Abstract
Juniperus seravschanica Kom. is a species that grows widely in the mountain ranges from Central Asia to Oman. It is an important tree for the formation of shrub–forest massifs in mountainous areas and for draining and fixing soils from middle to high altitudes. A comprehensive study of the species’ genetic diversity and population structure is a basic approach to understanding the current status of J. seravschanica resources for the development of future conservation strategies. Samples from 15 populations of J. seravschanica were collected from the mountain ranges of Uzbekistan, Kyrgyzstan, and Kazakhstan. The genetic diversity and population structure of 15 Central Asian populations of J. seravschanica were assessed using 11 polymorphic simple sequence repeat (SSR) markers. Genetic diversity parameters, including the number of alleles (na), the effective number of alleles (ne), Shannon’s information index (I), the percentage of polymorphic loci (PPL), Nei’s genetic diversity index (Nei), principal coordinate analysis (PCoA), etc., were evaluated. The analysis of 15 J. seravschanica populations based on 11 polymorphic SSRs detected 35 alleles. The average PIC value was 0.432, and the highest value (0.662) was found in the JT_40 marker. Nei’s genetic diversity index for the J. seravschanica populations was 0.450, ranging from 0.407 (population 14) to 0.566 (population 4). The analysis of molecular variance (AMOVA) showed that 90.3% of total genetic variation is distributed within the population. Using the alleles of all the populations, the gene flow (Nm) was found to be 4.654. Population structure analysis revealed poor clustering in the studied populations and confirmed our AMOVA results. The output of this work can be efficiently used for the maintenance of the species across the Central Asian region.
Keywords: Juniperus seravschanica, genetic diversity, population structure, simple sequence repeat (SSR), Central Asia
1. Introduction
Species of the genus Juniperus L., belonging to the Cupressaceae family, are common in the mountain ranges of Central Asian countries, including Kazakhstan (seven species), Kyrgyzstan (4), Tajikistan (4), and Uzbekistan (3) [1]. They grow as shrubs or trees and play an essential ecological role in forming shrub–forest massifs in mountainous areas and draining and fixing soils on mountain ranges. One common Juniperus species in this region is Juniperus seravschanica Kom. J. seravschanica is a tall, wind-pollinated, drought-resistant coniferous evergreen tree that is important as a medicinal plant. It is a source of cones, raw materials for producing essential oils (16.8% cedrol content), and leaves that are widely used in medicine [2]. The name of this species is associated with the region along the Zeravshan (Zarafshon) River in Uzbekistan and Tajikistan, where this tree was initially found, widely spread across the hills of the Pamir-Alay mountain system [3]. Therefore, it was assumed that J. seravschanica originated in that area and spread to the neighboring countries of Kyrgyzstan, Kazakhstan, Turkmenistan, Afghanistan, Pakistan, India, Iran, and Oman [4]. This species is the main forest-forming factor of the Western Tian Shan (Kazakhstan, Kyrgyzstan) and the Pamir-Alay (Kyrgyzstan, Tajikistan, Uzbekistan) mountain systems [5]. Juniper forests with J. seravschanica as the dominant species are typically found on dry foothills or at middle to high altitudes. This species is a key woodland component from 1000 to 3500 m above sea level [5]. J. seravschanica is often found in steep and sagebrush slopes, most often those with southern and western exposures, and frequently in rubbly–stony mountain gray-brown (xeromorphic) soils underlain by dense rocks. This species can grow on a wide variety of substrates, from steep (up to 70°) rocky outcrops to nutrient-rich brown soils with a significant (up to 16%) humus content. This property plays an essential ecological role in water and soil protection, preventing destructive mudflows on mountain ranges [6]. Despite the wide distribution of this species in the region, a decrease in the number of natural populations has been observed. The main factors in the species’ decline are environmental changes, the use of wood for construction, overgrazing, and fires [7]. Therefore, in order to preserve its natural population, this species was listed in the Red Book of Central Asia [8], including in Kazakhstan [9,10]. Therefore, the conservation of this species’ genetic resources and the reconstruction of its habitats are essential. A comprehensive assessment of the genetic diversity and genetic structure of this species’ natural populations will serve as a basic background for proposing conservation strategies and protecting J. servaschanica resources [4,11,12].
Taxonomically, J. seravschanica belongs to the Juniperus section Sabina, which was first described by Komarov in 1932 [13]. A distinctive feature of J. seravschanica is globular, blackish-blue fleshy 9–12 mm long fruits or cone-berries. The species J. excelsa, J. polycarpos K. Koch, and J. seravschanica Kom. have difficulties in identification due to their morphological characteristics. According to Adams et al. [14] J. excelsa is distributed from Greece to Turkey, J. polycarpos grows in Azerbaijan and Lebanon, and the distribution area of J. seravschanica stretches from Central Asia to Oman. Due to existing taxonomic uncertainties, it is important to study these species using polymorphic molecular markers [15]. Within the genus, a sufficiently large number of reports were dedicated to the evaluation of the genetic diversity of different Juniperus species [16,17,18,19,20,21,22,23,24,25,26]. Similarly, J. seravschanica was extensively studied for the determination of the area for core and peripheral populations [27], morphometric traits [13], chemical components [28], and essential oils [2,15]. Additionally, the complete chloroplast genome sequence of this species has been reported [29]. Nevertheless, there is a shortage of information on the evaluation of the genetic diversity and structure of the populations distributed in the Central Asian region.
Recent studies show that genetic diversity patterns significantly affect the viability and resistance of environmental conditions of local populations [30,31]. The importance of stochastic factors (gene flow, genetic drift, and founder events) and environmentally specific natural selection may change from a species’ core toward its boundary. Increased genetic drift caused by small population size and isolation at the species periphery leads to a loss of genetic variation, but may also promote population differentiation [32]. Thus, identifying patterns of intraspecific diversity within regions is vital for our understanding of ecological and evolutionary (e.g., speciation) processes [33], as well as for setting conservation priorities [34]. In addition, a comparative assessment of the genetic diversities of core and peripheral populations is relevant, as the original populations of a species often differ in the ecological conditions in which they live. Historically, the genetic diversity of the Juniperus genus populations was actively studied using different informative DNA markers, including RAPDs (randomly amplified polymorphisms of DNA) [35,36], AFLPs (amplified fragment length polymorphisms) [37], SSRs (simple sequence repeats) [21,26], etc. In particular, SSR markers, or microsatellites, can be very informative due to their wide distribution within genomes, high degree of polymorphism, good duplicability, and codominant inheritance [26,38,39,40,41,42]. However, the genetic diversity of J. seravschanica has only been assessed using a few types of DNA markers. For example, Adams [15], Sultangaziev, et al. [5], and Rahimian Boogar and Salehi [43] studied selected populations of the species using RAPD, PCR-RFLP, and ISSR markers, respectively. Naturally, assessing these populations using different types of DNA markers cannot provide a comprehensive platform for evaluating genetic diversity in Central Asia. Therefore, this study aimed to analyze the genetic diversity of J. seravschanica populations from mountain ranges in three Central Asian countries (Kazakhstan, Kyrgyzstan, and Uzbekistan) using polymorphic SSR markers. It was assumed that this investigation might help evaluate the variability of SSR markers in this species, and identify genetically diverse populations for the development of future strategies associated with the maintenance of J. seravschanica resources in Central Asia.
