Abstract
Four strains of an unknown coryneform bacterium were isolated in pure culture from females with urinary tract infections. Strong urease activity and the ability to slowly ferment maltose but not glucose were the most significant phenotypic features of this catalase-positive, nonmotile, nonlipophilic, rod-shaped bacterium which served to distinguish it from all other presently defined coryneform bacteria. Chemotaxonomic investigations demonstrated that the unknown bacterium belonged to the genus Corynebacterium. Comparative 16S rRNA gene sequence analysis revealed that the isolates were genealogically identical and represented a new subline within the genus Corynebacterium, for which the designation Corynebacterium riegelii sp. nov. is proposed. The type strain of Corynebacterium riegelii is CCUG 38180 (DSM 44326, CIP 105310).
For many years, the pathogenic potential of coryneform bacteria other than Corynebacterium diphtheriae has been grossly underestimated. It is only in the last decade that both clinical microbiologists and physicians became more appreciative both of the pathogenic potential of coryneform bacteria and of the great diversity of coryneform bacteria associated with human disease. This growing awareness, at least in part, stems from the implementation of more sophisticated schemes for the identification of coryneform bacteria, exploiting biochemical, chemotaxonomic, and molecular genetic methods. The application of such polyphasic approaches has led to the description of a plethora of new taxa of coryneform bacteria within recent years, and for some of them a clear disease association could be demonstrated (9). During an ongoing study on the differentiation of clinically significant coryneform bacteria isolated from human clinical specimens, workers in the Department of Medical Microbiology at the University of Zürich (DMMZ) isolated four strains from the urine of females with urinary tract infections. These isolates could not be assigned to any of the established taxa of coryneform bacteria. Therefore, we decided to study these four strains further using a polyphasic taxonomic approach including both phenotypic and molecular genetic methods. On the basis of the results of this investigation we propose a new Corynebacterium species, Corynebacterium riegelii sp. nov., for the strains associated with urinary tract infections in females.
MATERIALS AND METHODS
Strains and culture conditions.
Strains DMMZ 2415 (Culture Collection of the University of Göteborg [CCUG], Göteborg, Sweden; CCUG 38180), DMMZ 2582 (CCUG 38181), DMMZ 3128 (CCUG 38182), and DMMZ 3240 (CCUG 38242) were included in this study and were primarily isolated from dip-slide cystine-lactose-electrolyte-deficient agar (Orion Diagnostics, Espoo, Finland). All strains were subcultured on Columbia agar plates (Difco Laboratories, Detroit, Mich.) supplemented with 5% sheep blood for 24 h at 37°C in a 5% CO2 atmosphere. Staphylococcus aureus ATCC 25923 was used for analysis of the CAMP reaction.
Biochemical profiles.
The methods used for determination of the biochemical profiles have been described previously (7). API Coryne strips were read after 48 h, and API 50CH reactions performed with the 50 CHE medium (all from bioMérieux, Marcy l’Etoile, France) were read after 120 h of incubation at 37°C in ambient air.
Antimicrobial agent susceptibility patterns.
The MICs of 13 antimicrobial agents were determined with the Merlin Micronaut system (Merlin Diagnostics, Bornheim-Hersel, Germany) as outlined previously (5). MICs were interpreted according to the criteria for staphylococci established by the National Committee for Clinical Laboratory Standards (NCCLS) (12), although it is emphasized that NCCLS has not explicitly published criteria for coryneform bacteria.
Chemotaxonomic investigations.
Cellular fatty acid (CFA) patterns were determined with the Sherlock system (Microbial ID, Inc., Newark, Del.) as outlined previously (18). Techniques used for analyses of whole-cell hydrolysates for the presence of meso-diaminopimelic acid and mycolic acids were as described before (7).
Molecular genetic investigations.
