Abstract
Staphylococcus aureus is the cause of serious infections in humans, including endocarditis, deep-seated abscesses, and bacteremia, which lead to toxic and septic shock syndromes. Rapid and direct identification of this bacterium specifically and ubiquitously directly from clinical specimens would be useful in improving the diagnosis of S. aureus infections in the clinical microbiology laboratory. A wide variety of kits based on biochemical characteristics efficiently identify S. aureus, but the rapidity and the accuracy of each of these methods combined with testing of clinically relevant antibiotic resistance genes need to be improved. On the basis of hybridization assays with randomly selected clones from an S. aureus genomic library, we have identified a chromosomal DNA fragment which is specific for S. aureus and which detected all 82 S. aureus isolates tested. This 442-bp fragment was sequenced and was used to design a set of PCR amplification primers. The PCR assay was also specific and ubiquitous for the identification from bacterial cultures of 195 clinical strains of S. aureus isolated from a variety of anatomical sites and obtained from hospitals throughout the world. The PCR assay that we have developed is simple and can be performed in about 1 h. This DNA-based test provides a novel diagnostic tool for the diagnosis of S. aureus infections.
Staphylococci are frequently isolated as etiologic agents of infectious processes, with Staphylococcus aureus being the most important human pathogen of this group. S. aureus causes superficial, deep-skin, soft-tissue infections, endocarditis, and bacteremia with metastatic abscess formation and a variety of toxin-mediated diseases including gastroenteritis, staphylococcal scalded-skin syndrome, and toxic shock syndrome (11, 24). The isolation of the bacterium from the site of infection or blood culture is required to link S. aureus to a specific disease. In some cases, detection of the organism may be hampered by ongoing antimicrobial therapy.
The rapid identification of S. aureus is important so that the appropriate antibiotic therapy can be initiated, and this bacterial species must be differentiated from coagulase-negative staphylococci, which frequently appear as contaminants or as a separate cause of bacteremia and urinary tract infections. Several methods for the rapid identification of S. aureus have been evaluated. These methods include coagulase tests (9), a panel of commercial agglutination tests (7, 13, 34), a hybridization test for rRNA (8), and an enzymatic test for the detection of thermostable nuclease (18, 23). Although identification with the thermonuclease enzyme test has shown an excellent correlation with the conventional identification of S. aureus isolates (18, 23), false-positive results due to thermonuclease activity in some strains of coagulase-negative staphylococci may occur (6, 10, 17). In addition, some streptococcal isolates may give a positive thermonuclease test (20). Excellent specificity but variable sensitivity (39 to 80%) was noted with diagnostic kits based on agglutination for identification directly from clinical specimens (22). Furthermore, several of these kits may fail to detect methicillin-resistant staphylococci (12, 13, 21, 25, 34). The hybridization test for rRNA showed excellent specificity for S. aureus but demonstrated an evident lack of sensitivity for detection from blood cultures (8). Brakstad et al. (5) have developed a PCR assay specific for S. aureus by targeting the nuc gene coding for the staphylococcal thermonuclease. This assay was shown to be suitable for diagnostic purposes.
Although S. aureus is easy to cultivate and is not difficult to identify, there is still a need for the development of a rapid and sensitive DNA-based assay which is specific for S. aureus and which is suitable for the identification of S. aureus from blood cultures and for the direct detection of S. aureus from clinical specimens. In this study, we describe the development of a species-specific DNA-based assay for the identification of S. aureus. In order to select a genetic target suitable for diagnostic purposes, an S. aureus genomic library was screened by hybridization of S. aureus DNA to DNAs from an array of both gram-positive and gram-negative bacterial species. Using this strategy, we were able to identify a clone carrying a chromosomal DNA insert suitable as a probe for the identification of S. aureus. The genomic DNA fragment inserted into this clone was species specific and ubiquitous (i.e., it hybridized to DNA from any S. aureus strain) for the identification of S. aureus. This genomic DNA fragment was sequenced and was used to design PCR amplification primers for S. aureus-specific PCR assays. The simple and rapid PCR assay that we developed provides a new tool for the diagnosis of S. aureus infections, and the assay may be applied for the direct detection of S. aureus from blood cultures or clinical specimens.
MATERIALS AND METHODS
Bacterial strains.
The bacterial isolates used in this study were selected from the culture collection of the microbiology laboratories of the Centre Hospitalier Universitaire de Québec (Pavillon Centre Hospitalier de l’Université Laval [CHUL], Ste-Foy, Quebec, Canada), the Laval Hospital (Ste-Foy, Quebec, Canada), the Mount Sinai Hospital (Toronto, Ontario, Canada), the Shanghai Hospital (Shanghai, China), the Centers for Disease Control and Prevention (Atlanta, Ga.), the Institut Pasteur (Paris, France), the Laboratoire de Santé Publique du Québec (Sainte-Anne-de-Bellevue, Quebec, Canada), and the Universidad de Buenos Aires (Buenos Aires, Argentina). Eight S. aureus strains obtained from the American Type Culture Collection (ATCC; ATCC 13301, ATCC 25923, ATCC 29213, ATCC 27660, ATCC 33591, ATCC 33592, ATCC 33593, and ATCC 43300) were also used for this study. Duplicate isolates from the same patients, even if the sites of infection were different, were excluded from this study. Strains were cultured on sheep blood agar or in brain heart infusion (BHI) medium. Bacterial cultures were stored frozen (−80°C) in BHI medium containing 10% glycerol.