2. Results
2.1. Polymorphism of Tested SSR Markers
The initial sample screening with a set of 18 SSR markers demonstrated that 11 out of the 18 tested SSR markers were polymorphic and suitable for genetic analysis (Table 1).
Table 1.
Characteristics of 11 polymorphic microsatellite markers used in Juniperus seravschanica.
| Locus | na | ne | I | Nei | H | PIC |
|---|---|---|---|---|---|---|
| Jce03 | 3 | 1.8 | 0.666 | 0.442 | 0.485 | 0.410 |
| Jce04 | 4 | 1.7 | 0.625 | 0.442 | 0.495 | 0.393 |
| Jce05 | 2 | 1.1 | 0.132 | 0.077 | 0.081 | 0.077 |
| JT_04 | 3 | 2.0 | 0.779 | 0.511 | 0.564 | 0.499 |
| JT_30 | 3 | 2.3 | 0.921 | 0.586 | 0.606 | 0.538 |
| JT_33 | 2 | 1.7 | 0.578 | 0.417 | 0.440 | 0.343 |
| JT_34 | 2 | 1.3 | 0.292 | 0.191 | 0.207 | 0.185 |
| JT_37 | 5 | 2.9 | 1.170 | 0.688 | 0.705 | 0.653 |
| JT_40 | 5 | 2.6 | 1.028 | 0.620 | 0.712 | 0.662 |
| JT_46 | 3 | 2.0 | 0.729 | 0.466 | 0.615 | 0.537 |
| JS54 | 3 | 2.0 | 0.813 | 0.508 | 0.508 | 0.456 |
| Mean | 3.2 | 1.9 | 0.703 | 0.450 | 0.493 | 0.432 |
| SE | 0.069 | 0.051 | 0.027 | 0.016 | 0.195 | 0.180 |
Notes: na—number of alleles per locus; ne—effective number of alleles; I—Shannon’s Information Index; Nei—Nei’s genetic diversity index; H—heterozygosity value; PIC–polymorphism information content; SE—Standard error.
Genotyping of a total of 323 samples from 15 populations with 11 SSRs (Table S1) resulted in the identification of 35 alleles. The number of alleles per SSR marker ranged from 2 to 5, with an average of 3.2 alleles per locus. JT_37 and JT_40 markers amplified the ultimate number of alleles (5 alleles), while Jce05, JT_33, and JT_34 generated the lowest number of alleles (2 alleles) (Table 1). The average PIC value was 0.432, ranging from 0.077 (Jce05) to 0.662 (JT_40), and the lowest and highest level of heterozygosity 0.081 and 0.712 at loci Jce_05 and JT_40, respectively (Table 1).
2.2. Genetic Diversity in Collected Populations of J. seravschanica in Three Central Asian Countries
The average number of alleles amplified using 11 polymorphic SSR loci in the 15 studied populations was 2.6 and ranged from 2.2 (population 2) to 3 (population 12) (Table 2). The number of effective alleles (ne) ranged from 1.7 to 2.3, with an average value of 1.9. The average PPL was 91.5% and varied from 82% to 100%. The average number of observed heterozygosity (Ho) and expected heterozygosity (He) were 0.695, and 0.575, respectively. The mean Nei’s genetic diversity index for studied J. seravschanica populations was 0.450, ranging from 0.407 (population 14) to 0.566 (population 4) (Table 2). The highest average Nei’s index value was recorded in the populations collected in Uzbekistan (0.473), followed by those from South Kyrgyzstan and Kazakhstan (both 0.443). Interestingly, the assessment of the ne and PPL indices shows that the highest values were recorded for the populations from Kazakhstan (Table 2).
Table 2.
Assessment of the genetic diversity of studied Juniperus seravschanica populations.
| Pop | Size | Origin | na | ne | Ho | He | Nei | PPL |
|---|---|---|---|---|---|---|---|---|
| Pop 1 | 11 | Uzbekistan | 2.6 | 2.0 | 0.711 | 0.593 | 0.474 | 91% |
| Pop 2 | 10 | 2.2 | 1.7 | 0.700 | 0.562 | 0.418 | 91% | |
| Pop 3 | 11 | 2.4 | 1.9 | 0.802 | 0.605 | 0.433 | 82% | |
| Pop 4 | 12 | 2.7 | 2.3 | 0.682 | 0.639 | 0.566 | 91% | |
| Mean | 2.5 | 2.0 | 0.724 | 0.600 | 0.473 | 89% | ||
| Pop 5 | 13 | South Kyrgyzstan | 2.3 | 1.8 | 0.664 | 0.535 | 0.411 | 82% |
| Pop 6 | 21 | 2.8 | 2.2 | 0.684 | 0.592 | 0.497 | 100% | |
| Pop 7 | 21 | 2.5 | 1.8 | 0.680 | 0.562 | 0.414 | 91% | |
| Pop 8 | 20 | 2.5 | 2.0 | 0.700 | 0.581 | 0.452 | 91% | |
| Mean | 2.5 | 1.9 | 0.682 | 0.567 | 0.443 | 91% | ||
| Pop 9 | 20 | North Kyrgyzstan | 2.6 | 1.8 | 0.705 | 0.556 | 0.431 | 91% |
| Pop 10 | 20 | 2.5 | 1.9 | 0.668 | 0.555 | 0.430 | 91% | |
| Mean | 2.5 | 1.9 | 0.686 | 0.555 | 0.430 | 91% | ||
| Pop 11 | 41 | Kazakhstan | 2.8 | 1.8 | 0.694 | 0.568 | 0.419 | 100% |
| Pop 12 | 40 | 3.0 | 2.2 | 0.675 | 0.579 | 0.482 | 100% | |
| Pop 13 | 20 | 2.5 | 1.9 | 0.627 | 0.537 | 0.447 | 100% | |
| Pop 14 | 20 | 2.5 | 1.9 | 0.718 | 0.561 | 0.407 | 82% | |
| Pop 15 | 43 | 2.6 | 2.1 | 0.717 | 0.602 | 0.465 | 91% | |
| Mean | 2.7 | 2.0 | 0.686 | 0.569 | 0.443 | 94% | ||
| Total mean | 323 | 2.6 | 1.9 | 0.695 | 0.575 | 0.450 | 91.5% | |
| SE | 0.069 | 0.051 | 0.019 | 0.012 | 0.016 | 1.65% |
Notes: Pop—population; na—number of alleles per locus; ne—effective number of alleles; Ho—Observed Heterozygosity; He—Expected Heterozygosity; Nei—Nei’s genetic diversity index; PPL—the percentage of polymorphic loci; SE—Standard error.
The results of the AMOVA suggest that 90.3% of total genetic variation is distributed within, and 9.7% between population groups, respectively. The gene flow (Nm) was calculated using the Fst values and equals 4.654 migrants per generation (Table 3).
Table 3.
AMOVA results for 15 populations of Juniperus seravschanica based on 11 SSR markers.
| Source | df | SS | MS | % | p | Est. Var. | FST | Nm |
|---|---|---|---|---|---|---|---|---|
| Among populations | 14 | 113.302 | 8.093 | 9.7% | <0.001 | 0.266 | ||
| Within populations | 308 | 761.596 | 2.473 | 90.3% | <0.001 | 2.473 | ||
| Total | 322 | 874.898 | 100% | 2.738 | 0.050 * | 4.654 |
df—degrees of freedom; SS—sum of squares; MS—mean squared; Est. var.—estimates of variance; %—percentage of variation; FST—fixation index; Nm—gene flow (Nm) value. * p < 0.001; Nm = (1 − FST)/4FST.
