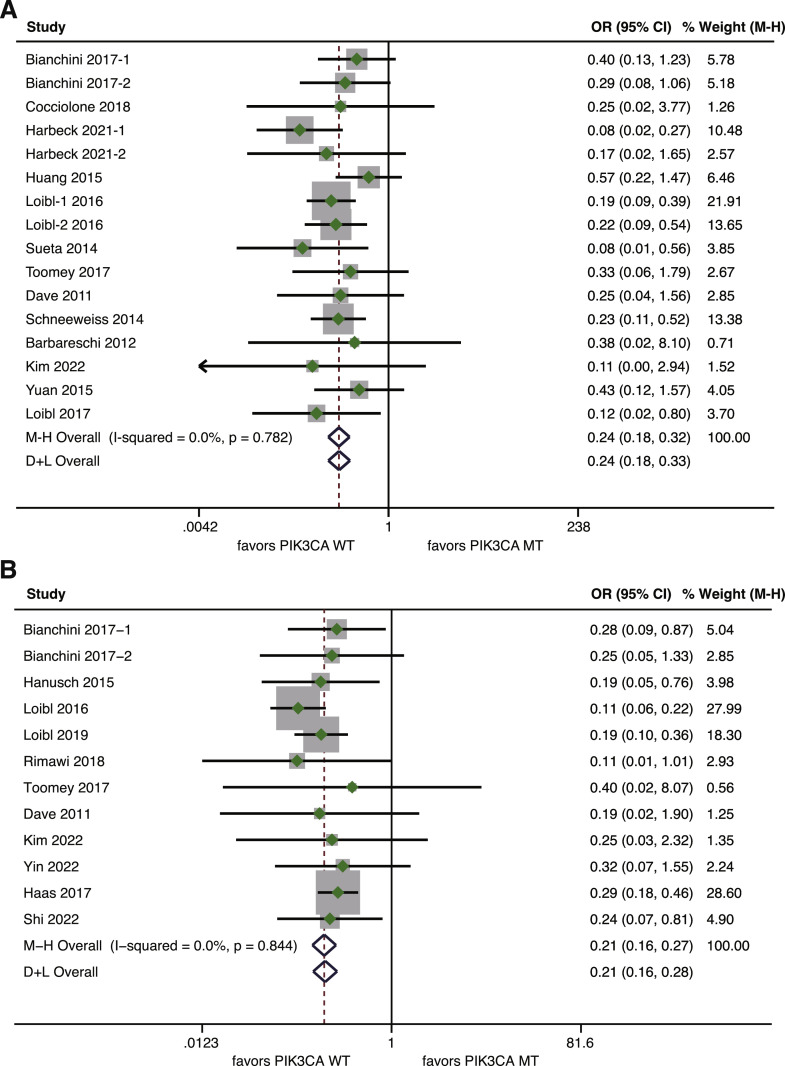
The association between PIK3CA mutation and pCR rate was further assessed in the context of single- or dual-agent anti-HER2 therapy. A total of 13 studies, encompassing 1378 patients treated with single-agent anti-HER2 therapy, had available pCR data according to PIK3CA status [8,9,20,30,32,34,35,[40], [41], [42], [43], [44],46]. Patients harboring PIK3CA mutation exhibited a reduced pCR rate compared to those with wild-type PIK3CA (OR=0.24, 95%CI 0.18–0.32, p<0.001; I2=0%, p = 0.782) (Fig. 3A). As for dual-agent anti-HER2 therapy, data on pCR rate was given in 11 studies involving 1540 patients [8,29,30,33,38,39,41,42,44,45,60]. Likewise, a statistically significant disadvantage in pCR rate was observed for PIK3CA mutated patients compared with wild-type patients (OR=0.21, 95%CI 0.16–0.27, p<0.001; I2=0%, p = 0.844) (Fig. 3B).
Fig. 3.
Forest plot of association between PIK3CA mutation and pCR for HER2-positve breast cancer patients treated with anti-HER2 therapy according to anti-HER2 agents. (A) Association between PIK3CA mutation and pCR for patients receiving neoadjuvant single-agent anti-HER2 therapy; (B) Association between PIK3CA mutation and pCR for patients receiving neoadjuvant dual-agent anti-HER2 therapy. pCR, pathological complete response; HER2, human epidermal growth factor receptor 2.
Effect of PIK3CA mutation on pCR according to the status of hormone receptor
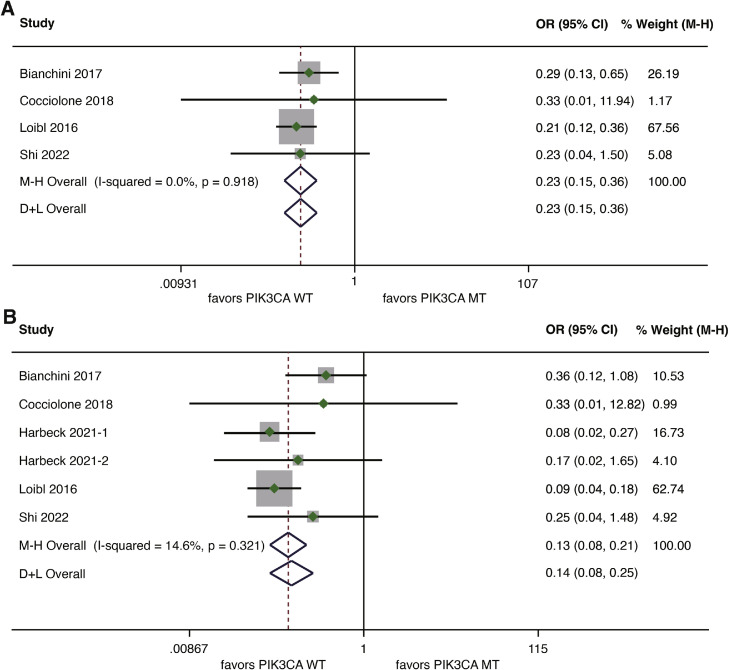
The association between PIK3CA mutation and pCR rate was also examined separately for hormone receptor-negative patients and hormone receptor-positive patients. Data on pCR rate were derived from four studies involving hormone receptor-negative patients [8,29,30,32], and from five studies covering hormone receptor-positive patients [8,29,30,32,34]. Among hormone receptor-negative patients, a significant association between PIK3CA mutation and a lower pCR rate was observed (OR=0.23, 95%CI 0.15–0.36, p<0.001; I2=0%, p = 0.918) (Fig. 4A). For hormone receptor-positive patients, the detrimental effect of PIK3CA mutation on pCR rate was more obvious, with a pooled OR of 0.13 (95%CI 0.08–0.21, p<0.001). No heterogeneity was demonstrated between studies (I2=14.6%, p = 0.321) (Fig. 4B).
Fig. 4.
Forest plot of association between PIK3CA mutation and pCR for HER2-positve breast cancer patients treated with anti-HER2 therapy according to hormone receptor status. (A) Association between PIK3CA mutation and pCR for hormone receptor-negative patients; (B) Association between PIK3CA mutation and pCR for hormone receptor-positive patients. pCR, pathological complete response; HER2, human epidermal growth factor receptor 2.
Effect of PIK3CA mutation on DFS and iDFS in the adjuvant setting
A total of four studies were available for assessing the effects of PIK3CA mutation on DFS in the adjuvant setting [11,13,31,48]. There was no significant difference in DFS between patients with PIK3CA mutation and those with wild-type PIK3CA (HR=0.84, 95%CI 0.66–1.08, p = 0.179; I2=26.5%, p = 0.245) (Supplementary Figure S2A). Only two studies evaluated the effect of PIK3CA mutation on iDFS [49,66]. No statistical association between PIK3CA status and iDFS was noted, with a pooled HR of 1.24 (95%CI 0.93–1.65, p = 0.141) and no evidence of heterogeneity (I2=0%, p = 0.732) (Supplementary Fig. S2B).
