Abstract
Four different standardization approaches based on a competitive reverse transcription (RT)-PCR assay were compared with a noncompetitive assay based on an external standard curve. Criteria for assessment were accuracy in quantitation, correctness of recovery, sensitivity, dynamic range, reproducibility, throughput, and convenience of sample handling. As a model system, we used the 5′-noncoding region of hepatitis C virus (HCV) for amplification in all quantitative RT-PCRs. A computer program that allowed parallel data processing was developed. Surprisingly, all methods were found suitable for accurate quantitation and comparable with respect to the criterion correctness of recovery. All results differed only by a factor of about 2. The reason for this finding might be that all of our mimics, as well as the wild-type genome of HCV, exhibited exactly the same amplification and hybridization efficacy. Moreover, minimal competition occurred in our experiments over a 5-log dynamic range. A further topic of our investigation was the comparison of two different competitive RNA fragments, mimics, with regard to their suitability as internal standards. One was a heterologous mimic, in which only the primer binding sites were identical to the wild type. The second one was a homologous mimic identical to the wild type except for a small region used for differential hybridization, which was replaced by a permutated sequence of the same length. Both the homologous and heterologous internal mimics were found appropriate for an accurate competitive RT-PCR assay, provided that amplification efficacy, as well as capture efficacy, is proven identical for both analyte and mimic.
Quantitation of nucleic acids has become an essential tool in molecular diagnostics. These quantitative determinations are helpful not only in understanding the progress of infectious diseases but also in monitoring antiviral drug therapy, e.g., for human immunodeficiency virus (HIV) or hepatitis C virus (HCV). In the past few years, there have been many publications dealing with the quantitation of PCR products. The first approaches were only semiquantitative and were based on limiting dilution of the analyte (25). Other methods used external standard curves for quantitation (27) or low-stringency PCR (4). None of these approaches overcame the problem of inhibition of individual probes. As a consequence, the next generation focused on amplification reactions that were internally controlled, either by coamplification of internal endogenous standards, such as housekeeping genes (5, 16), or by introduction of an artificial exogenous mimic fragment (2, 9, 26). For detailed reviews, see Clementi et al. (6, 7).
This last approach was finally established in the molecular diagnosis of many infectious disease parameters, either in commercially available tests or in in-house assays. A greater diversity can be found among standardization concepts. Frequently, a serial-dilution method (referred to here as method A) (Table 1), where either the analyte is diluted and coamplified with a constant amount of internal mimic or vice versa (16, 20, 22), is applied. Another common standardization format is based on the generation of an external standard curve, where known and increasing amounts of cloned wild-type fragments are coamplified with one constant amount of a mutated competitor mimic (method B) (Table 1). A third standardization method (method C) (Table 1) relies on a standard curve generated only by one mutated mimic template (18). A fourth standardization approach is even more simple and requires no standard curve (method D) (Table 1). In addition to the above internally controlled amplifications, an external standardization and/or quantitation approach based on a noncompetitive reverse transcription (RT)-PCR was also compared in our investigation (method E) (Table 1).
TABLE 1.
Characterization of the five standardization methods
| Method | Description | Internal mimic | No. of PCRs
|
|
|---|---|---|---|---|
| Per sample | Per standard curve | |||
| A | Constant amount of analyte spiked with a serial dilution of internal mimic | + | 5 | |
| B | External standard curve generated by coamplification of analyte and mimic in serial dilutions | + | 1 | 5 (Analyte including mimic) |
| C | Variant of method B; external standard curve is generated by amplifying only the mimic in serial dilutions | + | 1 | 5 (Mimic only) |
| D | No standard curve; quantitation is achieved by relating analyte signal to known amounts of mimic signal | + | 1 | |
| E | No internal mimic; quantitation on external standard curve generated with serial dilution of defined analyte material | − | 1 | 5 (Analyte only) |
The aim of the present study was to compare all five standardization approaches in one distinct and well-described format. This was done both in a model system using two cloned mimic fragments, pHCV-st1 and pHCV-wt1, and with clinical material (HCV-positive plasma samples). The second purpose of our investigation was to compare different RNA competitors with respect to their capability to mimic the overall RT-PCR efficacy. In vitro transcription and amplification has to be identical for both internal mimic and analyte in order to ensure accurate quantitation in a given dynamic range. This is usually considered for mimics of the same size as the wild-type template. But some have proposed that even the sequence itself and the nucleotide content of both templates play an important role in the above-mentioned efficacy (19). In order to clarify this, we cloned and compared two different mimics, both of the same length as the amplified wild-type region but differing in sequence.
MATERIALS AND METHODS
Patient samples.