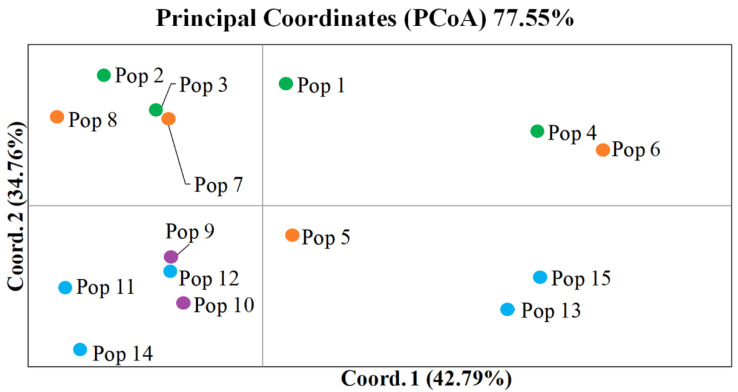
The PCoA plot shows the distances of the J. seravschanica populations using two principal coordinates: Coordinate 1 and Coordinate 2, which explain 42.79% and 34.76% of the total variation among populations, respectively (Figure 1). Coordinate 1 effectively separates populations 4, 6, 13, and 15 from the majority of the other populations. Coordinate 2 separates the populations of Uzbekistan and South Kyrgyzstan from the populations of North Kyrgyzstan and Kazakhstan, except for population 5, which was grouped together with populations from North Kyrgyzstan and Kazakhstan (Figure 1). The closest population to the intersection of eigenvalues 1 and 2 was population 5 from South Kyrgyzstan (Figure 1). The Mantel test for correlation between population genetic distance (PopGD) and geographic distance (GGD) matrices (r2) was equal to 0.0885 (p > 0.020). The correlation between Nei’s genetic distance (NeiP) and the geographic distance (GGD) was 0.0082 (p > 0.270).
Figure 1.
Principal coordinates analysis (PCoA) of 15 populations of Juniperus seravschanica from Central Asia based on pairwise population differentiation (PhiPT) values. Notes: Circles highlighted as green—Uzbekistan, orange—South Kyrgyzstan, purple—North Kyrgyzstan, blue—Kazakhstan.
The results of SSR analysis for the fifteen J. seravschanica populations were also evaluated using an unrooted dendrogram based on the unweighted pair-group method and the arithmetic means (UPGMA) method (Figure S1). The dendrogram separates the studied populations into three groups: I, II, and III (Figure S1). Groups I and II form the first clade, and populations in group III are separated into the second clade. The clustering results are concordant with the PCoA plot (Figure 1).
2.3. The Genetic Structure of J. seravschanica Based on the Analysis of Fifteen Populations from Central Asia
The analysis of the STRUCTURE output suggests poor clustering for the 15 studied populations collected from four different regions (Figure S2), which confirmed the AMOVA results for partitioning total genetic variation within and between populations (Table 3). The Structure Harvester analysis using the “elbow” method indicates that the most significant K steps were K3 and K5 (Figure S2). At the K3 step, cluster 3 mostly consisted of plants from three populations from Uzbekistan, one population from South Kyrgyzstan, two populations from North Kyrgyzstan, and three populations from Kazakhstan. At this K step, cluster 1 was mostly populated by plants from three populations from South Kyrgyzstan and two populations from Kazakhstan. Notably, population 4, from Uzbekistan, had an equal number of plants in both clusters (Table 4). A minor number of plants in all four regions represented cluster 2. At the K5 step, the plant ratio between clusters was slightly modified, as most plants were part of clusters 4 and 5 (Table 4). Cluster 3 was heavily represented by plants from population 4 (41.7%). The majority of plants in population 1 from Uzbekistan (45.5%) and population 7 from South Kyrgyzstan (52.4%) were part of cluster 2. Finally, cluster 1 was strongly represented by plants in population 5 from South Kyrgyzstan (30.8%). Notably, the plants in population 5 were equally represented in clusters 1, 4, and 5 (Table 4), which is in good agreement with the centralized position of this population in the PCoA plot (Figure 1).
Table 4.
The population structure of Juniperus seravschanica from four regions of Central Asia using the K3 and K5 steps of the STRUCTURE package.
| Population | Sample Size | Origin | K3 (%) | K5 (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cluster 1 | Cluster 2 | Cluster 3 | Cluster 1 | Cluster 2 | Cluster 3 | Cluster 4 | Cluster 5 | |||
| Pop 1 | 11 | UZ | 27.3 | 9.1 | 63.6 | 18.2 | 45.5 | 9.1 | 18.2 | 9.1 |
| Pop 2 | 10 | 20.0 | 0.0 | 80.0 | 10.0 | 10.0 | 10.0 | 60.0 | 10.0 | |
| Pop 3 | 11 | 18.2 | 9.1 | 72.7 | 0.0 | 0.0 | 9.1 | 72.7 | 18.2 | |
| Pop 4 | 12 | 50.0 | 0.0 | 50.0 | 0.0 | 8.3 | 41.7 | 0.0 | 50.0 | |
| Pop 5 | 13 | SKG | 53.8 | 0.0 | 46.2 | 30.8 | 7.7 | 0.0 | 30.8 | 30.8 |
| Pop 6 | 21 | 85.7 | 4.8 | 9.5 | 4.8 | 4.8 | 19.0 | 9.5 | 61.9 | |
| Pop 7 | 21 | 61.9 | 9.5 | 28.6 | 4.8 | 52.4 | 19.0 | 14.3 | 9.5 | |
| Pop 8 | 20 | 30.0 | 0.0 | 70.0 | 15.0 | 15.0 | 10.0 | 50.0 | 10.0 | |
| Pop 9 | 20 | NKG | 25.0 | 10.0 | 65.0 | 15.0 | 15.0 | 0.0 | 60.0 | 10.0 |
| Pop 10 | 20 | 15.0 | 15.0 | 70.0 | 20.0 | 15.0 | 5.0 | 55.0 | 5.0 | |
| Pop 11 | 41 | KZ | 19.5 | 12.2 | 68.3 | 24.4 | 29.3 | 4.9 | 39.0 | 2.4 |
| Pop 12 | 40 | 45.0 | 7.5 | 47.5 | 7.5 | 25.0 | 15.0 | 25.0 | 27.5 | |
| Pop 13 | 20 | 90.0 | 0.0 | 10.0 | 5.0 | 35.0 | 5.0 | 0.0 | 55.0 | |
| Pop 14 | 20 | 30.0 | 0.0 | 70.0 | 25.0 | 10.0 | 5.0 | 25.0 | 35.0 | |
| Pop 15 | 43 | 72.1 | 9.3 | 18.6 | 2.3 | 16.3 | 16.3 | 11.6 | 53.5 | |
Notes: UZ—Uzbekistan; SKG—South Kyrgyzstan; NKG—North Kyrgyzstan; KZ—Kazakhstan.