Effect of PIK3CA mutation on response and survival outcomes in the metastatic setting
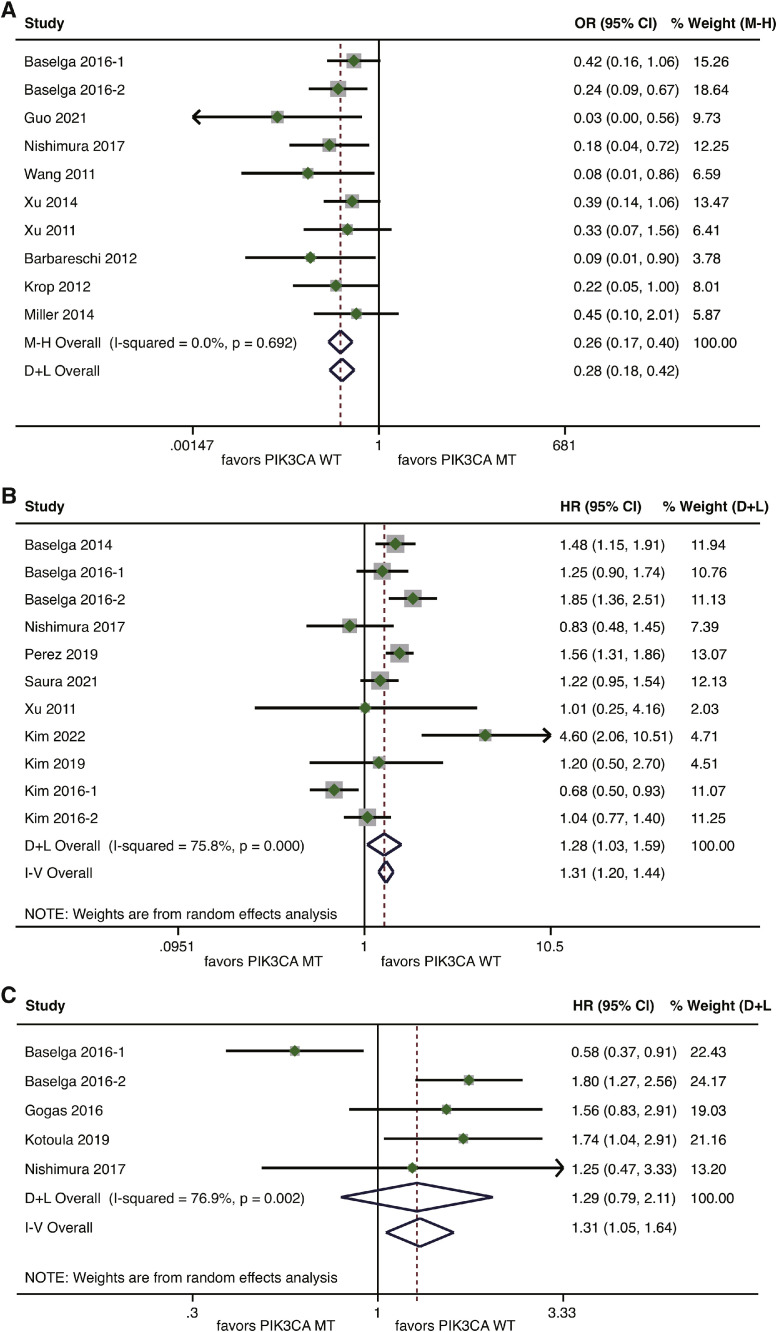
The analysis for ORR in the metastatic setting included nine studies covering 638 patients [15,18,43,51,53,[55], [56], [57], [58]]. The pooled analysis demonstrated that PIK3CA mutation was significantly associated with inferior ORR compared with wild-type PIK3CA (OR=0.26, 95%CI 0.17–0.40, p<0.001), without heterogeneity (I2=0%, p = 0.692) (Fig. 5A).
Fig. 5.
Forest plot of association between PIK3CA mutation and outcome measures for HER2-positve breast cancer patients treated with anti-HER2 therapy in the metastatic setting. (A) Association between PIK3CA mutation and ORR; (B) Association between PIK3CA mutation and PFS; (C) Association between PIK3CA mutation and OS. HER2, human epidermal growth factor receptor 2; ORR, objective response rate; PFS, progression-free survival; OS, overall survival.
For the meta-analysis of PFS, nine studies were included [[14], [15], [16], [17], [18],44,[53], [54],59]. Based on a random-effects model, patients with mutated PIK3CA had a significantly worse PFS than those harboring wild-type PIK3CA (HR=1.28, 95%CI 1.03–1.59, p = 0.024). There was a high level of heterogeneity (I2=75.8%, p<0.001) (Fig. 5B). Data on TTP according to the status of PIK3CA were available for only two studies [50,52]. Compared with wild-type PIK3CK, PIK3CK mutation was significantly associated with a higher risk of disease progression (HR=2.27, 95%CI 1.54–3.34, p<0.001; I2=0%, p = 0.864) (Supplementary Figure S3). As for the analysis of OS, data for comparison between mutated and wild-type PIK3CA were derived from four studies [15,50,52,53]. Compared with wild-type PIK3CA, PIK3CA mutation seemed to be associated with a higher risk of death, but without statistical significance according to a random-effects model (HR=1.29, 95%CI 0.79–2.11, p = 0.315; I2=76.9%, p = 0.002) (Fig. 5C).
Sensitivity analysis and publication bias
Sensitivity analyses were conducted to examine the influence of individual studies on pooled results by excluding one study at each time. For the pooled analysis of OS in the metastatic setting, the overall results would be altered after removing a sub-cohort (the T-DM1 arm) of the study by Baselga et al. which might contribute to the observed high heterogeneity [15](Supplementary Figure S4). After removing this, the pooled HR for OS comparing PIK3CA mutation versus wild type was 1.70 (95%CI 1.32–2.20, p<0.001), and there was no evidence of heterogeneity (I2=0%, p = 0.903) (Supplementary Figure S5). For other pooled analyses, the overall results were not significantly changed after excluding each study, demonstrating the stability and consistency of the pooled results (Supplementary Figure S6-S14).
The funnel plots for effect sizes in all pooled analyses, except for the HR for DFS in the neoadjuvant setting and the OR for ORR, were symmetric (Supplementary Figure S15-S24). According to the results of Begg's and Egger's test, there was no publication bias in all meta-analyses with the exception of the pooled analysis of DFS in the neoadjuvant setting (p = 0.734 for Begg's test, and p = 0.020 for egger's test), and ORR (p = 0.107 for Begg's test, and p = 0.034 for egger's test) which suggested there might be a potential publication bias (Supplementary Table S4).
