Abstract
Peroxisomes are metabolically active organelles that are known for exerting oxidative metabolism, but the precise mechanism remains unclear in diabetic nephropathy (DN). Here, we used proteomics to uncover a correlation between the antioxidant protein disulfide-bond A oxidoreductase-like protein (DsbA-L) and peroxisomal function. In vivo, renal tubular injury, oxidative stress, and cell apoptosis in high-fat diet plus streptozotocin (STZ)-induced diabetic mice were significantly increased, and these changes were accompanied by a "ghost" peroxisomal phenotype, which was further aggravated in DsbA-L-deficient diabetic mice. In vitro, the overexpression of DsbA-L in peroxisomes could improve peroxisomal phenotype and function, reduce oxidative stress and cell apoptosis induced by high glucose (HG, 30 mM) and palmitic acid (PA, 250 μM), but this effect was reversed by 3-Amino-1,2,4-triazole (3-AT, a catalase inhibitor). Mechanistically, DsbA-L regulated the activity of catalase by binding to it, thereby reducing peroxisomal leakage and proteasomal degradation of peroxisomal matrix proteins induced by HG and PA. Additionally, the expression of DsbA-L in renal tubules of patients with DN significantly decreased and was positively correlated with peroxisomal function. Taken together, these results highlight an important role of DsbA-L in ameliorating tubular injury in DN by improving peroxisomal function.
Keywords: Diabetic nephropathy, DsbA-L, Peroxisome, Catalase, Kidney, Oxidative stress
1. Introduction
Diabetic nephropathy (DN) is a serious complication of diabetes mellitus and the main cause of end-stage renal disease [1]. In the past, research on DN mainly focused on the glomerulus, but in recent years, the role of tubular injury in DN has been increasingly appreciated [[2], [3], [4], [5]]. Many studies have highlighted that the correlation between the progression of nephropathy and tubulointerstitial changes is stronger than that of glomerular changes [6,7]. The early changes of the proximal tubules (PTs) are the basis of oxidative stress, inflammation and tubulointerstitial fibrosis, which in turn promote the progression of DN [5,8]. These observations suggest that the proximal renal tubules play a crucial role in the pathophysiology of DN.
Peroxisomes, originally described in renal tubular cells by J. Rhodin, are small, single-membrane organelles [9]. These dynamic organelles perform essential functions in regulating numerous cellular processes, such as the synthesis of bile acids, the maintenance of cholesterol homeostasis and fatty acid β-oxidation [10]. Additionally, peroxisomes are also important organelles for redox reactions, contributing to both the generation and clearance of reactive oxygen species (ROS) [11]. Up to 20% of total oxygen consumption and 35% of total hydrogen peroxide (H2O2) generation may be attributed to peroxisomal respiration [12]. The high abundance of peroxisomes in the PTs strongly suggested an important role of peroxisomes in the kidney [13,14]. And proteomic results showed an extensive differential expression of peroxisomal proteins in the injured kidney [15,16]. In addition, it is known that peroxisome deficiency or dysfunction would lead to increased intracellular ROS, which contributes to oxidative stress-induced kidney injury [17,18]. However, limited studies have explored the contribution of peroxisomes in the development of DN oxidative damage, and the specific regulatory mechanism remains unclear.
Disulfide-bond A oxidoreductase-like protein (DsbA-L), originally named glutathione S-transferase (GST)-k, is a 25 kDa protein predominantly expressed in the proximal tubule cells (PTCs) of kidneys and localized mainly in the peroxisome and mitochondria [19,20]. DsbA-L exerts peroxidase activity and plays a crucial role in safeguarding cells against oxidative stress by facilitating the metabolism of exogenous substances, endogenous toxic metabolites, and free radicals [21]. Previous studies have shown that DsbA-L deficiency worsens oxidative stress and inflammation [22,23], while DsbA-L overexpression reduces ROS [24]. It is currently known that DsbA-L can protect PTCs by decreasing oxidative stress through improving mitochondrial function in DN [22], but the role of DsbA-L in peroxisome and the underlying mechanisms in DN remain to be elucidated.
This study revealed that diabetic mice with PT-specific knockout of the DsbA-L exhibited exacerbated albuminuria and tubular oxidative damage, co-occurring with a substantial decline in both peroxisomal number and function. Furthermore, in vitro studies indicated that overexpression of DsbA-L in HK-2 cells intensified catalase (CAT) activity through direct interaction with the protein, reduced the leakage of peroxisomal membrane, restored peroxisomal function, subsequently decreased the injury of tubular cells caused by high glucose (HG) and palmitic acid (PA).
2. Materials and methods
2.1. Antibodies and reagents
Antibodies against the following proteins/enzymes were used: DsbA-L (ab92819, PA5-80676, 14535-1-AP), Peroxisomal membrane protein 70 (PMP70, ab3421, SAB4200181), CAT (66765-1-Ig, 21260-1-AP, sc-271803), Peroxisomal acyl-coenzyme A oxidase 1 (ACOX1, 10957-1-AP), BAX (60267-I-Ig), BCL2 (26593-1-AP), Cleaved Caspase 3 (9661S), Caspase 9 (66169-1-Ig), 4-Hydroxynonenal (4-HNE, mab3249), 8-hydroxydeoxyguanosine (8-OHdG, sc-393871). 3-Amino-1,2,4-triazole (3-AT) was obtained from Selleck Chemicals. PA and streptozotocin (STZ) were obtained from Sigma-Aldrich. The DsbA-L plasmid was obtained from Dr. Liu Feng at Central South University, Changsha, China. Lipofectamine 3000 was purchased from Invitrogen (Waltham, MA). The high-fat diet (containing 35 kcal% carbohydrates, 45 kcal% fat, and 20 kcal% protein, D12451) was obtained from Research Diets, Inc (New Brunswick, NJ).
2.2. Generation of PT-specific DsbA-L-knockout mice
DsbA-Lflox/flox mice and PEPCK‐Cre mice were generously provided by Professor Liu Feng and Dr Tang from the Second Xiangya Hospital of Central South University, respectively [25]. To obtain the PT-specific DsbA-L-knockout (DsbA-LptKO) mice and their littermate controls (DsbA-Lctrl), we crossed DsbA-Lflox/flox mice with PEPCK-Cre mice. These mice were genotyped by polymerase chain reaction (PCR) analysis of genomic DNA isolated from tail tissues using a Mouse Direct PCR kit (Bimake) as described previously [26]. The sequences of the primers used were as follows: DsbA-L forward primer (5′–3′), CTGGATGGCTTCTGTTAGAG; DsbA-L reverse primer (5′–3′), AAGACTGCCACTCTGCAATG; Cre forward primer (5′–3′), ACCTGAAGATGTTCGCGATTATCT; Cre reverse primer (5′–3′), ACCGTCAGTACGTGAGATATCTT.
2.3. Mouse model
Male mice aged 6 weeks were randomly assigned to 4 groups: DsbA-Lctrl group, DsbA-LptKO group, high-fat diet (HFD)/STZ induced DsbA-Lctrl group and HFD/STZ induced DsbA-LptKO group. The DsbA-Lctrl and DsbA-LptKO groups were administered a normal diet, whereas the other two groups were given a HFD for four weeks and then injected intraperitoneally with STZ (100 mg/kg; Sigma-Aldrich) to generate hyperglycemia, as described in a previous study [26]. Mice with random blood glucose levels greater than 12 mmol/l were selected for the study three days after the injection, and they were maintained on the HFD for 16 weeks. The body weight and blood glucose were measured every two weeks. Urine was collected at 26 weeks of age and kidney tissues were collected for various experiments. The Medical Ethics Committee of Central South University approved all experiments (20220151).
2.4. General information on patients with DN
The clinical specimens used in this study were obtained from the Second Xiangya Hospital of Central South University. Patients diagnosed with DN via renal biopsy were included, and patients with glomerular minor lesion were selected as the control group. For these patients, routine clinical parameters, such as the estimated glomerular filtration rate (eGFR), as well as pathological data, including interstitial fibrosis and tubular atrophy (IFTA) were collected (Table S1). The protocol was approved by the Ethics Committee of The Second Xiangya Hospital, Central South University (XXF190105). All patients signed the informed consent in advance.
2.5. Measurement of urine albumin and creatinine
According to the kit's instructions, urine albumin and urinary creatinine levels were determined using a Mouse MAU (Microalbuminuria) ELISA Kit (Sangon Biotech) and CicaLiquid-N CRE (KANTO CHEMICAL CO., INC.), respectively.