3. Discussion
The genetic diversity and population structure of forests are shaped by past historical and current human-mediated processes. While its assessment is crucial to inform conservation and forestry management, the geographic scale at which it is quantified plays an essential role in developing species conservation strategies. We carried out a genetic diversity analysis of 15 populations of J. seravschanica originating from three Central Asian countries (Kazakhstan, Kyrgyzstan, and Uzbekistan) using 11 polymorphic SSR markers (Table 2). The estimated average number of alleles (3.2) identified in this study was comparable to those of J. cedrus reported by Rumeu et al. [20] and J. thurifera reported by Teixeira et al. [21], but lower than the J. sabina (6.70) reported by Lu et al. [26], J. excelsa (12.57) reported by Evren and Kaya [25] or J. communis (9–23 alleles) species reported by Michalczyk et al. [17]. The average Shannon’s diversity index (I) calculated using SSR markers was 0.703, higher than the index (0.428) calculated using RAPD markers [44]. Accordingly, 91.5% was the PPL percentage value determined utilizing SSR markers.
The mean genetic diversity for this species (0.450), including for peripheral populations in Kazakhstan (0.443), was relatively high (Table 2). A species’ resistance to environmental change is stronger the more genetic diversity it has [26,45]. The total genetic variation of J. seravschanica populations collected from mountain ranges of three Central Asian countries was divided into 90.3% within populations and 9.7% between populations (Table 3).
The values of Nm in conifers have a tendency to be much higher, with values of Nm ≥ 3 being the norm [46]. Our results indicate a high level of gene flow for this species-Nm-4.654), which is higher than the mean values of gene flow for other congeneric juniper species—J. communis—1.09 [47] and J. brevifolia—2.43 [48], suggesting a high level of historical gene flow between these populations. We believe that gene flow via wind-dispersed pollen is less likely to reach larger distances, as reported for J. tibetica by Opgenoorth [49], and does not exceed 2 km. Moreover, often isolated mountain slopes with deep valleys restrict pollen movement outside of that territory. Furthermore, frequent rains during the pollination period wash away the pollen; therefore, low levels of fertile seeds have been reported [50]. It is more likely that gene flow happens mainly via fleshy female cones, preferred mainly by birds from Turdus and Fringillidae, who feed on cones during the breeding season and disseminate seeds for a longer distance [51]. Similar patterns have been reported for other juniper species [52]. According to Slatkin [53], Nm of more than four migrants/ generation prevents genetic differentiation between populations, a pattern we observe for J. seravschanica in the Central Asian regions.
Our assessment of the STRUCTURE outputs (Table 4 and Table S2) for different K steps shows a very poor structuration of the populations in this species, which is in good agreement with our AMOVA results (Table 3). Nevertheless, the STRUCTURE plot during the K3 step suggests that the collected populations were separated into two groups. The first group consisted of three populations from Uzbekistan (populations 1–3), one from South Kyrgyzstan (population 8), two from North Kyrgyzstan (populations 9 and 10), and two from Kazakhstan (populations 12 and 14). The second group consisted of one population from Uzbekistan (population 4), three from South Kyrgyzstan (populations 5–7), and two from Kazakhstan (populations 14 and 15) (Table 4). This separation possibly indicates gene flow routes from the southern to northern territories of the region, as the highly populated Ferghana valley was found to be a significant barrier for gene flow between northern and southern sub-populations in Kyrgyzstan [5]. The gene flow is likely happening through the territory of Uzbekistan, more precisely via the Gissar and Western-Tianshan mountain ranges, where the highest average level of genetic diversity was registered (Table 2). Nevertheless, analysis of the plot from the K5 step suggests an outstanding role of population 5 in South Kyrgyzstan (Table 4), where plants were evenly distributed with high frequency in three out of five clusters. Additionally, the eigenvalues for population 5 are in the middle of those for Coordinates 1 and 2 in the PCoA plot (Figure 1). This result is unsurprising, as this population was collected in the Turkestan Range, just north of the Zeravshan River.
A high gene flow among populations probably influenced the relatively high level of genetic diversity in the populations from Kazakhstan. Although these populations grow on the edge of the area of J. seravschanica distribution, it was determined that their level of genetic diversity is equal to the values of populations from South Kyrgyzstan (Table 2). There are three main hypotheses in the literature regarding trends in genetic diversity along core–peripheral clines, each with different spatial implications [54]. The first hypothesis suggests that diversity increases from the periphery to the core, the “Carson hypothesis”; the second one suggests that diversity decreases from the periphery to the core, the “Fischer hypothesis”; and the third one suggests homogeneous diversity from the periphery to the core, the “Mayr hypothesis”. The results of this study support the third hypothesis, which, in this particular case, was based on (1) species distribution limited to the mountain chains of three neighboring countries in Central Asia, and (2) a high level of gene flow. A review of the literature indicates that the hypothetical region for the core populations of J. seravschanica is the mountain ranges of northern Pamir-Alay (Zaamin National Park, Zeravshan River) in Uzbekistan [12,55], although none of the populations from Tajikistan have been assessed so far. The results of this study suggest that the region for the core population of this species might be wider and may include the Turkestan Range in South Kyrgyzstan. The regions for the northern peripheral populations in Kazakhstan and North Kyrgyzstan collected in this work are possibly a natural northern border for the species distribution. Further north and northeast, no J. seravschanica populations were found, which indicates that Kazakhstan’s colder climate is unsuitable for this species’ growth. Thus, the population structure outputs and genetic diversity data in this work revealed a relatively high rate of gene flow [46,47,48] in the species and a high level of genetic diversity within the populations (90.3%). The results of this study can be used to develop species conservation strategies, as well as afforestation programs across the Central Asian region.
4. Materials and Methods
4.1. Sampling Area and Approaches
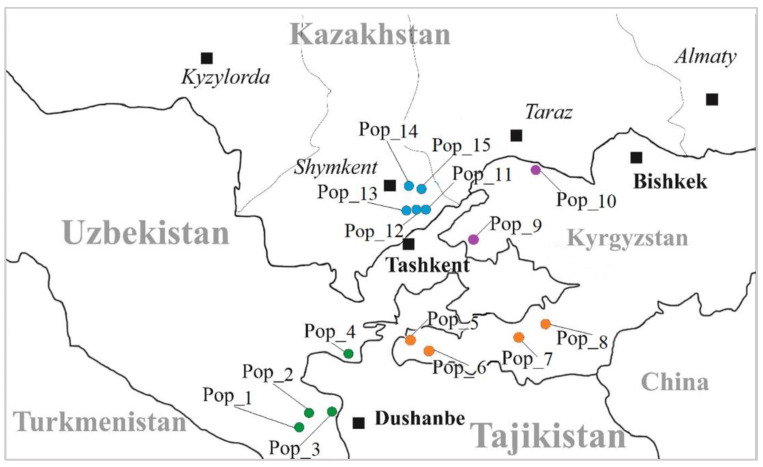
Across the three Central Asian countries (Kazakhstan, Kyrgyzstan, and Uzbekistan) 323 individuals of 15 J. seravschanica populations were sampled at altitudes from 1420 to 2293 m above sea level (Figure 2; Table 5). The number of samples in each population ranged from 10 (Population 1) to 43 (Population 15). The number of samples per population varied significantly due to difficulties related to the inaccessibility of terrains and population sizes.
Figure 2.
Location of sampled Juniperus seravschanica populations in Central Asia. Pop—population; enumeration according to Table 1. Notes: Circles highlighted as green—Uzbekistan, orange—South Kyrgyzstan, purple—North Kyrgyzstan, blue—Kazakhstan.
Table 5.