Mutational landscape of HER2-positive breast cancer according to PIK3CA mutation status
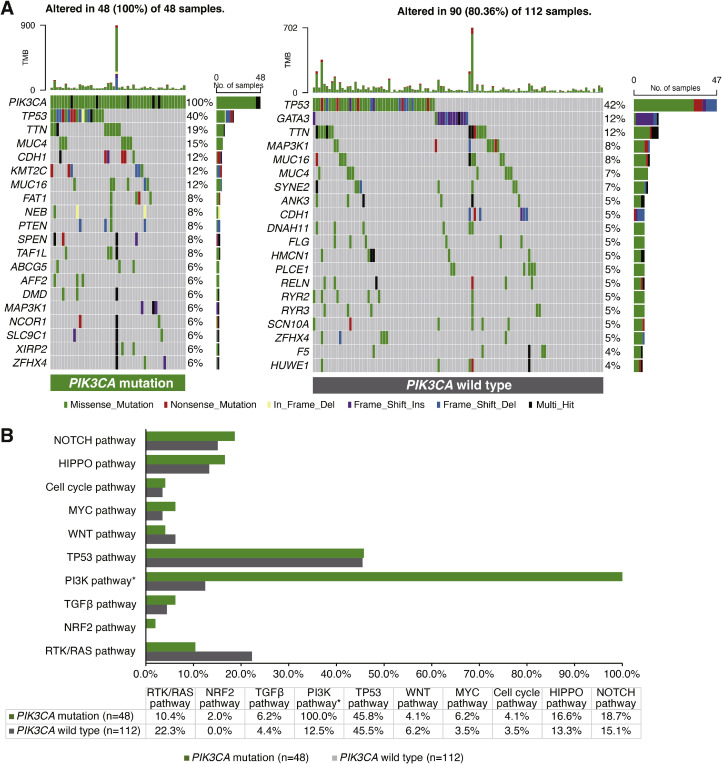
The mutational profile for 160 patients with HER2-positive breast cancer from TCGA was analyzed according to PIK3CA mutation status. The baseline characteristics for these patients are shown in Supplementary Table S5. Among 48 patients with PIK3CA mutation, the most common variation was TP53 mutation (40%), followed by mutations in TTN (19%), MUC4 (15%), CDH1 (12%), KMT2C (12%), MUC16 (12%), FAT1 (8%), NEB (8%), PTEN (8%), SPEN (8%) and TAF1L (8%) (Fig. 6A). Among 112 patients with wild-type PIK3CA, the top-ranking mutated genes were TP53 (42%), GATA (12%), TTN (12%), MAP3K1 (8%), MUC16 (8%), MUC4 (7%) and SYNE2 (7%) (Fig. 6A). Undoubtedly, the mutational frequencies in the PI3K pathway were significantly higher in the PIK3CA mutated group than those in the PIK3CA wild-type group (100% vs. 12.5%, P<0.001) (Fig. 6B). However, there was a tendency towards lower mutational frequencies in the RTK/RAS pathway in the PIK3CA mutated group compared to the wild-type group (10.4% vs. 22.3%, P = 0.122) (Fig. 6B).
Fig. 6.
Somatic mutations and signaling pathways in HER2-positive breast cancer from TCGA dataset. (A) The oncoprint of somatic mutations for PIK3CA mutated patients (left side, n = 48) and that for PIK3CA wild-type patients (right side, n = 112); (B) The bar-graph showing comparisons of ten canonical signaling pathways between PIK3CA mutated patients and PIK3CA wild-type patients. An asterisk (*) indicates the significant difference in mutational frequencies of the signaling pathway between PIK3CA mutated and wild-type patients. HER2, human epidermal growth factor receptor 2; TCGA, The Cancer Genome Atlas.
Discussion
This study demonstrated that PIK3CA mutation was associated with a lower pCR rate in HER2-positive breast cancer patients undergoing neoadjuvant anti-HER2 therapy. The detrimental impact of PIK3CA mutation on pCR remained consistent regardless of whether single-agent or dual-agent anti-HER2 therapy was administered. The deleterious effect of PIK3CA mutation on the pCR rate was particularly pronounced in hormone receptor-positive patients. However, there was no significant difference in DFS between patients with PIK3CA mutation and those with wild type, neither in the neoadjuvant nor the adjuvant setting. In the metastatic setting, PIK3CA mutation predicted for worse ORR, PFS and TTP, but not for OS. In addition, the bioinformatic analysis of TCGA breast cancer data revealed distinct mutational landscapes between PIK3CA mutated and wild-type HER2-positive breast cancer. To the best of our knowledge, this study represents the largest and most comprehensive meta-analysis assessing the effect of PIK3CA mutation on clinical outcomes in patients with HER2-positive breast cancer undergoing anti-HER2 therapy.
The results of current study were partly consistent with a previous meta-analysis by Ibrahim et al., which also examined the predictive and prognostic utility of PIK3CA mutation in HER2-postive breast cancer receiving anti-HER2 therapy [67]. However, pooled analyses for several outcome measures were not done in that study, including DFS for mutated versus wild-type PIK3CA in the adjuvant setting, and OS for mutated versus wild-type PIK3CA in the metastatic setting. In addition, the predictive effect of PIK3CA on pCR rates when taking consideration into single- or dual-agent anti-HER2 therapy and hormone receptor status was not addressed in that study, owing to limited number of included studies. We updated the systematic research and included additional studies that have been published since the last date for inclusion of the study by Ibrahim et al. [67]. This may largely explain the conflicting results regarding the association of PIK3CA mutation with ORR and PFS observed between our study and that study [67]. The present meta-analysis, with a much larger sample size, provides robust statistical evidence supporting the predictive and prognostic value of PIK3CA mutation in HER2-positive breast cancer patients receiving anti-HER2 therapy.
Aberrant activation of the PI3K/AKT/mTOR pathway, primarily driven by mutations in PIK3CA or loss of PTEN expression, leads to constitutive pathway activation downstream of HER2, which can result in the decreased sensitivity of anti-HER2 therapies [7]. Mutations in PI3K can enhance HER2-mediated transformation through promoting heregulin production and activating HER3 [68]. A preclinical study showed that PIK3CA accelerated HER2-mediated breast epithelial transformation and metastatic progression [69]. Moreover, PIK3CA mutations modified the intrinsic phenotype of HER2-overexpressing cancers, and induced resistance to anti-HER2 therapies [69]. HER2 and mutant PI3K collaborate to promote the establishment and metastatic progression of mammary tumors. Consequently, PIK3CA mutations may partly contribute to resistance to HER2-targeted agents.
This study demonstrated a significant association between PIK3CA mutation and a lower pCR rate, even in patients treated with dual-agent anti-HER2 therapy as neoadjuvant treatment. This finding further supports preclinical evidence that activating PIK3CA mutation may mediate resistance to trastuzumab alone, and in combination with lapatinib or pertuzumab [69], or lapatinib alone [70]. In contrast, T-DM1 exhibited potent activity in cell lines and xenograft models with PIK3CA mutation, probably due to its cytotoxic activity [15]. However, this observation does not align conclusively with the clinical findings from previous studies. In the ADAPT trial, a lower pCR rate (21.1% vs. 48.1%, p = 0.04) was observed in PIK3CA mutated patients than wild-type patients when treated with T-DM1-based therapy [34]. Similarly, in the KRISTINE trial, PIK3CA mutation was associated with numerically lower pCR rates (31.1% vs. 51.0%) in patients receiving T-DM1 plus pertuzumab [60]. These results suggest that PIK3CA mutation may be also responsible for resistance to T-DM1. However, the results in the metastatic setting appeared to be in contrast with data from the neoadjuvant setting. In the EMILIA trial, PIK3CA mutated patients had shorter median PFS and OS for the capecitabine plus lapatinib arm, but not for the T-DM1 arm, suggesting the activity of T-DM1 against PIK3CA-mutated metastatic breast cancer [15]. Likewise, the TH3RESA trial showed that PFS benefit was obtained with T-DM1 versus treatment of physician's choice irrespective of PIK3CA status [59]. Due to limited number of included studies, we were unable to perform pooled analyses specifically for T-DM1 treatment. It would be valuable to further assess the predictive effect of PIK3CA mutation on efficacy of anti-HER2 antibody-drug conjugates like T-DM1 or other novel drugs.
Although PIK3CA mutation was linked with a lower pCR rate after chemotherapy and anti-HER2 therapy, it was not the case for DFS. Moreover, the present study demonstrated that there was no association between PIK3CA mutation and DFS in the adjuvant setting, though this finding was not very conclusive owing to the small number of studies included. The NSABP B-31 and FinHER trials demonstrated that PIK3CA mutation could not predict reduced benefit from adjuvant trastuzumab [10,11]. Similarly, the ExteNET trial failed to identify PIK3CA alteration as a predictive marker of response to adjuvant neratinib in HER2-positive breast cancer [12]. Given these results, there may be a discrepancy in the relevance of PIK3CA mutation with pCR and long-term survival like DFS. However, the underlying mechanism for this discrepancy remains unclear. One possible explanation may be the hypothesis that there is a distinct role of PIK3CA mutation between macroscopic (neoadjuvant and metastatic) and microscopic disease [13]. Additionally, the association of PIK3CA with DFS may vary across molecular intrinsic subtypes, and PIK3CA mutation showed a favorable prognostic impact on DFS in the PAM50 HER2-enriched subtype [13]. More investigations are needed to assess the role of PIK3CA mutation in the adjuvant setting for HER2-positive breast cancer.