2.6. Real-time qPCR
Total RNA was extracted from renal cortex using the RNAiso Plus kit (TaKaRa), as previously described [27]. The extracted RNA was then converted to cDNA using the PrimeScript Reagent Kit (TaKaRa) and subsequently amplified using TB Green Premix Ex Taq II (TaKaRa) in the 7300 Real-Time PCR System (Applied Biosystems). The sequences of the primers used were as follows: DsbA-L forward primer (5′–3′), CTGGATGGCTTCTGTTAGAG; DsbA-L reverse primer (5′–3′), AAGACTGCCACTCTGCAATG; β-actin forward primer (5′–3′), CAAGCAGGAGTACGATGAGTC, β-actin reverse primer (5′–3′), AACGCAGCTCAGTAACAGTC.
2.7. Western blot
Total protein from cells and tissues was extracted using phosphatase inhibitors, protease inhibitors and RIPA buffer (CWBIO). The BCA Protein Assay Kit (Thermo Fisher Scientific) was used to quantify the protein concentrations. The protein was then subjected to the sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and transferred to polyvinylidene fluoride membranes. After an overnight incubation with the primary antibody at 4 °C, the blotted membrane was then incubated with appropriate secondary antibodies (Abcam). An enhanced chemiluminescence kit (Thermo Fisher Scientific) was used to detect the membrane blots, and band intensities were quantified by ImageJ software.
2.8. Proteomics analysis
The total protein extracted from the renal cortex of DsbA-Lctrl and DsbA-LptKO mice was obtained following the manufacturer’s provided instructions. Subsequently, the protein samples were sent to Luming Bio (Shanghai, China) (https://www.lumingbio.com/) for proteomics analysis.
2.9. Morphological analysis of the kidney
Fixed mouse kidney tissue using 4% paraformaldehyde was embedded in paraffin. Staining of the four-μm-thick paraffin sections with hematoxylin-eosin (HE), periodic acid Schiff (PAS) and Masson followed previously described methods [28]. The severity of tubulointerstitial injury was evaluated based on IFTA and interstitial inflammation. IFTA together as a percentage of the total involved area of interstitium and tubules: 0 = no IFTA, 1 = less than 25% IFTA, 2 = at least 25% but less than 50%, and 3 = at least 50% IFTA. Regarding interstitial inflammation, 0 = no interstitial infiltrates, 1 = infiltration limited to atrophic tubules, and 2 = inflammatory infiltrates in areas other than around atrophic tubules. Renal tubulointerstitial injury was observed under light microscope and 10 random visual fields were selected for scoring [29].
2.10. Immunohistochemistry (IHC)
The four-μm thick paraffin sections were dewaxed, hydrated, and antigen retrieved. Sections were then blocked with 5% bovine serum albumin (BSA), followed by overnight incubation of primary antibody at 4 °C. After washing, the sections were incubated with secondary antibodies, reacted with diaminobenzidine (Servicebio) and stained with hematoxylin to visualize the nuclei. Sections were observed by a light microscope. Proteins were quantified using Image-Pro plus 6.0.
2.11. Immunofluorescence (IF)
The paraffin sections were dewaxed, hydrated and antigen retrieved, then permeabilized and blocked with 5% BSA. Sections were incubated with primary antibody overnight at 4 °C. After PBS washing, the sections were exposed to Alexa Fluor-488 or Alexa Fluor-594-labeled secondary antibody (Abcam). Nuclei were stained with 4',6-diamidino-2-phenylindole (DAPI) (SouthernBiotech).
2.12. CAT and superoxide dismutase activity
Following the manufacturer's instructions, the superoxide dismutase (SOD) and CAT activity was measured using commercially available kits from Jiancheng Biotechnology.
2.13. Cell culture and treatments
The HK-2 cell line was obtained from ATCC and cultured. Transfection of cells with pcDNA3.1-DsbA-L plasmid was performed with a Lipofectamine 3000 reagent (Invitrogen) following the manufacturer's protocol. In this study, HK-2 cells were treated with different concentrations of d-glucose (5 mM or 30 mM) and PA (250 μM) as previously described [26]. And 3-AT (10 mM, Selleck Chemicals) were used to suppress CAT. MG132 (10 μM, Sigma-Aldrich) were used as a proteasome inhibitor.
2.14. HyPer-PTS1 biosensor and DsbA-L-PTS1
To monitor the H2O2 levels produced locally in peroxisomes, we employed a peroxisome-targeted and genetically encoded fluorescence probe, HyPer-PTS1 [30]. Subsequently, we established a line of HK-2 cells expressing the HyPer-PTS1 biosensor. Similarly, we added the SKL sequence after the DsbA-L sequence to targe DsbA-L into the peroxisome.
2.15. Determination of ROS in peroxisome
HK-2 cells stably transfected with HyPer-PTS1 were dispensed onto the black 96-well glass-bottom plates (Corning, 3603). To assay H2O2 production, fluorescence was immediately measured in each well using SpectraMax Multifunctional enzyme label with excitation at 420 nm and 500 nm; emission at 516 nm.
2.16. Cell immunofluorescence
After various treatments, the cells were fixed, permeabilized, and blocked. Next, the cells were incubated with primary antibodies overnight at 4 °C. After washing, the cells were incubated with AlexaFluor-conjugated secondary antibodies and stained with DAPI. Cells were visualized by an LSM 780 META laser scanning microscopy (Zeiss, Thornwood, NY). Colocalization analyses were performed using Image-Fiji.
2.17. Immunoprecipitation
The cells were lysed with immunoprecipitation lysis buffer (87787, Thermo Fisher Scientific) containing protease/phosphatase inhibitors and then incubated overnight at 4 °C with anti-DsbA-L, CAT and IgG (B900610, B900620, Proteintech). The precipitated materials were used for western blot analysis with a secondary antibody that does not interfere with denatured IgG (ab131366, Abcam).
2.18. Proteinase K protection assay
According to the experimental method of Mu et al. [31], the post nuclear supernatants were divided into three groups: Triton X-100 (GC204003, Servicebio), Proteinase K (G1237, Servicebio) and combination of Proteinase K with Triton X-100. The reaction was stopped after 30 min by adding PMSF, and the samples were further treated for Western blot assay.
2.19. Measurement of oxidative stress
Dihydroethidium (DHE) (Invitrogen) and dichloro-dihydro-fluorescein diacetate (DCFH-DA) were used to evaluate intracellular ROS in renal tissues and HK-2 cells, respectively.
2.20. Statistical analysis
All data were analyzed using GraphPad Prism9.0 software. The values were presented as means ± SEMs and analyzed by Student’s t-test or one-way analysis of variance (ANOVA). Pearson’s correlation analysis was performed to examine the correlations between two numerical variables. P < 0.05 was considered statistically significant.
3. Results
3.1. Proximal tubular cell-specific DsbA-L gene deficiency markedly exacerbated renal injury in diabetic mice
To gain insight into the role of DsbA-L in DN, we generated mice with a targeted deletion of the DsbA-L gene in PT. Specifically, we crossed DsbA-L exon 1,2,3-floxed mice (DsbA-Lflox/flox) with PEPCK-Cre mice to obtain the DsbA-LptKO strain (Fig. 1A).
Fig. 1.
PT-specific DsbA-L gene deficiency markedly exacerbated renal injury in diabetic mice. (A) Schematic of DsbA-L conditional knockout strategy. (B) PCR with genomic DNA from tail tissues as templates for verification the floxed mouse using primer pairs. (C) A representative western blot of DsbA-L expression in isolated PTCs from DsbA-LCtrl and DsbA-LptKO mice. (D) Real-time PCR analysis of DsbA-L in kidney cortices of mice (n=4). (E) Body weight, (F) blood glucose levels and (G) UACR of different groups of mice (n=4). (H) Morphological examinations of renal pathology by HE, PAS, Masson staining. Scale bars:20 μm. (I) Quantification of tubular interstitial damage score of the kidneys in each group (n=4). Data are presented as means±SEMs. *P < 0.05vs DsbA-Lctrl group; **P < 0.05vs HFD/STZ-DsbA-Lctrl group; ns, not significant. PTCs, proximal tubular cells; PCR, polymerase chain reaction; UACR; urine albumin creatine ratio.