Geographical coordinates of sampled Juniperus seravschanica populations in three Central Asian countries.
| Country | Population | N (Latitude) | E (Longitude) | Altitude (m) | Sample Size | Geographic Location |
|---|---|---|---|---|---|---|
| Uzbekistan | Pop 1 | 38.272472 | 67.291861 | 2120 | 11 | Southwestern Gissar range mountain of Baysuntau, Surkhandarya region |
| Pop 2 | 38.563444 | 67.479750 | 2210 | 10 | Basin of the Sangardak river, Surkhandarya region | |
| Pop 3 | 38.590361 | 67.970222 | 2200 | 11 | Basin of the Shargun, Surkhandarya region | |
| Pop 4 | 39.593611 | 68.465222 | 2267 | 12 | Basin of Kashkasu-Sai, Zaamin National Park, Turkestan Range | |
| South Kyrgyzstan | Pop 5 | 39.966694 | 69.642000 | 1800 | 13 | Sulukta, Leilek district |
| Pop 6 | 39.758083 | 69.949278 | 2273 | 21 | Baul, Leilek district | |
| Pop 7 | 40.055444 | 71.884389 | 2103 | 21 | Tamasha gorge, Batken Region | |
| Pop 8 | 40.204806 | 72.352278 | 2293 | 20 | Abshir-Sai gorge, Turkestan mountain Range | |
| North Kyrgyzstan | Pop 9 | 41.499278 | 70.965833 | 2160 | 20 | Jalal-Abad region, Chatkal gorge |
| Pop 10 | 42.763917 | 71.822817 | 1420 | 20 | Talas region, Kara-Archa gorge | |
| Kazakhstan | Pop 11 | 42.168389 | 70.381722 | 1701 | 41 | Turkestan region, Sairam-Ugam State National Nature Park (SNNP), Sairam gorge, right bank of the Sairamsu river |
| Pop 12 | 42.155694 | 70.235750 | 1817 | 40 | Turkestan region, Sairam-Ugam SNNP, Kaskasu gorge, left bank of the Sairamsu river | |
| Pop 13 | 42.179500 | 70.333556 | 1530 | 20 | Turkestan region, Sairam-Ugam SNNP, Saryaygyr gorge | |
| Pop 14 | 42.331250 | 70.372583 | 1605 | 20 | Turkestan region, Aksu-Zhabagly state nature reserve (SNR), Aksu canyon, right bank of the Aksu river | |
| Pop 15 | 42.416528 | 70.207417 | 2110 | 43 | Turkestan region, Tyulkubas district, Aksu-Zhabagly SNR, Mashat gorge |
The minimum distance of collected plant materials in the population ranged from 30 to 50 m between individuals, each individual was labeled and dried in silica gel and stored at room temperature until DNA was extracted. The geographical coordinates of each population are listed in Table 5. Due to the distinct geographic spread of J. seravschanica in Kyrgyzstan, the populations in the country were separated into two groups—North and South Kyrgyzstan (Figure 2, Table 5).
4.2. DNA Extraction and SSR Analysis
For each sample, total genomic DNA was extracted from 50 to 60 mg of dried leaves, which were ground to powder using two steel beads and a shaking mill (Retsch MM301, Haan, Germany), following the cetyltrimethylammonium bromide (CTAB) method [56]. The quality of the extracted DNA was checked using 1% Tris-borate-EDTA (TBE) agarose gels and NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The concentration of the DNA samples was normalized at 100 ng/μL and stored at –20 °C for further SSR analysis.
Eighteen primer pairs designed for the amplification of SSR loci for other Juniperus species, namely J. cedrus [20], J. thurifera [21], and J. sabina [26] were chosen to test their transferability for congeneric species—J. seravschanica. Information on the primer sequences, repeat types, and amplification conditions of the juniper microsatellites is provided in Table 6.
Table 6.
Characteristics of SSR primers used for the analysis of Juniperus seravschanica.
| № | Primers | Motif | Primer Sequences (5′-3′) | Expected Size (bp) |
Annealing T (°C) |
|---|---|---|---|---|---|
| 1 | Jce03 | (AATAC)6 | F: TGGTTTATTCATCAACGCCC R: TTCATCCAGTGTTAAGCATGTACC |
101–121 | 61 |
| 2 | Jce04 | (AAGAG)7 | F: TCTTTGCCTTGACTTCTCGG R: CAATAGCAGGAACACTAAACGG |
100–110 | 61 |
| 3 | Jce05 | (ATATC)6 | F: GCTGCTTTAGCTTCATGGACG R: CCAACTCTTGAACTCTAATCTGTACTGC |
173–198 | 63 |
| 4 | Jce08 | (ATAC)10 | F: TGGATTCTGAAATTTGTATGCAGC R: AAGCAATGACAAAGCAAAGGC |
176–202 | 60 |
| 5 | Jce09 | (ATAC)8 | F: TGTATATTTCTAGTCAAATGCCTTCC R: GATCTCCATTCATCTCTACAATCC |
120–155 | 61 |
| 6 | Jce12 | (TA)6...(CATA)8 | F: GCCAGATTCAAGGATAATTGG R: TTTAGCAACAGTACTATGCAAG |
157–163 | 55 |
| 7 | Jce13 | (CATA)12 | F: TGTTGTCATACCCCTGTGAGC R: GCAGTTGGATGAATTTTGTTG |
200–230 | 55 |
| 8 | JT_03 | (CTT)8 | F: ACCCTCTATAAGGATGCTACCATGA R: AAAAGATAGATTGATAAGTTGAAAGGG |
140 | 55 |
| 9 | JT_04 | (AGA)7 | F: CCAAGGAATGATCTAACCTTTGAA R: TGGGATGCATATCTTATCTTCCT |
174–219 | 55 |
| 10 | JT_30 | (TCT)10 | F: AATCCCCTATCCTTGCCAGT R: TCAACAATATCAGCAAGTAATGAGA |
128–220 | 57 |
| 11 | JT_33 | (CT)11 | F: GAGCTTCCTTTGTAGATTTTGGG R: GTAAGAAGACACCACTCAGTCGAT |
177–285 | 57 |
| 12 | JT_34 | (AG)12 | F: CATGCATGGGTTATAATAATAGAGATA R: TGGGCACAAATTTTAGTGTAATG |
110 | 55 |
| 13 | JT_37 | (GT)14 | F: GATGTTTGTATCATATCCTTGATTGG R: TCCACACCTATCGGGTTCAT |
123 | 55 |
| 14 | JT_38 | (AC)14 | F: CCAACAAGCCTCCACCCTAT R: CAAGTTTGGAAAGTGTGGTCA |
114 | 55 |
| 15 | JT_40 | (CA)20 | F: GGCCGCATGATCCATTACT R: TCGTAACGTAATGACATGTATAGTGC |
98–150 | 54 |
| 16 | JT_46 | (AGG) 7 | F: TGAGATCACCTACTTCCTAGTGGA R: CCACCAAGGGCATAGAGTTC |
174–237 | 54 |
| 17 | JS31 | (ATG)5 | F: TTGGCTAATGATGTGCTTGC R: ACCCAAGCTATGTGCAGGAT |
330–354 | 60 |
| 18 | JS54 | (CAT)7 | F: CTTGTGGTTAGTGGTTGGCA R: CACTCTCCCAGTGGTGGTTT |
255–279 | 60 |
The DNA was amplified in a thermal cycler (Thermo Fisher Scientific, Waltham, MA, USA) as follows: denaturation at 95 °C for 3 min; 35 cycles consisting of 30 s of denaturation at 95 °C; a 30 s annealing at temperatures ranging from 54 to 63 °C depending on the primer pair used; and a 90 s extension at 72 °C, followed by a final extension at 72 °C over 20 min. The PCR reactions were performed in a volume of 20 μL containing 1 × PCR buffer, 1.5 mM of MgCl2, 0.5 mM each of dNTPs, 0.2 μM of each primer, 1 unit of Taq DNA polymerase, and 100 ng of DNA template. QIAxcel Connect System capillary electrophoresis (QIAGEN, Hilden, Germany) with QIAxcel DNA High-Resolution Kit and QX Alignment Marker (15 bp/3 kb) were used for separating PCR products.