All plasma samples were from patients with historically proven hepatitis. Viral (HCV) status was determined by HCV antibody testing using the Abbott enzyme immunoassay and was confirmed by RT-PCR using the Amplicor HCV kit (Roche Molecular Systems).
Cloning and preparation of the mimics.
First, an unmodified wild-type standard, pHCV-wt1, was cloned based on amplification of wild-type HCV with primers KY80 (forward) and KY78 (reverse), both described by Young et al. (29), followed by blunt-end ligation into the vector pBluescript SK+ (Stratagene, Heidelberg, Germany).
Subsequently, the homologous standard, pHCV-st1, was obtained from pHCV-wt1 by site-directed mutagenesis as described in Ho et al. (12). The capture region was replaced by part of a sequence (21 bp) from a plastid-encoded gene (rbcL) of a green plant (11). This was done to avoid any cross-reactions between capture probe and human, animal, bacterial, or viral sequences. The 21-bp fragment (Ip102) has the same length, A+T content, and G+C content as the wild-type (p102) but differs from it in sequence.
The heterologous standard, pHCV-st2, also based on the above-mentioned primers, was constructed by using the PCR-mimic construction kit (Clontech, Palo Alto, Calif.). This fragment is derived from the v-erb B oncogene (24). The sequences of all standards were confirmed according to their ancestral sources. For RNA production, all standards were transcribed in vitro (1), treated twice with phenol and once with chloroform, and finally purified by gel filtration (Quick Spin columns; Boehringer Mannheim, Penzberg, Germany). The remaining DNA was digested twice with DNase I (Boehringer Mannheim), and RNA was checked for purity by PCR omitting the RT step and using Taq polymerase instead of Tth polymerase. The concentration of the RNA was determined photometrically by measuring the optical density at 260 nm. RNA was then serially diluted in water and frozen as aliquots at −80°C. These aliquots were stable for months without any degradation. The terms st1, st2, and wt1, used below, refer to the RNAs of the plasmids pHCV-st1, pHCV-st2, and pHCV-wt1, respectively.
Sample preparation.
Total RNA from HCV-positive plasma was extracted according to a method described in principle by Boom et al. (3) but slightly modified. In brief, virus particles present in 500 μl of plasma were lysed in a total volume of 1 ml in the presence of guanidinium isothiocyanate during a 10-min incubation at room temperature. Binding of both nucleic acids (genomic RNA and mimic RNA) to glass magnetic particles took place in a buffer containing chaotropic salts, isopropanol, and silica surfaces. Unspecifically bound material was removed by several washing steps. Finally, nucleic acids were eluted in a total volume of 100 μl at 80°C for 15 min. Ten microliters from the eluate was used for RT-PCR. In our model system, when only RNA competitor mimics were used, sample preparation was carried out in HCV-negative plasma in order to compensate for matrix effects.
RT-PCR assays.
The 5′-untranslated region of the HCV genome was used for amplification, since this region is known to be the most conserved among different HCV genotypes (14, 25). Primers used for in vitro transcription as well as amplification were KY80 and KY78; the latter was biotinylated, corresponding to our detection format (see below). Our RT-PCR protocol was performed as a one-step assay in a total volume of 100 μl containing 50 mM bicine, 115 mM potassium acetate, 8% (vol/vol) glycerol, 2.5 mM manganese acetate, 0.2 mM (each) dATP, dCTP, and dGTP, 0.6 mM dUTP, 0.3 μM each primer, and 10 U of Tth polymerase. In all amplifications, 2 U of heat-labile uracil-N-glycosylase (UNG) was used to control carryover of amplicons and to reduce background signals. All reagents were from Boehringer Mannheim GmbH. RT-PCR was performed in a PE 9600 thermocycler (Perkin-Elmer Applied Biosystems) according to the following procedure. A 10-min incubation at 37°C (which allows UNG to digest possible amplicon contaminations) was followed by a RT reaction for 30 min at 60°C. Subsequently, RNA-DNA heteroduplexes were denatured for 1 min at 95°C. PCR amplification proceeded with 35 cycles at 95°C for 15 s and 60°C for 20 s, followed by a final extension at 72°C for 10 min. PCR products were kept at 50°C on the thermocycler until detection in order to circumvent UNG renaturation.
Detection protocol.
For detection of amplified material, we used a very sensitive nonisotopic approach based on electrochemiluminescence (ECL). A ruthenium-tris(bipyridyl)-labeled oligonucleotide (capture probe) was hybridized specifically to the biotinylated denatured amplicon. Subsequent, this hybrid was bound to the surface of streptavidin-coated magnetic beads. After the beads were captured on an electrode by using a permanent magnet, the ECL reaction of the ruthenium label was triggered by voltage application. For details of the ECL detection process, see Hoyle et al. (13). The totally automated ECL detection was performed on an instrumental platform (preprototype of Elecsys 1010; Boehringer Mannheim GmbH).