A large fragment (ca. 1,500 bases) of the 16S rRNA genes of strains DMMZ 2415, DMMZ 2582, and DMMZ 3128 was amplified by PCR by using universal primers pA and pH* as described previously (11). The PCR products were purified by using a Prep-A-Gene kit (Bio-Rad, Hercules, Calif.) and were sequenced by using a Taq DyeDeoxy terminator cycle sequencing kit (Applied Biosystems, Inc., Foster City, Calif.) and a model 373A automatic sequencer (Applied Biosystems). The sequences determined were aligned with those of reference organisms (i.e., other actinomycetes with high G+C contents) obtained from the European Molecular Biology Laboratory (EMBL) Data Library by using the program PILEUP (3), and the alignment was corrected manually. An unrooted phylogenetic tree was constructed by using the neighbor-joining method (14). The stability of relationships was assessed by using the programs SEQBOOT, DNADIST, NEIGHBOR, and CONSENSE of the PHYLIP package (4).
Nucleotide sequence accession number.
The nucleotide sequence of the 16S rRNA of strain DMMZ 2415T (CCUG 38180T) has been deposited in the EMBL Data Library under accession no. Y14651.
RESULTS AND DISCUSSION
The four strains of coryneform bacteria were isolated in quantities of >105 CFU/ml from the urine of symptomatic female patients (between 21 and 62 years of age) with no underlying diseases. The unknown coryneforms grew in pure culture, and between 5 and 10 leukocytes/mm3 were seen on direct examination of the urine. Erythrocytes and proteins were not elevated, and crystals were not seen. There was no indication that the four outpatients were epidemiologically linked; the isolates had been recovered over a 1-year period, from August 1996 to July 1997.
All four strains grew as whitish, glistening, convex colonies with entire edges of up to 1.5 mm in diameter after 48 h of incubation. All strains were nonlipophilic. Two strains were of a creamy consistency, while the others exhibited a slightly sticky consistency. Gram stains showed typical club-shaped coryneform bacteria (indicative of true Corynebacterium spp.) of 1 to 3 μm in length, and they were arranged as single cells, in pairs, or in small clusters. The organisms were not partially acid fast.
The biochemical screening reactions of the four isolates by the scheme of von Graevenitz and Funke (17) were as follows: catalase positive; weak fermentative metabolism and weak anaerobic growth; nonmotile; nitrate reduction negative; strong urease activity (i.e., positive within 5 min in Christensen’s urea broth); esculin hydrolysis negative; CAMP reaction negative; and slow acid production from maltose (and ribose) but not from sucrose, mannitol, or xylose. Surprisingly, no acid formation from glucose was observed either in cystine Trypticase agar medium or in the API Coryne gallery (the corresponding numerical profiles were 0101224, 2001224, and 2101224 [two times]) and the API 50CH gallery. The authors are not aware of any other taxon belonging to the coryneform bacteria which produces acid from maltose but not from glucose. It was evident that the unknown coryneforms exhibited a biochemical pattern that was distinct from those of all other presently defined nonlipophilic, fermentative corynebacteria (Table 1). The unknown coryneform bacterium could be differentiated from the urea-splitting strains C. glucuronolyticum, C. pseudotuberculosis, and C. ulcerans by its negative CAMP reaction and from C. amycolatum by the unknown bacterium’s possession of mycolic acids (see below). All these other urea-splitting Corynebacterium species, however, also produce acid from glucose. Further testing of the enzymatic activities of the unknown bacterium revealed the presence of esterase (C4), esterase lipase (C8), leucine arylamidase, and cystine arylamidase.
TABLE 1.