4.3. Statistical Analysis
The number of alleles (na), effective number of alleles (ne), Shannon’s information index (I), observed heterozygosity (Ho), expected heterozygosity (He), percentage of polymorphic loci (PPL), Nei’s genetic diversity index (Nei), principal coordinate analysis (PCoA), and the analysis of molecular variance (AMOVA) were evaluated using the GenAlEx 6.5 package [57]. In addition, the Mantel R test (pair-wise geographical and genetic distances correlation among the populations) was performed using GenAlEx software [57]. The amount of gene flow (Nm) between gene pools was calculated based on FST estimates, Nm = [(1/FST) − 1]/4. The heterozygosity (H) and polymorphic information content (PIC) values were calculated using Gene-Calc software [58].
The unweighted pair-group method with arithmetic means (UPGMA) based on the genetic distance matrices was used for the reconstruction of the dendrogram of 15 populations in PAST software with 1000 bootstrap replications [59]. The STRUCTURE v2.3.4 program with the Bayesian clustering method was used to calculate the genetic structure of the 15 Central Asian J.seravschanica populations. The burn-in period and MCMC (Markov chain Monte Carlo) replications were set to 100,000, and the iteration number was set to 3. The results of the structure analysis were evaluated using the web-based program Structure Harvester [60], and the number of clusters (K) was obtained by employing Evanno et al. [61] and Jakobsson and Rosenberg’s [62] calculations in Structure Harvester.
5. Conclusions
In this study, 15 populations of J. seravschanica growing in the mountain ranges of four distinct Central Asian regions, including parts of Uzbekistan, South and North Kyrgyzstan, and Kazakhstan, were collected. The collected samples of 323 individuals from 15 natural populations were analyzed for genetic diversity and population structure using 11 polymorphic SSR markers. Genetic diversity results indicate that J. seravschanica in this region has a 0.450 value, with the lowest recorded value in North Kyrgyzstan (0.430) and the highest in Uzbekistan (0.473). The AMOVA results suggest that 90.3% of this genetic variation exists within populations and 9.7% between populations. The STRUCTURE outputs revealed a poor clustering in this species, confirming our AMOVA results. The Nm gene flow index was 4.654, suggesting a relatively high rate of gene flow. Population 5, collected in South Kyrgyzstan, was closest to the Zeravshan River and located in the middle of Coordinate 1 and Coordinate 2 eigenvalues of the PCoA plot, which indicates this population’s importance to the origin of this species. Accordingly, the obtained results on genetic diversity in J. seravschanica populations elucidated the populations with a high level of genetic diversity.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants12162961/s1, Table S1. Allelic status in eleven SSR markers for plants of fifteen populations of Juniperus seravschanica (bp). Table S2. The population structure for samples in Juniperus seravschanica collected in Kazakhstan, Kyrgyzstan, and Uzbekistan at K2, K3, and K5 steps based on STRUCTURE analysis. Figure S1. The UPGMA dendrogram of fifteen Juniperus seravschanica populations using eleven SSR markers and Nei’s genetic distance values. Figure S2. The optimal K steps for fifteen populations of Juniperus seravschanica using 11 SSR markers and the STRUCTURE Harvester package.
Author Contributions
Conceptualization, Y.T.; Methodology, S.A. (Shyryn Almerekova), O.T., O.S. and S.A. (Saule Abugalieva); Validation, S.A. (Shyryn Almerekova), S.A. (Saule Abugalieva), O.S. and Y.T.; Formal Analysis, M.Y., S.A. (Shyryn Almerekova) and S.A. (Saule Abugalieva); Investigation, M.Y., S.A. (Shyryn Almerekova), O.T. and O.S.; Resources, M.Y., S.A. (Shyryn Almerekova), O.T. and O.S.; Data Curation, S.A. (Shyryn Almerekova) and S.A. (Saule Abugalieva); Writing—Original Draft Preparation, M.Y. and Y.T.; Writing—Review & Editing, M.Y., S.A. (Saule Abugalieva), O.T., O.S. and Y.T.; Supervision, S.A. (Shyryn Almerekova) and Y.T.; Project Administration, S.A. (Shyryn Almerekova) and S.A. (Saule Abugalieva); Funding Acquisition, S.A. (Shyryn Almerekova). All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
All data are provided in the article.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by the Science Committee of the Ministry of Science and Higher Education of the Republic of Kazakhstan (Grant No. AP09259027).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Adams R.P. The Junipers of the World: The Genus Juniperus. 4th ed. Trafford Publ.; Victoria, BC, Canada: 2014. p. 415. [Google Scholar]
- 2.Özek G., Schepetkin I.A., Yermagambetova M., Özek T., Kirpotina L.N., Almerekova S.S., Abugalieva S.I., Khlebnikov A.I., Quinn M.T. Innate immunomodulatory activity of cedrol, a component of essential oils isolated from Juniperus species. Molecules. 2021;26:7644. doi: 10.3390/molecules26247644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Komarov V.L. Multi seed juniper species in Central Asia. Bot. J. 1932;17:474–482. (In Russian) [Google Scholar]
- 4.Al Farsi K.A., Lupton D., Hitchmough J.D., Cameron R.W. How fast can conifers climb mountains? Investigating the effects of a changing climate on the viability of Juniperus seravschanica within the mountains of Oman, and developing a conservation strategy for this tree species. J. Arid Environ. 2017;147:40–53. doi: 10.1016/j.jaridenv.2017.07.020. [DOI] [Google Scholar]
- 5.Sultangaziev O., Konrad H., Schueler S., Geburek T. North-south population subdivision of Juniperus seravschanica in Kyrgyzstan revealed through novel plastid DNA markers. J. Syst. Evol. 2012;50:411–421. doi: 10.1111/j.1759-6831.2012.00206.x. [DOI] [Google Scholar]
- 6.Yermagambetova M.M., Abugalieva S.I., Turuspekov Y.K., Almerekova S.S. Conspectus of the genus Juniperus L. growing in Kazakhstan. Proc. Appl. Bot. Genet. Breed. 2022;183:161–170. doi: 10.30901/2227-8834-2022-3-161-170. (In Russian with English Abstract) [DOI] [Google Scholar]
- 7.Chorfi K., Cornet J.G., Roesle G., Nadyrbekov A., Murzakmatova R., Sultangaziev O. The Social and Economic Context for Sustainable Management of Juniper Forests in South Kyrgyzstan: The View from Local Population. ENGR; Nancy, France: 2004. [Google Scholar]