Capture probes used for hybridization with the wild-type and different mimic amplicons were as follows: p102 (for the wild type standard, pHCV-wt1), 5′-GTCGTGCAGCCTCCAGGACCC-3′; Ip102 (for the homologous mimic, pHCV-st1); 5′-GGGGTAATGCGCCAGGTGCCG-3′; and probe 3 (for the heterologous mimic, pHCV-st2), 5′-CCACACCAGGGCTTTTTCAACTGC-3′. All capture probes were ruthenium labeled at their 5′ ends.
Standardization methods.
All RNA mimics were processed (unless otherwise stated) throughout the sample preparation with an initial concentration of 2 × 103 to 2 × 107 copies per ml. Assuming that no loss occurs after sample preparation, this corresponds to 102 to 106 copies of RNA per RT-PCR assay. Since no sample preparation process leads to a quantitative isolation of nucleic acids, the real copy number in our assay is lower than this.
(i) Method A—internal standard curve.
The sample to be determined (clinical plasma or wt1 RNA) was spiked with a serial dilution (102 to 106 copies per assay; see above) of competitor mimic st1. Subsequent, five RT-PCRs were carried out, and the amplicon of each reaction was split and hybridized with its corresponding specific probe. Either all signal values were plotted onto a double-log scale and quantitation was made graphically, or, for greater accuracy, signal values were processed mathematically with our software tool.
(ii) Method B—external standard curve with two RNAs.
Method B was first described by Gerna et al. (8) and was subsequently used for different applications (20, 23). First, a standard curve was generated by coamplification of increasing amounts of wt1 RNA (102 to 106 copies) and a constant amount of competitor mimic st1 (103 copies per assay). Signal ratios from differential hybridizations (wt1/st1) were plotted onto a double-log scale against the initial wt1 RNA concentration. Quantitation of individual HCV samples was then obtained by coamplifying each sample with the same amount of st1 RNA (103 copies) and then mathematically plotting the signal ratio between the sample and st1 RNA on the external standard curve.
(iii) Method C—external standard curve with one RNA.
An external standard curve was generated with st1 RNA in increasing amounts (102 to 106 copies). HCV samples were then coamplified with one internal st1 concentration (103 copies), followed by differential hybridization. Subsequently, the obtained signal from st1 was used to calibrate the standard curve individually for each sample by a factor derived from the difference between the expected and the measured signal. The signal of this sample was then read from the corrected standard curve.
(iv) Method D—without standard curve.
The simplest method lacks any standard curve and is based on coamplification of sample RNA and st1 RNA in one defined concentration (103 copies). Quantitation of the initial sample concentration was then derived from the following formula: initial sample concentration = (signal from sample/signal from st1) × initial st1 concentration.
(v) Method E—without competitive RT-PCR.
An external standard curve was generated by amplifying increasing amounts of wt1 RNA (102 to 106 copies). Afterwards, the sample signal was plotted on the standard curve. This approach served as an example of a noninfluenced RT-PCR, since no competitor mimic was coamplified.
Evaluation methods and software tool.
A special computer program which supports all mathematical algorithms needed for the evaluation of the five standardization approaches was developed. For each of the procedures, different curve-fitting algorithms are available, among them an especially adapted nonlinear curve fitting for the Rodbard model (21). Statistics were calculated based on sample measurements. This allowed a direct comparison of the different procedures. Assay-specific numbers—the critical level, the detection limit, and the minimal distinguishable difference—are also available. The software adheres to common standards for software running under Windows 95 and Windows NT.
To circumvent statistical phenomena, particularly at lower concentrations, multiple determinations were carried out as follows: fivefold determination for 10 copies of RNA, threefold determination for 100 copies of RNA, and twofold determination for every higher concentration. In addition, each individual experiment was carried out at least twice to confirm the results. In order to reduce interassay variations, most of our comparative studies were performed in the very same run. This means that most samples were eluted during one sample preparation and that all mimics used and quantified were derived from one aliquot after in vitro transcription. All RT-PCRs and most of the detection experiments were carried out in parallel in any case where data were directly compared. The interassay coefficient of variation was 25% for method A, 70% for method B, 45% for method C, 35% for method D, and 20% for method E.
RESULTS
Evaluation of wt1, st1, and st2 RNA.