The specificity of the DNA-based tests was verified by using a panel of clinical bacterial isolates consisting of 42 gram-negative and 51 gram-positive bacterial species (Table 1). This battery of bacterial strains includes isolates obtained from both ATCC and the Microbiology Laboratory of CHUL. The ubiquity (i.e., ability to detect all strains of S. aureus) of the DNA-based tests was verified by using a battery of 195 clinical isolates identified as S. aureus by standard biochemical methods and obtained from CHUL (n = 80), Laval Hospital (n = 24), Mount Sinai Hospital (n = 5), Shanghai Hospital (n = 21), ATCC (n = 8), Centers for Disease Control and Prevention (n = 4), the Institut Pasteur (n = 2), the Laboratoire de Santé Publique du Québec (n = 50), and the Universidad de Buenos Aires (n = 1). The identities of all strains were reconfirmed to be S. aureus with the Staphaurex Plus system (Murex Diagnostics Canada, Guelph, Ontario, Canada). The 80 S. aureus strains from CHUL were initially identified by using the MicroScan Autoscan-4 system equipped with the Positive BP Combo Panel Type 6 (Dade Diagnostics, Mississauga, Ontario, Canada).
TABLE 1.
Bacterial species used to test the specificity of selected probes and PCR primers
| Bacterial species (no. of species) | No. of strains tested |
|---|---|
| Gram-positive species (48) | |
| Bacillus cereus | 1 |
| Bacillus subtilis | 1 |
| Corynebacterium diphtheriae | 1 |
| Corynebacterium pseudodiphtheriticum | 1 |
| Enterococcus durans | 1 |
| Enterococcus faecalis | 4 |
| Enterococcus faecium | 2 |
| Lactobacillus acidophilus | 1 |
| Listeria grayi | 1 |
| Listeria innocua | 1 |
| Listeria ivanovii | 1 |
| Listeria monocytogenes | 1 |
| Listeria murrayi | 1 |
| Listeria seeligeri | 1 |
| Micrococcus luteus | 2 |
| Micrococcus lylae | 1 |
| Staphylococcus arlettae | 1 |
| Staphylococcus aureus subsp. anaerobius | 1 |
| Staphylococcus aureus subsp. aureus | 5 |
| Staphylococcus auricularis | 1 |
| Staphylococcus capitis subsp. capitis | 2 |
| Staphylococcus capitis subsp. ureolyticus | 1 |
| Staphylococcus caprae | 1 |
| Staphylococcus carnosus | 1 |
| Staphylococcus caseolyticus | 1 |
| Staphylococcus chromogenes | 1 |
| Staphylococcus cohnii subsp. cohnii | 1 |
| Staphylococcus cohnii subsp. urealyticum | 1 |
| Staphylococcus delphini | 1 |
| Staphylococcus epidermidis | 5 |
| Staphylococcus equorum | 1 |
| Staphylococcus felis | 1 |
| Staphylococcus gallinarum | 1 |
| Staphylococcus haemolyticus | 2 |
| Staphylococcus hominis | 2 |
| Staphylococcus intermedius | 1 |
| Staphylococcus kloosi | 1 |
| Staphylococcus lentus | 1 |
| Staphylococcus lugdunensis | 1 |
| Staphylococcus saprophyticus | 5 |
| Staphylococcus schleiferi subsp. coagulans | 1 |
| Staphylococcus sciuri subsp. sciuri | 1 |
| Staphylococcus simulans | 1 |
| Staphylococcus warneri | 1 |
| Staphylococcus xylosus | 1 |
| Streptococcus agalactiae | 2 |
| Streptococcus bovis | 1 |
| Streptococcus pneumoniae | 4 |
| Streptococcus pyogenes | 2 |
| Streptococcus salivarius | 2 |
| Viridans group streptococci | 2 |
| Gram-negative species (42) | |
| Acinetobacter calcoaceticus | 1 |
| Acinetobacter lwoffi | 1 |
| Bordetella pertussis | 2 |
| Burkholderia cepacia | 1 |
| Citrobacter diversus | 1 |
| Citrobacter freundii | 1 |
| Comamonas acidovorans | 1 |
| Enterobacter aerogenes | 1 |
| Enterobacter agglomerans | 1 |
| Enterobacter cloacae | 1 |
| Escherichia coli | 2 |
| Haemophilus aegyptius | 1 |
| Haemophilus haemolyticus | 1 |
| Haemophilus influenzae | 1 |
| Haemophilus parahaemolyticus | 1 |
| Haemophilus parainfluenzae | 1 |
| Hafnia alvei | 1 |
| Kingella indologenes | 1 |
| Klebsiella oxytoca | 1 |
| Klebsiella pneumoniae | 2 |
| Moraxella atlantae | 1 |
| Moraxella catarrhalis | 1 |
| Moraxella urethralis | 1 |
| Morganella morganii | 1 |
| Neisseria caviae | 1 |
| Neisseria mucosa | 1 |
| Neisseria subflava | 1 |
| Proteus mirabilis | 2 |
| Proteus vulgaris | 1 |
| Providencia rettgeri | 1 |
| Providencia rustigianii | 1 |
| Providencia stuartii | 1 |
| Pseudomonas aeruginosa | 2 |
| Pseudomonas fluorescens | 1 |
| Pseudomonas putida | 1 |
| Salmonella choleraesuis | 1 |
| Salmonella typhimurium | 1 |
| Serratia marcescens | 1 |
| Shigella flexneri | 1 |
| Shigella sonnei | 1 |
| Stenotrophomonas maltophilia | 1 |
| Yersinia enterocolitica | 1 |
Genomic DNA library construction.