- 8.Eastwood A., Lazkov G., Newton A.C. The Red List of Trees of Central Asia. Fauna and Flora International; Cambridge, UK: 2009. p. 27. [Google Scholar]
- 9.The Red Book of Kazakh USSR Rare and endangered animal and plant species. Alma-Ata. 1981;260 (In Russian) [Google Scholar]
- 10.The Red Book of Kazakhstan Volume 2. Part 1. Plants. Re-edition, completed and revised. Astana. 2014;446 [Google Scholar]
- 11.MacLaren C.A. Climate change drives decline of Juniperus seravschanica in Oman. J. Arid Environ. 2016;128:91–100. doi: 10.1016/j.jaridenv.2016.02.001. [DOI] [Google Scholar]
- 12.Seim A., Tulyaganov T., Omurova G., Nikolyai L., Botman E., Linderholm H.W. Dendroclimatological potential of three juniper species from the Turkestan range, northwestern Pamir-Alay Mountains, Uzbekistan. Trees. 2015;30:733–748. doi: 10.1007/s00468-015-1316-y. [DOI] [Google Scholar]
- 13.Sultangaziev O., Schueler S., Geburek T. Morphometric traits and sexual dimorphisms do not strongly differentiate populations of Zeravshan juniper (Juniperus seravschanica Kom.) in Kyrgyzstan. Flora-Morphol. Distrib. Funct. Ecol. Plants. 2010;205:532–539. doi: 10.1016/j.flora.2009.12.019. [DOI] [Google Scholar]
- 14.Adams R.P., Al-Farsi A., Schwarzbach A.E. Confirmation of the southern-most population of Juniperus seravschanica in Oman by DNA sequencing of nrDNA and four cpDNA regions. Phytologia. 2014;96:218–224. [Google Scholar]
- 15.Adams R.B. Geographic variation in leaf essential oils and RAPDs of Juniperus polycarpos K. Koch in Central Asia. Biochem. Syst. Ecol. 2001;29:609–619. doi: 10.1016/S0305-1978(00)00098-3. [DOI] [PubMed] [Google Scholar]
- 16.Man K.H., Hong W.H. Genetic Diversity and Population Structure of Juniperus rigida (Cupressaceae) and Juniperus coreana. Evol. Ecol. 2000;14:87–98. [Google Scholar]
- 17.Michalczyk I.M., Sebastiani F., Buonamici A., Cremer E., Mengel C., Ziegenhagen B., Vendramin G.G. Characterization of highly polymorphic nuclear microsatellite loci in Juniperus communis L. Mol. Ecol. Notes. 2006;6:346–348. doi: 10.1111/j.1471-8286.2005.01227.x. [DOI] [Google Scholar]
- 18.Qian Z., Yang Y.Z., Wu G.L., Zhang D.Y., Liu J.Q. Isolation and characterization of microsatellite DNA primers in Juniperus przewalskii Kom (Cupressaceae) Conserv. Genet. 2008;9:767–769. [Google Scholar]
- 19.Allphin L., Hunt A.F., Anderson V.J. Genetic diversity and low reproductive success in isolated populations of Utah juniper (Juniperus osteosperma, Cupressaceae) West. N. Am. Nat. 2013;67:323–337. doi: 10.3398/1527-0904(2007)67[323:GDALRS]2.0.CO;2. [DOI] [Google Scholar]
- 20.Rumeu B., Vargas P., Jaén-Molina R., Nogales M., Caujapé-Castells J. Phylogeography and genetic structure of the threatened Canarian Juniperus cedrus (Cupressaceae) Bot. J. Linn. Soc. 2014;175:376–394. doi: 10.1111/boj.12172. [DOI] [Google Scholar]
- 21.Teixeira H., Rodríguez-Echeverría S., Nabais C. Genetic diversity and differentiation of Juniperus thurifera in Spain and Morocco as determined by SSR. PLoS ONE. 2014;9:e88996. doi: 10.1371/journal.pone.0088996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adams R.P., Schwarzbach A.E., Tashev A.N. Chloroplast capture in Juniperus sabina var. balkanensis R. P. Adams and A. N. Tashev, from the Balkan peninsula: A new variety with a history of hybridization with J. thurifera. Phytologia. 2016;98:100–111. [Google Scholar]
- 23.Reim S., Lochschmidt F., Proft A., Tröber U., Wolf H. Genetic structure and diversity in Juniperus communis populations in Saxony, Germany. Biodivers. Res. Conserv. 2016;42:9–18. [Google Scholar]
- 24.Knyazeva S.G., Hantemirova E.V. Comparative Analysis of Genetic and Morpho-Anatomical Variability of Common Juniper (Juniperus communis L.) Russ. J. Genet. 2020;56:48–58. doi: 10.1134/S102279542001007X. [DOI] [Google Scholar]
- 25.Evren Ö.H., Kaya N. High genetic diversity within and low differentiation among Juniperus excelsa M. Bieb. populations: Molecular markers reveal their genetic structure patterns. Turk. J. Bot. 2021;45:192–202. doi: 10.3906/bot-2006-22. [DOI] [Google Scholar]
- 26.Lu D., Huang H., Wang A., Zhang G. Genetic Evaluation of Juniperus sabina L. (Cupressaceae) in Arid and Semi-Arid Regions of China Based on SSR Markers. Forests. 2022;13:231. doi: 10.3390/f13020231. [DOI] [Google Scholar]
- 27.Adams R.P., Hojjati F., Schwarzbach A.E. Taxonomy of Juniperus in Iran: DNA sequences of nr DNA plus three cpDNA reveal Juniperus polycarpos var. turcomanica and J. seravschanica in southern Iran. Phytologia. 2014;96:19–25. [Google Scholar]
- 28.Okasaka M., Takaishi Y., Kashiwada Y., Kodzhimatov O.K., Ashurmetov O., Lin A.J., Consentino L.M., Lee K.H. Terpenoids from Juniperus polycarpus var. seravschanica. Phytochemistry. 2006;67:2635–2640. doi: 10.1016/j.phytochem.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 29.Yermagambetova M., Abugalieva S., Turuspekov Y., Almerekova S. Illumina sequencing data of the complete chloroplast genome of rare species Juniperus seravschanica (Cupressaceae) from Kazakhstan. Data Brief. 2023;46:108866. doi: 10.1016/j.dib.2022.108866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Z., Naifa L., Zhou T. A comparative study of genetic diversity of peripheral and central populations of chukar partridge from Northwestern China. Biochem. Genet. 2005;43:613–621. doi: 10.1007/s10528-005-9118-3. [DOI] [PubMed] [Google Scholar]
- 31.Shulgina I., Yakubov B., Orlovsky N., Mendlinger S., Shulgina I., Volis S. Genetic (RAPD) Diversity Across Species Range: Core vs. Peripheral Populations of Wild Barley in Israel and Turkmenistan. Isr. J. Ecol. Evol. 2006;52:93–109. doi: 10.1560/IJEE_52_2_93. [DOI] [Google Scholar]
- 32.Volis S., Mendlinger S., Turuspekov Y., Esnazarov U., Abugalieva S., Orlovsky N. Allozyme variation in Turkmenian populations of wild barley, Hordeum spontaneum Koch. Ann. Bot. 2001;87:435–446. doi: 10.1006/anbo.2000.1354. [DOI] [Google Scholar]
- 33.Smith T.B., Wayne R.K., Girman D.J., Bruford M.W. A role for ecotones in generating rainforest biodiversity. Science. 1997;276:1855–1857. doi: 10.1126/science.276.5320.1855. [DOI] [Google Scholar]