The mice with the DsbA-Lflox/flox; PEPCK-Cre genotype were referred to as DsbA-LptKO, while their littermates without Cre amplification were used as controls, denoted as DsbA-LCtrl (Fig. 1B). The deletion of the DsbA-L gene was also confirmed by qPCR and western blot analysis of PTC isolated from the kidney (Fig. 1C and D). DN was then induced in both DsbA-LCtrl mice and DsbA-LptKO mice by administration of a HFD and STZ. Both groups displayed hyperglycemia after STZ treatment, and no significant differences in body weight and blood glucose were found (Fig. 1E and F). However, HFD/STZ-induced DsbA-LptKO mice showed further increased urine albumin creatine ratio (UACR) compared to HFD/STZ-induced DsbA-LCtrl mice (Fig. 1G). In addition, HE, PAS and Masson staining of diabetic mice kidneys revealed dilated cortical proximal tubules with loss of brush borders, as well as tubular basement membrane thickening and interstitial fibrosis, compared to the DsbA-LCtrl group. These lesions were further aggravated in diabetic DsbA-LptKO group (Fig. 1H and I), indicating that DsbA-L plays a crucial role in regulating tubular injury in DN.
3.2. The deficiency of DsbA-L resulted in peroxisomal dysfunction in the tubular cells of diabetic mice
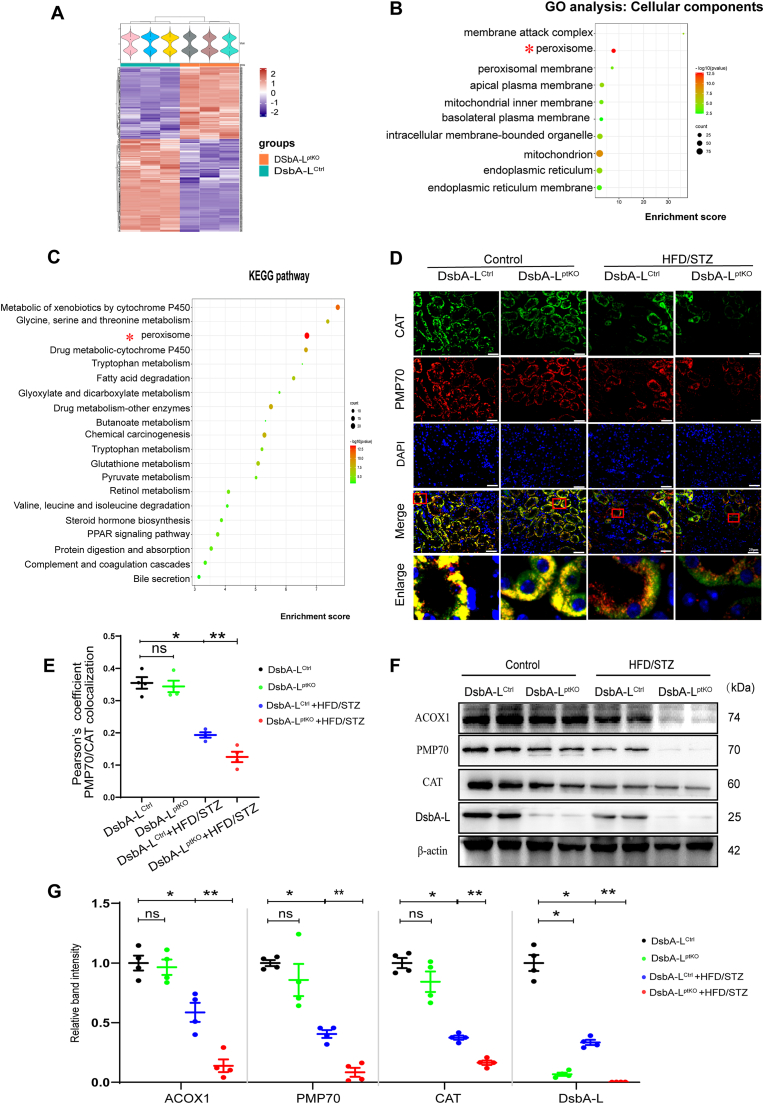
To investigate how DsbA-L affects the kidney, we extracted the renal cortex of DsbA-LCtrl and DsbA-LptKO mice for proteomic analysis. The analysis revealed 464 differentially expressed proteins between the two groups (Fig. 2A). Furthermore, the enrichment analysis of cellular components indicated the enrichment of DsbA-L in peroxisomes (Fig. 2B), while Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis demonstrated its significant role in peroxisome biology (Fig. 2C).
Fig. 2.
PTCs-specific-DsbA-L deficiency accelerated disruption of peroxisomal abundance and function in tubular cells of diabetic mice. (A) Clusters of differentially expressed proteins in the kidney tissues of DsbA-Lctrl and DsbA-LptKO mice. Each row represents a protein, and each column represents a mouse sample. Blue color indicates downregulation, and red shows upregulation. n = 3 mice per group. (B) Gene ontology (GO) analysis of differentially expressed proteins in the kidney of DsbA-LptKO with DsbA-LCtrl; the analysis was performed according to the cellular components. (C) Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of differentially expressed proteins in the kidney of DsbA-LptKO with DsbA-LCtrl. (D) Representative immunofluorescence images of CAT (green) and PMP70 (red) in kidney tissues from each group. The nuclei were counterstained by DAPI (blue). Overlap analyzed by the colocalization highlighter plugin (ImageJ) are shown in yellow. Scale bar: 20 μm. (E) Colocalization of CAT and PMP70 determined from the images represented in (D) using Pearson’s correlation coefficient (n=4). (F) Western blot analysis of ACOX1, PMP70, CAT and DsbA-L expression in kidney cortices (n=4). (G) Relative band intensity of ACOX1, PMP70, CAT and DsbA-L. Data are presented as means±SEMs. *P < 0.05vs DsbA-Lctrl group; **P < 0.05vs HFD/STZ-DsbA-Lctrl group; ns, not significant. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
To examine the impact of DsbA-L on peroxisomal function in the kidney, proteins associated with peroxisome in kidney were analyzed using immunofluorescence double staining in puncta co-stained with both PMP70 and CAT, which are identified as mature and functional peroxisomes [32]. Peroxisomal function disruption often presents a “ghost” peroxisomal phenotype where cells display defective CAT import but have peroxisomal bodies positive for PMP70 [33]. We detected such a peroxisomal phenotype in PTC. As shown in Fig. 2D and E, the co-staining with PMP70 and CAT demonstrated the HFD/STZ-induced DsbA-LCtrl mice had fewer peroxisomes and an increased "ghost" peroxisomal phenotype, while these indicators were further altered in the HFD/STZ-induced DsbA-LptKO mice. Furthermore, the western blot analysis of kidney cortex showed a significant decrease in CAT, PMP70, and ACOX1 expression in HFD/STZ-induced DsbA-LCtrl mice, which became more pronounced in HFD/STZ-induced DsbA-LptKO mice (Fig. 2F and G). Taken together, these data suggest that DsbA-L might be a key player in peroxisomal metabolism in PTC.
3.3. DsbA-L gene ablation in PTCs resulted in increased ROS production and cell apoptosis in diabetic mice
Peroxisomes are considered the primary site of oxidative reactions, and the accumulated dysfunctional peroxisomes is known to be significant sources of ROS and pro-apoptotic factors [17,34]. We detected the presence of ROS within kidney tissues through DHE staining, where we observed an overproduction of ROS in the tubules of HFD/STZ-induced DsbA-LCtrl mice, and the HFD/STZ-induced DsbA-LptKO mice showed significantly stronger signals (Fig. 3A and B). 4-HNE and 8-OHdG, markers of oxidative damage and lipid peroxidation, also accumulated to high levels in the kidney of the HFD/STZ-induced mice (Fig. 3A, C, 3D). And the activity level of vital antioxidants, CAT and SOD, were remarkably lower in the HFD/STZ-induced DsbA-LptKO mice (Fig. 3E and F). To observe changes in apoptosis, we detected alterations in some apoptotic factors and anti-apoptotic factors. Compared with DsbA-LCtrl mice, we measured up-regulated levels of Cleaved Caspase-3, Caspase-9, and BAX and down-regulated levels of BCL2 in the STZ/HFD-induced DsbA-LCtrl mice, all of which were further changed in STZ/HFD-induced DsbA-LptKO mice (Fig. 3G–K). Together, these observations indicate that DsbA-L deficiency leads to oxidative stress and cell death.
Fig. 3.
