Abstract
A rapid cytomegalovirus (CMV) pp65 antigenemia assay with direct erythrocyte lysis (DL) with 0.8% NH4Cl, followed by indirect immunofluorescence staining (IF), was evaluated with 82 blood samples from renal transplant recipients, and the results were compared to those of the conventional antigenemia assay with dextran sedimentation and two-cycle alkaline phosphatase, anti-alkaline phosphatase staining (DS-APAAP). The DL-IF modification gave a higher leukocyte yield compared to DS-APAAP (75.4 versus 54.9%; P < 0.05), with similar leukocyte viability rates of >95%. The DL-IF methodology involved fewer technical steps, and the assay time was shortened from 5 h to less than 3 h. Nineteen of the 82 samples concordantly tested positive for pp65 antigenemia by both assays, and the readings showed a good correlation (r = 0.996; P < 0.01). No discordant results were observed. We conclude that the CMV pp65 antigenemia assay by this novel DL-IF modification is technically simpler, cheaper, and less time-consuming but yields results comparable to those of the conventional DS-APAAP assay. The shortened assay time and increased capacity to handle more samples confer distinct advantages in the rapid diagnosis and prompt treatment of CMV disease in immunosuppressed patients.
Cytomegalovirus (CMV) infection accounts for much morbidity and mortality in immunocompromised patients. Early diagnosis and prompt therapy with antiviral agents such as ganciclovir and foscarnet are critical to ensuring a favorable clinical outcome (9). It is therefore essential to have an assay that allows the rapid and reliable diagnosis of CMV disease.
The CMV pp65 antigenemia assay, which quantitates the number of CMV-infected leukocytes in peripheral blood, has proven efficacy in the detection and monitoring of CMV infection in immunocompromised patients (6, 10, 12). The original methodology for CMV pp65 antigenemia assay by Van der Bij et al. (14) comprises three steps: isolation of leukocytes by dextran sedimentation (DS), fixation, and immunocytochemical detection by indirect immunoperoxidase staining. The assay time is approximately 5 h. Various modifications have been made to increase the sensitivity and specificity of this assay, such as the two-cycle alkaline phosphatase, anti-alkaline phosphatase (APAAP) staining method (2). However, few attempts have been made to shorten the assay time, reduce the technical complexities, or lower the assay cost.
We describe a direct erythrocyte lysis (DL) method for the isolation of leukocytes, coupled with antigen detection by indirect immunofluorescence staining (IF), which allows the assay to be completed in less than 3 h. The new DL-IF methodology offers sensitivity and specificity comparable to those of the DS-APAAP assay, while it reduces the reagent and labor costs compared to those for the conventional DS-APAAP assay.
(Part of the data were presented at the 6th International Cytomegalovirus Workshop, 5 to 9 March 1997, Orange Beach, Ala.)
MATERIALS AND METHODS
Blood samples.
Eighty-two fresh EDTA-anticoagulated blood samples from 60 renal transplant patients were studied. A total of 9 ml of blood was collected from each patient; 2 ml was used for total leukocyte and differential counts with a cell analyzer (Cobas Micro; Roche), 5 ml was used for CMV pp65 antigenemia assay by the conventional DS-APAAP method, and 2 ml was used for the novel DL-IF assay.
Leukocyte isolation by DS.
A total of 5 ml of EDTA-anticoagulated blood was mixed with 1.5 ml of a 6% dextran solution, and the mixture was incubated at 37°C for 30 min to allow aggregation and sedimentation of the erythrocytes. The leukocyte-rich supernatant was then mixed with 10 ml of phosphate-buffered saline (PBS), and the mixture was spun at 200 × g for 10 min at room temperature (RT). The cell pellet was suspended in 1 ml of 0.8% NH4Cl for 5 min to rupture the remaining erythrocytes. The cells were then washed once in PBS and centrifuged at 200 × g for 10 min. The final leukocyte pellet was suspended in 1 ml of 3% bovine serum albumin (BSA)-PBS. The cell yield and differential counts were determined with the cell analyzer. The former was defined as the percentage of leukocytes obtained compared to the maximum available percentage, as indicated by the total leukocyte count of the original blood sample. Leukocyte viability was examined by trypan blue staining. Cytospin slides were obtained by centrifugation of 2 × 105 leukocytes on glass slides at 500 rpm (Cytospin 3; Shandon, Cheshire, England) for 3 min.