Genomic DNA from S. aureus ATCC 25923 was extracted from 1.5 ml of an overnight S. aureus culture in BHI medium. The cells were pelleted by centrifugation (7,000 × g for 5 min). The cell pellet was suspended in 1 ml of a solution containing 200 μg of lysostaphin (Sigma Chemical Co., St. Louis, Mo.) per ml and 100 μg of lysozyme (Sigma) per ml, and the solution was then incubated for 30 min at 37°C. Subsequently, the cell preparation was treated with 100 μg of proteinase K (ICN Biochemicals, Costa Mesa, Calif.) per ml for 1 h at 37°C and was then extracted with phenol-chloroform. The purified genomic DNA, recovered by ethanol precipitation, was partially digested with the restriction enzyme Sau3AI (New England Biolabs Ltd., Mississauga, Ontario, Canada). The resulting DNA fragments were cloned into the BamHI site of the plasmid vector pGEM-7Zf (Promega Corp., Madison, Wis.) by using T4 DNA ligase (New England Biolabs). Recombinant plasmids were transformed into competent Escherichia coli DH5α cells by using standard procedures (1, 27).
Plasmid DNA isolation was done either by the method of Birnboim and Doly (4) for small-scale preparations or by using the Wizard Maxiprep kit (Promega) for large-scale preparations.
Hybridization.
Probe preparation and labeling were as described previously (19). For all staphylococcal species, genomic DNA was extracted from 1.5 ml of an overnight culture in BHI medium as described earlier for construction of the S. aureus genomic DNA library, except that the phenol-chloroform treatment, the RNase treatment, and the ethanol precipitation were not performed. The crude DNA preparation was denatured and was then spotted onto a nylon membrane with a dot blot apparatus (19). The genomic DNA extraction method used for bacterial species other than staphylococci (Table 1) was identical except that lysostaphin was not incorporated into the lysis solution. Hybridization conditions and posthybridization washes were as described earlier (19). In order to ensure that the bacterial cell lysis protocol was efficient for all species tested, hybridizations with a 16S rRNA universal DNA probe were also performed. For this purpose, a 241-bp DNA fragment amplified from a conserved region of the 16S rRNA gene and labeled with [α-32P]dATP was used as a probe (19).
DNA sequencing.
Both strands of genomic DNA fragments were sequenced by the dideoxynucleotide chain termination sequencing method with SP6 and T7 sequencing primers by using the Applied Biosystems 373A Automated DNA Sequencer with the PRISM Sequenase Terminator Double-Stranded DNA Sequencing Kit (Applied Biosystems Division, Perkin-Elmer Corp., Foster City, Calif.).
PCR amplification.
Oligonucleotide primers were synthesized with a model 391 DNA synthesizer (Perkin-Elmer Corp., Applied Biosystems Division). For all bacterial species, amplification was performed directly from a bacterial colony or from a bacterial suspension whose turbidity was adjusted to that of a 0.5 McFarland standard, which corresponds to approximately 1.5 × 108 bacteria per ml. A portion of an isolated colony or 1 μl of the standardized bacterial suspension was transferred directly to a 20-μl PCR mixture containing 50 mM KCl, 10 mM Tris-HCl (pH 9.0), 0.1% Triton X-100, 2.5 mM MgCl2, 0.4 μM (each) the two S. aureus-specific primers Sa442-1 (5′-AAT CTT TGT CGG TAC ACG ATA TTC TTC ACG-3′; positions 5 to 34) and Sa442-2 (5′-CGT AAT GAG ATT TCA GTA GAT AAT ACA ACA-3′; positions 83 to 112), 200 μM (each) the four deoxynucleoside triphosphates, and 0.5 U of Taq DNA polymerase (Promega). In order to reduce the formation of nonspecific extension products, a hot-start protocol was performed (19). The PCR mixtures were subjected to thermal cycling (3 min at 96°C and then 30 or 40 cycles of 1 s at 95°C for the denaturation step and 30 s at 55°C for the annealing-extension step) with a PTC-200 thermal cycler (MJ Research Inc., Watertown, Mass.). The quick lysis and the rapid cycling for PCR amplification required slightly less than 1 h for the 30-cycle PCR and approximately 70 min for the 40-cycle PCR.
Primer sequences derived from highly conserved regions of the bacterial 16S rRNA gene (19) were used to provide an internal control for all PCRs. These primers can amplify a 241-bp product from any bacterial species (universal bacterial amplification). The internal control was integrated into all amplification reactions to verify the efficiencies of the quick protocol for bacterial lysis and the PCR assays as well as to ensure that significant PCR inhibition was absent. The internal control and the S. aureus-specific amplifications were performed simultaneously in multiplex PCR assays as described previously (19).
Ten microliters of the PCR-amplified reaction mixture was resolved by electrophoresis through a 2% agarose gel containing 0.5 μg of ethidium bromide per ml in Tris-borate-EDTA buffer (89 mM Tris, 89 mM boric acid, 2 mM EDTA) at 170 V for 15 min. The gels were visualized under 254-nm UV light. The sizes of the amplification products were estimated by comparison with a 50-bp molecular size standard ladder. The total time for the PCR assays from the start of the assay with a bacterial colony or a standardized bacterial suspension was approximately 1 h.
For determination of the sensitivities of the PCR assays, a culture of S. aureus in the logarithmic phase of growth (optical density at 600 nm, ≈0.7 to 0.8) was diluted in phosphate-buffered saline. Each dilution (2 μl) was tested in PCR assays to determine the minimal number of CFU which can be detected. The number of CFU was estimated by standard plating procedures. A similar approach was applied to determine the minimal number of recombinant plasmid molecules which can be detected. Sensitivity assays were performed with recombinant plasmids linearized by digestion with the restriction endonuclease SacI.