Characteristics which differentiate C. riegelii from other fermenting, nonlipophilic Corynebacterium spp. encountered in human clinical specimensa
| Organism | Nitrate reduction | Urea hydrolysis | Esculin hydrolysis | Pyrazin- amidase activity | Alkaline phosphatase activity | Acid production from the following:
|
CAMP reaction | Other traits | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Glucose | Maltose | Sucrose | ||||||||
| C. riegelii | − | + | − | V | V | − | (+) | − | − | |
| C. amycolatum | V | V | − | + | + | + | V | V | − | Mycolic acid negative |
| C. argentoratense | − | − | − | + | V | + | − | − | − | Chymotrypsin positive |
| C. coyleae | − | − | − | + | + | (+) | − | − | + | |
| C. diphtheriae | V | − | − | − | − | + | + | − | − | Cystinase positive |
| C. glucuronolyticum | V | V | V | + | V | + | V | + | + | β-Glucuronidase positive |
| C. imitans | − | − | − | W | + | + | + | W | + | Tyrosine negative |
| C. matruchotii | + | − | V | + | − | + | + | + | − | “Whip handle” (Gram stain) |
| C. minutissimum | − | − | − | + | + | + | + | V | − | Tyrosine positive |
| C. pseudotuberculosis | − | + | − | − | V | + | + | V | REV | Glycogen negative |
| C. striatum | + | − | − | + | + | + | − | V | V | Tyrosine positive |
| C. ulcerans | − | + | − | − | + | + | + | − | REV | Glycogen positive |
| C. xerosis | V | − | − | + | + | + | + | + | − | Yellowish |
Determination of the MICs of various antimicrobial substances revealed that all four strains were susceptible to cephalothin (MIC, ≤0.125 μg/ml), chloramphenicol (MIC range, 1 to 4 μg/ml), ciprofloxacin (MIC, 0.125 μg/ml), fusidic acid (MIC, 0.03125 μg/ml), gentamicin (MIC range, 0.125 to 0.25 μg/ml), penicillin (MIC range, 0.0625 to 0.125 μg/ml), rifampin (MIC, ≤0.02 μg/ml), tetracycline (MIC range, 1 to 2 μg/ml), and vancomycin (MIC, 0.5 μg/ml) but were resistant to cefetamet (MIC range, 64 to >64 μg/ml), ceftibuten (MIC, 32 μg/ml), and fosfomycin (MIC, >256 μg/ml). The MICs of oxacillin were 1 to 4 μg/ml. Overall, the antimicrobial susceptibility patterns of the four unknown strains corresponded to those of most other nonlipophilic corynebacteria (9).
Chemotaxonomic investigations revealed that C16:0 (range, 48 to 56% of all CFAs), C18:1ω9c (range, 24 to 33%), and C18:0 (range, 7 to 11%) were the predominant CFAs, which was compatible with the assignment of the four strains to the genus Corynebacterium (1, 18). Additionally, thin-layer chromatographic analysis demonstrated meso-diaminopimelic acid as the cell wall diamino acid and the presence of short-chain mycolic acids, thereby confirming the identities of the isolates as members of the genus Corynebacterium.
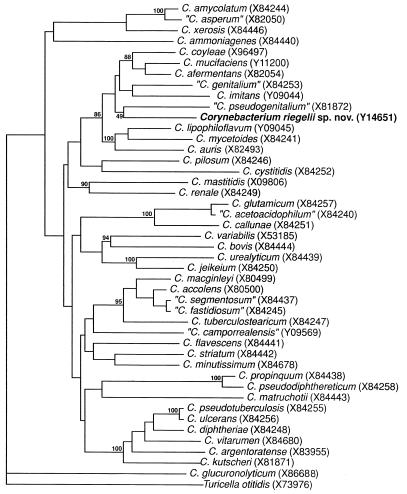
To determine the phylogenetic relatedness of the strains, their 16S rRNA genes were amplified by PCR and were subjected to sequence analysis. The almost complete 16S rRNA gene sequence (1,415 nucleotides) of strain DMMZ 2415 and partial 16S rRNA gene sequences of strains DMMZ 2582 and DMMZ 3128 (approximately 800 nucleotides) were determined. Comparative sequence analysis revealed no nucleotide differences between the isolates (100% sequence similarity), thereby demonstrating their genealogical homogeneity. Sequence searches of EMBL/GenBank libraries with the FASTA program revealed that the newly determined sequences were most closely related to those of the species of the genus Corynebacterium (16S rRNA sequence similarities, >92%; Table 2). Significantly lower levels of relatedness were shown with other gram-positive bacteria with high G+C contents (data not shown). A tree depicting the phylogenetic relationships of the unidentified bacterium (as exemplified by strain DMMZ 2415) within the genus Corynebacterium is presented in Fig. 1. The new bacterium showed a close phylogenetic affinity to the subcluster of species embracing “C. pseudogenitalium,” “C. genitalium,” C. imitans, C. coyleae, C. mucifaciens, C. afermentans, C. auris, C. mycetoides, and C. lipophiloflavum. It is evident from both sequence divergence values and the phylogenetic analysis that the unknown bacterium is not specifically related to any other species and that the >3% 16S rRNA sequence divergence unequivocally demonstrates that the bacterium represents a new Corynebacterium species (15).