Leukocyte isolation by DL.
A total of 2 ml of EDTA-anticoagulated blood was added to 25 ml of 0.8% NH4Cl solution in a 50-ml plastic centrifuge tube, roller mixed at high speed for 5 min at RT, and spun at 300 × g for 7 min. The leukocyte pellet was suspended in 1 ml of PBS and was transferred to a 2-ml V-bottom microcentrifuge tube (BioScience Inc.) and centrifuged for 20 s at 12,000 rpm (Cytospin 3; Shandon). After another wash in 1 ml of PBS, the leukocyte pellet was suspended in 1 ml of 3% BSA–PBS. The viability test was performed as described above, and cell yield was determined as described above.
Fixation of cytospin slides.
Leukocytes on cytospin slides prepared by both the DS and the DL methods were fixed in 5% formaldehyde for 10 min at RT. After washing the slides three times in PBS, the cells were permeabilized with 0.5% Nonidet P-40 for 5 min at RT and then air dried for 15 min before staining for pp65 antigen.
Detection of CMV pp65 antigen by IF.
After cell fixation, each slide was covered with 30 μl of 1/8-diluted Clonab CMV monoclonal antibody (Clonab clone C10/C11, mouse immunoglobulin G [IgG] type; Biotest, Dreieich, Germany) and incubated for 30 min at 37°C. After washing three times in PBS, the slides were incubated with 30 μl of 1/20-diluted fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG antibody (Clonab Ig-FITC; Biotest) for 30 min at 37°C. After washing, the numbers of cells with green fluorescence were scored under a UV microscope at ×400 magnification. A positive assay result was defined by the presence of at least 1 positively stained leukocyte on the slide, and the result was expressed as the number of CMV pp65-positive cells per 2 × 105 leukocytes.
Detection of CMV pp65 antigen by two-cycle APAAP staining.
The CMV pp65 antigen was detected by APAAP staining as described previously (2). After cell fixation, each slide was incubated with 30 μl of 1/8-diluted Clonab CMV monoclonal antibody for 30 min at 37°C. After washing three times in PBS, the slides were incubated with 30 μl of 1/50-diluted rabbit anti-mouse immunoglobulin (Dako, Glostrup, Denmark) for 45 min at RT. Following another three washes in PBS, they were incubated with 30 μl of 1/100-diluted APAAP complex (Serotec, Oxford, United Kingdom) for 45 min at RT. After another wash in PBS, the rabbit anti-mouse immunoglobulin and APAAP complex incubation steps were repeated at RT, each for 10 min. Nephthol-AS-B1 phosphate in new fuchsin stain (Sigma, St. Louis, Mo.) was then added for 30 min for color development. The total numbers of positively stained leukocytes with bright red nuclei were counted by light microscopy.
Statistics.
Nonparametric data were compared by the Mann-Whitney test. Correlation of the results of the DL-IF and DS-APAAP assays was examined with the Spearman rank correlation coefficient.