Nucleotide sequence accession number.
The nucleotide sequence of the S. aureus genomic fragment is available from GenBank as accession no. AFO33191.
RESULTS
Identification of S. aureus-specific genomic DNA fragments.
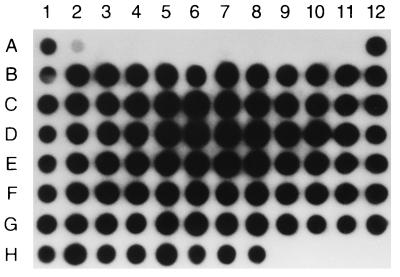
The S. aureus genomic library was selected for clones carrying species-specific DNA inserts by hybridization of S. aureus DNA to DNAs from an array of bacterial species (Table 1). A total of 12 randomly selected clones had to be tested in order to obtain a species-specific probe. For each clone, the genomic DNA insert was excised from the vector by digestion with the restriction endonucleases SacI and ClaI. The resulting 33 genomic DNA fragments with sizes ranging from 100 bp to 1.2 kbp were individually gel purified, labeled, and tested in hybridization assays. Using this strategy, we isolated a fragment of genomic DNA of 442 bp which was shown to be specific for S. aureus (Fig. 1). This probe hybridized only to DNAs from the S. aureus isolates, and no hybridization signal was observed with the other bacterial species listed in Table 1. Negative hybridization signals were not attributable to the inefficiency of the cell lysis protocol because hybridization with the 241-bp DNA probe amplified from the 16S rRNA gene showed a uniform hybridization signal for all bacterial species tested, thereby indicating adequate bacterial cell lysis (data not shown). Ubiquity tests performed with the S. aureus-specific probe with an array of 82 clinical strains of S. aureus (CHUL [n = 80] and ATCC [n = 2]) showed that DNAs from all strains hybridized specifically with the probe, thereby demonstrating 100% ubiquity (Fig. 2). The identification of all 82 of these strains as S. aureus was initially performed by using the MicroScan Autoscan-4 system and was subsequently reconfirmed by using the Staphaurex Plus system.
FIG. 1.
Test for specificity by dot blot hybridization by using DNAs from a variety of bacterial species (Table 1) as targets. (A) Test for specificity with the 32P-labeled, S. aureus-specific, 442-bp DNA fragment as a probe. (B) Example of a test for specificity performed with a nonspecific S. aureus genomic DNA probe. For panels A and B, a variety of gram-positive bacterial species (Table 1) were used as targets (locations 1A to 8D). DNAs from S. aureus isolates are at locations 9D (ATCC 43300), 10D (ATCC 33593), 11D (ATCC 29213), and 12D (ATCC 25923) in panels A and B. (C) Test for specificity with the 32P-labeled, S. aureus-specific, 442-bp DNA fragment as a probe and DNAs from a variety of gram-negative bacterial species (Table 1) as targets (locations 1A to 11D). Genomic DNA from an S. aureus isolate (ATCC 25923) was spotted at location 12D.
FIG. 2.
Ubiquity test by dot blot hybridization by using the 32P-labeled, S. aureus-specific, 442-bp DNA fragment as a probe and DNAs from 82 clinical isolates of S. aureus as targets. DNAs from the S. aureus strains are at locations 1A (ATCC 43300), 12A (ATCC 25923), and 1B to 8H (80 clinical strains from CHUL). A battery of eight different staphylococcal species including S. epidermidis, S. saprophyticus, S. simulans, S. lugdunensis, S. haemolyticus, S. hominis, S. warneri, and S. schleiferi were used as negative controls (locations 3A to 10A, respectively). Micrococcus luteus was also used as a negative control (location 11A). DNA from transformed E. coli DH5α carrying the recombinant plasmid pGEM-Sa442 was spotted at location 2A. No DNA was spotted at locations 9H to 12H.
Subcloning and sequencing of the S. aureus-specific probe.
The S. aureus-specific 442-bp genomic DNA fragment probe was subcloned into pGEM-7Zf. The resulting recombinant plasmid, pGEM-Sa442, was then used to transform E. coli DH5α. Subsequently, the sequences of both strands of the S. aureus genomic DNA insert were determined. Searches of this sequence in various data banks did not reveal any significant homologies with known sequences. From the sequence of this DNA insert, which hybridized specifically to S. aureus DNA, we attempted to design a pair of PCR primers that could be used in amplification assays for the detection of S. aureus. One set of optimal PCR primers derived from this sequence (see above) was designed with the help of Oligo, version 4.0, Primer Analysis software (National Biosciences, Plymouth, Minn.).
PCR assays.