TABLE 2.
Levels of 16S rRNA sequence similarity between C. riegelii sp. nov. and other Corynebacterium species
| Species (EMBL accession no.)a | % 16S rRNA sequence similarity with C. riegelii sp. nov. |
|---|---|
| C. accolens (X80500) | 94.7 |
| “C. acetoacidophilum” (X84240) | 93.1 |
| C. afermentans subsp. afermentans (X82054) | 96.4 |
| C. ammoniagenes (X84440) | 94.0 |
| C. amycolatum (X84244) | 94.7 |
| C. argentoratense (X83955) | 94.6 |
| C. auris (X82493) | 95.5 |
| C. bovis (X84444) | 92.5 |
| C. callunae (X84251) | 93.3 |
| “C. camporrealensis” (Y09569) | 93.2 |
| C. coyleae (X96497) | 96.9 |
| C. cystitidis (X84252) | 93.3 |
| C. diphtheriae (X84248) | 94.6 |
| “C. fastidiosum” (X84245) | 94.1 |
| C. flavescens (X84441) | 93.5 |
| “C. genitalium” (X84253) | 96.9 |
| C. glucuronolyticum (X86688) | 92.1 |
| C. glutamicum (X84257) | 92.9 |
| C. imitans (Y09044) | 96.0 |
| C. jeikeium (X84250) | 93.9 |
| C. kutscheri (X81871) | 94.2 |
| C. lipophiloflavum (Y09045) | 96.3 |
| C. macginleyi (X80499) | 94.6 |
| C. mastitidis (Y09806) | 93.7 |
| C. matruchotii (X84443) | 93.6 |
| C. minutissimum (X84678) | 94.5 |
| C. mucifaciens (Y11200) | 96.4 |
| C. mycetoides (X84241) | 95.9 |
| C. pilosum (X84246) | 94.0 |
| C. propinquum (X84438) | 94.2 |
| C. pseudodiphtheriticum (X84258) | 94.0 |
| “C. pseudogenitalium” (X81872) | 96.4 |
| C. pseudotuberculosis (X84255) | 94.3 |
| C. renale (X84249) | 94.7 |
| “C. segmentosum” (X84437) | 94.0 |
| C. striatum (X84442) | 93.9 |
| “C. tuberculostearicum” (X84247) | 94.1 |
| C. ulcerans (X84256) | 94.2 |
| C. urealyticum (X84439) | 93.4 |
| C. variabilis (X53185) | 93.6 |
| C. vitarumen (X84680) | 93.8 |
| C. xerosis (X84446) | 94.0 |
The numbers in parentheses are EMBL 16S rRNA nucleotide sequence accession numbers.
FIG. 1.
Unrooted tree showing the phylogenetic relationships of C. riegelii sp. nov. and other members of the genus Corynebacterium. The tree, constructed by using the neighbor-joining method, was based on a comparison of approximately 1,320 nucleotides. Bootstrap values, expressed as percentages of 500 replications, are given at the branch points. EMBL sequence accession numbers are given in parentheses.
We explain that the Corynebacterium species described in this report has not been reported in the literature before by the fact that coryneform bacteria from urine samples, even when growing in pure cultures, were considered contaminants and were not further identified by many clinical laboratories for many years. However, as has been demonstrated for C. urealyticum (9), true corynebacteria may cause urinary tract infections. Our data indicate that the newly described Corynebacterium may not be recovered from patients with urinary tract infections as often as C. urealyticum is (four patients with infections caused by the newly described bacterium versus nine patients with C. urealyticum infections diagnosed by DMMZ during a 12-month period from August 1996 to July 1997). C. urealyticum and the new Corynebacterium both exhibit strong urease activity, which has been demonstrated as a pathogenicity factor for C. urealyticum as well as for other genitourinary bacterial pathogens (e.g., Proteus mirabilis) and which may also be the case for the new Corynebacterium.