- 34.Kark S., Volis S., Novoplansky A. Biodiversity along core-periphery distribution clines. Biodivers. Drylands. 1999;3:30–56. [Google Scholar]
- 35.Adams R.P., Turuspekov Y. Taxonomic reassessment of some Central Asian and Himalayan scale-leaved taxa of Juniperus (Cupressaceae) supported by random amplification of polymorphic DNA. Taxon. 1998;47:75–83. doi: 10.2307/1224021. [DOI] [Google Scholar]
- 36.Dzialuk A., Mazur M., Boratyńska K., Montserrat J.M., Romo A., Boratyński A. Population genetic structure of Juniperus phoenicea (Cupressaceae) in the western Mediterranean Basin: Gradient of diversity on a broad geographical scale. Ann. For. Sci. 2011;68:1341–1350. doi: 10.1007/s13595-011-0150-7. [DOI] [Google Scholar]
- 37.Juan A., Fay M.F., Pastor J., Juan R., Fernández I., Crespo M.B. Genetic structure and phylogeography in Juniperus oxycedrus subsp. macrocarpa around the Mediterranean and Atlantic coasts of the Iberian Peninsula, based on AFLP and plastid markers. Eur. J. For. Res. 2012;131:845–856. doi: 10.1007/s10342-011-0558-5. [DOI] [Google Scholar]
- 38.Ellegren H. Microsatellites: Simple sequences with complex evolution. Nat. Rev. Genet. 2004;5:435–445. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
- 39.Selkoe K.A., Toonen R.J. Microsatellites for ecologists: A practical guide to using and evaluating microsatellite markers. Ecol. Lett. 2006;9:615–629. doi: 10.1111/j.1461-0248.2006.00889.x. [DOI] [PubMed] [Google Scholar]
- 40.Turuspekov Y., Nakamura K., Yoshikawa R., Tuberosa R. Genetic diversity of Japanese barley cultivars based on SSR analysis. Breed. Sci. 2001;51:215–218. doi: 10.1270/jsbbs.51.215. [DOI] [Google Scholar]
- 41.Almerekova S., Favarisova N., Turuspekov Y., Abugalieva S. Cross-Genera Transferability of Microsatellite Markers and Phylogenetic Assessment of Three Salsola Species from Western Kazakhstan. Proc. Latv. Acad. Sci. Sect. B Nat. Exact Appl. Sci. 2020;74:325–334. doi: 10.2478/prolas-2020-0049. [DOI] [Google Scholar]
- 42.Yermagambetova M.M., Almerekova S.S., Krekova Y., Abugalieva S.I., Turuspekov Y.K. Genetic variation in populations of Picea schrenkiana Fisch. et CA Mey. based on simple sequence repeat markers. Mosc. Univ. Biol. Sci. Bull. 2022;77:76–83. doi: 10.3103/S0096392522020134. [DOI] [Google Scholar]
- 43.Rahimian Boogar A., Salehi H. ISSR-based genetic diversity assessment of five populations of Juniperus polycarpos K. Koch in southern habitats of Iran. Flower Ornam. Plants. 2021;6:1–12. doi: 10.52547/flowerjournal.6.1.1. [DOI] [Google Scholar]
- 44.Zhang G., Zhang X., Wang L., Liang X., Wen G., Hong Y. RAPD analysis on genetic differentiation of Sabina vulgaris populations in Inner Mongolia area. J. Arid Land Resour. Environ. 2005;19:193–198. [Google Scholar]
- 45.Hughes A.R., Inouye B.D., Johnson M.T., Underwood N., Vellend M. Ecological consequences of genetic diversity. Ecol. Lett. 2008;11:609–623. doi: 10.1111/j.1461-0248.2008.01179.x. [DOI] [PubMed] [Google Scholar]
- 46.Ledig F.T. Genetic Variation in Pinus. In: Richardson D.M., editor. Ecology and Biology of Pinus. Cambridge University Press; Cambridge, UK: 1998. pp. 251–280. [Google Scholar]
- 47.Provan J., Beatty G.E., Hunter A.M., McDonald R.A., McLaughlin E., Preston S.J., Wilson S. Restricted gene flow in fragmented populations of a wind-pollinated tree. Conserv. Genetic. 2008;9:1521–1532. doi: 10.1007/s10592-007-9484-y. [DOI] [Google Scholar]
- 48.Bettencourt S.X., Mendonça D., Lopes M.S., Rocha S., Monjardino P., Monteiro L., da Câmara Machado A. Genetic diversity and population structure of the endemic Azorean juniper, Juniperus brevifolia (Seub.) Antoine, inferred from SSRs and ISSR markers. Biochem. Syst. Ecol. 2015;59:314–324. doi: 10.1016/j.bse.2015.02.003. [DOI] [Google Scholar]
- 49.Opgenoorth L. Identification and characterization of microsatellite marker in the tetraploid Juniperus tibetica Kom. using next generation sequencing. Conserv. Genet. Resour. 2009;1:253–255. doi: 10.1007/s12686-009-9062-3. [DOI] [Google Scholar]
- 50.Aleksandrovsky E.S., Abdurazakova U.T. In: Nauchnye Osnovy Lesomelioratsiiv Uzbekistane. (Scientific Bases of Forest Amelioration in Uzbekistan) Kayimov A.K., editor. UZNIILH; Tashkent, Uzbekistan: 1996. pp. 131–189. (In Russian) [Google Scholar]
- 51.Sultangaziev O. Population differentiation of Juniperus seravschanica Kom. in Kyrgyzstan. Akamediker Verlag. Nat. Sci. Ser. 2017;108 [Google Scholar]
- 52.Rumeu B., Caujape-Castells J., Blanco-Pastor J.L., Ja en-Molina R., Nogales M., Elias R.B., Vargas P. The colonization history of Juniperus brevifolia (Cupressaceae) in the Azores islands. PLoS ONE. 2011;6:e27697. doi: 10.1371/journal.pone.0027697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Slatkin M. Gene flow and the geographical structure of natural populations. Science. 1987;236:787–792. doi: 10.1126/science.3576198. [DOI] [PubMed] [Google Scholar]
- 54.Kark S. Within-Population Diversity in the Distribution Range: Partridges as a Research Model. Department of Evolution, Systematics and Ecology, The Hebrew University of Jerusalem; Jerusalem, Israel: The Desertification and Restoration Ecology Research Center; Negev, Israel: 1999. [Google Scholar]
- 55.Mustafaev I.M., Islomiddinov Z.S., Iminova M.M., Ortiqov I.Z. Distribution of species of the genus Gymnosporangium (Pucciniales) in Uzbekistan. Ukr. Bot. J. 2021;78:39–46. doi: 10.15407/ukrbotj78.01.039. [DOI] [Google Scholar]
- 56.Doyle J.J., Doyle J.L. A Rapid DNA Isolation Procedure for Small Quantities of Fresh Leaf Tissue. Phytochem. Bull. 1987;19:11–15. [Google Scholar]
- 57.Peakall R., Smouse P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics. 2012;28:2537–2539. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bińkowski J., Miks S. Gene-Calc [Computer Software] 2018. [(accessed on 8 August 2023)]. Available online: www.gene-calc.pl.
- 59.Hammer Ø., Harper D.A., Ryan P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001;4:4–9. [Google Scholar]
- 60.Earl D.A., vonHoldt B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet Resour. 2012;4:359–361. doi: 10.1007/s12686-011-9548-7. [DOI] [Google Scholar]
- 61.Evanno G., Regnaut S., Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 62.Jakobsson M., Rosenberg N.A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are provided in the article.