RESULTS
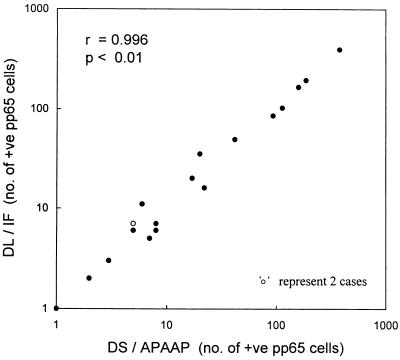
The level of recovery of leukocytes was 75.4% by DL and 54.9% by DS (P < 0.05). Leukocyte viability was >95% by both methods. Differential counts of isolated leukocytes were also similar by both methods. All 82 samples were tested by both the DL-IF and the DS-APAAP methodologies. Nineteen samples concordantly tested positive by both methodologies, with a good correlation (r = 0.996; P < 0.01) (Fig. 1). DL-IF gave mean and median values of 58 and 11 (range, 1 to 391) CMV pp65-positive cells per 2 × 105 leukocytes, respectively, while the corresponding values for DS-APAAP were 57 and 8 (range, 1 to 380) CMV pp65-positive cells per 2 × 105 leukocytes. Preparation of leukocyte cytospin slides by DL was 1 h quicker than that by DS, and the assay time was less than 3 h by DL-IF, whereas it was 5 h by DS-APAAP (Table 1). Compared to DS-APAAP, the cost of reagents for DL-IF was 36% lower. In addition to the requirement for fewer reagents, the DL-IF methodology was also simpler because of the reduced number of steps.
FIG. 1.
Correlation of results for the 19 samples concordantly testing positive for CMV pp65 antigenemia by both the DS-APAAP and the DL-IF methodologies.
TABLE 1.
Comparison of DL followed by IF and DS followed by two-cycle APAAP for the measurement of CMV pp65 antigenemia
| Parameter | DL-IF | DS-APAAP |
|---|---|---|
| Leukocyte isolation | ||
| Vol (ml) of EDTA-anticoagulated blood used | 2 | 5 |
| Mean leukocyte recovery ratea (% [range]) | 75.4 (68.9–79.9) | 54.9 (48.7–63.9) |
| Leukocyte viability (%) | >95 | >95 |
| Time to prepare a cytospin slide | 25 min | 1 h and 30 min |
| No. of assay procedures | ||
| Sedimentation | 0 | 1 |
| Lysing | 1 | 1 |
| Incubation | 2 | 4 |
| Color development | 0 | 1 |
| No. of principle reagents | 3 | 11 |
| Reagent cost per sample (US$) | 3.2 | 5.1 |
| Assay time (h) per sample | <3 | 5 |
| No. of samples processed per technician-day | 16 | 8 |
P < 0.05.
DISCUSSION
CMV infection is common among organ allograft recipients. Despite the presence of effective antiviral agents, CMV disease still accounts for considerable morbidity and mortality, which often relate to delay in therapy due to unsatisfactory diagnostic assays. Diagnosis by viral culture (shell vial assay) or serological response is time-consuming, is often retrospective, and is generally not sensitive. Detection of the CMV genome by PCR is highly sensitive, but its positive predictive value for symptomatic disease in seropositive patients is low, ranging from 25 to 45% (1). Rapid diagnosis of CMV disease is now possible with the CMV pp65 antigenemia assay, in which a monoclonal antibody is used to detect a 65-kDa lower-matrix phosphoprotein (pp65) that is expressed in the host cell nuclei during the early phase of the CMV replication cycle. The sensitivity, specificity, and relative rapidity of this assay have been established (5, 8, 10, 12–14). In addition, the quantitative nature of the pp65 antigenemia assay help discriminate between patients with latent CMV infection and those with symptomatic disease (10). In this study, we evaluated a modified methodology for the pp65 antigenemia assay by DL followed by IF, and our results indicate that this modified assay is simple, reliable, less time-consuming, and cheaper than the conventional pp65 assay methodology.