Only a very few prokaryotes (e.g., Ruminobacter amylophilus [16]) have been reported to produce acid from maltose (which is composed of two glucose molecules) but not from glucose. Although it is not the purpose of this paper to address the mechanism responsible for this observation, it seems not unlikely that the new Corynebacterium species contains an ATP binding cassette transporter system for maltose uptake (10) but lacks a glucose phosphotransferase system for glucose uptake (13). However, irrespective of the precise mechanism, it is important to reemphasize that to our knowledge the newly described Corynebacterium species is the only coryneform bacterium known to date to possess this physiological attribute, thereby making it easily recognizable in the routine laboratory.
It is obvious that the new Corynebacterium is probably a rarely encountered microorganism, but it is most likely that other clinical microbiologists will also recognize it once it has been described. We therefore emphasize the importance of identifying coryneform bacteria to the species level whenever those are recovered in pure culture from clinical specimens (9).
On the basis of the results of the phenotypic and molecular genetic findings, we propose that the unknown coryneform bacterium described above be classified as a new species of the genus Corynebacterium, for which the name Corynebacterium riegelii sp. nov. is proposed.
C. riegelii sp. nov.
Corynebacterium riegelii (rie.gel′ii. N.L. gen. n. riegelii, of Riegel, to honor contemporary French microbiologist Philippe Riegel for his contributions to the taxonomy of the genus Corynebacterium as well as to the clinical microbiology of coryneform bacteria). The description of the characteristics given below is based on the results of studies with the four strains.
Cells are gram positive, non-spore forming, and nonmotile. They are typically club-shaped rods which appear as single cells, in pairs, or in small clusters. Colonies are whitish, circular with entire edges, convex, glistening, of up to 1.5 mm in diameter after 48 h of incubation, and of a creamy or a slightly sticky consistency. They have weak anaerobic growth. The organism is catalase positive. Acid is produced from maltose, ribose, trehalose, d-tagatose, and 5-ketogluconate, but acid is not produced from glucose, sucrose, mannitol, xylose, glycerol, erythritol, arabinose, adonitol, β-methylxyloside, galactose, d-fructose, d-mannose, l-sorbose, rhamnose, dulcitol, inositol, sorbitol, α-methyl-d-mannoside, α-methyl-d-glucoside, N-acetylglucosamine, amygdalin, arbutin, salicin, cellobiose, lactose, melibiose, inulin, melezitose, d-raffinose, amidon, glycogen, xylitol, β-gentiobiose, d-turanose, d-lyxose, fucose, arabitol, gluconate, or 2-ketogluconate. Nitrate is not reduced. Urea hydrolysis is strongly positive. Esculin is not hydrolyzed. The CAMP reaction is negative. The activities of esterase (C4), esterase lipase (C8), leucine arylamidase, and cystine arylamidase are detected, whereas the activities of pyrazinamidase and alkaline, as well as that of acid phosphatase, are variable. The activities of pyrrolidonylarylamidase, lipase (C14), valine arylamidase, trypsin, chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase are not detected.
The cell wall contains meso-diaminopimelic acid. Mycolic acids are present. The main straight-chain saturated fatty acids are palmitic and stearic acids; oleic acid is the predominant unsaturated fatty acid. The organism is isolated from human clinical specimens. The type strain, strain DMMZ 2415, has been deposited in CCUG as strain CCUG 38180, in the German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany, as strain DSM 44326, and in the Collection of the Institute Pasteur, Paris, France, as strain CIP 105310. It has the features described above except that the activity of alkaline phosphatase but not that of pyrazinamidase or acid phosphatase is detected.