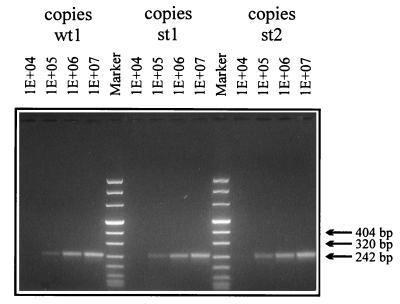
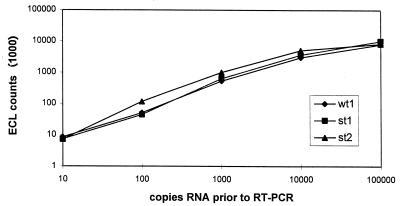
A well defined model system requires that all mimics be amplified and captured with the same efficacy. To prove this, amplification efficacy was determined by amplifying different starting copy numbers independently for each mimic. Here, direct detection by agarose gel electrophoresis was preferred in order to exclude any hybridization influence (Fig. 1). Afterwards, different capture probes were evaluated on each mimic in order to ensure the same capture efficacy on their corresponding templates (data not shown). For each mimic, a capture probe which totally fulfilled this requirement was chosen (Fig. 2). As can be seen in Fig. 2, each specific capture probe generated a signal in the same range for a given concentration. Copy numbers higher than 104 to 105 led to a saturation of either amplification or hybridization, as the curves in Fig. 2 indicate. For capture probe 3, which hybridizes with the st2 amplicon, signals were slightly higher. Hence, most of our competitive RT-PCRs were performed either with st1 and clinical material, or with st1 and wt1 when our model system was used.
FIG. 1.
Amplification efficacies of different competitive mimics. The length marker is no. VIII from Boehringer Mannheim GmbH. The amplicon length is 244 bp.
FIG. 2.
Illustration of the capture efficacies of different mimics hybridized with their corresponding capture probes. wt1, wild-type RNA; st1, homologous mimic RNA; st2, heterologous mimic RNA.
Since all signal values in Fig. 2 were blank corrected, one can also see that our analytical sensitivity is consistently in the range of about 10 copies of RNA per amplification assay.
Competitive RT-PCR with homologous (st1) and heterologous (st2) mimics.
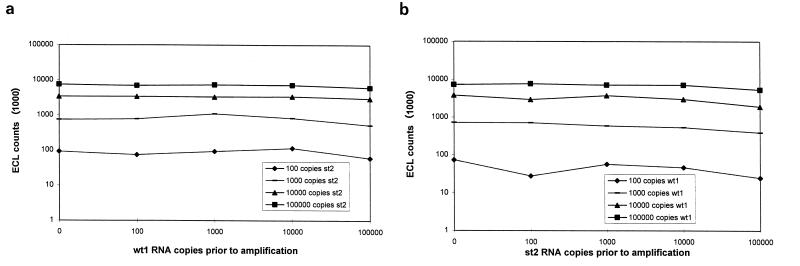
All competitive RT-PCRs were performed either in our model system with wt1 as a wild-type analog and st1 or st2 as the counterpart or with clinical material and st1. In order to assess competition between both templates, two experiments were designed; in the first, serial dilutions of wt1 were spiked with a constant amount of st2 (Fig. 3a), and in the second, serial dilutions of st2 were spiked with a constant amount of wt1 (Fig. 3b). As illustrated in Fig. 3, nearly no competition was seen over a 5-log dynamic range, indicating that our system is very stable and suitable for comparing different standardization approaches (see below). Only at the extreme target/competitor ratio of 105:102 could a very weak competition be observed. The same results were obtained when both experiments were repeated with st1 as the counterpart to clinical material or to wt1 (data not shown). Hence, our competitive RT-PCR is independent of the kind of mimic introduced, homologous or heterologous.
FIG. 3.
Competitive RT-PCR assay. (a) A constant amount of st2 RNA was coamplified with increasing amounts of wt1 RNA, followed by hybridization with an st2-specific capture probe. (b) A constant amount of wt1 RNA was coamplified with increasing amounts of st2 RNA, followed by hybridization with a wt1-specific capture probe. No competition could be seen in either case over a 5-log dynamic range.
Quantitation according to different standardization approaches.
In order to cover a wide dynamic range in those experiments which require one internal mimic amount (methods B, C, and D), we have chosen a mean concentration of competitor mimic of 20,000 copies per ml of plasma. Provided that no loss of RNA occurs during sample preparation, this would lead to 1,000 copies per RT-PCR assay.
In addition to our model system (wt1-st1), we also quantified five different HCV-positive plasma samples, referred to as no. 025, 043, 100, 114, and 122, undiluted and 10-fold diluted three times (1:10; 1:100; and 1:1,000) in HCV-negative plasma.
(i) Method A.