DS is a standard methodology for leukocyte isolation prior to staining for the pp65 antigen. Alternatively, leukocyte extraction with Polymorphprep leukocyte separation medium (Nycomed Pharma AS, Majorstua, Norway) has been reported to yield pp65 antigenemia results comparable to those of DS. However, this modification is more expensive and offers no technical advantage, and the viability rate and the differential count of the leukocytes thus isolated remain unclear (4). Leukocyte isolation by the conventional DS method involves the use of 1 to 3 ml of 0.8% NH4Cl to lyse contaminating erythrocytes in the leukocyte pellet. With our modification, a greater volume (25 ml) of 0.8% NH4Cl is added directly to 2 ml of EDTA-anticoagulated whole blood to lyse the erythrocytes, which is completed in 5 min. The process of leukocyte isolation is simplified since the modified assay does not involve dextran sedimentation. Washing of the isolated leukocytes after erythrocyte lysis can be done within 20 s with a 2-ml V-bottom microcentrifuge tube instead of 10 min in a 15-ml centrifuge tube following DS. Our data indicate that the DL modification has an improved leukocyte recovery rate of 75%, whereas it is 55% by DS, and gives percentages of granulocytes, monocytes, and lymphocytes in isolated leukocyte populations similar to those given by DS. The latter is important since CMV predominantly infects granulocyte and monocyte populations (7, 11). In addition, leukocyte isolation by DL requires only 2 ml of blood sample, whereas 5 ml is required for DS. Leukocyte isolation by DL is thus faster and is technically easier to perform.
We used 5% formaldehyde after trying different cell fixatives, since following permeabilization with Nonidet P-40 it gives a more definitive staining of CMV pp65-positive cells and better reproducibility of positive cell yield than the use of methanol and acetone as cell fixatives (3). Immunoperoxidase staining is used in the original assay described by Van der Bij et al. (14). Other staining techniques such as staining with the avidin-biotin peroxidase complex, immunofluorescence staining, and two-cycle APAAP have been reported to confer increased sensitivity, reduced levels of false-positive or high background staining due to endogenous leukocyte peroxidase (2, 5). In particular, Gerna et al. (5) reported better results with IF staining than with immunoperoxidase, avidin-biotin peroxidase, or APAAP staining (5). We have found similar staining results with IF and two-cycle APAAP. However, the latter involves more reagents and incubation steps and is thus more cumbersome and prone to interassay variations.
We conclude that while assays for CMV pp65 antigenemia by the DL-IF and DS-APAAP methodologies give comparable results, the DL-IF modification is preferred since it is less tedious, involving fewer steps and reagents, and costs about one-third less. Of particular importance is the fact that the DL-IF modification can give the result within 3 h. The rapidity of diagnosis and the capacity for the technician to handle more samples with the DL-IF modification offer distinct advantages for the clinical management of patients with suspected CMV disease.
ACKNOWLEDGMENTS
The work was supported by the Renal Research Fund (360/041/0599) of the Department of Medicine, The University of Hong Kong.
We thank A. Chan, C. Tang, and R. Wong for assistance in technical steps and statistical analyses.
REFERENCES
- 1.Abecassis M M, Koffron A J, Kaplan B, Buckingham M, Muldoon J P, Cribbins A J, Kaufman D B, Fryer J P, Stuart J, Stuart F P. The role of PCR in the diagnosis and management of CMV in solid organ recipients: what is the predictive value for the development of disease and should PCR be used to guide antiviral therapy? Transplantation. 1997;63:275–279. doi: 10.1097/00007890-199701270-00017. [DOI] [PubMed] [Google Scholar]