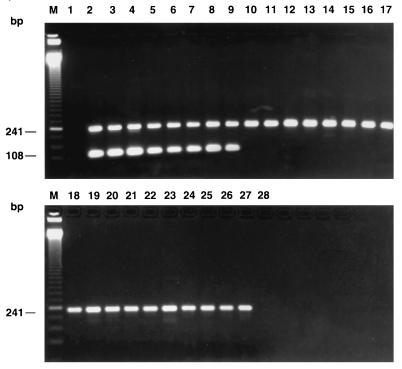
Specificity tests performed with the panel of gram-positive and gram-negative bacterial species listed in Table 1 indicated that the selected PCR primer pair amplified only DNA from clinical isolates of S. aureus. In order to ensure that the negative PCR results obtained with the bacterial species other than the target species were not attributable to PCR inhibitors or to the inadequacy of the PCR assay, all cell lysates were simultaneously amplified in a multiplex PCR assay with both the S. aureus-specific primers and the universal primers specific for the highly conserved bacterial 16S rRNA gene. The results indicated that all bacterial species were efficiently amplified by the universal primers, thereby showing the absence of PCR inhibitors and the suitability of the PCR assays for amplifying DNAs from the variety of bacterial species tested (Table 1 and Fig. 3). It is important that the S. aureus-specific PCR assay did not yield any specific amplification product with 25 staphylococcal species other than S. aureus (Fig. 3). This confirmed the specificity of this PCR assay for S. aureus. Increasing the number of amplification cycles from 30 to 40 does not appear to affect the specificity of the S. aureus-specific PCR assay because all species of coagulase-negative staphylococci tested (listed in Table 1) could not be amplified by the 40-cycle PCR assay (data not shown).
FIG. 3.
Multiplex PCR amplification with the S. aureus-specific PCR primer pair and the universal primers, which were used to provide an internal control. PCR assays were performed with 1 μl of a bacterial suspension whose turbidity was adjusted to that of a 0.5 McFarland standard prepared from a variety of reference strains or from clinical isolates from CHUL. Lanes: 2, S. aureus ATCC 33591; 3, S. aureus ATCC 33592; 4, S. aureus ATCC 33593; 5, S. aureus ATCC 43300; 6, S. aureus ATCC 25923; 7, S. aureus ATCC 13301; 8, S. aureus ATCC 29213; 9, S. aureus ATCC 27660; 10, S. saprophyticus ATCC 15305; 11, S. epidermidis ATCC 14990; 12, S. haemolyticus ATCC 29970; 13, S. hominis ATCC 27844; 14, S. simulans ATCC 27848; 15, S. lugdunensis ATCC 43809; 16, S. capitis subsp. ureolyticus ATCC 49326; 17, S. schleiferi subsp. coagulans ATCC 49545; 18, S. auricularis ATCC 33753; 19, S. cohnii subsp. urealyticum DSM 20260; 20, S. warneri ATCC 27836; 21, S. sciuri subsp. sciuri ATCC 29060; 22, S. xylosus LSPQ 2517; 23, M. luteus ATCC 9341; 24, S. pneumoniae ATCC 27336; 25, S. pyogenes ATCC 19615; 26, E. faecalis ATCC 29212; 27, E. faecium ATCC 51559. Lanes 1 and 28, controls to which no DNA was added; lanes M, 50-bp molecular size standard ladder.
The S. aureus-specific PCR assay was also tested for its ubiquity by performing PCR assays with the 82 clinical isolates of S. aureus previously used for the hybridization assays. The ubiquity test showed that DNAs from all isolates were specifically amplified by this PCR assay, thereby showing a perfect correlation with standard identification methods. Subsequently, the ubiquity of this PCR assay was further confirmed by testing an additional 113 strains of S. aureus originating from various geographical locations (Table 2). DNAs from all strains tested were also shown to be efficiently amplified, thereby demonstrating a 100% ubiquity (Table 2).
TABLE 2.
Sources of the clinical isolates of S. aureus used to test the ubiquity of the S. aureus PCR assay
| Source | No. of strains positive by PCR/no. of strains tested |
|---|---|
| 1. Microbiology Laboratory of CHUL | 80/80 |
| 2. Laboratoire de Santé Publique du Québec | 50/50 |
| 3. Microbiology Laboratory of Laval Hospital | 24/24 |
| 4. Microbiology Laboratory of the Shanghai Hospital | 21/21 |
| 5. ATCC | 8/8 |
| 6. Microbiology Laboratory of the Mount Sinai Hospital | 5/5 |
| 7. Centers for Disease Control and Prevention | 4/4 |
| 8. Institut Pasteur | 2/2 |
| 9. Universidad de Buenos Aires | 1/1 |
| Total | 195/195 |
We determined the sensitivity of our S. aureus-specific 30-cycle PCR assay. We found a detection limit of approximately 750 copies of the linearized recombinant plasmid pGEM-Sa442 or 500 copies of the S. aureus genome. In terms of the numbers of CFU, the detection limit with logarithmically growing S. aureus cultures was approximately 1.0 × 103 CFU. In order to enhance the sensitivity of the assay, we have increased the number of cycles. For PCR assays with 40 cycles, the sensitivity was increased to 25 copies of the linearized plasmid, 50 copies of the S. aureus genome, or 45 CFU of S. aureus, and the time for completion was increased by approximately 10 min.
DISCUSSION
Although S. aureus is not difficult to grow and is easy to identify, there is a need for the development of rapid and sensitive DNA-based assays which are suitable for the direct detection of S. aureus from clinical specimens to improve the rapidity and the accuracy of the diagnosis of S. aureus infections. Nucleic acid amplification by PCR has several applications in the detection of bacteria and other infectious agents in the clinical microbiology laboratory. In the present study, we have developed a rapid PCR-based assay to improve the ability to diagnose S. aureus infections. Initially, an S. aureus genomic library was screened randomly by hybridization in order to isolate a genomic DNA fragment which is species specific and ubiquitous for the identification of S. aureus. Using this strategy, we were able to obtain such a genomic DNA fragment, a probe of 442 bp. This hybridization assay, which was performed with samples from bacterial cultures, was found to be adequate for the confirmation of the presence of S. aureus in a culture. In order to simplify the assay as well as to improve its rapidity and sensitivity, the sequence of the 442-bp DNA probe, whose coding potential is unknown, was used to develop a PCR assay suitable for the rapid and accurate diagnosis of S. aureus infections. The PCR assay, which was performed directly with samples from bacterial colonies or a standardized bacterial suspension, was designed and optimized to be simple and performed in approximately 1 h. Our data indicate that the PCR assay (with 30 or 40 cycles of amplification) that we developed is also specific and ubiquitous for S. aureus. This simple and rapid PCR assay represents an alternative to currently used methods, especially when it is used in combination with other PCR assays developed by our group and which are specific for other clinically important bacterial species as well as for associated antibiotic resistance genes (data not shown). These assays can be combined in multiplex PCRs because all of our PCR assays are performed under uniform amplification conditions.