This study demonstrates PIK3CA mutation may identify a subset of patients who are resistant or have a worse prognosis when treated with anti-HER2 therapies. These results have clinical implications suggesting that combining anti-HER2 therapy with PI3K inhibitors may be a better treatment option for HER2-positive patients carrying PIK3CA mutation. Preclinical data have shown anti-HER2 drug resistance induced by PI3K may be partially reversed by the addition of PI3K inhibitors [69,70]. Encouragingly, clinical development of new drugs targeting the PI3K pathway is emerging. Several clinical trials investigating anti-HER2 therapy in combination with PIK3 inhibitors for patients with HER2-positive breast cancer are ongoing or have been completed (Table 2). Preliminary clinical activity has been shown for these combinations, however, it has not been determined from these studies whether this activity is confined to patients with PIK3CA mutation or PIK3 activation (Supplementary Table S5). In the NeoPHOEBE trial, no significant difference in pCR rates were observed between the neoadjuvant buparlisib plus trastuzumab and paclitaxel arm and the placebo plus trastuzumab and paclitaxel arm in HER2-positive breast cancer [46]. The small number of patients with PIK3CA mutation (n = 8) hindered the ability to detect differences in pCR rates between the PIK3CA mutated cohort and the wild-type cohort [46]. The BOLERO-1 and BOLERO-3 trials evaluated the addition of everolimus to trastuzumab and chemotherapy in advanced HER2-positive breast cancer [71,72]. Pooled exploratory biomarker analysis of these two trials found that PFS benefit from the addition of everolimus was only confined to patients harboring PIK3CA mutation, PTEN loss, or hyperactive PI3K pathway [73]. These results indicate a potential role for PIK3 inhibitors in HER2-positive breast cancer, and meantime, highlight the importance of identifying predictive biomarkers.
Table 2.
Clinical trials of PI3K inhibitors combined with anti-HER2 therapy in HER2-positive breast cancer.
| Identifier | Study center | Study design | Treatment setting | Treatment | Biomarker selected* | No. | Status |
|---|---|---|---|---|---|---|---|
| NCT04208178 | Multicenter | Part 1: Open-label, safety run-in part; Part 2: Randomized, double-blind, placebo-controlled, phase III | Metastatic, maintenance therapy | Part 1: Alpelisibz + H + P; Part 2: Alpelisib + H + P vs. Placebo + H + P | PIK3CA MT | 511 | Recruiting |
| NCT05306041 | Multicenter | Randomized, open-label, phase II | Neoadjuvant | Inavolisib + H+ P + ET vs. H+ P + ET | PIK3CA MT | 170 | Not yet recruiting |
| NCT04108858 | Multicenter | Randomized, open-label, phase Ib/II | Metastatic, maintenance therapy | Phase Ib: Copanlisib + H + P; Phase II: Copanlisib + H + P vs. H + P | PI3K-activated | 12 | Recruiting |
| NCT05230810 | Multicenter | Single arm, open-label, phase Ib/II | Metastatic | Alpelisib + tucatinib ± fulvestrant | PIK3CA MT | 40 | Recruiting |
| NCT01816594 | Multicenter | Randomized, double-blind, placebo-controlled, phase II | Neoadjuvant | Buparlisib (BKM120) + H + paclitaxel vs. Placebo + H + paclitaxel | Unselected | 50 | Completed |
| NCT05063786 | Multicenter | Randomized, open-label, phase II | Metastatic, previously treated | Alpelisib + H ± fulvestrant vs.H + CT | PIK3CA MT | 300 | Recruiting |
| NCT02947685 | Multicenter | Randomized, open-label, phase III | Metastatic, maintenance therapy | Palbociclib + H+ P + ET vs. H+ P + ET | Unselected | 496 | Active, not recruiting |
| NCT01589861 | Multicenter | Single arm, open-label, phase Ib/II | Metastatic, previously treated | Buparlisib (BKM120) + lapatinib | PI3K-activated | 106 | Suspended |
| NCT04253561 | Multicenter | Single arm, open-label, phase Ib | Metastatic, maintenance therapy | Ipatasertib + H + P | PIK3CA MT | 25 | Recruiting |
| NCT03767335 | Multicenter | Open-label, dose-escalation, phase Ib | Metastatic, previously treated | MEN1611 + H ± Fulvestrant | PIK3CA MT | 62 | Active, not recruiting |
| NCT02038010 | Multicenter | Single arm, open-label, phase I | Metastatic, previously treated | Alpelisib (BYL719) + T-DM1 | Unselected | 17 | Completed |
| NCT01132664 | Multicenter | Single arm, open-label, phase Ib/IIa | Metastatic, previously treated | Buparlisib (BKM120) + H ± capecitabine | Unselected | 72 | Completed |
| NCT03765983 | Single center | Single arm, open-label, phase II | Metastatic, previously treated | GDC-0084 + H | Unselected | 47 | Recruiting |
| NCT01471847 | Multicenter | Phase Ib: Single arm, open-label; Phase II: Randomized, open-label | Metastatic, previously treated | Phase Ib: BEZ235 + H; Phase II: BEZ235 + H vs. Lapatinib + capecitabine | Unselected | 5 | Completed |
| NCT01042925 | Multicenter | Non-randomized, open-label, phase 1/2 | Metastatic, previously treated | Pilaralisib (XL147) + H (arm 1), or pilaralisib + H + paclitaxel (arm 2) | Unselected | 42 | Completed |
| NCT00736970 | Multicenter | Single arm, open-label, phase II | Metastatic, previously treated | Ridaforolimus + H | Unselected | 34 | Completed |
| NCT04736589 | Multicenter | Randomized, open-label, phase III | Metastatic, previously treated | Inetetamab + rapamycin + CT vs. pyrotinib + CT | PI3K-activated | 270 | Not yet recruiting |
| NCT00876395 | Multicenter | Randomized, double-blind, placebo-controlled, phase II | Metastatic, first-line | Everolimus + paclitaxel + H vs. Placebo + paclitaxel + H | Unselected | 719 | Completed |
| NCT01007942 | Multicenter | Randomized, double-blind, placebo-controlled, phase II | Metastatic, previously treated | Everolimus + vinorelbine + H vs. Placebo + vinorelbine + H | Unselected | 569 | Completed |
| NCT02705859 | Multicenter | Single arm, open-label, phase Ib/II | Metastatic, previously treated | Copanlisib + H | Unselected | 26 | Completed |
| NCT01305941 | Multicenter | Single-arm, open-label phase II | Metastatic, previously treated | Everolimus + H + vinorelbine | Unselected | 32 | Completed |
| NCT00674414 | Multicenter | Randomized, open-label, phase II | Neoadjuvant | H + everolimus vs. H | Unselected | 82 | Completed |
This refers to whether the study included PIK3CA mutation or PI3K pathway activation as a biomarker for patient selection at enrollment.
Abbreviations: PI3K, phosphoinositide 3-kinase; HER2, human epidermal growth factor receptor; MT, mutation; H, trastuzumab; P, pertuzumab; ET, endocrine therapy; CT, chemotherapy; T-DM1, trastuzumab emtansine; No., Number.
Several limitations should be acknowledged. Firstly, the anti-HER2 agents administrated in each study and methods of assessment of PIK3CA mutation were non-uniform, which may lead to bias. Secondly, the number of included studies for several pooled analyses is small, which limited the power of statistical analysis. For example, only four studies were available for DFS analysis in both the neoadjuvant and adjuvant settings, and only four studies were included for OS analysis in the metastatic setting. Therefore, validation with larger sample sizes is needed. Additionally, due to limited data, we did not perform subgroup analyses according to PIK3CA exons of mutation (exon 9 vs. exon 20). Therefore, whether different PIK3CA exons of mutation render differential predictive and prognostic effect remains unknown. Moreover, although we examined the mutational landscape according to PIK3CA mutation status, the potential impact of these differences on the clinical relevance of PIK3CA remains unknown. The molecular mechanisms underlying the observed associations between PIK3CA mutation and clinical outcomes were not investigated in this study. Despite these limitations, the large number of samples enabled us to comprehensively explore the predictive and prognostic relevance of PIK3CA status across different treatment settings. These results support the potential clinical importance of PIK3CA assessment for patients with HER2-positive breast cancer, providing a rationale for investigation of PI3K inhibitors in this subset of patients.