PTCs-specific-DsbA-L deficiency resulted in increased ROS production and cell apoptosis in diabetic mice. (A) The upper layer is DHE staining (Scale bar: 50 μm), and the middle and lower layer represents the immunohistochemical results of 4-HNE and 8-OHdG (Scale bar: 20 μm), respectively. (B) Relative mean fluorescence intensity of DHE (n=4). (C) Relative immunostaining density/area of 4-HNE (n=4). (D) Relative immunostaining density/area of 8-OHdG (n=4). (E) CAT activity was detected (n=4). mgprot, milligram protein. (F) Total SOD activity was detected (n=4). (G–K) Representative western blot bands (G) and relative band intensity (H–K) of Caspase-9, BCL2, BAX and Cleaved Caspase-3 protein in the kidney cortices (n=4). Data are presented as means±SEMs. *P < 0.05vs DsbA-Lctrl group; **P < 0.05vs HFD/STZ-DsbA-Lctrl group; ns, not significant.
3.4. The overexpression of DsbA-L in HK-2 cells prevented peroxisomal dysfunction induced by HG and PA through enhancing CAT activity by interacting with CAT directly
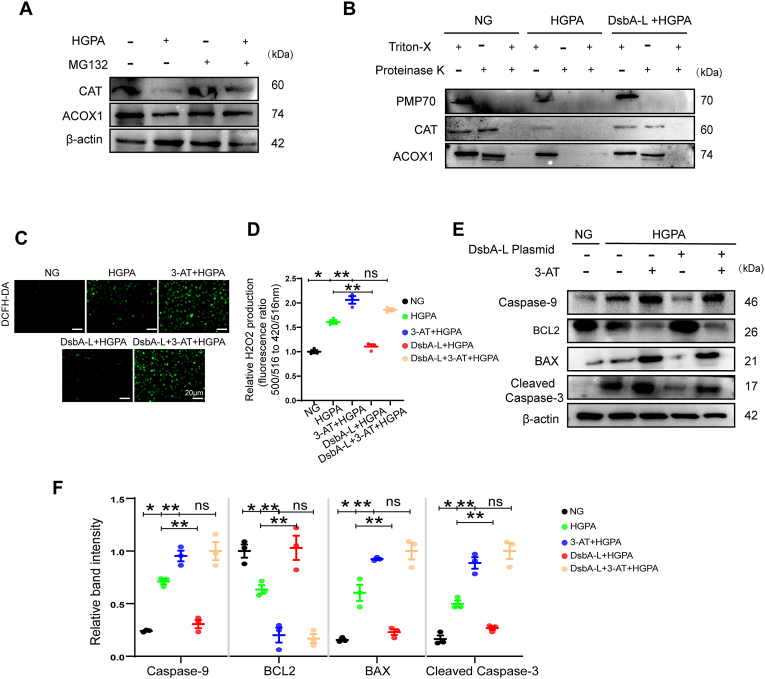
We conducted a series of in vitro experiments to investigate whether the overexpression of DsbA-L could rescue HG and PA-induced effects in HK-2 cells and explore its potential mechanisms. Firstly, we checked the expression and subcellular localization of DsbA-L in tubular cells. Amounts of DsbA-L proteins were co-localized with PMP70 in HK-2 cells, indicating that some of the DsbA-L proteins localize in the peroxisome (Fig. 4A). Secondly, based on the analysis of the website database, DsbA-L appeared to interact with multiple molecules. Among them, CAT is located in the peroxisome and is closely related to the oxidative reaction (Fig. 4B). Next, we explored whether DsbA-L improves peroxisome abundance and function via CAT. The results showed that mature and functional peroxisomes and the expression of ACOX1, PMP70, and CAT decreased in the HG and PA environment but were partially restored by overexpression of DsbA-L, and the effect was abolished by the treatment of 3-AT, a CAT inhibitor (Fig. 4C–F). CAT, a peroxisomal matrix protein, was also observed to leak into the cytoplasm under HG and PA conditions, and the overexpression of DsbA-L partially improved the leakage of CAT into the cytoplasm (Fig. 4C). These findings suggest that DsbA-L may regulate peroxisomal function through CAT.
Fig. 4.
DsbA-L enhanced CAT activity by directly interacting with CAT, and then alleviated HG and PA-induced peroxisomal dysfunction in HK-2 cells. (A) Confocal images of endogenous DsbA-L (green) and peroxisomal marker PMP70 (red) in HK-2 cells. Nuclei were counterstained with DAPI (blue). Overlap analyzed by the colocalization highlighter plugin (ImageJ) are shown in yellow. Scale bar: 10 μm. (B) Protein interaction made using Cytoscape (data source STRING). (C) HK-2 cells were treated with DsbA-L overexpression plasmid, normal glucose (NG, 5 mM), high glucose (HG, 30 mM) and palmitic acid (PA, 250 μM) or 3-AT and immunostained for CAT (green) and PMP70 (red). Nuclei were counterstained with DAPI (blue). Overlaps analyzed by the colocalization highlighter plugin (ImageJ) are shown in yellow. Scale bar: 10 μm. (D) Colocalization of CAT and PMP70 determined from the images represented in (C) using Pearson’s correlation coefficient (n=3). (E) Representative western blot images of ACOX1, PMP70, CAT and DsbA-L expression in HK-2 cells under various treatment conditions. (F) Relative band intensity of ACOX1, PMP70 and CAT in HK-2 cells (n=3). (G) Cell lysates were subjected to immunoprecipitation (IP) with control IgG and anti-DsbA-L antibody (upper panel) or control IgG and anti-CAT antibody (lower panel). The immune-precipitates and whole-cell lysates (input) were analyzed by Western blotting with the indicated antibodies. IB, immunoblotting. (H) Representative images showing colocalization of CAT (red) and DsbA-L (green) in HK-2 cells. Nuclei were counterstained with DAPI (blue). Overlaps analyzed by the colocalization highlighter plugin (ImageJ) are shown in yellow. Scale bar: 20 μm. (I) The predicted interaction mode between DsbA-L (green) and CAT (blue). CAT active site signature (FDRERIPERVVHAKGAGA) was yellow. (J) CAT activity was detected in HK-2 cells (n=3). mgprot, milligram protein. Data are presented as means±SEMs. *P < 0.05vs NG; **P < 0.05vs HG and PA; ns, not significant; ACOT2, Acyl-coenzyme A thioesterase 2; AGXT, Alanine--glyoxylate aminotransferase; DECR2, Peroxisomal 2,4-dienoyl-CoA reductase; EPHX2, Bifunctional epoxide hydrolase 2. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Furthermore, it has previously been reported that DsbA-L may interact with CAT [35]. To verify the interaction between DsbA-L and CAT, we performed immunoprecipitation with HK-2 cells (Fig. 4G). The interaction between DsbA-L and CAT was further validated through immunofluorescence, and the results showed that overexpression of DsbA-L increased binding under HG and PA-induced conditions (Fig. 4H). Next, by analyzing the protein sequence of the two molecules and using GRAMM for prediction, we discovered that DsbA-L binds to the active site of CAT [36] (Fig. 4I, Fig. S1). And we also found that DsbA-L could regulate CAT activity under the condition of HG and PA (Fig. 4J). Taken together, these results suggest that DsbA-L may regulate the activity of CAT by interacting with it, thereby improving the abundance and function of the peroxisome.
3.5. Overexpression of DsbA-L alleviated leakage of peroxisome membrane, proteasomal degradation of peroxisomal matrix proteins, peroxisomal H2O2 and apoptosis in HK-2 cells induced by HG and PA
Our results demonstrated that peroxisomal matrix proteins could be released into the cytoplasm under HG and PA conditions. And peroxisomal ROS generation promotes peroxisomal leakage and proteasomal degradation of peroxisomal matrix proteins [31]. Therefore, we hypothesized that the elevated peroxisomal ROS generation under HG and PA conditions may cause leaky peroxisome, which subsequently results in the leakage of matrix contents followed by proteasomal degradation. To verify HG and PA in the proteasomal degradation of peroxisomal matrix proteins, HK-2 cells were treated with a proteasome inhibitor, MG132, along with HG and PA. As shown in Fig. 5A, the addition of MG132 has been shown to inhibit HG and PA mediated degradation of peroxisome matrix proteins, including CAT and ACOX1. To confirm the degradation of peroxisome matrix proteins in the cytosol, Proteinase K protection assay was carried out. In Fig. 5B, PMP70 was degraded by proteinase K without Triton X-100, while peroxisomal matrix proteins, including CAT and ACOX1, were only degraded by a combination of proteinase K and Triton X-100 in control cells. These findings suggest that in control cells, the degradation of matrix proteins by proteinase K requires the matrix protein to leak into the cytoplasm via pores induced by Triton X-100 on the peroxisomal membrane. ACOX1 and CAT were degraded by protease K under the intervention of HG and PA, indicating that the peroxisomal membrane has leaked. And these effects were partially restored by overexpression of DsbA-L(Fig. 5B).