The S. aureus-specific PCR assay developed in this study was specific because it did not amplify DNAs from a variety of gram-positive and gram-negative bacterial species including 25 staphylococcal species other than S. aureus. Furthermore, this assay was shown to be 100% ubiquitous on the basis of the testing of 195 clinical S. aureus isolates from various countries, and 58 (29.7%) of these isolates were methicillin resistant. All 195 strains were initially reconfirmed as being S. aureus by using the Staphaurex Plus system, thereby showing a perfect correlation with the identifications by the S. aureus-specific PCR assay. Furthermore, there was no discordance between the S. aureus identifications obtained by the MicroScan Autoscan-4 system and those obtained by our PCR assay for the clinical isolates from CHUL. Therefore, the S. aureus genomic target of unknown coding potential selected for the PCR assay appears to be present in all S. aureus strains and is also well conserved in this species at the nucleotide level but is either absent from or distinct in other bacterial species including coagulase-negative staphylococci.
Others have developed PCR amplification assays targeting various S. aureus genes, including mecA coding for methicillin resistance (14, 31–33), as well as genes coding for toxins which are produced by many S. aureus isolates such as the exfoliative toxin (16, 26), the toxic shock syndrome toxin (16), and enterotoxins (16, 29, 35). However, all of these targets are not ubiquitously found in the species S. aureus, and consequently, these PCR assays are not suitable for the detection and identification of S. aureus. The coa gene coding for the coagulase protein has also been considered a candidate for the development of DNA-based diagnostic assays for S. aureus. However, this gene is highly polymorphic and cannot permit a ubiquitous identification of all strains of S. aureus (30). PCR protocols suitable for the specific detection and identification of S. aureus were designed to detect the femA gene in blood samples (32, 33). However, Jayaratne and Rutherford (15) have recently shown that these PCR assays may be unsuitable for diagnostic purposes because of polymorphism in the femA gene. PCR amplification of the nuc gene coding for the staphylococcal thermonuclease gene appears to be suitable for diagnostic purposes (5). This nuc gene PCR assay was shown to be specific and ubiquitous for S. aureus on the basis of testing of samples from positive blood culture bottles (5). Finally, another strategy oriented toward the identification of S. aureus by using a single-base-pair mismatch at the 3′ end of the primer in the 16S rRNA gene was optimized by testing 28 staphylococcal and nonstaphylococcal strains, and only S. aureus strains gave a positive reaction (28).
In this study, we have used a different approach to the elaboration of S. aureus-specific DNA-based diagnostic tests. A genomic DNA probe which was specific and ubiquitous for the identification of S. aureus was used to derive optimal PCR primers. The sequence of the 442-bp DNA probe did not show any significant homology with the sequences available in various data banks. Our goal was to develop a simple and rapid (about 1 h) PCR assay which is specific and ubiquitous for S. aureus and which can be applied to detection directly with samples from bacterial cultures or a variety of clinical specimens. We have designed a simple lysis protocol and a rapid thermal cycling which allow the assay to be performed in about 1 h. Even though this PCR protocol includes only 30 cycles of amplification, our results indicate that the levels of sensitivity achieved (i.e., about 1,000 CFU, 500 copies of the S. aureus genome, or 750 copies of linearized pGEM-Sa442) are sufficient for culture confirmation assays. This PCR assay was applied for the identification of S. aureus directly with samples from blood cultures, and preliminary data indicate that it is also suitable for that purpose. However, this application needs to be confirmed in a larger study to validate the procedure (work is in progress).
Increased levels of sensitivity of the PCR will be required for the direct detection of S. aureus directly from clinical specimens, in which the number of target cells can be much lower than in a bacterial colony or in blood cultures. It is possible to efficiently increase the sensitivity of a PCR assay by simply increasing the number of amplification cycles. For example, sensitivity assays performed with 40-cycle amplifications showed that the detection limit was increased to about 45 CFU, 50 copies of the S. aureus genome, or 25 copies of linearized pGEM-Sa442. Although the lysis protocol is rapid, it appears to lyse S. aureus cells relatively efficiently since the levels of sensitivity measured by titrations of CFU counts and linearized recombinant plasmids or numbers of S. aureus genomes were similar.
The shotgun approach that we have used in this study to generate S. aureus DNA probes and primers has also been useful for generating DNA-based tests for Moraxella catarrhalis (2) and Staphylococcus epidermidis (19). We have also developed specific and ubiquitous PCR assays for the detection of 16 other species and three genera of the most frequently encountered bacterial pathogens which account for approximately 85% of the bacteria routinely isolated in the microbiology laboratory (unpublished data). We have also developed PCR assays specific for 25 clinically relevant antibiotic resistance genes associated with these bacterial pathogens (unpublished data). The PCR assay for S. aureus reported here will be combined in multiplex with these PCR assays as well as with others which are under development. All of these assays can be performed under uniform amplification conditions and will be adapted for the direct detection of organisms from a variety of clinical specimens. A direct impact of such diagnostic tests is that they should allow the faster establishment of effective antibiotic therapy and reduce the level of use of empirical treatments with broad-spectrum antibiotics which are associated with high costs and toxicity (3). The consequent reduction of antibiotic use should reduce the emergence of resistance.