ACKNOWLEDGMENTS
A. von Graevenitz is acknowledged for careful review of the manuscript.
This study was supported in part by grant BIO2-CT943098 awarded by the European Community and by the Swiss National Science Foundation (grant 3100-050648.97/1). G.F. is a recipient of a European Society for Clinical Microbiology and Infectious Diseases research fellowship.
REFERENCES
- 1.Bernard K A, Bellefeuille M, Ewan E P. Cellular fatty acid composition as an adjunct to the identification of asporogenous, aerobic gram-positive rods. J Clin Microbiol. 1991;29:83–89. doi: 10.1128/jcm.29.1.83-89.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.bioMérieux. API Coryne analytical profile index 2.0. Marcy l’Etoile, France: bioMérieux; 1997. [Google Scholar]
- 3.Devereux J, Haeberli P, Smithies D. A comprehensive set of sequence analysis programmes for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Felsenstein J. PHYLIP-phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 5.Funke G, Alvarez N, Pascual C, Falsen E, Akervall E, Sabbe L, Schouls L, Weiss N, Collins M D. Actinomyces europaeus sp. nov., isolated from human clinical specimens. Int J Syst Bacteriol. 1997;47:687–692. doi: 10.1099/00207713-47-3-687. [DOI] [PubMed] [Google Scholar]
- 6.Funke G, Efstratiou A, Kuklinska D, Hutson R A, de Zoysa A, Engler K H, Collins M D. Corynebacterium imitans sp. nov. isolated from patients with suspected diphtheria. J Clin Microbiol. 1997;35:1978–1983. doi: 10.1128/jcm.35.8.1978-1983.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Funke G, Martinetti Lucchini G, Pfyffer G E, Marchiani M, von Graevenitz A. Characteristics of CDC group 1 and group 1-like coryneform bacteria isolated from clinical specimens. J Clin Microbiol. 1993;31:2907–2912. doi: 10.1128/jcm.31.11.2907-2912.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Funke G, Pascual Ramos C, Collins M D. Corynebacterium coyleae sp. nov., isolated from human clinical specimens. Int J Syst Bacteriol. 1997;47:92–96. doi: 10.1099/00207713-47-1-92. [DOI] [PubMed] [Google Scholar]
- 9.Funke G, von Graevenitz A, Clarridge III J E, Bernard K A. Clinical microbiology of coryneform bacteria. Clin Microbiol Rev. 1997;10:125–159. doi: 10.1128/cmr.10.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Higgins C F. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 11.Hutson R A, Thompson D E, Collins M D. Genetic interrelationships of saccharolytic Clostridium botulinum types B, E and F and related clostridia as revealed by small-subunit rRNA gene sequences. FEMS Microbiol Lett. 1993;108:103–110. doi: 10.1111/j.1574-6968.1993.tb06081.x. [DOI] [PubMed] [Google Scholar]
- 12.National Committee for Clinical Laboratory Standards. Minimum inhibitory concentration (MIC) interpretive standards (μg/ml) for organisms other than Haemophilus spp., Neisseria gonorrhoeae, and Streptococcus spp. NCCLS document M7-A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 13.Postma P W, Lengeler J W, Jacobson G R. Phosphoenolpyruvate: carbohydrate phosphotransferease systems of bacteria. Microbiol Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 15.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 16.Stackebrandt E, Hippe H. Transfer of Bacteroides amylophilus to a new genus Ruminobacter gen. nov., nom. rev. as Ruminobacter amylophilus comb. nov. Syst Appl Microbiol. 1986;8:204–207. [Google Scholar]
- 17.von Graevenitz A, Funke G. An identification scheme for rapidly and aerobically growing gram-positive rods. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg Abt 1 Orig. 1996;284:246–254. doi: 10.1016/s0934-8840(96)80100-9. [DOI] [PubMed] [Google Scholar]
- 18.von Graevenitz A, Osterhout G, Dick J. Grouping of some clinically relevant gram-positive rods by automated fatty acid analysis. APMIS. 1991;99:147–154. doi: 10.1111/j.1699-0463.1991.tb05132.x. [DOI] [PubMed] [Google Scholar]