Fig. 5.
Overexpression of DsbA-L alleviated leakage of peroxisomal membrane, proteasomal degradation of peroxisomal matrix proteins, peroxisomal H2O2and apoptosis in HK-2 cells induced by HG and PA. (A) Cells were treated with NG, HG and PA, or MG132 (10 μM) for 24 h and followed by immunoblotting. (B) HK-2 Cells were treated with NG, HG and PA, or DsbA-L plasmid, followed by Proteinase K protection assay, and immunoblotting for PMP70, CAT and ACOX1. (C) HK-2 cells were treated with DCFH-DA to determine the ROS and examined under fluorescence microscope. (D) Peroxisomal H2O2 production was measured by HyPer-PTS1(n=3). (E) Representative western blot bands of Caspase-9, BCL2, BAX and Cleaved Caspase-3 protein in HK-2 cells. (F) Relative band intensity of Caspase-9, BCL2, BAX and Cleaved Caspase-3 protein in HK-2 cells (n=3). Data are presented as means±SEMs. *P < 0.05vs NG; **P < 0.05vs HG and PA; ns, not significant.
Furthermore, intracellular ROS level was measured using DCFH-DA, and we found that overexpression of DsbA-L in HK-2 cells could reduce HG and PA-induced ROS, but this alleviating effect could be inhibited by 3-AT (Fig. 5C). Similar results were also observed with H2O2 in peroxisomes (Fig. 5D). Consistent with the improvement of peroxisomal ROS, apoptosis in HK-2 cells exposed to HG and PA as revealed was also inhibited with overexpression of DsbA-L (Fig. 5E and F).
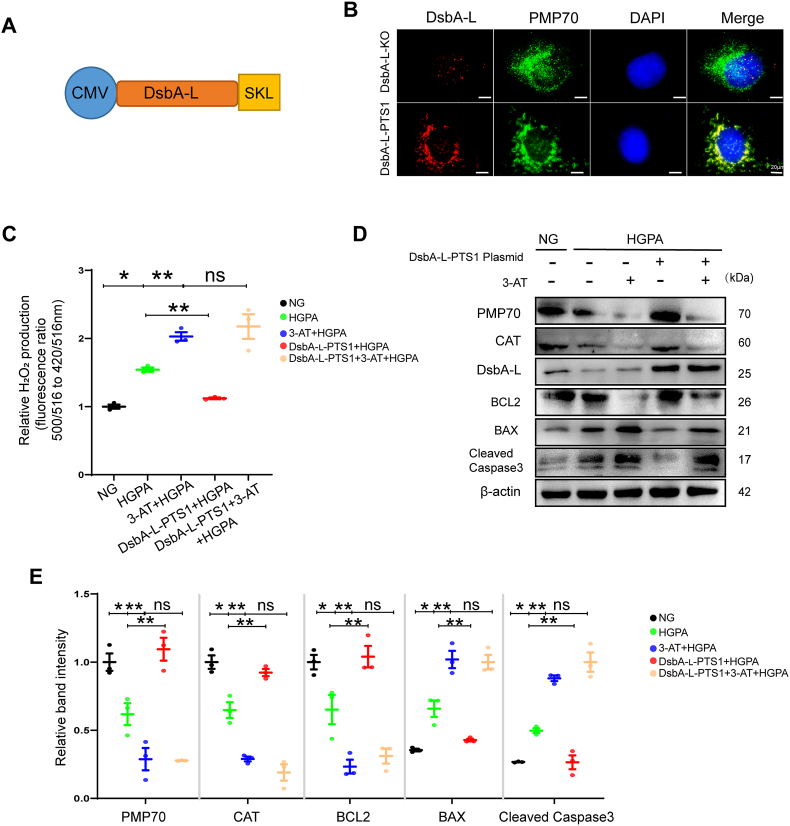
3.6. Peroxisomal localization of DsbA-L improved peroxisomal function and cell injury by CAT
The protein DsbA-L is located in both mitochondria and peroxisomes. We were interested in whether peroxisomal or mitochondrial DsbA-L is responsible for the CAT function. To investigate this, we generated organelle-specific targeting of DsbA-L by genetically fusing it with SKL, which was named DsbA-L-PTS1 (Fig. 6A). Immunofluorescence was used to confirm the specific localization of DsbA-L-PTS1 within the peroxisomes (Fig. 6B). DsbA-L-PTS1 blocked the reduction of peroxisomal abundance and dysfunction, prevented peroxisomal H2O2 and cell apoptosis in HK-2 cells induced by HG and PA (Fig. 6C–E). Therefore, based on these results, it seems that peroxisomal DsbA-L is responsible for peroxisomal function as well as kidney injury.
Fig. 6.
Peroxisomal localization of DsbA-L improved peroxisomal function and apoptosis. (A) Construction of DsbA-L plasmid directed to peroxisome. (B) The DsbA-L knockout stable transfection strain (DsbA-L-KO) was constructed, and DsbA-L-PTS1 was transfected in the DsbA-L-KO stable transfection strain. Panel shows the DsbA-L as red fluorescence, the peroxisomes showed as PMP-70 positive green staining, and the co-localization of DsbA-L with PMP-70 is shown as yellow fluorescence puncta. Nuclear DNA is stained with DAPI and shown in blue. Scale bar: 20 μm. (C) Peroxisomal H2O2 production was measured by HyPer-PTS1 (n=3). (D) Representative western blot bands of PMP70, CAT, DsbA-L, BCL2, BAX and Cleaved Caspase-3 protein in HK-2 cells. (E) Relative band intensity of PMP70, CAT, BCL2, BAX and Cleaved Caspase-3 protein in HK-2 cells (n=3). Data are presented as means ± SEMs. *P < 0.05vs NG; **P < 0.05vs HG and PA; ns, not significant. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.7. Decreased DsbA-L expression and altered peroxisomal function are associated with kidney injury in DN patients
Obviously decreased expression of DsbA-L was observed in the renal tubular cells of patients with DN compared with those of controls as confirmed by immunofluorescence, and this finding was consistent with the pathological grade (Fig. 7A). Moreover, the expression of DsbA-L in the tubules of diabetic kidney was positively correlated with eGFR (Fig. 7B), and negatively correlated with degrees of IFTA(Fig. 7C). Similar result was obtained for CAT and peroxisomal function (Fig. 7D–H). In addition, we also found that the expression of DsbA-L had a positive correlation with peroxisomal function (Fig. 7I).
Fig. 7.
Decreased DsbA-L expression and altered peroxisomal function are associated with kidney injury in DN patients. (A) IF staining showed the expression of DsbA-L in renal biopsy specimens of patients with diabetic nephropathy (DN) and control. Scale bar: 20 μm. (B, C) Correlation between DsbA-L expression and eGFR (n=13) (B) and IFTA (n=13) (C) in patients with DN. (D) Representative immunofluorescence images of CAT (green) and PMP70 (red) in kidney tissues. The nuclei were counterstained by DAPI (blue). Overlap analyzed by the colocalization highlighter plugin (ImageJ) are shown in yellow. Scale bar: 20 μm. (E, F) Correlation between CAT expression and eGFR (n=13) (E) and IFTA (n=13) (F) in patients with DN. (G, H) Colocalization of CAT and PMP70 determined from the images (D) represented in using Pearson’s correlation coefficient. Correlation between Pearson’s correlation coefficient and eGFR (n=13) (G) and IFTA (n=13) (H) in patients with DN. (I) Correlation between DsbA-L expression and Pearson’s correlation coefficient (n=8). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
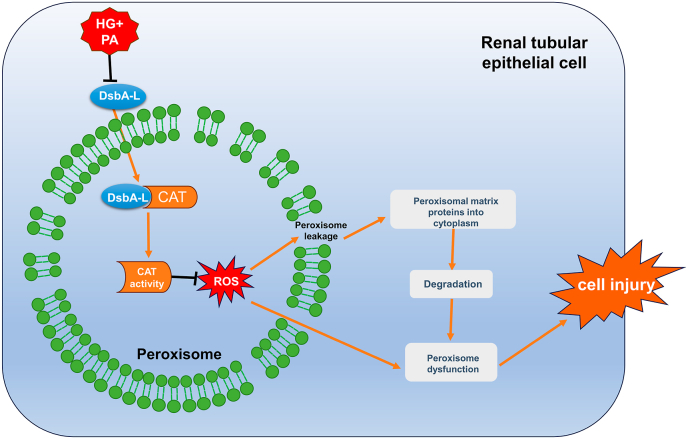
In this study, we found that knockout of DsbA-L gene in PTC further worsened peroxisomal function and tubular oxidative damage in the kidney of DN mice. DsbA-L is localized in peroxisomes, and upregulation of DsbA-L expression in the peroxisomes helps to regulate the activity of CAT by binding to it, alleviate leakage of peroxisomal membranes, improve peroxisome function, and thereby improving tubular cell damage inducted by HG and PA (Fig. 8). These results suggest that DsbA-L plays a key role in the regulation of peroxisomal function and ameliorates tubular cell damage in DN.