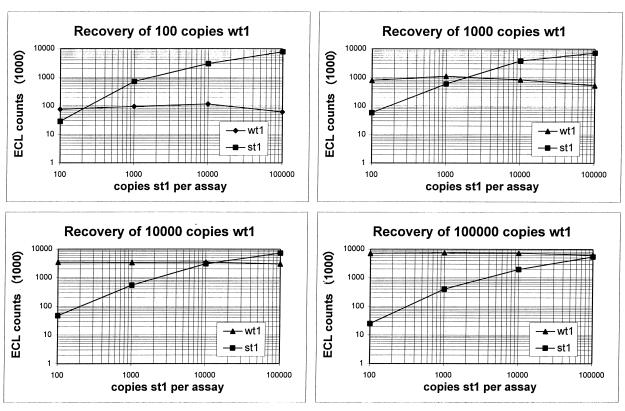
In the first experiments, the serial dilution method was used in our model system without sample preparation, in which a constant amount of wt1 was coamplified with increasing amounts of st1. With this method, 100 copies of wt1 RNA were quantified (recovered) with approximately 200 copies of st1, 1,000 copies were found with approximately 1,900 copies, 10,000 copies were found with approximately 14,000 copies, and 100,000 copies could be found with nearly the given nominal value (Fig. 4). As seen in former experiments, only very weak competition was found across the entire tested dynamic range (Fig. 4). If the mimics were processed throughout sample preparation, recovery appeared slightly reduced, indicating little loss of RNA (in the range of 10% [data not shown]). In the following quantitative experiments, we applied this method on HCV-positive plasma. Again, only minimal competition was seen. Data derived from these experiments are partly summarized in Table 2.
FIG. 4.
Accuracy of quantitation according to method A. In all experiments, a competitive RT-PCR was carried out with constant amounts of wt1 and increasing amounts of st1 RNA. The recovery of wt1 RNA in each experiment is depicted and can be read at the point of crossing of the two curves.
TABLE 2.
Comparison of different standardization/quantitation approaches applied on several HCV-positive plasma samples
| Plasma sample no. and dilution | No. of copies with:
|
||||
|---|---|---|---|---|---|
| Method A | Method B | Method C | Method D | Method E | |
| 100 | 7.0 × 105 | 3.3 × 105 | 4.8 × 105 | 6.0 × 105 | 5.1 × 105 |
| 1:10 | NDa | 4.0 × 104 | 6.2 × 104 | 5.1 × 104 | 9.5 × 104 |
| 1:100 | ND | <2 × 103 | <2 × 103 | <2 × 103 | <2 × 103 |
| 1:1000 | ND | <2 × 103 | <2 × 103 | <2 × 103 | <2 × 103 |
| 122 | 3.6 × 106 | 9.0 × 105 | 1.5 × 106 | 2.0 × 106 | 1.2 × 106 |
| 1:10 | ND | 1.2 × 105 | 1.8 × 105 | 1.8 × 105 | 2.7 × 105 |
| 1:100 | ND | 2.2 × 104 | 2.2 × 104 | 2.4 × 104 | 4.1 × 104 |
| 1:1000 | ND | <2 × 103 | <2 × 103 | <2 × 103 | <2 × 103 |
| 114 | 4.0 × 106 | 9.1 × 105 | 1.8 × 106 | 2.1 × 106 | 1.8 × 106 |
| 1:10 | ND | 2.4 × 105 | 3.5 × 105 | 4.2 × 105 | 4.5 × 105 |
| 1:100 | ND | 5.5 × 104 | 8.4 × 104 | 7.2 × 104 | 1.2 × 105 |
| 1:1000 | ND | 1.2 × 104 | 1.4 × 104 | 1.0 × 104 | 3.2 × 104 |
| 043 | ND | >2 × 107 | >2 × 107 | >2 × 107 | 1.5 × 107 |
| 1:10 | ND | 4.3 × 106 | 1.0 × 107 | 8.3 × 106 | 5.1 × 106 |
| 1:100 | ND | 4.7 × 105 | 6.8 × 105 | 9.1 × 105 | 8.1 × 105 |
| 1:1000 | ND | 7.4 × 104 | 1.1 × 105 | 1.0 × 105 | 1.4 × 105 |
| 025 | 1.2 × 107 | 2.4 × 106 | 4.6 × 106 | 5.4 × 106 | 4.1 × 106 |
| 1:10 | ND | 4.7 × 105 | 7.8 × 105 | 9.7 × 105 | 7.2 × 105 |
| 1:100 | ND | 6.0 × 104 | 9.0 × 104 | 7.9 × 104 | 1.1 × 105 |
| 1:1000 | ND | 1.8 × 104 | 2.5 × 104 | 1.6 × 104 | 3.6 × 104 |
ND, not determined.