ACKNOWLEDGMENTS
We thank Louise Côté, director of the Microbiology Laboratory of CHUL, for free access to the laboratory and for providing the S. aureus and other clinical isolates. We thank Martin Gagnon, Caroline Paquet, Jean-Luc Simard, and Gisèle Chassé for technical help, which is highly appreciated. We also thank Louise Jetté (Laboratoire de Santé Publique du Québec), Pierre Auclair (Laval Hospital), Wang Fu (Shanghai Hospital), Donald E. Low (Mount Sinai Hospital), Fred C. Tenover (Centers for Disease Control and Prevention), Nevine El Sohl (Institut Pasteur), and Daniela Centron-Garcia (Universidad de Buenos Aires) for providing S. aureus strains. We thank Christian Ménard for critical comments regarding the manuscript.
Francis Martineau is a scholar from the Fonds de la Recherche en Santé du Québec. Marc Ouellette is a scholar from the Fonds de la Recherche en Santé du Québec and is the recipient of a Burroughs Wellcome Fund New Investigator Award in Molecular Parasitology. This study was supported by Infectio Diagnostic (I.D.I.) Inc., Sainte-Foy, Quebec, Canada.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. 1st ed. New York, N.Y: Green Publishing Associates and Wiley Interscience; 1987. [Google Scholar]
- 2.Beaulieu D, Bergeron M G, Roy P H. Development of a species-specific DNA probe for Moraxella (Branhamella) catarrhalis. Mol Cell Probes. 1991;5:37–48. doi: 10.1016/0890-8508(91)90036-j. [DOI] [PubMed] [Google Scholar]
- 3.Bergeron M G, Ouellette M. Diagnosing bacterial infectious diseases in one hour: an essential upcoming revolution. Infection. 1995;23:69–72. doi: 10.1007/BF01833867. [DOI] [PubMed] [Google Scholar]
- 4.Birnboim H C, Doly D. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brakstad O G, Aasbakk K, Maeland J A. Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. J Clin Microbiol. 1992;30:1654–1660. doi: 10.1128/jcm.30.7.1654-1660.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chesneau O, Allignet J, El Sohl N. Thermonuclease gene as a target nucleotide sequence for specific recognition of S. aureus. Mol Cell Probes. 1993;7:301–310. doi: 10.1006/mcpr.1993.1044. [DOI] [PubMed] [Google Scholar]
- 7.Croizé J, Gialannella P, Monnet D, Okada J, Orsi A, Voss A, Merlin S. Improved identification of Staphylococcus aureus using a new agglutination test. Results of an international study. APMIS. 1993;101:487–491. [PubMed] [Google Scholar]
- 8.Davis T E, Fuller D D. Direct identification of bacterial isolates in blood cultures by using a DNA probe. J Clin Microbiol. 1991;29:2193–2196. doi: 10.1128/jcm.29.10.2193-2196.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis T E, Fuller D D, Aeschleman E C. Rapid, direct identification of Staphylococcus aureus and Streptococcus pneumoniae from blood cultures using commercial immunological kits and modified conventional tests. Diagn Microbiol Infect Dis. 1992;15:295–300. doi: 10.1016/0732-8893(92)90014-k. [DOI] [PubMed] [Google Scholar]
- 10.Faruki H, Murray P. Medium dependence for rapid detection of thermonuclease in blood culture broths. J Clin Microbiol. 1986;24:482–483. doi: 10.1128/jcm.24.3.482-483.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fidalgo S, Vasquez F, Mendoza M C, Perez F, Mendez F J. Bacteremia due to Staphylococcus epidermidis: microbiological, epidemiologic, clinical, and prognostic features. Rev Infect Dis. 1990;12:520–528. doi: 10.1093/clinids/12.3.520. [DOI] [PubMed] [Google Scholar]
- 12.Fournier J-M, Boutonnier A, Bouvet A. Staphylococcus aureus strains which are not identified by rapid agglutination methods are of capsular polysaccharide serotype 5. J Clin Microbiol. 1989;27:1372–1374. doi: 10.1128/jcm.27.6.1372-1374.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fournier J-M, Bouvet A, Mathieu D, Nato F, Boutonnier A, Gerbal R, Brunengo P, Saulnier C, Sagot N, Slizewicz B, Mazie J-C. New latex reagent using monoclonal antibodies to capsular polysaccharide for reliable identification of both oxacillin-susceptible and oxacillin-resistant Staphylococcus aureus. J Clin Microbiol. 1993;31:1342–1344. doi: 10.1128/jcm.31.5.1342-1344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geha D L, Uhl J R, Gustaferro C A, Persing D H. Multiplex detection for identification of methicillin-resistant staphylococci in the clinical laboratory. J Clin Microbiol. 1994;32:1768–1772. doi: 10.1128/jcm.32.7.1768-1772.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jayaratne P, Rutherford C. Abstracts of the 97th General Meeting of the American Society for Microbiology 1997. Washington, D.C: American Society for Microbiology; 1997. Association of femA gene heterogeneity with unusual phenotypic characteristics of methicillin-resistant Staphylococcus aureus (MRSA), abstr. D-159; p. 236. [Google Scholar]