Fig. 8.
Proposed schematic model for the role of DsbA-L in improving HG and PA-induced PTC injury. Under the induction of HG and PA, the expression of DsbA-L is down-regulated, the activity of CAT decreases, and the production of ROS increases, which in turn destroys the peroxisomal membrane structure and causes the peroxisomal proteins to leak into the cytoplasm and be degraded, resulting in impaired peroxisome function and increased cell injury. The overexpression of DsbA-L can interact with CAT to increase its activity, thereby improving peroxisomal function and tubular damage.
DsbA-L is a vital antioxidant protein mainly located in the peroxisome and mitochondria [20,26]. Previous studies have shown that DsbA-L mediates renal tubular oxidative damage in DN [22], but the precise mechanisms remain unclear. In this study, we also found that specific knockout of DsbA-L gene in PTC could lead to diabetic kidney damage induced by HFD and STZ (Fig. 1). To further identify the relevant mechanisms, proteomic was carried out and indicated that DsbA-L was closely associated with peroxisomal function, and DsbA-L deletion in diabetic mice further worsened peroxisomal function and aggravated oxidative damage compared with diabetic DsbA-Lctrl mice (Figs. 2 and 3). Moreover, overexpression of DsbA-L partially reversed peroxisomal function and oxidative damage in HG and PA-induced HK-2 cells (Fig. 4), which is consistent with the restoration of peroxisomal function being able to protect the kidney from STZ-induced renal oxidative damage in diabetic mice [18]. These results suggest that DsbA-L depletion under diabetic conditions leads to renal oxidative damage, partly due to impaired peroxisomal function. Additionally, the changes of oxidative stress, apoptosis, and peroxisomal function were not obvious in DsbA-LptKO mice. Similar observations were previously noted in DsbA-L full knockout and liver gene-specific knockout mice [22,25]. Based on these observations, we hypothesized that DsbA-L is a stress-responsive protein. DsbA-L deficient mice display no anomalies under basal conditions, but the deficiency of DsbA-L would disturb the redox homeostasis during stress condition such as diabetes. ACOX1 is the first and rate-limiting enzyme in peroxisomal β-oxidation of very-long-chain fatty acids (VLCFAs), and ACOX1 activity leads to H2O2 production, which is then transformed into water and oxygen by CAT [37]. Our study revealed a decrease in ACOX1 expression but an increase in ROS under HG and PA, which was consistent with the findings of You et al. [37,38]. This finding may be attributed to the decrease in ACOX1, which results in the accumulation of VLCFAs. The decrease in CAT and the accumulation of VLCFAs induce ROS production [[37], [38], [39]].
The next question is how DsbA-L affects the morphology and function of peroxisome in DN. First of all, our data further confirmed that DsbA-L is predominantly located in the peroxisomes of tubular epithelial cells. The database revealed that CAT may be involved in the downstream effector mechanisms of DsbA-L (Fig. 4). It has been known that CAT is predominantly located in the peroxisomes, where it accounts for 10%–25% of peroxisomal protein [40]. Additionally, humans with inherited CAT deficiency (for acatalasemia, hypocatalasemia) are at greater risk of developing age-related disorders such as diabetes, atherosclerosis, and cancer [41]. A previous study indicated that CAT deficiency leads to ineffective ROS clearance [32]. And CAT could protect the kidney from oxidative stress damage in STZ-induced diabetic mice by maintaining peroxisomal function [18]. Moreover, Liu et al. reported a potential interaction between DsbA-L and CAT through affinity purification mass spectrometry and complementary proximity labeling [35]. It is suggested that DsbA-L may protect DN tubular cells from oxidative damage attributed to the effects of CAT on peroxisomal morphology and function. Here we discovered co-localization and interaction between DsbA-L and CAT. And under the condition of HG and PA, the expression of CAT and the function of peroxisome decreased. Overexpression of DsbA-L in vitro restored CAT expression, alleviated peroxisomal ROS production and cell apoptosis in HG and PA treated HK-2 cells, which could be reversed by CAT inhibitor 3-AT (Figs. 4 and 5). In addition, in order to rule out that DsbA-L is the mitochondrial pathway to improve the oxidative damage of HK-2 cells induced by HG and PA, we constructed the DsbA-L-PTS1 plasmid that was localized in peroxisome, and found that overexpressed DsbA-L-PTS1 improved the function of peroxisome through CAT (Fig. 6). These data strongly support that DsbA-L can mediate direct interactions with CAT, thus enhancing peroxisomal function to ameliorate the damage of DN tubular cells.
Furthermore, it is unclear how the reduction of CAT activity in DN results in decrease of peroxisomal matrix proteins and impaired peroxisomal function. Some studies have shown that inhibition of CAT activity will trigger a decrease in autophagy and peroxisomal density [42,43]. Deficiency or inactivation of CAT can result in the inhibition of peroxisomal biosynthesis [44,45]. Studies have also shown that the inhibition of CAT activity will lead to accumulation of peroxisomal ROS and the leakage of proteins in the peroxisome to the cytoplasm, which in turn causes the selective degradation of peroxisomal matrix proteins, thereby damaging the function of the peroxisome [31]. But it has also been shown that peroxisomes can selectively inhibit the import of CAT under conditions of oxidative stress, leading to elevated levels of CAT in the cytoplasm [46], which significantly protects the cytoplasm from H2O2-induced damage [47]. The difference mentioned above could relate to cell type and intervention duration. Our study indicates that CAT is mislocated to the cytoplasm in HG and PA-induced HK-2 cells in addition to the decrease in its activity, and DsbA-L overexpression can reverse this phenomenon (Fig. 4). So, we speculate that DsbA-L may regulate CAT activity, improve peroxisome membrane leakage and degradation of peroxisomal matrix proteins to maintain peroxisome function and improve oxidative damage in renal tubular cells. The results in Fig. 5 also confirmed our hypothesis. An interesting question remains to be elucidated in this study is that how DsbA-L regulates CAT activity. Studies have shown that when mammalian cells are exposed to external H2O2 or other ROS stimulus, CAT can be phosphorylated [48,49], succinylated [50,51], monomethylated [52],or ubiquitinated [53,54], thereby altering CAT activity. Further experiments are required to explore whether the regulation of CAT activity by DsbA-L in DN is associated with these post-translational modifications.
Collectively, our study reveals the importance of DsbA-L in maintaining peroxisomal function in DN. We also demonstrated that DsbA-L improves peroxisome abundance and function by regulating CAT activity, which provides new insights into renal tubular damage in DN and provides a new potential target for the treatment of DN.
Funding
This work was supported by Natural Science Foundation of Hunan Province (2021JC0003) and National Natural Science Foundation of China (No. 81730018 and 82000697).
Author contributions
Y.L. conducted the experiments and composed the manuscript. W. C., C.L., L.L., M.Y., N.J., S.L., Y.X., C.L., Y.H., H.Z. and S.L. provided technical support for and participated in discussions about this study. S. Y., X.Z., L.X., and L.S. designed this study and edited this manuscript. All authors have approved the final version of the manuscript. L.S. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Declaration of competing interest
The authors declare no potential conflicts of interest.