Conclusions
In conclusion, this study reveals a significant association between PIK3CA mutation and a lower pCR rate in HER2-positive breast cancer patients treated with neoadjuvant anti-HER2 therapy. This association remained significant irrespective of the type of anti-HER2 therapy (single-agent or dual-agent) and hormone receptor status. In the metastatic setting, PIK3CA mutation was associated with worse clinical outcomes in terms of ORR, PFS and TTP, whereas it was not predictive of OS. Distinct mutational landscapes were observed in HER2-positive breast cancer between individuals with PIK3CA mutations and those with wild-type PIK3CA. These results suggest the potential clinical importance of PIK3CA mutation status assessment for patients with HER2-positive breast cancer, and there is an opportunity to develop PI3K inhibitors for these patients. Further studies examining the molecular mechanisms underlying the associations between PIK3CA mutation and clinical outcomes in HER2-positive patients are warranted.
Ethics approval and consent to participate
Not applicable because this work was a meta-analysis.
Consent for publication
Not applicable.
Availability of data and materials
This study used publicly available data from published studies. All data and material analyzed during this study are included in this article.
Authors’ contributions
HRY, YFY, HZC and XBH contributed to the study design. HZC and XBH performed the systematic search. HZC, XBH, DQW and YW contributed to selection of eligible studies, data extraction and assessment of study quality. HZC and XBH did the statistical analysis, interpreted the data, and drafted the manuscript. HRY and YFY interpreted the data and revised the manuscript. All authors have reviewed and approved the final version of the manuscript.
Declaration of Competing Interest
All authors declare no potential conflicts of interest.
Acknowledgments
Funding
This study was supported by grants 82273204 and 81972471 from the National Natural Science Foundation of China, grants 2023A1515012412 and 2023A1515011214 GuangDong Basic and Applied Basic Research Foundation, grants 2023A03J072, 202206010078 and 202201020574 from the Guangzhou Science and Technology Project, grant 2018007 from the Sun Yat-Sen University Clinical Research 5010 Program, grant SYS-C-201801 from the Sun Yat-Sen Clinical Research Cultivating Program, grant A2020558 from the Guangdong Medical Science and Technology Program, grant 7670020025 from Tencent Charity Foundation, grant YXQH202209 from the Scientific Research Launch Project of Sun Yat-Sen Memorial Hospital.
Acknowledgements
Not applicable.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2023.101738.
Contributor Information
Yunfang Yu, Email: yuyf9@mail.sysu.edu.cn.
Herui Yao, Email: yaoherui@mail.sysu.edu.cn.
Appendix. Supplementary materials
References
- 1.Slamon D.J., Clark G.M., Wong S.G., Levin W.J., Ullrich A., McGuire W.L. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/NEU oncogene. Science. 1987;235(4785):177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 2.Loibl S., Gianni L. HER2-positive breast cancer. Lancet. 2017;389(10087):2415–2429. doi: 10.1016/S0140-6736(16)32417-5. [DOI] [PubMed] [Google Scholar]
- 3.Liu P., Cheng H., Santiago S., Raeder M., Zhang F., Isabella A., Yang J., Semaan D.J., Chen C., Fox E.A., et al. Oncogenic PIK3CA-driven mammary tumors frequently recur via PI3K pathway-dependent and PI3K pathway-independent mechanisms. Nat. Med. 2011;17(9):1116–1120. doi: 10.1038/nm.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer genome atlas N: comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zardavas D., Te Marvelde L., Milne R.L., Fumagalli D., Fountzilas G., Kotoula V., Razis E., Papaxoinis G., Joensuu H., Moynahan M.E., et al. Tumor PIK3CA genotype and prognosis in early-stage breast cancer: a pooled analysis of individual patient data. J. Clin. Oncol. 2018;36(10):981–990. doi: 10.1200/JCO.2017.74.8301. [DOI] [PubMed] [Google Scholar]
- 6.Andre F., Ciruelos E., Rubovszky G., Campone M., Loibl S., Rugo H.S., Iwata H., Conte P., Mayer I.A., Kaufman B., et al. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N Engl. J. Med. 2019;380(20):1929–1940. doi: 10.1056/NEJMoa1813904. [DOI] [PubMed] [Google Scholar]
- 7.Berns K., Horlings H.M., Hennessy B.T., Madiredjo M., Hijmans E.M., Beelen K., Linn S.C., Gonzalez-Angulo A.M., Stemke-Hale K., Hauptmann M., et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12(4):395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 8.Loibl S., Majewski I., Guarneri V., Nekljudova V., Holmes E., Bria E., Denkert C., Schem C., Sotiriou C., Loi S., et al. PIK3CA mutations are associated with reduced pathological complete response rates in primary HER2-positive breast cancer: pooled analysis of 967 patients from five prospective trials investigating lapatinib and trastuzumab. Ann. Oncol. 2016;27(8):1519–1525. doi: 10.1093/annonc/mdw197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schneeweiss A., Chia S., Hegg R., Tausch C., Deb R., Ratnayake J., McNally V., Ross G., Kiermaier A., Cortes J. Evaluating the predictive value of biomarkers for efficacy outcomes in response to pertuzumab- and trastuzumab-based therapy: an exploratory analysis of the TRYPHAENA study. Breast. Cancer Res. 2014;16(4):R73. doi: 10.1186/bcr3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loi S., Michiels S., Lambrechts D., Fumagalli D., Claes B., Kellokumpu-Lehtinen P.L., Bono P., Kataja V., Piccart M.J., Joensuu H., et al. Somatic mutation profiling and associations with prognosis and trastuzumab benefit in early breast cancer. J. Natl. Cancer Inst. 2013;105(13):960–967. doi: 10.1093/jnci/djt121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pogue-Geile K.L., Song N., Jeong J.H., Gavin P.G., Kim S.R., Blackmon N.L., Finnigan M., Rastogi P., Fehrenbacher L., Mamounas E.P., et al. Intrinsic subtypes, PIK3CA mutation, and the degree of benefit from adjuvant trastuzumab in the NSABP B-31 trial. J. Clin. Oncol. 2015;33(12):1340–1347. doi: 10.1200/JCO.2014.56.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chia S.K.L., Martin M., Holmes F.A., Ejlertsen B., Delaloge S., Moy B., Iwata H., von Minckwitz G., Mansi J., Barrios C.H., et al. PIK3CA alterations and benefit with neratinib: analysis from the randomized, double-blind, placebo-controlled, phase III ExteNET trial. Breast. Cancer Res. 2019;21(1):39. doi: 10.1186/s13058-019-1115-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guarneri V., Dieci M.V., Bisagni G., Brandes A.A., Frassoldati A., Cavanna L., Musolino A., Giotta F., Rimanti A., Garrone O., et al. PIK3CA mutation in the ShortHER randomized adjuvant trial for patients with early HER2(+) breast cancer: association with prognosis and integration with PAM50 subtype. Clin. Cancer Res. 2020;26(22):5843–5851. doi: 10.1158/1078-0432.CCR-20-1731. [DOI] [PubMed] [Google Scholar]
- 14.Baselga J., Cortés J., Im S.A., Clark E., Ross G., Kiermaier A., Swain S.M. Biomarker analyses in CLEOPATRA: a phase III, placebo-controlled study of pertuzumab in human epidermal growth factor receptor 2-positive, first-line metastatic breast cancer. J. Clin. Oncol. 2014;32(33):3753–3761. doi: 10.1200/JCO.2013.54.5384. [DOI] [PubMed] [Google Scholar]