(ii) Method B.
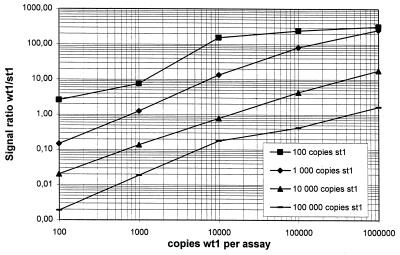
For determining the optimal concentration of st1 RNA, 102 to 106 copies of wt1 RNA were coamplified with varying amounts of st1 (102 to 105 copies). Standard curves were generated by plotting the signal ratio of wt1/st1 against the initial wt1 concentration on a logarithmic scale (Fig. 5). As seen in Fig. 5, the linearity of the standard curves, which is a prerequisite for accurate quantitation, is almost independent of the concentration of coamplified st1; deviations were observed only with the lowest concentration of st1. We therefore decided to use 104 copies of st1 for further coamplification experiments. This concentration gives an excellent linearity of the standard curve and also avoids possible competition effects at higher concentrations or high variations due to statistical distributions of st1 at lower concentrations.
FIG. 5.
Standard curves according to method B. In a competitive RT-PCR, serial dilutions of wt1 RNA were coamplified with a constant amount of st1 RNA. This was done for four different st1 RNA concentrations (100 to 100,000 copies). The signal ratio of each individual assay is depicted, showing linearity over the entire range.
(iii) Other methods.
For all other methods, data were not depicted in detail but were processed with our software tool and are summarized in Table 2. As can be seen, all quantitative results are in the same range and differ only by a factor of about 2. Sample readings greater than 2 × 107 or smaller than 2 × 103 (Table 2) are not facilitated by our software tool in the current version.
DISCUSSION
PCR has become an important tool in the diagnosis of infectious diseases because of its high sensitivity. Particularly in those cases where monitoring of therapy is required (e.g., in HCV viremia), quantitative determinations are mandatory. The need for accuracy and correctness of recovery is obvious, but beyond that and for routine applications, quantitative PCR should offer maximal convenience and high sample throughput (28). In order to achieve these goals, different quantitation approaches have been used in the past. Presently, competitive RT-PCR is considered most reliable and reproducible (7), since this approach is solely able to recognize inhibitors in individual samples. Besides the necessity of controlling those tube-to-tube variations during amplification, a further important requirement in a routine molecular diagnostic laboratory is to standardize all processes, including sample preparation, amplification, and detection. This can be achieved by introducing an RNA mimic for RT-PCR assays directly into the sample preparation. It is highly imprecise to use DNA mimics for RNA quantitation and to calculate reverse transcription efficacy, since the efficacy of RT-PCR is controlled more by reverse transcription into cDNA than by the amplification of this cDNA (7).
These findings are widely accepted and taken into account, but great diversity can be found among standardization concepts. In order to compare different standardization approaches in parallel, we developed a well-defined model system based on the amplification of RNA mimics. In addition to this model system, we applied these standardization methods to clinical plasma samples from HCV-infected patients in order to confirm our findings. Plasma samples were used because significant RNA loss could sometimes be observed in serum samples (17).
The serial-dilution approach (method A), often found in research laboratories (2, 10, 15), is proposed to be most accurate in absolute quantitation but is very cumbersome and has a poor throughput with regard to routine applications. Every sample has to be spiked with increasing amounts of RNA mimic, usually four to five concentrations, and run throughout the entire process. After amplification, each competitive reaction has to be split into two hybridization reactions, ending up with eight to ten detections per sample.
In contrast to this internal standardization, different external approaches exist to increase throughput and convenience. In these approaches, an external standard curve is generated by coamplification either of two different mimics (method B) (8, 20, 23) or of only one mimic (method C) (18). The theoretical advantage of method B is that the hybridization efficacies of the wild type and the internal mimic might be different, but the ratio between the two curves is the same in both the external standard curve and the sample. Method C further simplifies calibration by using only one mimic and is therefore even more convenient. Finally, method D completely omits any standard curves and calculates the sample concentration just from the signal ratios of the internal mimic and the analyte multiplied by the initial mimic concentration. In addition to these internally controlled assays, a noncompetitive RT-PCR quantified with an external standard curve (method E) was also implemented in our comparison in order to investigate a noninfluenced (no mimic was coamplified) RT-PCR.