Acknowledgements
We appreciate all the members of our study team from Department of Nephrology of Second Xiangya Hospital Affiliated to Central South University for their assistance in completing this project.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102855.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Tuttle K.R., Bakris G.L., Bilous R.W., Chiang J.L., de Boer I.H., Goldstein-Fuchs J., Hirsch I.B., Kalantar-Zadeh K., Narva A.S., Navaneethan S.D., Neumiller J.J., Patel U.D., Ratner R.E., Whaley-Connell A.T., Molitch M.E. Diabetic kidney disease: a report from an ADA Consensus Conference. Am. J. Kidney Dis. 2014;64:510–533. doi: 10.1053/j.ajkd.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 2.Gilbert R.E. Proximal tubulopathy: prime mover and key therapeutic target in diabetic kidney disease. Diabetes. 2017;66:791–800. doi: 10.2337/db16-0796. [DOI] [PubMed] [Google Scholar]
- 3.Nowak N., Skupien J., Niewczas M.A., Yamanouchi M., Major M., Croall S., Smiles A., Warram J.H., Bonventre J.V., Krolewski A.S. Increased plasma kidney injury molecule-1 suggests early progressive renal decline in non-proteinuric patients with type 1 diabetes. Kidney Int. 2016;89:459–467. doi: 10.1038/ki.2015.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duan S., Lu F., Song D., Zhang C., Zhang B., Xing C., Yuan Y. Current challenges and future perspectives of renal tubular dysfunction in diabetic kidney disease. Front. Endocrinol. 2021;12 doi: 10.3389/fendo.2021.661185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bagby S.P. Diabetic nephropathy and proximal tubule ROS: challenging our glomerulocentricity. Kidney Int. 2007;71:1199–1202. doi: 10.1038/sj.ki.5002286. [DOI] [PubMed] [Google Scholar]
- 6.Bonventre J.V. Can we target tubular damage to prevent renal function decline in diabetes? Semin. Nephrol. 2012;32:452–462. doi: 10.1016/j.semnephrol.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalla Vestra M., Saller A., Bortoloso E., Mauer M., Fioretto P. Structural involvement in type 1 and type 2 diabetic nephropathy. Diabetes Metab. 2000;26(Suppl 4):8–14. [PubMed] [Google Scholar]
- 8.Vallon V. The proximal tubule in the pathophysiology of the diabetic kidney. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011;300:R1009–R1022. doi: 10.1152/ajpregu.00809.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rhodin J. Thesis Kalolinska Instutet; 1954. Correlation of Ultrastructural Organization and Function in Normal and Experimentally Changed Proximal Convoluted Tubule Cells of the Mouse Kidney. [Google Scholar]
- 10.Zalckvar E., Schuldiner M. Beyond rare disorders: a new era for peroxisomal pathophysiology. Mol. Cell. 2022;82:2228–2235. doi: 10.1016/j.molcel.2022.05.028. [DOI] [PubMed] [Google Scholar]
- 11.Bonekamp N.A., Volkl A., Fahimi H.D., Schrader M. Reactive oxygen species and peroxisomes: struggling for balance. Biofactors. 2009;35:346–355. doi: 10.1002/biof.48. [DOI] [PubMed] [Google Scholar]
- 12.De Duve C., Baudhuin P. Peroxisomes (microbodies and related particles) Physiol. Rev. 1966;46:323–357. doi: 10.1152/physrev.1966.46.2.323. [DOI] [PubMed] [Google Scholar]
- 13.Mi J., Kirchner E., Cristobal S. Quantitative proteomic comparison of mouse peroxisomes from liver and kidney. Proteomics. 2007;7:1916–1928. doi: 10.1002/pmic.200600638. [DOI] [PubMed] [Google Scholar]
- 14.Vasko R. Peroxisomes and kidney injury. Antioxidants Redox Signal. 2016;25:217–231. doi: 10.1089/ars.2016.6666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tserga A., Pouloudi D., Saulnier-Blache J.S., Stroggilos R., Theochari I., Gakiopoulou H., Mischak H., Zoidakis J., Schanstra J.P., Vlahou A., Makridakis M. Proteomic analysis of mouse kidney tissue associates peroxisomal dysfunction with early diabetic kidney disease. Biomedicines. 2022;10 doi: 10.3390/biomedicines10020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burton J.B., Silva-Barbosa A., Bons J., Rose J., Pfister K., Simona F., Gandhi T., Reiter L., Bernhardt O., Hunter C.L., Goetzman E.S., Sims-Lucas S., Schilling B. Substantial downregulation of mitochondrial and peroxisomal proteins during acute kidney injury revealed by data-independent acquisition proteomics. bioRxiv. 2023 doi: 10.1101/2023.02.26.530107. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y., Zhang X., Yao H., Chen X., Shang L., Li P., Cui X., Zeng J. Peroxisome-generated succinate induces lipid accumulation and oxidative stress in the kidneys of diabetic mice. J. Biol. Chem. 2022;298 doi: 10.1016/j.jbc.2022.101660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hwang I., Lee J., Huh J.Y., Park J., Lee H.B., Ho Y.S., Ha H. Catalase deficiency accelerates diabetic renal injury through peroxisomal dysfunction. Diabetes. 2012;61:728–738. doi: 10.2337/db11-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morel F., Rauch C., Petit E., Piton A., Theret N., Coles B., Guillouzo A. Gene and protein characterization of the human glutathione S-transferase kappa and evidence for a peroxisomal localization. J. Biol. Chem. 2004;279:16246–16253. doi: 10.1074/jbc.M313357200. [DOI] [PubMed] [Google Scholar]
- 20.Thomson R.E., Bigley A.L., Foster J.R., Jowsey I.R., Elcombe C.R., Orton T.C., Hayes J.D. Tissue-specific expression and subcellular distribution of murine glutathione S-transferase class kappa. J. Histochem. Cytochem. 2004;52:653–662. doi: 10.1177/002215540405200509. [DOI] [PubMed] [Google Scholar]
- 21.Morel F., Aninat C. The glutathione transferase kappa family. Drug Metab. Rev. 2011;43:281–291. doi: 10.3109/03602532.2011.556122. [DOI] [PubMed] [Google Scholar]
- 22.Gao P., Yang M., Chen X., Xiong S., Liu J., Sun L. DsbA-L deficiency exacerbates mitochondrial dysfunction of tubular cells in diabetic kidney disease. Clin. Sci. (Lond.) 2020;134:677–694. doi: 10.1042/CS20200005. [DOI] [PubMed] [Google Scholar]
- 23.Bai J., Cervantes C., Liu J., He S., Zhou H., Zhang B., Cai H., Yin D., Hu D., Li Z., Chen H., Gao X., Wang F., O'Connor J.C., Xu Y., Liu M., Dong L.Q., Liu F. DsbA-L prevents obesity-induced inflammation and insulin resistance by suppressing the mtDNA release-activated cGAS-cGAMP-STING pathway. Proc. Natl. Acad. Sci. U.S.A. 2017;114:12196–12201. doi: 10.1073/pnas.1708744114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oniki K., Nohara H., Nakashima R., Obata Y., Muto N., Sakamoto Y., Ueno-Shuto K., Imafuku T., Ishima Y., Watanabe H., Maruyama T., Otake K., Ogata Y., Suico M.A., Kai H., Shuto T., Saruwatari J. The DsbA-L gene is associated with respiratory function of the elderly via its adiponectin multimeric or antioxidant properties. Sci. Rep. 2020;10:5973. doi: 10.1038/s41598-020-62872-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen H., Bai J., Dong F., Fang H., Zhang Y., Meng W., Liu B., Luo Y., Liu M., Bai Y., Abdul-Ghani M.A., Li R., Wu J., Zeng R., Zhou Z., Dong L.Q., Liu F. Hepatic DsbA-L protects mice from diet-induced hepatosteatosis and insulin resistance. FASEB J. 2017;31:2314–2326. doi: 10.1096/fj.201600985R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X., Han Y., Gao P., Yang M., Xiao L., Xiong X., Zhao H., Tang C., Chen G., Zhu X., Yuan S., Liu F., Dong L.Q., Liu F., Kanwar Y.S., Sun L. Disulfide-bond A oxidoreductase-like protein protects against ectopic fat deposition and lipid-related kidney damage in diabetic nephropathy. Kidney Int. 2019;95:880–895. doi: 10.1016/j.kint.2018.10.038. [DOI] [PubMed] [Google Scholar]
- 27.Li C., Li L., Yang M., Yang J., Zhao C., Han Y., Zhao H., Jiang N., Wei L., Xiao Y., Liu Y., Xiong X., Xi Y., Luo S., Deng F., Chen W., Yuan S., Zhu X., Xiao L., Sun L. PACS-2 ameliorates tubular injury by facilitating endoplasmic reticulum-mitochondria contact and mitophagy in diabetic nephropathy. Diabetes. 2022;71:1034–1050. doi: 10.2337/db21-0983. [DOI] [PubMed] [Google Scholar]