- 15.Baselga J., Lewis Phillips G.D., Verma S., Ro J., Huober J., Guardino A.E., Samant M.K., Olsen S., de Haas S.L., Pegram M.D. Relationship between tumor biomarkers and efficacy in EMILIA, a phase III study of trastuzumab emtansine in HER2-positive metastatic breast cancer. Clin. Cancer Res. 2016;22(15):3755–3763. doi: 10.1158/1078-0432.CCR-15-2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saura C., Matito J., Oliveira M., Wildiers H., Brufksy A.M., Waters S.H., Hurvitz S.A., Moy B., Kim S.B., Gradishar W.J., et al. Biomarker analysis of the phase III NALA Study of Neratinib + Capecitabine versus Lapatinib + Capecitabine in patients with previously treated metastatic breast cancer. Clin. Cancer Res. 2021;27(21):5818–5827. doi: 10.1158/1078-0432.CCR-21-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim S.B., Do I.G., Tsang J., Kim T.Y., Yap Y.S., Cornelio G., Gong G., Paik S., Lee S., Ng T.Y., et al. BioPATH: a biomarker study in asian patients with HER2+ advanced breast cancer treated with lapatinib and other Anti-HER2 Therapy. Cancer Res. Treat. 2019;51(4):1527–1539. doi: 10.4143/crt.2018.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu B.H., Jiang Z.F., Chua D., Shao Z.M., Luo R.C., Wang X.J., Liu D.G., Yeo W., Yu S.Y., Newstat B., et al. Lapatinib plus capecitabine in treating HER2-positive advanced breast cancer: efficacy, safety, and biomarker results from Chinese patients. Chin. J. Cancer. 2011;30(5):327–335. doi: 10.5732/cjc.010.10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moher D., Liberati A., Tetzlaff J., Altman D.G., Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. 2009;339:b2535. doi: 10.1136/bmj.b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan H., Chen J., Liu Y., Ouyang T., Li J., Wang T., Fan Z., Fan T., Lin B., Xie Y. Association of PIK3CA mutation status before and after neoadjuvant chemotherapy with response to chemotherapy in women with breast cancer. Clin. Cancer Res. 2015;21(19):4365–4372. doi: 10.1158/1078-0432.CCR-14-3354. [DOI] [PubMed] [Google Scholar]
- 21.Tierney J.F., Stewart L.A., Ghersi D., Burdett S., Sydes M.R. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur. J. Epidemiol. 2010;25(9):603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 23.Sanchez-Vega F., Mina M., Armenia J., Chatila W.K., Luna A., La K.C., Dimitriadoy S., Liu D.L., Kantheti H.S., Saghafinia S., et al. Oncogenic signaling pathways in the cancer genome atlas. Cell. 2018;173(2):321–337. doi: 10.1016/j.cell.2018.03.035. e310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mantel N., Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J. Natl. Cancer Inst. 1959;22(4):719–748. [PubMed] [Google Scholar]
- 25.DerSimonian R., Laird N. Meta-analysis in clinical trials. Control Clin. Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 26.Higgins J.P., Thompson S.G., Deeks J.J., Altman D.G. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Begg C.B., Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–1101. [PubMed] [Google Scholar]
- 28.Egger M., Davey Smith G., Schneider M., Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi Q., Xuhong J., Luo T., Ge J., Liu F., Lan Y., Chen Q., Tang P., Fan L., Chen L., et al. PIK3CA mutations are associated with pathologic complete response rate to neoadjuvant pyrotinib and trastuzumab plus chemotherapy for HER2-positive breast cancer. Br. J. Cancer. 2022;128(1):121–129. doi: 10.1038/s41416-022-02021-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bianchini G., Kiermaier A., Bianchi G.V., Im Y.H., Pienkowski T., Liu M.C., Tseng L.M., Dowsett M., Zabaglo L., Kirk S., et al. Biomarker analysis of the NeoSphere study: pertuzumab, trastuzumab, and docetaxel versus trastuzumab plus docetaxel, pertuzumab plus trastuzumab, or pertuzumab plus docetaxel for the neoadjuvant treatment of HER2-positive breast cancer. Breast Cancer Res. 2017;19(1):16. doi: 10.1186/s13058-017-0806-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cizkova M., Dujaric M.E., Lehmann-Che J., Scott V., Tembo O., Asselain B., Pierga J.Y., Marty M., de Cremoux P., Spyratos F., et al. Outcome impact of PIK3CA mutations in HER2-positive breast cancer patients treated with trastuzumab. Br. J. Cancer. 2013;108(9):1807–1809. doi: 10.1038/bjc.2013.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cocciolone V., Cannita K., Tessitore A., Mastroiaco V., Rinaldi L., Paradisi S., Irelli A., Baldi P.L., Sidoni T., Ricevuto E., et al. Neoadjuvant chemotherapy in breast cancer: a dose-dense schedule in real life and putative role of PIK3CA mutations. Oncotarget. 2018;9(44):27380–27396. doi: 10.18632/oncotarget.25270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanusch C., Schneeweiss A., Loibl S., Untch M., Paepke S., Kümmel S., Jackisch C., Huober J., Hilfrich J., Gerber B., et al. Dual blockade with AFatinib and trastuzumab as NEoadjuvant treatment for patients with locally advanced or operable breast cancer receiving taxane-anthracycline containing chemotherapy-DAFNE (GBG-70) Clin. Cancer Res. 2015;21(13):2924–2931. doi: 10.1158/1078-0432.CCR-14-2774. [DOI] [PubMed] [Google Scholar]
- 34.Harbeck N., von Schumann R., Kates R.E., Braun M., Kuemmel S., Schumacher C., Potenberg J., Malter W., Augustin D., Aktas B., et al. Immune markers and tumor-related processes predict neoadjuvant therapy response in the WSG-ADAPT HER2-positive/hormone receptor-positive trial in early breast cancer. Cancers (Basel) 2021;13(19) doi: 10.3390/cancers13194884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang L., Chen S., Yang W., Xu B., Huang T., Yang H., Zheng H., Wang Y., Song E., Zhang J., et al. Efficacy and safety analysis of trastuzumab and paclitaxel based regimen plus carboplatin or epirubicin as neoadjuvant therapy for clinical stage II-III, HER2-positive breast cancer patients: a phase 2, open-label, multicenter, randomized trial. Oncotarget. 2015;6(21):18683–18692. doi: 10.18632/oncotarget.4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Irelli A., Parisi A., D'Orazio C., Sidoni T., Rotondaro S., Patruno L., Pavese F., Bafile A., Resta V., Pizzorno L., et al. Anthracycline-free neoadjuvant treatment in patients with HER2-positive breast cancer: real-life use of pertuzumab, trastuzumab and taxanes association with an exploratory analysis of PIK3CA mutational status. Cancers (Basel) 2022;14(12) doi: 10.3390/cancers14123003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li K., Liao N., Chen B., Zhang G., Wang Y., Guo L., Wei G., Jia M., Wen L., Ren C., et al. Genetic mutation profile of Chinese HER2-positive breast cancers and genetic predictors of responses to Neoadjuvant anti-HER2 therapy. Breast Cancer Res. Treat. 2020;183(2):321–332. doi: 10.1007/s10549-020-05778-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loibl S., Treue D., Budczies J., Weber K., Stenzinger A., Schmitt W.D., Weichert W., Jank P., Furlanetto J., Klauschen F., et al. Mutational diversity and therapy response in breast cancer: a sequencing analysis in the Neoadjuvant GeparSepto trial. Clin. Cancer Res. 2019;25(13):3986–3995. doi: 10.1158/1078-0432.CCR-18-3258. [DOI] [PubMed] [Google Scholar]