- 16.Johnson W M, Tyler S D, Ewan E P, Ashton F E, Pollard D R, Rozee K R. Detection of genes for enterotoxins, exfoliative toxins, and toxic shock syndrome toxin 1 in Staphylococcus aureus by the polymerase chain reaction. J Clin Microbiol. 1991;29:426–430. doi: 10.1128/jcm.29.3.426-430.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kloos W E, Bannerman T L. Update on clinical significance of coagulase-negative staphylococci. Clin Microbiol Rev. 1994;7:117–140. doi: 10.1128/cmr.7.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madison B M, Baselski V S. Rapid identification of Staphylococcus aureus in blood cultures by thermonuclease testing. J Clin Microbiol. 1983;18:722–724. doi: 10.1128/jcm.18.3.722-724.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martineau F, Picard F J, Roy P H, Ouellette M, Bergeron M G. Species-specific and ubiquitous DNA-based assays for rapid identification of Staphylococcus epidermidis. J Clin Microbiol. 1996;34:2888–2893. doi: 10.1128/jcm.34.12.2888-2893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park C E, De Melo Serrano M, Landgraf M, Huang J C, Stankiewicz Z, Rayman M K. A survey of microorganisms for thermonuclease production. Can J Microbiol. 1980;26:532–535. doi: 10.1139/m80-089. [DOI] [PubMed] [Google Scholar]
- 21.Piper J, Hadfield T, McCleskey F, Evans M, Friedstrom S, Lauderdale P, Winn R. Efficacies of rapid agglutination tests for identification of methicillin-resistant staphylococcal strains as Staphylococcus aureus. J Clin Microbiol. 1988;26:1907–1909. doi: 10.1128/jcm.26.9.1907-1909.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rappaport T, Sawyer K P, Nachamkin I. Evaluation of several commercial and immunologic methods for rapid identification of gram-positive cocci directly from blood cultures. J Clin Microbiol. 1988;21:1335–1338. doi: 10.1128/jcm.26.7.1335-1338.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ratner H B, Stratton C W. Thermonuclease test for same-day identification of Staphylococcus aureus in blood cultures. J Clin Microbiol. 1985;21:995–996. doi: 10.1128/jcm.21.6.995-996.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roberts F J, Geere I W, Coldman A. A three-year study of positive blood cultures, with emphasis on prognosis. Rev Infect Dis. 1991;13:34–46. doi: 10.1093/clinids/13.1.34. [DOI] [PubMed] [Google Scholar]
- 25.Ruane P J, Morgan M A, Citron D M, Mulligan M E. Failure of rapid agglutination methods to detect oxacillin-resistant Staphylococcus aureus. J Clin Microbiol. 1986;24:490–492. doi: 10.1128/jcm.24.3.490-492.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakurai S, Suzuki H, Machida K. Rapid identification by polymerase chain reaction of staphylococcal exfoliative toxin serotype A and B genes. Microbiol Immunol. 1995;39:379–386. doi: 10.1111/j.1348-0421.1995.tb02216.x. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Saruta K, Hoshina S, Machida K. Genetic identification of Staphylococcus aureus by polymerase chain reaction using single-base-pair mismatch in 16S ribosomal RNA gene. Microbiol Immunol. 1995;39:839–844. doi: 10.1111/j.1348-0421.1995.tb03280.x. [DOI] [PubMed] [Google Scholar]
- 29.Schumacher Perdreau F, Akatova A, Pulverer G. Detection of staphylococcal enterotoxin B and toxic shock syndrome toxin: PCR versus conventional methods. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg Abt 1 Orig. 1995;282:367–371. doi: 10.1016/s0934-8840(11)80706-1. [DOI] [PubMed] [Google Scholar]
- 30.Schwarzkopf A, Karch H. Genetic variation in Staphylococcus aureus coagulase genes: potential and limits for use as epidemiological marker. J Clin Microbiol. 1994;32:2407–2412. doi: 10.1128/jcm.32.10.2407-2412.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ubukata K, Nakagami S, Nitta A, Yamane A, Kawakami S, Sugiura M, Konno M. Rapid detection of the mecA gene in methicillin-resistant staphylococci by enzymatic detection of polymerase chain reaction products. J Clin Microbiol. 1992;30:1728–1733. doi: 10.1128/jcm.30.7.1728-1733.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ünal S, Hoskins J, Flokowitsch J E, Wu C Y E, Preston D A, Skatrud P L. Detection of methicillin-resistant staphylococci by using the polymerase chain reaction. J Clin Microbiol. 1992;30:1685–1691. doi: 10.1128/jcm.30.7.1685-1691.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vannuffel P, Gigi J, Ezzedine H, Vandercam B, Delmee M, Wauters G, Gala J L. Specific detection of methicillin-resistant Staphylococcus species by multiplex PCR. J Clin Microbiol. 1995;33:2864–2867. doi: 10.1128/jcm.33.11.2864-2867.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilkerson M, McAllister S, Miller J M, Heiter B J, Bourbeau P P. Comparison of five agglutination tests for identification of Staphylococcus aureus. J Clin Microbiol. 1997;35:148–151. doi: 10.1128/jcm.35.1.148-151.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson I G, Cooper J E, Gilmour A. Detection of enterotoxigenic Staphylococcus aureus in dried skimmed milk: use of the polymerase chain reaction for amplification and detection of staphylococcal enterotoxin genes entB and entC1 and the thermonuclease gene nuc. Appl Environ Microbiol. 1991;57:1793–1798. doi: 10.1128/aem.57.6.1793-1798.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]