- 28.Han Y., Xu X., Tang C., Gao P., Chen X., Xiong X., Yang M., Yang S., Zhu X., Yuan S., Liu F., Xiao L., Kanwar Y.S., Sun L. Reactive oxygen species promote tubular injury in diabetic nephropathy: the role of the mitochondrial ros-txnip-nlrp3 biological axis. Redox Biol. 2018;16:32–46. doi: 10.1016/j.redox.2018.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tervaert T.W., Mooyaart A.L., Amann K., Cohen A.H., Cook H.T., Drachenberg C.B., Ferrario F., Fogo A.B., Haas M., de Heer E., Joh K., Noel L.H., Radhakrishnan J., Seshan S.V., Bajema I.M., Bruijn J.A., Renal Pathology S. Pathologic classification of diabetic nephropathy. J. Am. Soc. Nephrol. 2010;21:556–563. doi: 10.1681/ASN.2010010010. [DOI] [PubMed] [Google Scholar]
- 30.Elsner M., Gehrmann W., Lenzen S. Peroxisome-generated hydrogen peroxide as important mediator of lipotoxicity in insulin-producing cells. Diabetes. 2011;60:200–208. doi: 10.2337/db09-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mu Y., Maharjan Y., Kumar Dutta R., Wei X., Kim J.H., Son J., Park C., Park R. Pharmacological inhibition of catalase induces peroxisome leakage and suppression of LPS induced inflammatory response in Raw 264.7 cell. PLoS One. 2021;16 doi: 10.1371/journal.pone.0245799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mao X., Bharti P., Thaivalappil A., Cao K. Peroxisomal abnormalities and catalase deficiency in hutchinson-gilford progeria syndrome. Aging (Albany NY) 2020;12:5195–5208. doi: 10.18632/aging.102941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baldwin H.A., Wang C., Kanfer G., Shah H.V., Velayos-Baeza A., Dulovic-Mahlow M., Bruggemann N., Anding A., Baehrecke E.H., Maric D., Prinz W.A., Youle R.J. VPS13D promotes peroxisome biogenesis. J. Cell Biol. 2021;220 doi: 10.1083/jcb.202001188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang C., Okazaki T. Control of mitochondrial dynamics and apoptotic pathways by peroxisomes. Front. Cell Dev. Biol. 2022;10 doi: 10.3389/fcell.2022.938177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu X., Salokas K., Tamene F., Jiu Y., Weldatsadik R.G., Ohman T., Varjosalo M. An AP-MS- and BioID-compatible MAC-tag enables comprehensive mapping of protein interactions and subcellular localizations. Nat. Commun. 2018;9:1188. doi: 10.1038/s41467-018-03523-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dash B., Phillips T.D. Molecular characterization of a catalase from Hydra vulgaris. Gene. 2012;501:144–152. doi: 10.1016/j.gene.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.You L., Chen J., Liu W., Xiang Q., Luo Z., Wang W., Xu W., Wu K., Zhang Q., Liu Y., Wu J. Enterovirus 71 induces neural cell apoptosis and autophagy through promoting ACOX1 downregulation and ROS generation. Virulence. 2020;11:537–553. doi: 10.1080/21505594.2020.1766790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baarine M., Andreoletti P., Athias A., Nury T., Zarrouk A., Ragot K., Vejux A., Riedinger J.M., Kattan Z., Bessede G., Trompier D., Savary S., Cherkaoui-Malki M., Lizard G. Evidence of oxidative stress in very long chain fatty acid--treated oligodendrocytes and potentialization of ROS production using RNA interference-directed knockdown of ABCD1 and ACOX1 peroxisomal proteins. Neuroscience. 2012;213:1–18. doi: 10.1016/j.neuroscience.2012.03.058. [DOI] [PubMed] [Google Scholar]
- 39.Doria M., Nury T., Delmas D., Moreau T., Lizard G., Vejux A. Protective function of autophagy during VLCFA-induced cytotoxicity in a neurodegenerative cell model. Free Radic. Biol. Med. 2019;137:46–58. doi: 10.1016/j.freeradbiomed.2019.04.016. [DOI] [PubMed] [Google Scholar]
- 40.Reumann S., Ma C., Lemke S., Babujee L. AraPerox. A database of putative Arabidopsis proteins from plant peroxisomes. Plant Physiol. 2004;136:2587–2608. doi: 10.1104/pp.104.043695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goth L., Nagy T. Inherited catalase deficiency: is it benign or a factor in various age related disorders? Mutat. Res. 2013;753:147–154. doi: 10.1016/j.mrrev.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 42.Nitta Y., Muraoka-Hirayama S., Sakurai K. Catalase is required for peroxisome maintenance during adipogenesis. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2020;1865 doi: 10.1016/j.bbalip.2020.158726. [DOI] [PubMed] [Google Scholar]
- 43.Lee J.N., Dutta R.K., Maharjan Y., Liu Z.Q., Lim J.Y., Kim S.J., Cho D.H., So H.S., Choe S.K., Park R. Catalase inhibition induces pexophagy through ROS accumulation. Biochem. Biophys. Res. Commun. 2018;501:696–702. doi: 10.1016/j.bbrc.2018.05.050. [DOI] [PubMed] [Google Scholar]
- 44.Koepke J.I., Wood C.S., Terlecky L.J., Walton P.A., Terlecky S.R. Progeric effects of catalase inactivation in human cells. Toxicol. Appl. Pharmacol. 2008;232:99–108. doi: 10.1016/j.taap.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 45.Wang B., Van Veldhoven P.P., Brees C., Rubio N., Nordgren M., Apanasets O., Kunze M., Baes M., Agostinis P., Fransen M. Mitochondria are targets for peroxisome-derived oxidative stress in cultured mammalian cells. Free Radic. Biol. Med. 2013;65:882–894. doi: 10.1016/j.freeradbiomed.2013.08.173. [DOI] [PubMed] [Google Scholar]
- 46.Fujiki Y., Bassik M.C. A new paradigm in catalase research. Trends Cell Biol. 2021;31:148–151. doi: 10.1016/j.tcb.2020.12.006. [DOI] [PubMed] [Google Scholar]
- 47.Lismont C., Revenco I., Fransen M. Peroxisomal hydrogen peroxide metabolism and signaling in health and disease. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20153673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rafikov R., Kumar S., Aggarwal S., Hou Y., Kangath A., Pardo D., Fineman J.R., Black S.M. Endothelin-1 stimulates catalase activity through the PKCdelta-mediated phosphorylation of serine 167. Free Radic. Biol. Med. 2014;67:255–264. doi: 10.1016/j.freeradbiomed.2013.10.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cao C., Leng Y., Kufe D. Catalase activity is regulated by c-Abl and Arg in the oxidative stress response. J. Biol. Chem. 2003;278:29667–29675. doi: 10.1074/jbc.M301292200. [DOI] [PubMed] [Google Scholar]
- 50.Park J., Chen Y., Tishkoff D.X., Peng C., Tan M., Dai L., Xie Z., Zhang Y., Zwaans B.M., Skinner M.E., Lombard D.B., Zhao Y. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol. Cell. 2013;50:919–930. doi: 10.1016/j.molcel.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baker A., Lin C.C., Lett C., Karpinska B., Wright M.H., Foyer C.H. Catalase: a critical node in the regulation of cell fate. Free Radic. Biol. Med. 2023;199:56–66. doi: 10.1016/j.freeradbiomed.2023.02.009. [DOI] [PubMed] [Google Scholar]
- 52.Larsen S.C., Sylvestersen K.B., Mund A., Lyon D., Mullari M., Madsen M.V., Daniel J.A., Jensen L.J., Nielsen M.L. Proteome-wide analysis of arginine monomethylation reveals widespread occurrence in human cells. Sci. Signal. 2016;9:rs9. doi: 10.1126/scisignal.aaf7329. [DOI] [PubMed] [Google Scholar]
- 53.Kim W., Bennett E.J., Huttlin E.L., Guo A., Li J., Possemato A., Sowa M.E., Rad R., Rush J., Comb M.J., Harper J.W., Gygi S.P. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Akimov V., Barrio-Hernandez I., Hansen S.V.F., Hallenborg P., Pedersen A.K., Bekker-Jensen D.B., Puglia M., Christensen S.D.K., Vanselow J.T., Nielsen M.M., Kratchmarova I., Kelstrup C.D., Olsen J.V., Blagoev B. UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat. Struct. Mol. Biol. 2018;25:631–640. doi: 10.1038/s41594-018-0084-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.