- 39.Rimawi M.F., De Angelis C., Contreras A., Pareja F., Geyer F.C., Burke K.A., Herrera S., Wang T., Mayer I.A., Forero A., et al. Low PTEN levels and PIK3CA mutations predict resistance to neoadjuvant lapatinib and trastuzumab without chemotherapy in patients with HER2 over-expressing breast cancer. Breast Cancer Res. Treat. 2018;167(3):731–740. doi: 10.1007/s10549-017-4533-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sueta A., Yamamoto Y., Yamamoto-Ibusuki M., Hayashi M., Takeshita T., Yamamoto S., Iwase H. An integrative analysis of PIK3CA mutation, PTEN, and INPP4B expression in terms of trastuzumab efficacy in HER2-positive breast cancer. PLoS ONE. 2014;9(12) doi: 10.1371/journal.pone.0116054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toomey S., Eustace A.J., Fay J., Sheehan K.M., Carr A., Milewska M., Madden S.F., Teiserskiene A., Kay E.W., O'Donovan N., et al. Impact of somatic PI3K pathway and ERBB family mutations on pathological complete response (pCR) in HER2-positive breast cancer patients who received neoadjuvant HER2-targeted therapies. Breast Cancer Res. 2017;19(1):87. doi: 10.1186/s13058-017-0883-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dave B., Migliaccio I., Gutierrez M.C., Wu M.F., Chamness G.C., Wong H., Narasanna A., Chakrabarty A., Hilsenbeck S.G., Huang J., et al. Loss of phosphatase and tensin homolog or phosphoinositol-3 kinase activation and response to trastuzumab or lapatinib in human epidermal growth factor receptor 2-overexpressing locally advanced breast cancers. J. Clin. Oncol. 2011;29(2):166–173. doi: 10.1200/JCO.2009.27.7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barbareschi M., Cuorvo L.V., Girlando S., Bragantini E., Eccher C., Leonardi E., Ferro A., Caldara A., Triolo R., Cantaloni C., et al. PI3KCA mutations and/or PTEN loss in Her2-positive breast carcinomas treated with trastuzumab are not related to resistance to anti-Her2 therapy. Virchows Arch. 2012;461(2):129–139. doi: 10.1007/s00428-012-1267-2. [DOI] [PubMed] [Google Scholar]
- 44.Kim J.W., Lim A.R., You J.Y., Lee J.H., Song S.E., Lee N.K., Jung S.P., Cho K.R., Kim C.Y., Park K.H. PIK3CA mutation is associated with poor response to HER2-targeted therapy in breast cancer patients. Cancer Res. Treat. 2023;55(2):531–541. doi: 10.4143/crt.2022.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yin W., Wang Y., Wu Z., Ye Y., Zhou L., Xu S., Lin Y., Du Y., Yan T., Yang F., et al. Neoadjuvant trastuzumab and pyrotinib for locally advanced HER2-positive breast cancer (NeoATP): primary analysis of a phase II study. Clin. Cancer Res. 2022;28(17):3677–3685. doi: 10.1158/1078-0432.CCR-22-0446. [DOI] [PubMed] [Google Scholar]
- 46.Loibl S., de la Pena L., Nekljudova V., Zardavas D., Michiels S., Denkert C., Rezai M., Bermejo B., Untch M., Lee S.C., et al. Neoadjuvant buparlisib plus trastuzumab and paclitaxel for women with HER2+ primary breast cancer: a randomised, double-blind, placebo-controlled phase II trial (NeoPHOEBE) Eur J Cancer. 2017;85:133–145. doi: 10.1016/j.ejca.2017.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carey L.A., Berry D.A., Cirrincione C.T., Barry W.T., Pitcher B.N., Harris L.N., Ollila D.W., Krop I.E., Henry N.L., Weckstein D.J., et al. Molecular heterogeneity and response to neoadjuvant human epidermal growth factor receptor 2 targeting in CALGB 40601, a randomized phase III trial of paclitaxel plus trastuzumab with or without lapatinib. J. Clin. Oncol. 2016;34(6):542–549. doi: 10.1200/JCO.2015.62.1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fountzilas G., Giannoulatou E., Alexopoulou Z., Zagouri F., Timotheadou E., Papadopoulou K., Lakis S., Bobos M., Poulios C., Sotiropoulou M., et al. TP53 mutations and protein immunopositivity may predict for poor outcome but also for trastuzumab benefit in patients with early breast cancer treated in the adjuvant setting. Oncotarget. 2016;7(22):32731–32753. doi: 10.18632/oncotarget.9022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jensen J.D., Knoop A., Laenkholm A.V., Grauslund M., Jensen M.B., Santoni-Rugiu E., Andersson M., Ewertz M. PIK3CA mutations, PTEN, and pHER2 expression and impact on outcome in HER2-positive early-stage breast cancer patients treated with adjuvant chemotherapy and trastuzumab. Ann. Oncol. 2012;23(8):2034–2042. doi: 10.1093/annonc/mdr546. [DOI] [PubMed] [Google Scholar]
- 50.Gogas H., Kotoula V., Alexopoulou Z., Christodoulou C., Kostopoulos I., Bobos M., Raptou G., Charalambous E., Tsolaki E., Xanthakis I., et al. MYC copy gain, chromosomal instability and PI3K activation as potential markers of unfavourable outcome in trastuzumab-treated patients with metastatic breast cancer. J. Transl. Med. 2016;14(1):136. doi: 10.1186/s12967-016-0883-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guo L.W., Li X.G., Yang Y.S., Lu X.X., Han X.C., Lang G.T., Chen L., Shao Z.M., Hu X. Large-scale genomic sequencing reveals adaptive opportunity of targeting mutated-PI3Kα in early and advanced HER2-positive breast cancer. Clin. Transl. Med. 2021;11(11):e589. doi: 10.1002/ctm2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kotoula V., Tsakiri K., Koliou G.A., Lazaridis G., Papadopoulou K., Giannoulatou E., Tikas I., Christodoulou C., Chatzopoulos K., Bobos M., et al. Relapsed and De novo metastatic HER2-positive breast cancer treated with trastuzumab: tumor genotypes and clinical measures associated with patient outcome. Clin. Breast Cancer. 2019;19(2):113–125. doi: 10.1016/j.clbc.2018.10.014. e114. [DOI] [PubMed] [Google Scholar]
- 53.Nishimura R., Toh U., Tanaka M., Saimura M., Okumura Y., Saito T., Tanaka T., Teraoka M., Shimada K., Katayama K., et al. Role of HER2-related biomarkers (HER2, p95HER2, HER3, PTEN, and PIK3CA) in the efficacy of lapatinib plus capecitabine in HER2-positive advanced breast cancer refractory to trastuzumab. Oncology. 2017;93(1):51–61. doi: 10.1159/000468521. [DOI] [PubMed] [Google Scholar]
- 54.Perez E.A., de Haas S.L., Eiermann W., Barrios C.H., Toi M., Im Y.H., Conte P.F., Martin M., Pienkowski T., Pivot X.B., et al. Relationship between tumor biomarkers and efficacy in MARIANNE, a phase III study of trastuzumab emtansine ± pertuzumab versus trastuzumab plus taxane in HER2-positive advanced breast cancer. BMC Cancer. 2019;19(1):517. doi: 10.1186/s12885-019-5687-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang L., Zhang Q., Zhang J., Sun S., Guo H., Jia Z., Wang B., Shao Z., Wang Z., Hu X. PI3K pathway activation results in low efficacy of both trastuzumab and lapatinib. BMC Cancer. 2011;11:248. doi: 10.1186/1471-2407-11-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu B., Guan Z., Shen Z., Tong Z., Jiang Z., Yang J., DeSilvio M., Russo M., Leigh M., Ellis C. Association of phosphatase and tensin homolog low and phosphatidylinositol 3-kinase catalytic subunit alpha gene mutations on outcome in human epidermal growth factor receptor 2-positive metastatic breast cancer patients treated with first-line lapatinib plus paclitaxel or paclitaxel alone. Breast Cancer Res. 2014;16(4):405. doi: 10.1186/s13058-014-0405-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krop I.E., LoRusso P., Miller K.D., Modi S., Yardley D., Rodriguez G., Guardino E., Lu M., Zheng M., Girish S., et al. A phase II study of trastuzumab emtansine in patients with human epidermal growth factor receptor 2-positive metastatic breast cancer who were previously treated with trastuzumab, lapatinib, an anthracycline, a taxane, and capecitabine. J. Clin. Oncol. 2012;30(26):3234–3241. doi: 10.1200/JCO.2011.40.5902. [DOI] [PubMed] [Google Scholar]