Besides the criteria of convenience, throughput and accuracy, we also assessed reproducibility, dynamic range, sensitivity, and correctness of recovery. Interestingly, and in contrast to our expectation, we found all methods comparable with regard to the latter set of criteria. All results differed approximately by a factor of 2 (Table 2). In our understanding there are three main reasons for this finding. First, all of our mimics, as well as the analyte, reveal exactly the same amplification and hybridization efficacy, an important prerequisite for any accurate quantitative test. The observation that minimal competition occurs in our experiments over a 5-log dynamic range could be due either to this very same efficacy or to the fact that our RT-PCR was not in any saturated condition because of our highly sensitive ECL detection. Second, as can be concluded from method E, we had no inhibition of individual samples, and therefore it was not necessary to correct (calibrate) the standard curves. The finding that no inhibition was observed could be due either to the homogeneous specimen (plasma) or to effective separation of all inhibitors during sample preparation. Besides this, the lack of sample inhibition might also be due to the limited number of plasma samples tested in our investigation. Artificial spiking with strong inhibitors, such as ethanol or hemin, revealed the limitation of method E where an internal amplification control is missing, whereas the other methods gave comparable results because any inhibition was indicated by the internal control used in each of these methods (data not shown). Third, the equivalence of our standardization methods may also be due to our particular system. Other sample preparation methods, more cycles, or another detection process might lead to other results.
Beyond that, our results further indicate that equal amplification and capture efficacy are independent from the kind of mimic introduced, homologous or heterologous. Hence, both mimics are suitable for competitive RT-PCR assays.
If the above-mentioned conditions are ensured, then method D, the easiest standardization/quantitation concept, is as good as any other approach and is therefore the method of choice.
ACKNOWLEDGMENTS
We are very grateful to G. Kagerer, S. Mitzel, and I. Egger for technical assistance and to C. Berding and G. Ziegler for stimulating discussions regarding statistical analysis.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1995. [Google Scholar]
- 2.Besnard N C, Andre P M. Automated quantitative determination of hepatitis C virus viremia by reverse transcription-PCR. J Clin Microbiol. 1994;32:1887–1893. doi: 10.1128/jcm.32.8.1887-1893.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caballero O L, Villa L L, Simpson A J G. Low stringency-PCR (LS-PCR) allows entirely internally standardized DNA quantitation. Nucleic Acids Res. 1995;23:192–193. doi: 10.1093/nar/23.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chelly J, Montarras D, Pinset C, Berwald-Netter Y, Kaplan J-C. Quantitative estimation of minor mRNAs by cDNA-polymerase chain reaction: application to dystrophin mRNA in cultured myogenic and brain cells. Eur J Biochem. 1990;187:691–698. doi: 10.1111/j.1432-1033.1990.tb15355.x. [DOI] [PubMed] [Google Scholar]
- 6.Clementi M, Menzo S, Bagnarelli P, Manzin A, Valenza A, Varaldo P E. Quantitative PCR and RT-PCR in virology. PCR Methods Applic. 1993;2:191–196. doi: 10.1101/gr.2.3.191. [DOI] [PubMed] [Google Scholar]
- 7.Clementi M, Menzo S, Manzin A, Bagnarelli P. Quantitative molecular methods in virology. Arch Virol. 1995;140:1523–1539. doi: 10.1007/BF01322527. [DOI] [PubMed] [Google Scholar]
- 8.Gerna G, Balfanti F, Sarasini A, Furione M, Percivalle E, Revello M G, Zipeto D, Zella D The Italian Foscarnet Study Group. Effect of foscarnet induction treatment on quantitation of human cytomegalovirus (HCMV) DNA in peripheral blood polymorphonuclear leukocytes and aqueous humor of AIDS patients with HCMV retinitis. Antimicrob Agents Chemother. 1994;38:38–44. doi: 10.1128/aac.38.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilliland G, Perrin S, Blanchard K, Bunn H F. Analysis of cytokine mRNA and DNA: detection and quantitation by competitive polymerase chain reaction. Proc Natl Acad Sci USA. 1990;87:2725–2729. doi: 10.1073/pnas.87.7.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gretch D, Corey L, Wilson J, dela Rosa C, Willson R, Carithers R, Jr, Busch M, Hart J, Sayers M, Han J. Assessment of hepatitis C virus RNA levels by quantitative competitive RNA polymerase chain reaction: high-titer viremia correlates with advanced stage of disease. J Infect Dis. 1994;169:1219–1225. doi: 10.1093/infdis/169.6.1219. [DOI] [PubMed] [Google Scholar]