- 2.Bein G, Bitsch A, Hoyer J, Kirchner H. The detection of human cytomegalovirus immediate early antigen in peripheral blood leukocytes. J Immunol Methods. 1991;137:175–180. doi: 10.1016/0022-1759(91)90022-8. [DOI] [PubMed] [Google Scholar]
- 3.Boeckh M, Woogerd P M, Stevens-Ayers T, Ray C G, Bowden R A. Factors influencing detection of quantitative cytomegalovirus antigenemia. J Clin Microbiol. 1994;32:832–834. doi: 10.1128/jcm.32.3.832-834.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia A, Niubo J, Benitez M A, Viqueira M, Perez J. Comparison of two leukocyte extraction methods for cytomegalovirus antigenemia assay. J Clin Microbiol. 1996;34:182–184. doi: 10.1128/jcm.34.1.182-184.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerna G, Revello M G, Percivalle E, Morini F. Comparison of different immunostaining techniques and monoclonal antibodies to the lower matrix phosphoprotein (pp65) for optimal quantitation of human cytomegalovirus antigenemia. J Clin Microbiol. 1992;30:1232–1237. doi: 10.1128/jcm.30.5.1232-1237.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerna G, Revello M G, Percivalle E, Zavattoni M, Parea M, Battaglia M. Quantification of human cytomegalovirus viremia by using monoclonal antibodies to different viral proteins. J Clin Microbiol. 1990;28:2681–2688. doi: 10.1128/jcm.28.12.2681-2688.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerna G, Zipeto D, Percivalle E, Parea M, Revello M G, Maccario R, Peri G, Milanesi G. Human cytomegalovirus infection of the major leukocyte subpopulations and evidence for initial viral replication in polymorphonuclear leukocytes from viremic patients. J Infect Dis. 1992;166:1236–1244. doi: 10.1093/infdis/166.6.1236. [DOI] [PubMed] [Google Scholar]
- 8.Halwachs G, Zach R, Pogglitsch H, Holzer H, Tiran A, Iberer F, Wasler A, Tscheliessnigg H P, Lanzer G, Folsch B. A rapid immunocytochemical assay for CMV detection in peripheral blood of organ-transplanted patients in clinical practice. Transplantation. 1993;56:338–342. doi: 10.1097/00007890-199308000-00016. [DOI] [PubMed] [Google Scholar]
- 9.Hrebinko R, Jordan M L, Dummer J S, Hickey D P, Shapiro R, Vivas C, Starzl T E, Simmons R L. Ganciclovir for invasive cytomegalovirus infection in renal allograft recipients. Transplant Proc. 1991;23:1346–1347. [PMC free article] [PubMed] [Google Scholar]
- 10.Murray B M, Brentjens J, Amsterdam D, Myers J, Gray V, Pawlowski I, Schewegler K, Singh J P, Venuto R C. The cytomegalovirus-antigenemia assay in the diagnosis of posttransplant cytomegalovirus infection. J Am Soc Nephrol. 1994;4:1615–1622. doi: 10.1681/ASN.V481615. [DOI] [PubMed] [Google Scholar]
- 11.Taylor-Wiedeman J, Sissons J G P, Borysiewiez L K, Sinclair J H. Monocytes are a major site of persistence of human cytomegalovirus in peripheral blood mononuclear cells. J Gen Virol. 1991;72:2059–2064. doi: 10.1099/0022-1317-72-9-2059. [DOI] [PubMed] [Google Scholar]
- 12.The T H, Van der Ploeg M, Van den Berg A P, Vlieger A M, Van der Giessen M, Van Son W J. Direct detection of cytomegalovirus in peripheral blood leukocytes—a review of the antigenemia assay and polymerase chain reaction. Transplantation. 1992;54:193–198. doi: 10.1097/00007890-199208000-00001. [DOI] [PubMed] [Google Scholar]
- 13.Van den Berg A P, van der Bij W, van Son W J, Anema J, van der Giessen M, Schirm J, Tegzess A M, The T H. Cytomegalovirus antigenemia as a useful marker of symptomatic cytomegalovirus infection after renal transplantation—a report of 130 consecutive patients. Transplantation. 1989;48:991–995. doi: 10.1097/00007890-198912000-00019. [DOI] [PubMed] [Google Scholar]
- 14.Van der Bij W, Torensma R, Van Son W J, Anema J, Tegzess A M, The T H. Rapid immunodiagnosis of active cytomegalovirus infection by monoclonal antibody staining of blood leukocytes. J Med Virol. 1988;25:179–188. doi: 10.1002/jmv.1890250208. [DOI] [PubMed] [Google Scholar]