- 58.Miller K.D., Dieras V., Harbeck N., Andre F., Mahtani R.L., Gianni L., Albain K.S., Crivellari D., Fang L., Michelson G., et al. Phase IIa trial of trastuzumab emtansine with pertuzumab for patients with human epidermal growth factor receptor 2-positive, locally advanced, or metastatic breast cancer. J. Clin. Oncol. 2014;32(14):1437–1444. doi: 10.1200/JCO.2013.52.6590. [DOI] [PubMed] [Google Scholar]
- 59.Kim S.B., Wildiers H., Krop I.E., Smitt M., Yu R., Lysbet de Haas S., Gonzalez-Martin A. Relationship between tumor biomarkers and efficacy in TH3RESA, a phase III study of trastuzumab emtansine (T-DM1) vs. treatment of physician's choice in previously treated HER2-positive advanced breast cancer. Int. J. Cancer. 2016;139(10):2336–2342. doi: 10.1002/ijc.30276. [DOI] [PubMed] [Google Scholar]
- 60.de Haas S., Hurvitz S., Martin M., Kiermaier A., Lewis Phillips G., Xu J., Helms H., Slamon D., Press M. Abstract P6-07-09: biomarker analysis from the neoadjuvant KRISTINE study in HER2-positive early breast cancer (EBC) Cancer Res. 2017;77(4_Supplement) P6-07-09-P06-07-09. [Google Scholar]
- 61.Metzger O., Lambertini C., Krop I., Phillips G.L., Perou C., Symmans F., Melero I., Harbeck N., Winer E. Im S: 42O Biomarker analysis from KAITLIN, a randomised phase III study of adjuvant trastuzumab emtansine (TDM-1; K) plus pertuzumab (P) versus trastuzumab (H) plus taxane (T) plus P after anthracyclines (AC) for high-risk HER2-positive early breast cancer (EBC) Ann. Oncol. 2021;32:S37–S38. [Google Scholar]
- 62.Loibl S., Darb-Esfahani S., Huober J., Klimowicz A., Furlanetto J., Lederer B., Hartmann A., Eidtmann H., Pfitzner B., Fasching P.A., et al. Integrated analysis of PTEN and p4EBP1 protein expression as predictors for pCR in HER2-positive breast cancer. Clin. Cancer Res. 2016;22(11):2675–2683. doi: 10.1158/1078-0432.CCR-15-0965. [DOI] [PubMed] [Google Scholar]
- 63.Loibl S., Denkert C., Schneeweis A., Paepke S., Lehmann A., Rezai M., Zahm D., Sinn P., Khandan F., Eidtmann H. Abstract S4-06: PIK3CA mutation predicts resistance to anti-HER2/chemotherapy in primary HER2-positive/hormone-receptor-positive breast cancer–Prospective analysis of 737 participants of the GeparSixto and GeparQuinto studies. Cancer Res. 2013;73(24_Supplement) S4-06-S04-06. [Google Scholar]
- 64.Shi W., Jiang T., Nuciforo P., Hatzis C., Holmes E., Harbeck N., Sotiriou C., Pena L., Loi S., Rosa D.D., et al. Pathway level alterations rather than mutations in single genes predict response to HER2-targeted therapies in the neo-ALTTO trial. Ann. Oncol. 2017;28(1):128–135. doi: 10.1093/annonc/mdw434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guarneri V., Dieci M.V., Frassoldati A., Maiorana A., Ficarra G., Bettelli S., Tagliafico E., Bicciato S., Generali D.G., Cagossi K., et al. Prospective biomarker analysis of the randomized CHER-LOB study evaluating the dual anti-HER2 treatment with trastuzumab and lapatinib plus chemotherapy as neoadjuvant therapy for HER2-positive breast cancer. Oncologist. 2015;20(9):1001–1010. doi: 10.1634/theoncologist.2015-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Metzger O., Lambertini C., Krop I.E., Lewis Phillips G., Perou C.M., Symmans F., Melero I., Harbeck N., Winer E.P., et al. 42O Biomarker analysis from KAITLIN, a randomised phase III study of adjuvant trastuzumab emtansine (TDM-1; K) plus pertuzumab (P) versus trastuzumab (H) plus taxane (T) plus P after anthracyclines (AC) for high-risk HER2-positive early breast cancer (EBC) Ann. Oncol. 2021;32:S37–S38. [Google Scholar]
- 67.Ibrahim E.M., Kazkaz G.A., Al-Mansour M.M., Al-Foheidi M.E. The predictive and prognostic role of phosphatase phosphoinositol-3 (PI3) kinase (PIK3CA) mutation in HER2-positive breast cancer receiving HER2-targeted therapy: a meta-analysis. Breast Cancer Res. Treat. 2015;152(3):463–476. doi: 10.1007/s10549-015-3480-6. [DOI] [PubMed] [Google Scholar]
- 68.Chakrabarty A., Rexer B.N., Wang S.E., Cook R.S., Engelman J.A., Arteaga C.L. H1047R phosphatidylinositol 3-kinase mutant enhances HER2-mediated transformation by heregulin production and activation of HER3. Oncogene. 2010;29(37):5193–5203. doi: 10.1038/onc.2010.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hanker A.B., Pfefferle A.D., Balko J.M., Kuba M.G., Young C.D., Sanchez V., Sutton C.R., Cheng H., Perou C.M., Zhao J.J., et al. Mutant PIK3CA accelerates HER2-driven transgenic mammary tumors and induces resistance to combinations of anti-HER2 therapies. Proc. Natl. Acad. Sci. U S A. 2013;110(35):14372–14377. doi: 10.1073/pnas.1303204110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eichhorn P.J., Gili M., Scaltriti M., Serra V., Guzman M., Nijkamp W., Beijersbergen R.L., Valero V., Seoane J., Bernards R., et al. Phosphatidylinositol 3-kinase hyperactivation results in lapatinib resistance that is reversed by the mTOR/phosphatidylinositol 3-kinase inhibitor NVP-BEZ235. Cancer Res. 2008;68(22):9221–9230. doi: 10.1158/0008-5472.CAN-08-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hurvitz S.A., Andre F., Jiang Z., Shao Z., Mano M.S., Neciosup S.P., Tseng L.M., Zhang Q., Shen K., Liu D., et al. Combination of everolimus with trastuzumab plus paclitaxel as first-line treatment for patients with HER2-positive advanced breast cancer (BOLERO-1): a phase 3, randomised, double-blind, multicentre trial. Lancet Oncol. 2015;16(7):816–829. doi: 10.1016/S1470-2045(15)00051-0. [DOI] [PubMed] [Google Scholar]
- 72.Andre F., O'Regan R., Ozguroglu M., Toi M., Xu B., Jerusalem G., Masuda N., Wilks S., Arena F., Isaacs C., et al. Everolimus for women with trastuzumab-resistant, HER2-positive, advanced breast cancer (BOLERO-3): a randomised, double-blind, placebo-controlled phase 3 trial. Lancet Oncol. 2014;15(6):580–591. doi: 10.1016/S1470-2045(14)70138-X. [DOI] [PubMed] [Google Scholar]
- 73.Andre F., Hurvitz S., Fasolo A., Tseng L.M., Jerusalem G., Wilks S., O'Regan R., Isaacs C., Toi M., Burris H., et al. Molecular alterations and everolimus efficacy in human epidermal growth factor receptor 2-overexpressing metastatic breast cancers: combined exploratory biomarker analysis From BOLERO-1 and BOLERO-3. J. Clin. Oncol. 2016;34(18):2115–2124. doi: 10.1200/JCO.2015.63.9161. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study used publicly available data from published studies. All data and material analyzed during this study are included in this article.