- 11.Haberhausen G, Zetsche K. Nucleotide sequence of the rbcL gene and the intergenic promoter region between the divergently transcribed rbcL and atpB genes of Ipomoea purpurea (L.) Plant Mol Biol. 1992;18:823–825. doi: 10.1007/BF00020029. [DOI] [PubMed] [Google Scholar]
- 12.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 13.Hoyle N R, Eckert B, Kraiss S. Electrochemiluminescence: leading edge technology for automated immunoassay analyte detection. Clin Chem. 1996;42:1576–1578. [Google Scholar]
- 14.Kleter G E M, van Doorn L-J, Brouwer J T, Schalm S W, Heijtink R A, Quint W G V. Sequence analysis of the 5′ untranslated region in isolates of at least four genotypes of hepatitis C virus in The Netherlands. J Clin Microbiol. 1994;32:306–310. doi: 10.1128/jcm.32.2.306-310.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar U, Thomas H C, Monjardino J. Serum HCV RNA levels in chronic HCV hepatitis measured by quantitative PCR assay; correlation with serum AST. J Virol Methods. 1994;47:95–102. doi: 10.1016/0166-0934(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 16.Mallet F, Hebrard C, Livrozet J M, Lees O, Tron F, Touraine J L, Mandrand B. Quantitation of human immunodeficiency virus type 1 DNA by two PCR procedures coupled with enzyme-linked oligosorbent assay. J Clin Microbiol. 1995;33:3201–3208. doi: 10.1128/jcm.33.12.3201-3208.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manzin A, Bagnarelli P, Menzo S, Giostra F, Brugia M, Francesconi R, Bianchi F B, Clementi M. Quantitation of hepatitis C virus genome molecules in plasma samples. J Clin Microbiol. 1994;32:1939–1944. doi: 10.1128/jcm.32.8.1939-1944.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mulder J, McKinney N, Christopherson C, Sninsky J, Greenfield L, Kwok S. Rapid and simple PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma: application to acute retroviral infection. J Clin Microbiol. 1994;32:292–300. doi: 10.1128/jcm.32.2.292-300.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nedelmann J, Heagerty P, Lawrence C. Quantitative PCR: procedures and precisions. Bull Math Biol. 1992;54:477–502. [Google Scholar]
- 20.Ravaggi A, Zonaro A, Mazza C, Albertini A, Cariani E. Quantification of hepatitis C virus RNA by competitive amplification of RNA from denatured serum and hybridization on microtiter plates. J Clin Microbiol. 1995;33:265–269. doi: 10.1128/jcm.33.2.265-269.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodbard D. Statistical quality control and routine data processing for radioimmunoassay (RIA) and immunoradiometric assays (IRMA) Clin Chem. 1974;20:1255–1270. [PubMed] [Google Scholar]
- 22.Roth W K, Lee J-H, Rüster B, Zeuzem S. Comparison of two quantitative hepatitis C virus reverse transcriptase PCR assays. J Clin Microbiol. 1996;34:261–264. doi: 10.1128/jcm.34.2.261-264.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Secchiero P, Zella D, Crowley R W, Gallo R C, Lusso P. Quantitative PCR for human herpesviruses 6 and 7. J Clin Microbiol. 1995;33:2124–2130. doi: 10.1128/jcm.33.8.2124-2130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siebert P D, Larrick J W. PCR MIMICS: competitive DNA fragments for use as internal standards in quantitative PCR. BioTechniques. 1993;14:244–249. [PubMed] [Google Scholar]
- 25.Simmonds P, McOmish F, Yap P L, Chan S-W, Lin C K, Dusheiko G, Saeed A A, Holmes E C. Sequence variability in the 5′ non-coding region of hepatitis C virus: identification of a new virus type and restrictions on sequence diversity. J Gen Virol. 1993;74:661–668. doi: 10.1099/0022-1317-74-4-661. [DOI] [PubMed] [Google Scholar]
- 26.Wang A M, Doyle M V, Mark D F. Quantitation of mRNA by the polymerase chain reaction. Proc Natl Acad Sci USA. 1989;86:9717–9721. doi: 10.1073/pnas.86.24.9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whitby K, Garson J A. Optimisation and evaluation of a quantitative chemiluminescent polymerase chain reaction assay for hepatitis C virus RNA. J Virol Methods. 1995;51:75–88. doi: 10.1016/0166-0934(94)00144-6. [DOI] [PubMed] [Google Scholar]
- 28.White T J. The future of PCR technology: diversification of technologies and applications. Trends Biotechnol. 1996;14:478–483. doi: 10.1016/s0167-7799(96)10068-8. [DOI] [PubMed] [Google Scholar]
- 29.Young K Y, Resnick R M, Myers T W. Detection of hepatitis C virus RNA by combined reverse transcription-polymerase chain reaction assay. J Clin Microbiol. 1993;31:882–886. doi: 10.1128/jcm.31.4.882-886.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]